Abstract
Arabidopsis plants possess a composite AtLKR/SDH locus encoding two different polypeptides involved in lysine catabolism: a bifunctional lysine-ketoglutarate reductase/saccharopine dehydrogenase (LKR/SDH) enzyme and a monofunctional SDH enzyme. To unravel the physiological significance of these two enzymes, we analyzed their subcellular localization and detailed biochemical properties. Sucrose gradient analysis showed that the two enzymes are localized in the cytosol and therefore may operate at relatively neutral pH values in vivo. Yet while the physiological pH may provide an optimum environment for LKR activity, the pH optima for the activities of both the linked and non-linked SDH enzymes were above pH 9, suggesting that these two enzymes may operate under suboptimal conditions in vivo. The basic biochemical properties of the monofunctional SDH, including its pH optimum as well as the apparent Michaelis constant (Km) values for its substrates saccharopine and nicotinamide adenine dinucleotide at neutral and basic pH values, were similar to those of its SDH counterpart that is linked to LKR. Taken together, our results suggest that production of the monofunctional SDH provides Arabidopsis plants with enhanced levels of SDH activity (maximum initial velocity), rather than with an SDH isozyme with significantly altered kinetic parameters. Excess levels of this enzyme might enable efficient flux of lysine catabolism via the SDH reaction in the unfavorable physiological pH of the cytosol.
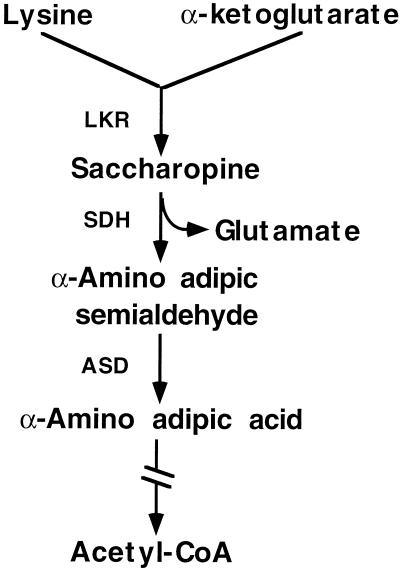
In both plant and animal cells, excess Lys is catabolized into Glu and acetyl-coenzyme A via the α-amino adipic acid pathway (Fig. 1) (Markovitz et al., 1984; Galili, 1995). The first enzyme in the Lys catabolic pathway, Lys-ketoglutarate reductase (LKR), condenses Lys and α-ketoglutarate into saccharopine. The second enzyme, saccharopine dehydrogenase (SDH), converts saccharopine into α-amino adipic semi-aldehyde and Glu (Markovitz et al., 1984; Goncalves-Butruille et al., 1996). Lys catabolism plays an important physiological role in both mammals and plants. In humans, the genetic disease familial hyperlysinemia, which causes mental retardation, is associated with reduced LKR activity (Markovitz et al., 1984). The molecular basis for this defect is yet unknown, but LKR activity was shown to be significantly up-regulated during embryonic development of rat brain (Rao et al., 1992). The level of LKR in plants was shown to be significantly up-regulated in inflorescence tissues and developing seeds, as well as in response to osmotic stress (Karchi et al., 1994; Karchi et al., 1995; Tang et al., 1997; Deleu et al., 1999). LKR activity is also stimulated by excess intracellular Lys, both in mammals and plants (Foster et al., 1993; Karchi et al., 1994; Karchi et al., 1995; Kemper et al., 1999). The stimulation of LKR activity in plants was shown to be regulated by an intracellular signaling cascade involving Ca2+ and protein phosphorylation (Karchi et al., 1995).
Figure 1.
Schematic diagram of the Lys catabolism pathway. ASD, Aminoadipic acid semialdehyde dehydrogenase. The broken arrow represents six enzymatic reactions leading to acetyl-coenzyme A (CoA) synthesis.
The control of metabolite flux via the LKR and SDH enzymes is still unclear. In both animals and plants, these enzymes are linked on a single bifunctional polypeptide (Markovitz et al., 1984; Markovitz and Chuang, 1987; Brochetto-Braga et al., 1992; Goncalves-Butruille et al., 1996; Epelbaum et al., 1997; Gaziola et al., 1997; Tang et al., 1997; Miron et al., 2000). Moreover, despite their physical linkage LKR and SDH possess significantly different pH optima; physiological pH values are optimal for LKR but not for SDH activity, suggesting that the in vivo metabolite flux via the SDH enzyme of LKR/SDH may be inefficient.
Metabolite flux via the α-amino adipic acid pathway may not be solely regulated by the bifunctional LKR/SDH enzyme. We have recently demonstrated that Arabidopsis plants possess in addition to a bifunctional LKR/SDH, a monofunctional SDH enzyme (Tang et al., 1997). Moreover, we have also shown that these two enzymes are produced by a single composite locus and that the monofunctional SDH possesses identical amino acid sequence to the SDH domain of the bifunctional LKR/SDH (Tang et al., 1997, 2000). The importance of the linkage between LKR and SDH and the significance of the monofunctional SDH is not clear. Kemper et al. (1998) have provided evidence suggesting that the activity of LKR may be influenced by its linked SDH domain, but whether the linkage between the two enzymes also affects SDH activity is still unknown. To study this issue further, in the present report we have compared the subcellular localization and biochemical properties of the two SDH enzymes produced by the composite LKR/SDH locus of Arabidopsis. We found that both the AtLKR/SDHp and AtSDHp polypeptides are localized in the cytosol and that the monofunctional SDH possesses similar biochemical properties to its counterpart enzyme that is linked to LKR.
RESULTS
Intracellular Localization of the Bifunctional LKR/SDH and Monofunctional SDH in Arabidopsis Cells
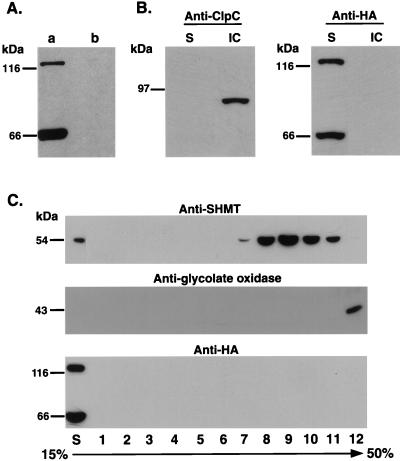
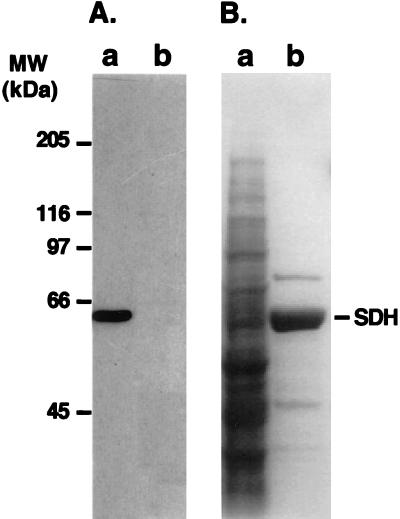
To unravel the physiological environment for the activity of the bifunctional AtLKR/SDHp and monofunctional AtSDHp, which are encoded by a single composite locus (Tang et al., 2000), we analyzed their subcellular localization. To address this, we used transgenic Arabidopsis plants expressing a recombinant construct of the Arabidopsis composite AtLKR/SDH locus (with its own promoter and introns) to which a DNA encoding three copies of the hemagglutinin (HA) epitope were fused in frame just prior to the translation stop codon. The details of these transgenic Arabidopsis plants, which produce both HA-tagged LKR/SDH and HA-tagged monofunctional SDH, are described elsewhere (Tang et al., 2000). Extracts from stem sections containing inflorescences and developing siliques from transgenic plants harboring this recombinant construct produced both an HA-tagged bifunctional AtLKR/SDHp and an HA-tagged monofunctional AtSDHp, which were detected with the commercial anti-HA monoclonal antibodies (Fig. 2A, lane a). Control non-transformed plants showed no cross-reactive bands with these antibodies, confirming their specificity to AtLKR/SDHp and AtSDHp (Fig. 2A, lane b).
Figure 2.
Subcellular localization of AtLKR/SDHp and AtSDHp. A, Expression of HA-tagged AtLKR/SDHp and AtSDHp in transgenic Arabidopsis. Protein extracts from a transgenic Arabidopsis plant expressing these HA-tagged proteins (Tang et al., 2000) (lane a) and control non-transformed plants (lane b) were reacted in a western blot with anti-HA antibodies. B, Tissue homogenate was used for isolation of intact chloroplasts as described in “Materials and Methods.” A PVDF membrane, containing proteins from the supernatant (S) and intact chloroplast fraction (IC), was subjected to two sequential western-blot analyses with antibodies to the chloroplast stroma marker protein, ClcP (left), and to the HA epitope (right). C, Tissue homogenate was used for fractionation of mitochondria and peroxisomes on 15% to 50% linear Suc gradient as described in “Materials and Methods.” A PVDF membrane, containing proteins from the supernatant (S) and from individual fractions of the Suc gradient, was subjected to three sequential western-blot analyses with antibodies to the mitochondrial matrix marker protein Ser hydroxymethyltransferase (SHMT) (top), the peroxisomal marker protein glycolate oxidase (middle), and the HA epitope (bottom). The sizes of molecular mass protein markers are indicated on the left.
The AtLKR/SDH and AtSDHp do not possess any amino acid sequence that resembles targeting peptides to plastids, mitochondria, or peroxisomes, suggesting that they may reside in the cytosol. To confirm this hypothesis, we prepared fractions containing intact chloroplasts (pelleted via a 40% percol cushion), as well as mitochondria and peroxisome (Suc gradient fractionation) (see “Materials and Methods”) from stems containing inflorescences and developing siliques of the transgenic plants expressing the HA-tagged AtLKR/SDHp and AtSDHp. Subcellular fractionation generally results in relatively low yield of intact organelles. Therefore, to detect AtLKR/SDHp and AtSDHp in any of the organelles, we used 200-fold enriched fraction of isolated chloroplasts (40% percol pellet) and 100-fold enriched fraction containing mitochondria and peroxisomes (12,000-g pellet) than that of the supernatant. Organelle enrichment was based on normalization to the original tissue weight. As shown in Figure 2B, the chloroplast stroma protein marker ClcP (Halperin and Adam, 1996; Ostersetzer and Adam, 1996) was detected only in intact chloroplasts but not in the supernatant. In contrast, AtLKR/SDHp and AtSDHp were detected only in the supernatant but not in the intact chloroplasts, showing that they are not localized in this organelle. As shown in Figure 2C, both the mitochondrial matrix protein marker, Ser hydroxymethyltransferase (Bourguignon et al., 1988) and the peroxisomal protein marker glycolate oxidase (Volokita, 1991) were highly enriched in the respective fractions 7 to 11 and fraction 12 of the Suc gradient, which are the expected densities for mitochondria and peroxisomes. Still, AtLKR/SDH and AtSDH were highly enriched in the supernatant, confirming that they were not localized in any of these two organelles.
Biochemical Properties of the Arabidopsis LKR/SDH Enzyme
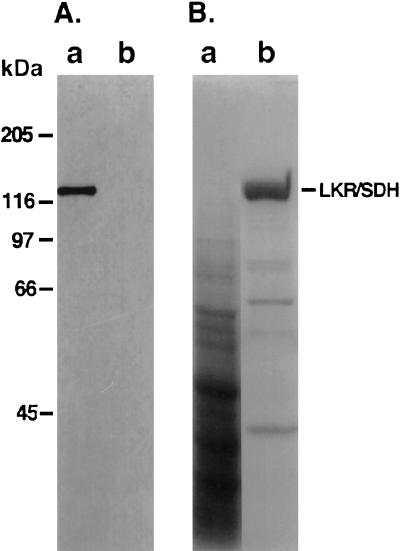
To study the metabolic significance of the two AtLKR/SDHp and AtSDHp polypeptides encoded by the Arabidopsis composite LKR/SDH locus, we wished to compare the biochemical properties of these two enzymes. Because of the small size of Arabidopsis, it is quite difficult to purify sufficient levels of LKR/SDH and monofunctional SDH enzymes from this plant to allow accurate kinetic measurements. Therefore, we used yeast as an heterologous system for expression of the Arabidopsis enzymes. We first tested whether yeast is a suitable system for production of active Arabidopsis AtLKR/SDHp. To address this, yeast cells were transformed with plasmids encoding three different forms of AtLKR/SDHp. One is a wild-type polypeptide and the other two are fused at either the N terminus or the C terminus to a His-tag epitope (see “Materials and Methods”). Purified His-tagged AtLKR/SDHp and partially purified extracts from yeast cells expressing the natural AtLKR/SDHp showed comparable catabolic LKR and SDH activities (data not shown), suggesting that: (a) the AtLKR/SDHp was folded correctly in the yeast cells to produce an active enzyme and (b) the His tags did not interfere with the LKR and SDH activities of AtLKR/SDHp. Next, the His-tagged AtLKR/SDHp enzymes were purified using a nickel column. Whereas AtLKR/SDH possessing the N-terminal His tag was easily purified in large amounts, its counterpart enzyme possessing the C-terminal His tag did not, suggesting that the C-terminal His tag was masked in this bifunctional protein. Thus, for subsequent analysis of the biochemical properties of AtLKR/SDHp, we used nearly purified preparations of AtLKR/SDHp containing the N-terminal His tag. The production of the approximately 120-kD His-tagged AtLKR/SDHp in yeast cells, as well as its purification using a nickel column, are illustrated in the western-blot (using anti-His-tag antibodies) and Coomassie Blue-stained gel shown in Figure 3, panels A and B, respectively.
Figure 3.
Expression of AtLKR/SDHp fused to N-terminal His tag in yeast and its purification. A, Protein extracts from yeast cells harboring the pVT-At-His-LKR/SDH plasmid or control vector alone (lanes a and b, respectively) were reacted in a western blot with anti-His-tag antibodies. B, Protein extracts from yeast cells harboring the pVT-At-His-LKR/SDH plasmid either before purification (lane a) or after purification on a nickel column (lane b) were fractionated on SDS polyacrylamide gel, and the proteins were stained with Coomassie Blue R-250. The position of AtLKR/SDHp is indicated on the right. The migrations of molecular mass protein markers are shown on the left.
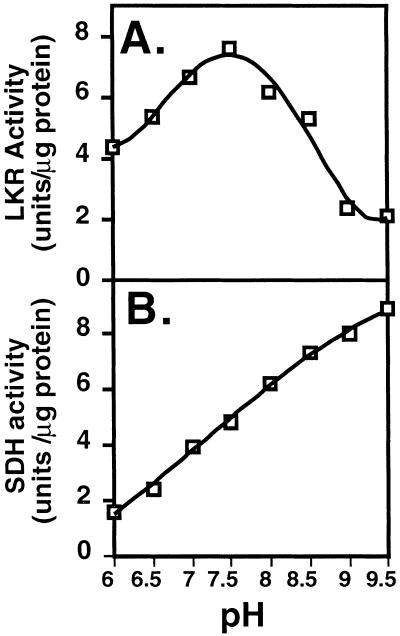
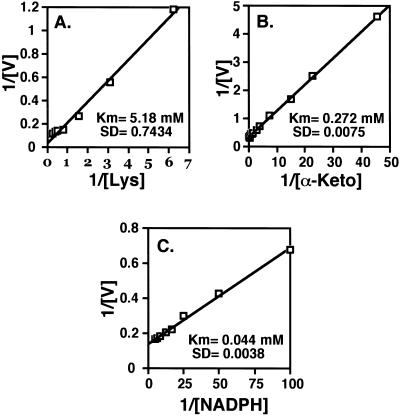
LKR and SDH activities of bifunctional LKR/DH enzymes from various plant species possess different pH optima (Brochetto-Braga et al., 1992; Goncalves-Butruille et al., 1996; Gaziola et al., 1997; Miron et al., 2000). Thus, we first tested the LKR and SDH activities of AtLKR/SDHp at different pH values under conditions of excess substrate concentrations. As shown in Figure 4, the optimal pH for LKR activity of AtLKR/SDHp was approximately pH 7.5, whereas that for SDH activity of AtLKR/SDHp was above pH 9, similarly to the maize, rice, and soybean LKR/SDH enzymes. Next, we tested the apparent Km values of the linked LKR and SDH enzyme of AtLKR/SDHp for their substrates. Since AtLKR/SDHp is localized in the cytosol (Fig. 2) and apparently functions at neutral pH values in vivo, we tested the apparent Km values of LKR for its substrates at its optimal pH value of 7.5. In addition, we tested the apparent Km values of SDH for its substrates at both pH values of 7 (near physiological pH) and 9 (near optimal pH of activity in vitro). As shown in Figure 5, the LKR enzyme of AtLKR/SDHp possessed a relatively high apparent Km of 5.180 mm for Lys, a lower apparent Km of 0.272 mm for α-ketoglutarate, and the lowest apparent Km of 0.044 mm for NADPH (Fig. 5, A, B, and C, respectively). These values were very similar to the values observed for natural LKR/SDH enzymes purified from maize, rice, and soybean (Brochetto-Braga et al., 1992; Goncalves-Butruille et al., 1996; Gaziola et al., 1997; Miron et al., 2000). For example, the LKR/SDH of rice, on which a detailed apparent Km analysis for all substrates was performed, possesses apparent Km values of 4.5 mm, 0.8 mm, and 0.032 mm for Lys, α-ketoglutarate, and NADPH, respectively (Gaziola et al., 1997).
Figure 4.
Determination of the optimal pH values for LKR and SDH activity of AtLKR/SDHp. The kinetics of LKR (A) and SDH (B) activities were assayed spectrophotometrically under conditions of excess substrate concentrations in reaction mixtures with increasing pH levels as indicated in the figures. These experiments were replicated three times with similar results.
Figure 5.
Determination of apparent Km values for LKR activity of AtLKR/SDHp toward its substrates. Apparent Km values were obtained by double reciprocal plots of the initial velocities of LKR with Lys (A), α-ketoglutarate (B), and NADPH (C) as the variable substrates. The calculated apparent Km values are given in A, B, and C. These experiments were replicated three times with similar results.
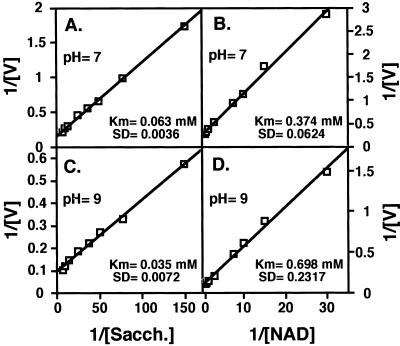
The apparent Km values of SDH of AtLKR/SDHp for its substrates saccharopine and NAD were calculated to be 0.063 mm and 0.374 mm, respectively at pH 7 (Fig. 6, A and B), as well as 0.035 mm and 0.698 mm, respectively at pH 9 (Fig. 6, C and D). The values at pH 9 were also very similar to the apparent Km values obtained for maize, rice, and soybean LKR/SDH enzymes, which were generally analyzed at pH 8.5 (Brochetto-Braga et al., 1992; Goncalves-Butruille et al., 1996; Gaziola et al., 1997; Miron et al., 2000). Again, the rice LKR/SDH, on which detailed analysis of SDH activity was performed at pH 8.5, possesses apparent Km values of 0.130 and 0.490 mm for saccharopine and NAD, respectively (Gaziola et al., 1997).
Figure 6.
Determination of apparent Km values for SDH activity of AtLKR/SDHp toward its substrates. Apparent Km values were obtained by double reciprocal plots of the initial velocities of SDH with saccharopine (A and C) and NAD (B and D) as the variable substrates. Activity assays were performed either at pH 7 (A and B) or at pH 9 (C and D). The calculated apparent Km values are given in A, B, C, and D. These experiments were replicated three times with similar results.
The Two SDH Enzymes Encoded by the Composite AtLKR/SDH Locus Possess Similar Biochemical Properties
The SDH domain of AtLKR/SDHp and the monofunctional AtSDHp are encoded by the same open reading frame of the AtLKR/SDH locus with AtSDHp translation being initiated from an internal ATG codon (Tang et al., 2000). To test whether the SDH domain of AtLKR/SDHp and the monofunctional AtSDHp possess similar or distinct biochemical properties, we have expressed in yeast two recombinant constructs encoding only the SDH open reading frame of AtLKR/SDH. One of these constructs encoded a wild-type AtSDH polypeptide, whereas the second encoded an AtSDH containing a C-terminal His tag. Partially purified extracts containing these two enzymes possessed comparable catabolic SDH activity (data not shown), suggesting that as in the case of AtLKR/SDHp, the His tag did not interfere with the activity of AtSDH. Notwithstanding, as opposed to AtLKR/SDHp, AtSDHp with the C-terminal His tag could be easily purified to large amounts using a nickel column. Thus, for subsequent analysis of the biochemical properties of AtSDHp, we used nearly purified preparations of this His-tagged AtSDHp. The production of the approximately 63-kD His-tagged monofunctional AtSDHp in the yeast cells, as well as its purification using a nickel column are illustrated the western-blot (using anti-His-tag antibodies) and Coomassie Blue-stained gel shown in Figure 7, A and B, respectively.
Figure 7.
Expression of AtSDHp fused to a C-terminal His tag in yeast and its purification. A, Protein extracts from yeast cells harboring the pVT-AtSDH-His plasmid or control vector alone (lanes a and b, respectively) were reacted in a western blot with anti-His-tag antibodies. B, Protein extracts from yeast cells harboring the pVT-AtSDH-His plasmid either before purification (lane a) or after purification on a nickel column (lane b) were fractionated on SDS-polyacrylamide gel, and the proteins were stained with Coomassie Blue R-250. The position of AtSDHp is indicated on the right. The migrations of molecular mass protein markers are shown on the left.
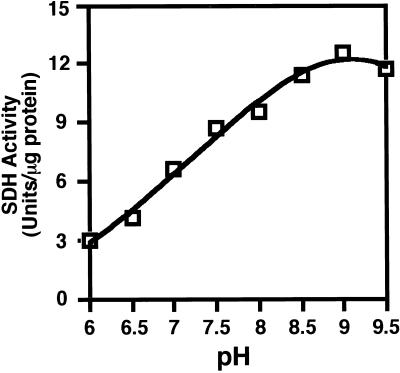
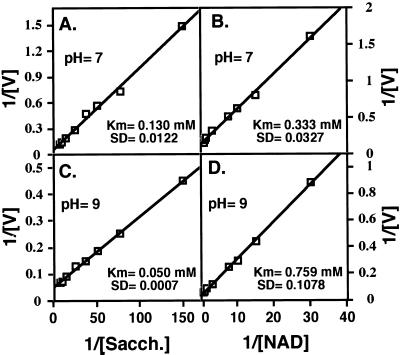
At the next stage, we tested the pH optimum for activity of the monofunctional AtSDHp under excess concentrations of substrates. As shown in Figure 8, activity of the recombinant AtSDHp was highest at approximately pH 9, and its activity was progressively reduced with reduction in pH values. The pH-dependent activity of the monofunctional AtSDHp was very similar to that of the SDH enzyme that is linked to LKR in AtLKR/SDHp (compare with Figs. 4B and Fig. 8). We also tested the apparent Km values of AtSDHp for saccharopine and NAD at pH values of 7 and 9. As shown in Figure 9, these apparent Km values were calculated to be 0.130 and 0.333 mm, respectively, at pH 7, (Fig. 9, A and B), as well as 0.05 and 0.759 mm, respectively, at pH 9 (Fig. 9, C and D). These values were comparable to the values obtained with the bifunctional AtLKR/SDHp (compare with Figs. 6 and 9).
Figure 8.
Determination of the optimal pH values for SDH activity of AtSDHp. The kinetics of SDH activity was assayed spectrophotometrically under conditions of excess substrate concentrations in reaction mixtures with increasing pH levels as indicated in the figure. These experiments were replicated three times with similar results.
Figure 9.
Determination of apparent Km values for SDH activity of AtSDHp toward its substrates. Apparent Km values were obtained by double reciprocal plots of the initial velocities of SDH with saccharopine (A and C) and NAD (B and D) as the variable substrates. Activity assays were performed either at pH 7 (A and B) or at pH 9 (C and D). The calculated apparent Km values are given in A, B, C, and D. These experiments were replicated three times with similar results.
DISCUSSION
AtLKR/SDH and AtSDH Are Localized in the Cytosol and Possess Comparable in Vitro Biochemical Properties
We have previously shown that Arabidopsis possesses a composite LKR/SDH locus encoding both a bifunctional AtLKR/SDHp as well as an additional monofunctional AtSDHp (Tang et al., 1997, 2000). To unravel the significance of the Arabidopsis monofunctional SDH, we compared its subcellular localization with that of AtLKR/SDHp and also compared its biochemical properties with those of its counterpart, which is linked to LKR. Since the small size of Arabidopsis renders this plant useless for obtaining large quantities of purified AtLKR/SDHp and AtSDHp, we used yeast as a heterologous expression system for production of these purified enzymes. Our finding that yeast can produce active AtLKR/SDHp and AtSDHp is in accordance with previous reports showing that yeast is a very useful heterologous system for biochemical and molecular characterization of large repertoire of eukaryotic proteins, including plant proteins (for example, see Schaller and Bleecker, 1995; Su et al., 1997).
By subcellular fractionation, we showed that both AtLKR/SDHp and AtSDHp are localized neither in intact chloroplasts nor in mitochondria and peroxisomes and are therefore likely to be localized in the cytosol. This is in agreement with the fact that targeting sequences to these three organelles were found neither in AtLKR/SDHp nor in AtSDHp. Our results are also in agreement with those reported for maize LKR/SDH enzyme, which was also shown to reside in the cytosol (Kemper et al., 1999). The cytosolic localization of plant LKR/SDH enzymes may be different from the mammalian LKR/SDH enzymes, which were suggested to be localized in the mitochondria (Blemings et al., 1994). The significance of this differential localization has yet to be elucidated.
Our biochemical analyses showed that the monofunctional SDH possesses similar biochemical properties to those of its counterpart SDH enzyme derived from AtLKR/SDHp. These biochemical properties include similar pH optima under excess substrate concentrations (Vmax) as well as similar apparent Km values for their substrates saccharopine and NAD both at the physiological and near optimal pH values of 7 and 9, respectively. These results imply that: (a) the basic activity of AtSDH may not be largely influenced by its link LKR domain, and (b) production of the monofunctional SDH essentially enhances the total level of SDH activity (Vmax) in the plant rather than providing the plant with a SDH enzyme having significantly different kinetic properties than its counterpart-linked SDH enzyme.
The SDH Activities of AtLKR/SDHp and AtSDHp May Operate Suboptimally at Physiological pH
The consequence of the different pH optima of LKR and SDH (pH approximately 7.5 for LKR and pH 9 or above for SDH) in the regulation of Lys catabolism is not clear. Although the cytosolic microenvironment in which AtLKR/SDHp and AtSDHp operate in vivo may allow them to be more efficient that it appears for our in vitro results, it is likely that the cytosol offers suboptimal conditions for the two SDH enzymes than for LKR. This hypothesis is supported by previous studies (Falco et al., 1995; Mazur et al., 1999) showing that transgenic Lys-overproducing soybean plants, which lack a monofunctional SDH enzyme (Tang et al., 1997; Miron et al., 2000), accumulate saccharopine as an intermediate product of Lys catabolism (Fig. 1). The postulated operation of SDH at suboptimal conditions in vivo is also supported by the results of the present work, which provides for the first time detailed analysis of SDH activity at its near physiological pH. Since the SDH substrate saccharopine is derived from LKR, which is a relatively inefficient enzyme (apparent Km of approximately 5 mm for Lys at the physiological pH; see Fig. 5), it is likely that this metabolite represents a major limiting substrate for SDH activity. Our kinetic analysis showed that apparent Km values of the two Arabidopsis SDH enzymes for saccharopine were nearly 75% higher at pH 7 than at pH 9 (Figs. 6 and 9). In addition, the specific activity of these enzymes under conditions of excess concentrations of both substrates (Vmax) were nearly 50% lower at pH 7 than at pH 9 (Figs. 4 and 8).
The in vivo physiological significance of the monofunctional SDH enzyme of Arabidopsis is still not clear. Despite its operation at suboptimal pH, this enzyme is produced in excess over the bifunctional LKR/SDH in Arabidopsis plants (Fig. 2). Thus, the relatively high expression level of this enzyme may overcome its suboptimal function resulting in enhanced flux of Lys catabolism. This presumption is supported by previous studies (Falco et al., 1995; Mazur et al., 1999) in which transgenic Lys-overproducing canola plants, which possess both a bifunctional LKR/SDH and monofunctional SDH enzymes (Tang et al., 1997; Deleu et al., 1999), accumulate α-amino adipic acid (a downstream metabolite to SDH; Fig. 1).
A Hypothesis for the Regulatory Significance of the Arabidopsis Composite AtLKR/SDH Locus
The regulatory significance of the linkage between LKR/SDH and the presence of an additional monofunctional SDH enzyme in some plant species are still ill defined. Kemper et al. (1998) have recently shown that the activity of maize LKR is inhibited by peptides derived from its linked SDH domain. Moreover, the activities of the maize and Arabidopsis LKR are stimulated by the ionic strength of the incubation medium (Kemper et al., 1998; X. Zhu, G. Tang, G. Galili, manuscript in preparation), suggesting that LKR activity is highly sensitive to conformational modulations of the LKR/SDH polypeptide. Thus, it is likely that the linkage between LKR and SDH was evolved for regulatory properties in which LKR activity is regulated by “cross-talk” interactions with its linked SDH domain. Nevertheless, the bifunctional LKR/SDH enzyme of plants is still not an optimal enzyme since it provides relatively limited levels of SDH activity to enable efficient flux of Lys catabolism. Taken together, we hypothesize that composite AtLKR/SDH locus, which encodes both a bifunctional AtLKR/SDHp and a monofunctional AtSDHp, provides a novel mechanism of metabolic regulation, enabling both a highly controlled but efficient flux of Lys catabolism.
MATERIALS AND METHODS
Materials
Wild-type and transgenic Arabidopsis plants expressing a HA epitope-tagged AtLKR/SDHp and AtSDHp (Tang et al., 1997) were grown in greenhouse conditions as previously described (Shaul et al., 1999). The Saccharomyces cerevisiae strain 8989c (ura3, lys9) was kindly provided by Dr. Andre Pierard. Anti-pea ClcP antibodies were kindly provided by Dr. Zach Adam, anti-pea Ser hydroximethyltransferase were kindly provided by R. Douce, and anti-spinach glycolate oxidase antibodies were kindly provided by Dr. Micha Volokita. Anti-HA and anti-His monoclonal antibody was purchased from Sigma (St. Louis).
Plasmid Construction
The full-length cDNA encoding the Arabidopsis bifunctional LKR/SDH was originally cloned into the pBluescript SK− (Stratagene, La Jolla, CA) vector to generate SK-AtLKR/SDH. For construction of a chimeric gene encoding AtLKR/SDHp fused at its N terminus to a tag of six His (His tag), a double-stranded oligonucleotide comprising the following two oligonucleotides 5′CTAGAATGCACCACCACCACCACCACATG3′, and 3′TTACGTGGTGGTGGTGGTGGTGTACTTAA5′ was used to replace the Xbal-EcoRI fragment of SK-AtLKR/SDH. The resulting clone was designated SK-At-His-LKR/SDH. For construction of a chimeric gene encoding AtLKR/SDHp fused at its C terminus to a His tag (SK-AtLKR/SDH-His), the double-stranded oligonucleotide comprising the following two oligonucleotides: 5′CCCCATCATCATCATCATCATG3′ and 3′GGGGTAGTAGTAGTAGTAGTACAGCT5′ was used to replace the Saml-SalI fragment of SK-AtLKR/SDH. A construct encoding a monofunctional AtSDHp was obtained by PCR to introduce a XbaI site at the end of the inter-domain of SK-AtLKR/SDH or SK-AtLKR/SDH-His. The PCR products were subcloned as the Xbal-Xhol sites of SK-AtLKR/SDH to generate SK-AtSDH and SK-AtSDH-His, respectively.
All constructs were subcloned from pBluescript into the yeast expression vector pVT-102 u (Vernet et al., 1987) to generate pVT-102 u-AtLKR/SDH, pVT-102 u-At-His-LKR/SDH, pVT-102 u-AtLKR/SDH-His, pVT-102 u-AtSDH, and pVT-102 u-At.SDH-His.
Yeast Transformation and Complementation
The expression plasmids were transformed into the yeast lys9 mutant cells by lithium acetate as previously described (Ito et al., 1983). Uracil+ transformants were identified on the synthetic complete uracil-free plates.
Subcellular Fractionation
Subcellular fractionation of intact chloroplasts, mitochondria, and peroxisomes was performed essentially as previously described (Gualberto et al., 1995). Transgenic Arabidopsis plants were kept in the dark for 3 d before harvesting of stems containing inflorescences and developing siliques. For isolation of intact chloroplasts, the tissue was cut into small pieces and mixed with precooled chloroplast extraction buffer {50 mm HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid]-KOH, pH 7.3, 0.33 m sorbitol, 2 mm EDTA, 0.1% [w/v] bovine serum albumin} at a ratio of 10 mL/g tissue. Homogenization was conducted in a Polytron (Kinematic, Lucerne, Switzerland) set at 70% power by three 5-s bursts. The homogenate was filtered through two layers of Miracloth (Calbiochem, La Jolla, CA) into precooled polycarbonate centrifuge tubes. Chloroplasts were pelleted by centrifugation at 3,000g for 5 min at 4°C, and the pellet was gently suspended in the remaining buffer with a fine brush. The volume was increased to 4 mL, layered on top of a 16-mL cushion containing 40% (v/v) Percoll in chloroplast extraction buffer, and centrifuged at 3,000g for 5 min. The pellet containing intact chloroplasts was collected.
For subcellular fractionation of mitochondria and peroxisomes, tissue was homogenized as described above in a buffer containing 50 mm Tris-HCl, pH 7.5, 0.4 m Suc, 2 mm EDTA, 0.1% (w/v) bovine serum albumin, and 4 mm β-mercaptoethanol. The homogenate was centrifuged at 3,000g for 5 min at 4°C to remove debris and plastids, and the supernatant was recentrifuged at 12,000g for 20 min at 4°C to obtain a pellet containing mitochondria and peroxisomes. The pellet was gently suspended in 10 mL of extraction buffer, homogenized with a Dounce homogenizer with a loose pestle, and recentrifuged at 3,000g and 12,000g as described above. The washed pellet was gently suspended in a 1-mL extraction buffer and layered onto a 11-mL continuous 15% to 50% (w/v) Suc gradients prepared in Tris buffer (50 mm Tris, pH 7.5, 2 mm EDTA, and 0.1% [w/v] bovine serum albumin). The gradient was centrifuged for 5 h at 4°C at 250,000g in a SW40Ti rotor (Beckman Instruments, Fullerton, CA), fractionated into 1-mL fractions, and individual fractions were subjected to trichloroacetic acid precipitation.
SDS PAGE and Western-Blot Analysis
Protein extraction from Arabidopsis plants and the yeast cells, fractionation on SDS PAGE, transfer to polyvinylidene difluoride (PVDF) membrane, staining of the membrane with Coomassie Blue R, and western-blot analysis were performed essentially as previously described (Tang et al., 2000; Miron et al., 2000). Anti-HA and anti-His monoclonal antibodies (commercially available) were used as primary antibodies for detecting the expression of epitope-tagged AtLKR/SDHp and AtSDHp in plants and yeast, respectively. Anti-pea ClpC (a regulatory subunit of the chloroplast Clp protease that is located in the chloroplast stroma) (Halperin and Adam, 1996; Ostersetzer and Adam, 1996), anti-pea Ser hydroxymethyltransferase (a mitochondrial matrix enzyme) (Bourguignon et al., 1988), and anti-spinach glycolate oxidase (a peroxisomal enzyme) were used for detection of the organelles possessing these proteins.
Purification of Recombinant LKR/SDH and SDH
The purification of LKR/SDH and SDH was performed as previously described (Su et al., 1997). Frozen yeast cells (2 g) were suspended in 8 mL of 25 mm potassium phosphate buffer, pH 7.5, containing 1 mm EDTA, 5% (v/v) glycerol, and 1 mm phenylmethylsulfonyl fluoride and were broken by vortexing with glass beads for 30 min at 4°C. The crude extract was either fractionated through a G50 column and used directly for enzymatic analysis (partially purified extracts) or purified using the His tag. For this affinity purification, the extracts were incubated with 4 mL of 80% (v/v) CL-6B sepharose (Pharmacia Biotech, Piscataway, NJ) at 4°C for 30 min and then centrifuged for 5 min at 18,000g. The supernatant was then incubated at 4°C for 30 min with 1 mL of a 50% (v/v) slurry of nickel-nitrilotriacetate agarose (Qiagen USA, Valencia, CA) in nickel-nitrilotriacetate-gel buffer (25 mm potassium phosphate buffer, pH 7.5, 20 mm imidazole, 10 mm 2-mercaptoethanol, and 5% [v/v] glycerol) and centrifuged at 2,000g for 0.5 min. The column resin was washed with 20 mL of 25 mm potassium phosphate buffer, pH 7.0, 1 mm EDTA, 5% (v/v) glycerol, 300 mm NaCl, and 10 mm 2-mercaptoethanol, resuspended in 5 mL of MOPS buffer {50 mm MOPS [3-(N-morpholino)propanesulfonic acid], pH 7.5, 1 mm EDTA, and 5% [v/v] glycerol}, and transferred to a 5-mL column. The column was washed with 5 mL of MOPS buffer, and LKR/SDH and SDH were eluted with 4 mL of MOPS buffer-containing 200 mm imidazole. Column fractions with maximal LKR activity or SDH activity were pooled.
Analysis of Protein Levels and LKR and SDH Activities
Protein levels were determined by the method of Bradford (1976). The kinetics of LKR activity were measured spectrophotometrically by determining the rate of NADPH oxidation at 340 nm for 10 min at 30°C. The activity assays included 30 μg of protein from the partially purified extracts or 0.1 μg of protein from the nearly purified preparations in 0.3 mL of 0.1 m potassium phosphate buffer (pH 6.0–7.5), Tris buffer (pH 8.0–9.0), or Gly buffer (pH 9.5), containing 20 mm Lys, 14 mm α-ketoglutarate, and 0.4 mm NADPH. One unit of LKR was defined as the amount of enzyme that catalyzes the oxidation of 1 nmol of NADPH per min at 30°C.
The kinetics of SDH activity was measured spectrophotometrically by determining the rate of NAD+ reduction at 340 nm for 10 min at 30°C. The activity assays included 30 μg of protein from the partially purified extracts or 0.1 μg of protein from the nearly purified preparation in 0.3 mL of buffers with different pH values as described above, containing 2 mm saccharopine and 2 mm NAD+. One unit of SDH was defined as the amount of enzyme that catalyzes the reduction 1 nmol of NAD+ per min at 30°C.
In all of the activity assays, controls lacking one of the substrates (Lys, α-ketoglutarate or NADPH for LKR, and saccharopine or NAD+ for SDH) were run in parallel. All of these controls showed nearly zero baseline activity. Experiments studying the apparent Km of a given substrate used different concentration of this specific substrate as indicated in the figures. The apparent Km values were determined graphically from double-reciprocal plots of activity against the variable substrate concentration.
ACKNOWLEDGMENTS
We thank Zach Adam for his kind help in the isolation of intact plastids, for probing our western blots with anti-pea Ser hydroximethyltransferase antibodies, and for providing the anti-ClcP antibodies. We also thank Dr. Andre Pierard for the Saccharomyces cerevisiae strain 8989c and Dr. Micha Volokita for the anti-spinach glycolate oxidase antibodies.
Footnotes
This work was supported by the FrameWork Program of the Commission of the European Communities, by the Israel Academy of Sciences and Humanities, National Council for Research and Development, Israel (grant no. BIO4–CT97–2182), and by a Leon and Kathe Fallek scholarship (to G.T.). G.G. is an incumbent of the Bronfman Chair of Plant Sciences.
LITERATURE CITED
- Blemings KP, Crenshaw TD, Swick RW, Benevenga NJ. Lysine-α-ketoglutarate reductase and saccharopine dehydrogenase are located only in the mitochondrial matrix in rat liver. J Nutr. 1994;124:1215–1221. doi: 10.1093/jn/124.8.1215. [DOI] [PubMed] [Google Scholar]
- Bourguignon J, Neuburger M, Douce R. Resolution and characterization of the glycine-cleavage reaction in pea leaf mitochondria: properties of the forward reaction catalyzed by glycine decarboxylase and serine hydroxymethyltransferase. Biochem J. 1988;255:169–178. doi: 10.1042/bj2550169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brochetto-Braga M, Leite A, Arruda P. Partial purification and characterization of lysine-ketoglutarate reductase in normal and opaque-2 maize endosperms. Plant Physiol. 1992;98:1139–1147. doi: 10.1104/pp.98.3.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleu C, Coustaut M, Niogert M-F, Larpher F. Three new osmotic stress-regulated cDNAs identified by differential display polymerase chain reaction in rapeseed leaf discs. Plant Cell Environ. 1999;22:979–988. [Google Scholar]
- Epelbaum S, McDevitt R, Falco SC. Lysine-ketoglutarate reductase and saccharopine dehydrogenase from Arabidopsis thaliana: nucleotide sequence and characterization. Plant Mol Biol. 1997;35:735–748. doi: 10.1023/a:1005808923191. [DOI] [PubMed] [Google Scholar]
- Falco SC, Guida T, Locke M, Mauvais J, Sandres C, Ward RT, Webber P. Transgenic canola and soybean seeds with increased lysine. Biotechnol. 1995;13:577–582. doi: 10.1038/nbt0695-577. [DOI] [PubMed] [Google Scholar]
- Foster AR, Scislowski PWD, Harris CI, Fuller MF. Metabolic response of liver lysine α-ketoglutarate reductase activity in rats fed lysine limiting or lysine excessive diets. Nutr Res. 1993;13:1433–1443. [Google Scholar]
- Galili G. Regulation of lysine and threonine synthesis. Plant Cell. 1995;7:899–906. doi: 10.1105/tpc.7.7.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaziola A, Teixeira CMG, Lugli J, Sodek L, Azevedo RA. The enzymology of lysine catabolism in rice seeds: isolation, characterization, and regulatory properties of a lysine 2-oxoglutarate reductase/saccharopine dehydrogenase bifunctional polypeptide. Eur J Biochem. 1997;247:364–371. doi: 10.1111/j.1432-1033.1997.00364.x. [DOI] [PubMed] [Google Scholar]
- Goncalves-Butruille M, Szajner P, Torigoi E, Leite A, Arruda P. Purification of characterization of the bifunctional enzyme lysine-ketoglutarate reductase-saccharopine dehydrogenase from maize. Plant Physiol. 1996;110:765–771. doi: 10.1104/pp.110.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualberto JM, Handa H, Grienenberger JM. Isolation and fractionation of plant mitochondria and chloroplasts: specific examples. Methods Cell Biol. 1995;50:161–175. doi: 10.1016/s0091-679x(08)61029-8. [DOI] [PubMed] [Google Scholar]
- Halperin T, Adam Z. Degradation of mistargeted OEE33 in the chloroplast stroma. Plant Mol Biol. 1996;30:925–933. doi: 10.1007/BF00020804. [DOI] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karchi H, Miron D, Ben-Yaacov S, Galili G. The lysine-dependent stimulation of lysine catabolism in tobacco seeds requires calcium and protein phosphorylation. Plant Cell. 1995;7:1963–1970. doi: 10.1105/tpc.7.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karchi H, Shaul O, Galili G. Lysine synthesis and catabolism are coordinately regulated during tobacco seed development. Proc Natl Acad Sci USA. 1994;91:2577–2581. doi: 10.1073/pnas.91.7.2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper EL, Cord-Neto G, Capella AN, Goncalves-Butruile M, Azevedo RA, Arruda P. Structure and regulation of the bifunctional enzyme lysine-oxoglutarate reductase-saccharopine dehydrogenase in maize. Eur J Biochem. 1998;253:720–729. doi: 10.1046/j.1432-1327.1998.2530720.x. [DOI] [PubMed] [Google Scholar]
- Kemper EL, Neto GC, Papes F, Moraes KC, Leite A, Arruda P. The role of Opaque2 in the control of lysine-degrading activities in developing maize endosperm. Plant Cell. 1999;11:1981–1994. doi: 10.1105/tpc.11.10.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovitz PJ, Chuang DT. The bifunctional aminoadipic semialdehyde synthase in lysine degradation-separation of reductase and dehydrogenase domains by limited proteolysis and column chromatography. J Biol Chem. 1987;262:9353–9358. [PubMed] [Google Scholar]
- Markovitz PJ, Chuang DT, Cox RP. Familial hyperlysinemias: purification and characterization of the bifunctional aminoadipic semialdehyde synthase with lysine-ketoglutarate reductase and saccharopine dehydrogenase activities. J Biol Chem. 1984;259:11643–11646. [PubMed] [Google Scholar]
- Mazur B, Krebbers E, Tingey S. Gene discovery and product development for grain quality traits. Science. 1999;285:372–375. doi: 10.1126/science.285.5426.372. [DOI] [PubMed] [Google Scholar]
- Miron D, Ben-Yaacov S, Reches C, Schupper A, Galili G. Purification and characterization of bifunctional lysine-ketoglutarate reductase/saccharopine dehydrogenase from developing soybean seeds. Plant Physiol. 2000;123:655–663. doi: 10.1104/pp.123.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostersetzer O, Adam Z. Effects of light and temperature on expression of ClpC, the regulatory subunit of chloroplastic clp protease in pea seedlings. Plant Mol Biol. 1996;31:673–676. doi: 10.1007/BF00042238. [DOI] [PubMed] [Google Scholar]
- Rao VV, Pan X, Chang YF. Developmental changes in l-lysine-ketoglutarate reductase in rat brain and liver. Comp Biochem Physiol. 1992;103B:221–224. doi: 10.1016/0305-0491(92)90435-t. [DOI] [PubMed] [Google Scholar]
- Schaller GE, Bleecker AB. Ethylene-binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science. 1995;270:1809–1811. doi: 10.1126/science.270.5243.1809. [DOI] [PubMed] [Google Scholar]
- Shaul O, Hilgemann D, Engler J, Van Montagu M, Inze D, Galili G. Cloning and characterization of a novel Mg2+/H+ exchanger. EMBO J. 1999;18:3973–3980. doi: 10.1093/emboj/18.14.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W, Mertens JA, Kanamaru K, Campbell WH, Crawford NM. Analysis of wild-type and mutant plant nitrate reductase expressed in the methylotrophic yeast Pichia pastoris. Plant Physiol. 1997;115:1135–1143. doi: 10.1104/pp.115.3.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Miron D, Zhu-Shimoni JX, Galili G. Regulation of lysine catabolism through lysine-ketoglutarate reductase and saccharopine dehydrogenase in Arabidopsis. Plant Cell. 1997;9:1305–1316. doi: 10.1105/tpc.9.8.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Zhu X, Tang X, Galili G. A novel composite locus of Arabidopsis encoding simulatenously two polypeptides with metabolically related but distinct functions in lysine catabolism. Plant J. 2000;23:195–204. doi: 10.1046/j.1365-313x.2000.00770.x. [DOI] [PubMed] [Google Scholar]
- Vernet T, Dignard D, Thomas DY. A family of yeast expression vectors containing the phage f1 intergenic region. Gene. 1987;52:225–233. doi: 10.1016/0378-1119(87)90049-7. [DOI] [PubMed] [Google Scholar]
- Volokita M. Carboxy-terminal end of glycolate oxidase directs a foreign protein into tobacco leaf peroxisomes. Plant J. 1991;1:361–366. doi: 10.1046/j.1365-313x.1991.t01-4-00999.x. [DOI] [PubMed] [Google Scholar]