Abstract
Protein phosphorylation is the most common reversible posttranslational modification in eukaryotes. Humans have over 500 protein kinases, of which more than a dozen are established targets for anti-cancer drugs. All kinases share a structurally similar catalytic domain, yet each one is uniquely positioned within signaling networks controlling essentially all aspects of cell behavior. Kinases are distinguished from one another based on their modes of regulation and their substrate repertoires. Coupling specific inputs to the proper signaling outputs requires that kinases phosphorylate a limited number of sites to the exclusion of hundreds of thousands of off-target phosphorylation sites. Here, we review recent progress in understanding mechanisms of kinase substrate specificity and how they function to shape cellular signaling networks.
Keywords: Protein kinase, protein phosphorylation, enzyme specificity, linear sequence motif, protein interactions
Principles of protein kinase substrate specificity
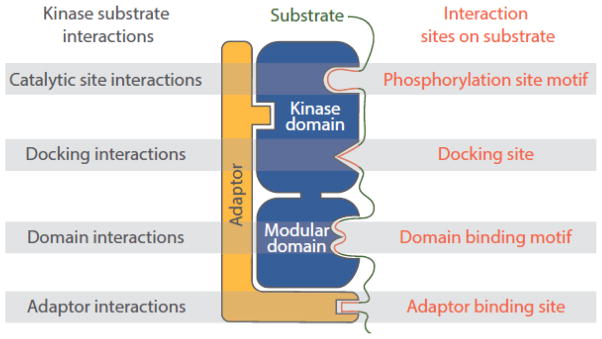
Protein kinases selectively target specific substrates through several types of physical interactions (Figure 1) [1, 2]. For example, it is self-evident that the phosphorylated amino acid residue must interact at least transiently with the active site of the kinase. Eukaryotic protein kinases are generally subdivided into tyrosine kinases (TyrKs), serine-threonine kinases (STKs) and dual-specificity kinases based on their favored substrate phosphoacceptor residues, which are determined by conserved features of the kinase active site unique to each class [3]. As with other protein-modifying enzymes, kinases have broad catalytic clefts that accommodate multiple residues flanking the site of phosphorylation, leading to specificity at the level of phosphorylation site sequence [4, 5]. However, as a rule, catalytic site interactions alone are insufficient to mediate selection of protein substrates. Kinase recognition motifs typically consist of only one to three residues that are critical for efficient phosphorylation (Table 1). As a consequence, essentially all proteins will harbor sites matching the simplest of these motifs, and there will be thousands of occurrences of more stringent motifs within a proteome. Furthermore, related kinases can have identical phosphorylation site motifs, yet mediate different cellular functions through phosphorylation of distinct substrates [6]. Phosphorylation site interactions may therefore be primarily important in determining which specific residues within a substrate protein are phosphorylated, and in cooperating with additional interactions to select substrates.
Figure 1.
Overview of the major types of kinase-substrate interactions. Kinases target their substrates through a combination of catalytic domain interactions both proximal and distal to the active site, interactions of short linear sequence motifs with protein interaction modules, and indirect interactions mediated by adaptor or scaffold proteins.
Table 1.
Protein kinase phosphorylation site motifs.
| Kinase | Consensus sequence | Refs | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −5 | −4 | −3 | −2 | −1 | 0 | +1 | +2 | +3 | +4 | |||
| Basophilic | PKA | R | − R/K | − x | − S | − ϕ | [1] | |||||
|
| ||||||||||||
| PAK | R/K | − R | − x | − S | − ϕ | [88] | ||||||
|
| ||||||||||||
| PKC | R/K | − R/K | − R/K | − G/R | − S/T | − ϕ | − R/K | − R/K | [18] | |||
|
| ||||||||||||
| PIM/S6K | R | − x | − R | − x | − x | − S/T | [53, 57, 89] | |||||
|
| ||||||||||||
| AKT/RSK | R | − x | − R | − x | − x | − S/T | − ϕ | [53, 89] | ||||
|
| ||||||||||||
| CAMK/PKD | ϕ | − x | − R | − Q | − x | − S | − ϕ | [18, 53] | ||||
|
| ||||||||||||
| AMPK/MARK | ϕ | − x | − R | − x | − x | − S | − x | − x | − x | − I/L | [60, 62] | |
|
| ||||||||||||
| Haspin | A/V | − R | − T | − K | [14] | |||||||
|
| ||||||||||||
| Pro-directed | MAPK | P/ϕ | − x | − S/T | − P | [18] | ||||||
|
| ||||||||||||
| CDK | S/T | − P | − x | − K/R | [18] | |||||||
|
| ||||||||||||
| DYRK | R | − x | − x | − S/T | − P | [90] | ||||||
|
| ||||||||||||
| Acidophilic | CK1 | D/E | − D/E | − D/E | − x | − x | − S/T | − ϕ | [18] | |||
| pS/pT | − x | − x | − S/T | − ϕ | ||||||||
|
| ||||||||||||
| CK2 | S/T | − D/E | − x | − D/E | [18, 35] | |||||||
| S/T | − D/E | − x | − pS | |||||||||
|
| ||||||||||||
| GSK3β | S | − x | − x | − x | − pS | [18] | ||||||
|
| ||||||||||||
| PLK | D/E | − x | − S/T | − ϕ | [9] | |||||||
|
| ||||||||||||
| FAM20C | S | − x | − E/pS | [91] | ||||||||
|
| ||||||||||||
| Other | ULK | L/M | − x | − x | − S/T | − ϕ | − ϕ | [92] | ||||
|
| ||||||||||||
| NEK | L/F | − x | − x | − S/T | − ϕ | [9, 93] | ||||||
|
| ||||||||||||
| LKB1 | L/I | − x | − T | [57] | ||||||||
|
| ||||||||||||
| ATM/ATR/DNAPK | S/T | − Q | [94] | |||||||||
|
| ||||||||||||
| Tyrosine | EGFR | E/D | − E/D | − E/D | − Y | − pY/F | − x | − ϕ | [43] | |||
|
| ||||||||||||
| SRC | E | − E | − I/V | − Y | − E/G | − x | − ϕ | [1] | ||||
|
| ||||||||||||
| ZAP70 | D/E | − Y | − E/ϕ | − x | − ϕ | [10] | ||||||
|
| ||||||||||||
| ABL | I | Y | − A | − x | − P | [1] | ||||||
Preferences for acidic (colored red, pS = phosphoserine; pT = phosphothreonine; pY = phosphotyrosine), basic (colored blue), hydrophobic (represented by ϕ), and proline residues are indicated. The underlined residue is the phosphoacceptor.
Substrate affinity and specificity can be enhanced by docking interactions, in which regions distal to the site of phosphorylation bind to grooves, pockets or surfaces outside of the kinase catalytic cleft [1, 2, 5, 6]. Like catalytic site interactions, docking interactions can involve recognition of short linear sequence motifs, for example through peptide-binding modules such as SH2 and SH3 domains. Docking interactions in some cases mediate processivity, facilitating phosphorylation of multiple distinct sites on a substrate. Kinases can also be recruited to their substrates through indirect interactions mediated by adaptor and scaffold proteins. Adaptor proteins that bind to both kinase and substrate promote phosphorylation through induced proximity. In addition, adaptor proteins can direct kinases to their substrates by controlling their subcellular localization independently of direct interactions with substrates. Scaffolds, which form stable complexes with multiple proteins, often serve as hubs for kinase regulation [2]. Kinases are frequently organized into cascades, and in these cases scaffolds can be important for channeling upstream kinases to activate specific downstream kinases. Scaffolds can also promote substrate specificity by localizing an active pool of the kinase in proximity to its substrates and, in some cases, causing conformational changes in substrates that promote phosphorylation [2].
The mechanisms of substrate targeting illustrated above are not mutually exclusive, and authentic kinase-substrate pairs likely require multiple interactions to achieve efficient phosphorylation in vivo. Recent insight into the biochemical and structural principles underlying these mechanisms has provided a more complete picture of how kinases interact with their substrates, moving beyond classical concepts involving recognition of simple consensus sequences. This detailed understanding has in turn influenced models of how kinases function within complex biological systems.
Recognition of phosphorylation site sequence motifs
Early biochemical studies suggested that protein kinases phosphorylated substrates in the context of specific sequence motifs. In co-crystal structures, peptide substrates generally bind to the kinase in an extended conformation (Figure 2A). The peptide makes β-sheet-like hydrogen bonding interactions with a portion of the kinase activation loop, a conformationally flexible region important for regulation. Residues within the catalytic cleft define its shape and biophysical characteristics [4, 5] to determine phosphorylation site specificity, which varies substantially among kinases. Examples of both classical and more recently established kinase consensus sequences are provided in Table 1.
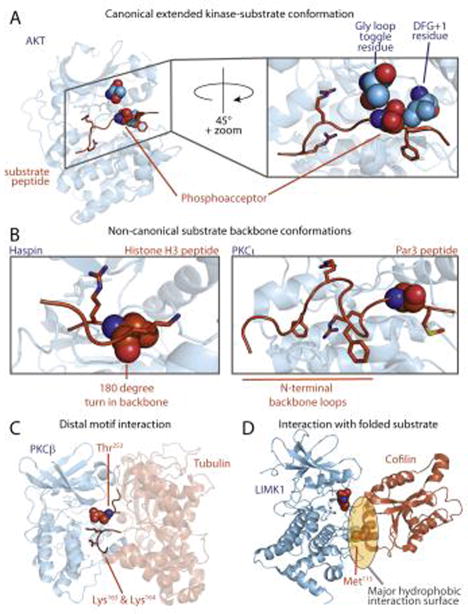
Figure 2.
Canonical and non-canonical kinase-substrate interactions. (A) Canonical peptide binding mode (PDB:1O6K). Substrate peptide binds in an extended conformation, with amino acid side-chains accessing distinct pockets. Kinase residues that determine phosphoacceptor residue specificity are shown in space fill, including the phosphorylatable residue in the Gly loop [8] (green) and the DFG+1 residue [7] in the activation loop (purple). (B) Atypical interactions between the kinase catalytic cleft and peptide substrates. The Haspin-histone H3 peptide complex, in which the peptide backbone makes a 180° turn near the phosphoacceptor, is shown at left (PDB: 4OUC). In the PKCι-Par3 peptide complex shown at right (PDB: 4DC2) the peptide backbone makes two β-turns N-terminal to the phosphoacceptor residue. (C) A model for PKCβ bound to tubulin, in which basic residues in the substrate distal in primary sequence are close to the phosphoacceptor residue in the 3D structure. (D) LIMK interacts with cofilin through a hydrophobic interaction centered on the αG helix of the kinase, in which placement of the phosphoacceptor requires a structured substrate (PDB: 5HVK). The Met115 residue on cofilin is critical for LIMK recognition.
Recent structural and biochemical studies have provided new insight into phosphorylation site specificity, including how kinases recognize the phosphoacceptor residue itself (Figure 2A). For example, it has been long appreciated that some STKs have substantial preference for either Ser or Thr as the phosphoacceptor residue. Chen et al. identified the residue immediately downstream of the conserved DFG motif found in the kinase activation loop (DFG+1) as a major determinant of Ser-Thr phosphoacceptor specificity [7]. Large hydrophobic DFG+1 residues promote Ser phosphorylation, whereas smaller β-branched residues confer specificity for Thr. Phosphoacceptor identity did not appear to affect substrate binding to the kinase. Rather, in kinase-peptide co-crystal structures, the DFG+1 residue appeared to position the phosphoacceptor residue in either a productive or non-productive conformation for catalysis. Intriguingly, a separate study found that phosphorylation of protein kinase C (PKC)-δ could alter its Ser-Thr phosphoacceptor specificity. Phosphorylation of the kinase at a site (Ser359) within the Gly-rich loop, a conserved region proximal to the ATP binding site [8] (Figure 2A), conferred a strong preference for a Ser phosphoacceptor, while the dephosphorylated form was non-selective. As a phosphorylatable residue is present at the analogous position in ~30% of human kinases, phosphorylation at this site may be a common mechanism for dynamic regulation of kinase phosphoacceptor specificity.
While phosphorylation site motifs are typically described in terms of residues that promote phosphorylation, negatively selected residues can also be an important component of substrate recognition. Such “forbidden” resides can act as a filter to prevent phosphorylation of a site by the “wrong” kinase, which can help establish the correct order and timing of phosphorylation events [9]. This concept has been illustrated recently in a study using a novel method in which genetically encoded peptide libraries are displayed on the surface of bacteria. Peptide-expressing bacteria were treated with a TyrK, and bacteria harboring phosphorylated substrates were labeled with a fluorophore-coupled anti-phosphotyrosine (pTyr) antibody for isolation by fluorescence-activated cell sorting (FACS). This method revealed that the TyrK ZAP70, which plays a key role in transducing signals from the T cell receptor, preferred acidic residues at multiple positions and had little tolerance for basic residues at any position [10]. This “electrostatic filter” both insulates ZAP70 substrates from other kinases and prevents the kinase from activating itself through autophosphorylation. Similar analysis of the upstream TyrK LCK explained its ability to phosphorylate itself as well as ZAP70. Thus, features of substrate selectivity enforce ordered phosphorylation of kinases and the compartmentalization of substrates in the T-cell activation cascade.
Roles for secondary and tertiary structure in kinase substrates
Known sites of phosphorylation on substrates are enriched in unstructured regions found outside of defined protein domains, which are likely to be more accessible to interact with kinases in the canonical, extended conformation (Figure 2A) [11–13]. Recently several kinase substrates have been observed to bind in alternative conformations, suggesting that kinases may in some cases recognize elements of secondary structure. One recent example involves Haspin, an atypical kinase lacking recognizable sequence similarity to most eukaryotic protein kinases. Kettenbach et al. analyzed the substrate specificity of Haspin using a novel type of peptide library comprising a dephosphorylated proteolytic digest of HeLa cell extract [14]. Following treatment of the peptide mixture with a kinase, phosphopeptides were purified and identified using high throughput tandem mass spectrometry (LC-MS/MS). This analysis found Haspin to preferentially phosphorylate sites near peptide N termini and defined a stringent sequence motif (Table 1) conforming to the sole known Haspin site, Thr3 of histone H3. Subsequent X-ray crystallography studies revealed that Haspin adopts a canonical bi-lobed kinase fold, but its activation loop assumes a distinct, largely helical conformation [15]. This conformation would preclude a substrate from adopting the typical extended binding mode. In a co-crystal structure, a bound histone H3 peptide made a sharp turn downstream of the phosphoacceptor to project outward toward solvent (Figure 2B). This arrangement allows for close contact between the sidechains of three residues flanking the phosphoacceptor and specific pockets in the kinase, explaining its unusually stringent sequence specificity and its preference for N-terminal sequences. Interestingly, these flanking residues, Arg2 and Lys4, are hotspots for acetylation and methylation. Modification of these residues would likely abolish phosphorylation by Haspin, rendering its activity responsive to epigenetic signals.
Several recent studies of PKC isozymes have uncovered novel modes of substrate interaction in which residues within the substrate are selected based on their arrangement within a folded structure rather than their position within a linear sequence. Unlike most “basophilic” kinases that have strict positional selectivity, PKCs prefer basic residues at multiple positions both upstream and downstream of the phosphorylation site (Table 1). While basic residues C-terminal to the phosphorylation site appear to promote catalytic efficiency, possibly by helping to position the γ-phosphate of ATP, N-terminal basic residues contribute to substrate binding [16]. An X-ray co-crystal structure of PKCι with a fragment of its substrate Par3 has revealed a unique mode of interaction with residues located upstream of the phosphorylation site [17] (Figure 2B). A hydrophobic pocket unique to PKCι (and its closest relative PKCζ) is created by an insertion within the kinase C-terminal lobe. This pocket anchors a Phe residue at the −5 position in Par3, promoting an unusual conformation involving two β-turns in the substrate backbone. This conformation allows a basic residue positioned far upstream of the phosphorylation site to engage an acidic pocket on the kinase that typically binds to more proximal residues. In this case, a sequence that is presumably disordered in the absence of the kinase adopts a specific conformation upon binding. A similar phenomenon may explain earlier observations that more distally positioned residues can be essential for phosphorylation [18]. An alternative model, in which the kinase recognizes a substrate within the context of pre-formed secondary structure, has been proposed for the interaction between PKCβ and α-tubulin [19] (Figure 2C). In this model, basic residues typically found within the PKC consensus motif are instead quite distal to the phosphorylation site (~90 residues upstream) in the primary sequence. However, in the folded α-tubulin structure, these residues are located proximal to the phosphosite (Figure 2C). This type of “structural consensus” may explain other instances where a substrate phosphorylation site does not conform to the simple linear sequence motif of the kinase (see Outstanding Questions box).
Substrate interactions outside of the catalytic cleft
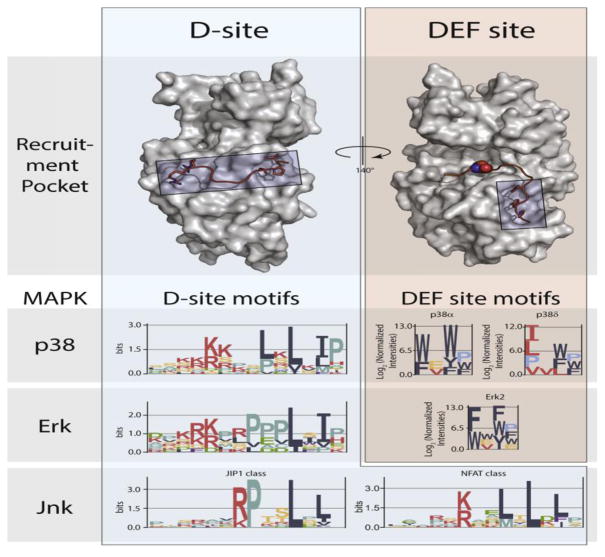
As for other types of protein-protein interactions, kinase-substrate docking interactions can occur through large binding interfaces or through recognition of short linear sequence motifs. For kinases with a large number of substrates, the use of short motifs for substrate targeting makes sense from an evolutionary standpoint, as substrates can be lost or gained through a small number of point mutations (see Box 1 – Evolution of kinase-substrate interactions). Recent studies of mitogen-activated protein kinases (MAPKs) have provided insight into how linear docking motifs can mediate selective targeting by closely related members of a kinase family. MAPKs share a common phosphorylation site consensus (S/T-P), but members of the different subfamilies (ERK, p38 and JNK) target a largely distinct set of substrates [6]. Two different regions of MAPK catalytic domains are known to interact with substrate docking motifs (Figure 3). DEF (docking site for ERK, FxF) motifs (also called F-sites) are generally Ω-x-Ω sequences (where Ω is an aromatic residue) that engage a small hydrophobic pocket proximal to the catalytic cleft of ERK and p38 family MAPKs [20, 21]. Distinct DEF site sequence preferences among p38 isoforms have been rationalized based primarily on the size of this pocket. D-site motifs consist of a basic patch separated by a short linker from a ϕ-x-ϕ sequence (where ϕ is a hydrophobic residue [5]). The D-site motif, which is found in both substrates and regulators of MAPKs, interacts with a groove within the kinase C-lobe that is located on the opposite face from the active site (Figure 3). Because this groove is structurally similar among the various MAPKs, how it might bind to specific sequences to mediate selective targeting has remained obscure. The Reményi group recently reported a series of X-ray crystal structures of MAPKs in complex with D-site peptides. Comparison with previously reported structures revealed that variations in the linker sequence gave rise to distinct conformations that promoted selective targeting of JNK vs. the p38 and ERK MAPKs [22]. In contrast, the Bardwell group reported that the identity of the hydrophobic residues drove selective interactions with JNK MAPK [23]. By leveraging unique structural or sequence features of the D-site, both groups have been able to computationally predict and verify new MAPK substrates [24, 25]. Intriguingly, a number of the predicted and established JNK D-sites overlap with a docking site for the phosphatase calcineurin, suggesting a mechanism for coordinate regulation of substrate phosphorylation and dephosphorylation through a common motif [26]. Other recent work has indicated that in some cases MAPK regulators, including the ERK5 activator MEK5 [27] and the p38 phosphatase HePTP [28], make additional contacts with the catalytic domain outside of the canonical D-site interaction groove. These additional interactions serve to enhance binding affinity and likely mediate precise targeting of specific MAPKs to prevent potentially deleterious crosstalk between pathways.
Box 1. Evolution of kinase-substrate interactions.
Changes in phosphorylation networks have been proposed as a source of biological diversity, acting as a driving force for evolution [77]. Phosphorylation sites can be gained or lost through mutation of both kinases and substrate proteins. The small fraction (2–6%) of phosphorylation sites highly conserved across species are construed to be ancient (>500 million years old) and likely functional [11, 13, 78]. However, as phosphoacceptor residues and kinase recognition motifs can be gained and lost through a small number of point mutations, phosphorylation sites tend to evolve rapidly. Indeed, the use of short linear motifs for both phosphorylation site and docking interactions provides a straightforward mechanism for rapidly expanding the substrate repertoire for kinases that phosphorylate numerous substrates. One general mechanism for biological diversification is through gene duplication followed by functional specification of the resulting paralogs. Both gain or loss of phosphorylation sites, or changes to sequence motifs surrounding conserved sites, can place homologous proteins under control of distinct signaling pathways [61]. Following the evolutionary history of substrates of kinases with known docking motifs has suggested that phosphorylation sites are generally introduced early, with docking sites appearing later [25, 47]. In some cases, late introduction of docking motifs suggests that they are not absolutely required for phosphorylation, but confer robustness to perturbation by enhancing substrate quality (see “Some substrates are better than others” in the main text) [47]. In other cases, for example with MAPKs, late-evolving docking motifs are critical for selective substrate targeting by distinct members of the family (Figure 3) [22, 25].
While kinase substrates tend to evolve rapidly, changes to the kinases themselves, which have the capacity to dramatically perturb phosphorylation networks, tend to occur on a slower time scale. Gain or loss of specific kinase families along various yeast lineages, for example, leads to increased phosphorylation of sites corresponding their cognate phosphorylation site motifs [13]. The diversity of sequence motifs targeted by extant protein kinases of similar structure and primary sequence indicates that kinase specificity must also change during evolution, likely following duplication events. Ancestral resurrection of “extinct” protein kinases suggested that extant kinases with stringent phosphorylation site specificity can arise from more promiscuous precursors [79]. Furthermore, substantial changes to phosphorylation site specificity can occur with a single amino acid substitution [7, 79]. Attempts to rationally re-engineer kinase specificity, however, suggest that the introduction of multiple point mutations effects more substantial changes in specificity, and that mutations within the catalytic cleft can result in large decreases in catalytic activity [80]. These observations suggest that kinases evolve new specificity through accumulation of multiple mutations, likely including mutations outside the catalytic cleft that cooperate with specificity determining residues in maintaining enzyme activity [81]. The evolution of kinases with new specificity could be deleterious through the generation of large numbers of new phosphorylation sites. One mechanism for adaptation to this type of perturbation is the evolution of the substrate pool. For example, the introduction of TyrKs in the metazoan lineage was associated with a significant decrease in the occurrence of tyrosine residues within the proteome [82]. In the case of the yeast meiotic kinase Ime2, however, a mutant with distinct specificity lost biological activity, yet was not toxic when present alongside the wild-type kinase [79]. These observations suggest that organisms can tolerate substantial changes to the phosphoproteome or alternatively that additional substrate targeting mechanisms must evolve to promote phosphorylation of new substrates.
Once evolved, kinase phosphorylation site specificity can be remarkably conserved across distant lineages. Human kinases, for example, almost invariably have identical specificity as their closest budding yeast homologs [59]. Indeed, budding yeast kinase mutants can frequently be complemented by their mammalian counterparts [83], suggesting that they have maintained the capacity to target critical substrates despite their vast evolutionary distance.
The evolutionary plasticity of kinase signaling networks may also be relevant to human tumors, an evolving system in which specific mutations confer fitness in the context of competitive growth and survival. Several recent studies predict that kinase signaling networks are frequently rewired in cancer cells through a variety of mechanisms. An analysis of point mutants in The Cancer Genome Atlas predicted that almost 20% of mutations within seven residues of a phosphorylation site may have an effect on kinase targeting [84], in some cases to change the likely phosphorylating kinase. Cancer-associated mutations in linear motifs can also rewire signaling networks by introducing or eliminating a kinase phosphorylation site. Interestingly, phosphorylation site mutations are enriched in known oncogenes and tumor suppressors, suggesting that at least some are likely to be functionally relevant. An algorithm designed to predict determinants of kinase specificity [81] suggested that >10% of kinase mutations map to putative specificity-determining residues, potentially changing the substrate repertoire [85]. There is experimental support that cancer-associated mutations indeed change the phosphorylation site specificity of several kinases, including PKD1, PKC-γ, and PKA [85, 86], and mutations in substrate targeting SH2 and SH3 domains of PTK6 can change its substrate repertoire [87]. Taken together, these studies suggest that evolution of kinase-substrate networks may be an important factor in cancer pathology.
Figure 3.
MAPK docking interactions. Substrate D-site motifs (left) bind to a groove located on the opposite face of the kinase from the catalytic cleft. D-sites contain a cluster of basic residues upstream of two or three hydrophobic residues, the spacing of which can select for distinct MAPKs. ERK and p38 MAPKs can also bind DEF-sites (right) through a pocket adjacent to the active site, with different isozymes targeting distinct motifs. Logos for D-sites and DEF-sites were generated from published data [20, 25], using EnoLogos. ERK2 structures (top) were based on the co-crystal structure with the HePTP D-site peptide (PDB: 2FYS) and an HDX-MS model of a bound DEF-site peptide [21].
In contrast to their less selective counterparts, kinases that phosphorylate only a small number of proteins tend to have larger interaction surfaces that confer stringent specificity [29, 30]. A recently studied example is the interaction between LIM kinase (LIMK) and its major substrate, the actin depolymerizing factor cofilin. LIMK phosphorylates cofilin family proteins at a single site near their N termini (Ser2). The recently solved structure of the LIMK1 catalytic domain in complex with cofilin revealed an extensive binding interface involving a region of the kinase C-terminal lobe adjacent to the catalytic cleft [31] (Figure 2D). This region includes a conserved structural feature of the kinase (helix αG) that has previously been observed to engage in kinase docking interactions [29]. The docking interaction appears to guide the phosphorylation site residue into the catalytic center without making canonical main chain interactions with the activation loop [31]. Instead, the N-terminal region binds in a manner perpendicular to the typical substrate binding mode. Presumably the additional binding energy provided by the interaction surface can compensate for the loss of main chain hydrogen bonds typically thought to be essential for substrate binding.
G protein-coupled receptor kinases (GRKs) are another group of kinases whose limited substrate repertoire is associated with an extensive binding interface. GRKs phosphorylate activated GPCRs at multiple sites, promoting binding of arrestin proteins to mediate receptor desensitization and G protein-independent signaling [32]. Both localization of GRKs to the plasma membrane as well as direct physical interactions with the GPCR contribute to specific targeting. In part because they form transient, state-dependent, low affinity complexes, structural insight into the nature of GRK-GPCR interactions has been limited. Recent studies using chemical crosslinking and hydrogen-deuterium exchange mass spectrometry have mapped points of interaction between GRK5 and its substrate β2-adrenergic receptor (B2AR) [33]. These studies suggest a structural model in which the GRK catalytic domain and its associated regulator of G-protein signaling homology (RH) domain directly contact multiple intracellular loops of the GPCR in addition to the C-terminal tail that harbors sites of phosphorylation. In addition to promoting recruitment of the kinase to its substrate, interaction with a GPCR also increases GRK catalytic activity. Prior crystallographic analyses of GRKs have suggested that the RH domain holds the catalytic domain in an open, inactive conformation [34]. Interaction with the B2AR appears to promote an elongated conformation in GRK5 in which specific autoinhibitory interactions between the RH and catalytic domains are broken. Activation of the kinase only upon direct binding to its substrate provides a mechanism for enforcing tight control of substrate specificity.
Multisite phosphorylation: signal integration, amplification and attenuation
A large majority of phosphoproteins are phosphorylated at multiple sites, often by distinct protein kinases. While multiple kinases can phosphorylate a substrate independently of each other, multisite phosphorylation can occur in a hierarchical manner, in which prior phosphorylation generates a recognition motif for a kinase to phosphorylate other sites [35]. This phenomenon of substrate “priming” is a common mechanism for signal integration and amplification in eukaryotic signaling pathways [36]. Recent studies have expanded the scope of priming-dependent phosphorylation and provided insight into its structural basis. Classically, substrate priming occurs close to the site of subsequent phosphorylation. For some kinases, such as casein kinase 2, phosphorylated residues can substitute for negatively charged Asp or Glu residues found at multiple positions near its phosphorylation sites [18, 37] (Table 1). By contrast, glycogen synthase kinase 3β (GSK3β) is an obligate priming-dependent kinase with strict positional selectivity, requiring a phosphoserine (pSer) residue at the +4 position in its substrates (Table 1). Prior biochemical studies had suggested that the priming phosphate group binds GSK3β at a site analogous to that occupied by a phosphorylated residue within the activation loop of other kinases, leading to allosteric activation of the kinase [38]. This model has been confirmed through a series of X-ray co-crystal structures of GSK3 bound to peptides harboring pSer, which offered additional details of how priming phosphorylation promotes substrate binding [39].
While generally associated with STKs, in some cases TyrKs can also recognize primed substrates through catalytic site interactions [40, 41]. Recent studies have investigated hierarchical phosphorylation by the TyrKs BMX [42] and epidermal growth factor receptor (EGFR) [43]. EGFR has a preference for a pTyr residue at the +1 position, mediating signal integration with the non-receptor TyrK SRC through dual phosphorylation of the SHC adapter protein. Co-crystal structures with EGFR revealed that primed substrates have a binding mode distinct from unprimed substrates: the +1 pTyr residue interacts with a Lys residue in the catalytic cleft that interacts with residues upstream of the phosphorylation site in non-primed substrates. These observations suggest the overall EGFR recognition motif may differ between primed and unprimed substrates, or that substrate priming may override the necessity for a canonical consensus sequence. As the analogous catalytic cleft residue is either Lys or Arg in all human receptor TyrKs, priming-dependent phosphorylation may prove to be a general phenomenon among kinases in this group.
Priming phosphorylation can also generate a binding site for some protein-interaction domains and adaptor proteins that recruit kinases to phosphorylate more distal sites [36]. Well characterized phosphoprotein-interacting domains include pTyr-binding SH2 domains present in many non-receptor TyrKs and the pSer/pThr-interacting polobox domain found in polo-like kinases. These domains typically bind to phosphoproteins in the context of linear motifs unrelated to known kinase phosphorylation motifs. As a consequence, many different kinases can theoretically generate an interaction site for a given phosphobinding domain, allowing for flexibility in crosstalk between pathways. In contrast, two adaptor proteins were recently found to interact with phosphopeptide sequences similar to known kinase phosphorylation site motifs. One such protein, the cyclin-dependent kinase (CDK) adaptor Cks1, binds phosphopeptides within a sequence context (ϕ-x-pT-P) that overlaps the CDK phosphorylation site consensus (S/T-P-x-K/R) [44]. Cks1 can thereby act as a processivity factor, allowing a single CDK phosphorylation site to prime for subsequent phosphorylation at multiple sites on the same substrate [45]. Another example involves MOB1 adapter proteins, which are obligate activators of LATS family kinases that act in the tumor suppressive Hippo pathway [46, 47]. Interestingly, the phosphopeptide binding specificity of MOB1 conforms to the phosphorylation site consensus of MST1 and 2, kinases which activate LATS through direct phosphorylation [48]. In this case, MST1/2 autophosphorylation generates MOB1 binding sites, recruiting LATS to its upstream kinase to facilitate its activation [49]. In this way, a limited level of MST1/2 activity would be sufficient to activate the Hippo pathway without off-target phosphorylation of other proteins.
Multisite phosphorylation often arises from a single kinase acting on multiple sites within a substrate in a non-processive manner [35]. In some cases, sites are phosphorylated sequentially, with the order dictated by the relative catalytic efficiency of the kinase for each site [50]. When this occurs, high efficiency sites that are phosphorylated first can act as decoys to inhibit phosphorylation of low efficiency sites. This phenomenon has been argued to be a source of switch-like responses to graded stimuli at saturating levels of substrate [51]. A recent study used time-resolved NMR analysis to investigate the dynamics of multisite phosphorylation of the transcription factor Elk-1 by ERK MAPK [52]. Competition between two ERK docking motifs promoted different rates of phosphorylation at eight sites within the Elk-1 activation domain. Intriguingly, while rapidly phosphorylated sites promoted transcriptional activation by Elk-1, sites phosphorylated more slowly led to inactivation. These results demonstrate that multisite phosphorylation dynamics can provide a mechanism for signal attenuation in the absence of counteracting phosphatases, which may facilitate differential timing of the various signaling outputs of a kinase.
Some substrates are better than others: consequences of differential substrate quality
Classically protein kinases were viewed as having stringent consensus sequences that dictated, in a binary manner, whether or not a specific site would be a substrate [18]. In a number of cases, new substrates for kinases have been discovered based on the presence of such consensus sequences, but these efforts are generally challenging due to motif degeneracy and redundancy. The presence of both a phosphorylation site and docking site sequence motif appears to be predictive of true substrates of some kinases [47]. Substrate prediction can also be improved by considering the contribution of residues other than those strictly required for phosphorylation. Peptide library approaches [14, 53–56] have facilitated comprehensive quantitative analysis of substrate specificity, in which the impact on phosphorylation rate of each of the 20 amino acids can be assessed for multiple positions within the peptide. Computational tools can use such quantified data to identify candidate substrates predicted to be phosphorylated most efficiently by the kinase [57, 58]. This approach can take advantage of subtle differences between related kinases to identify their unique substrates [59–63]. As substrates harboring suboptimal sites (discussed below) will escape detection, other approaches such as chemical genetic tagging [64] and time-resolved phosphoproteomics analysis [65, 66] have emerged to globally identify direct kinase substrates.
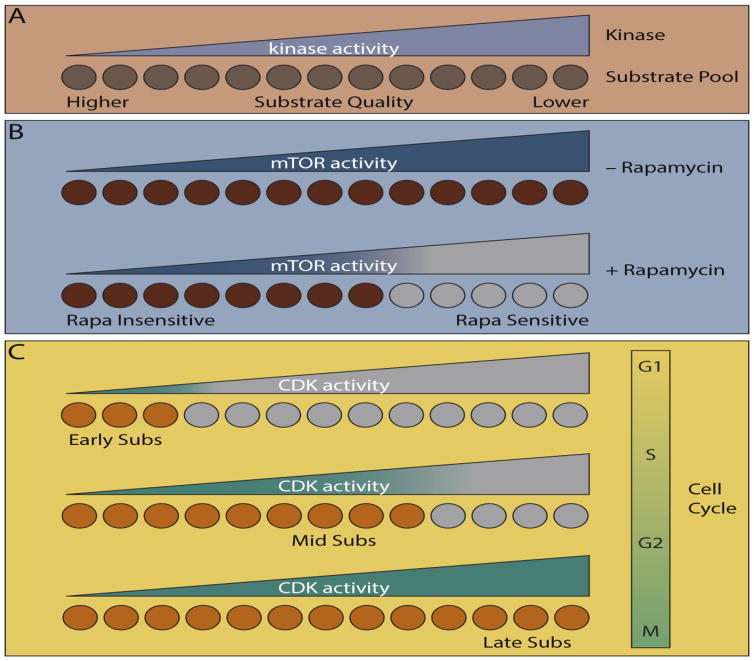
A more quantitative view of kinase specificity suggests a continuum of phosphorylation rates for the various substrates of a particular kinase (Figure 4A). Such differences in “substrate quality” can arise from variations in phosphorylation site or docking sequences and may explain why the timing or sensitivity to perturbation can vary among substrates of the same kinase. This concept was illustrated recently for substrates of the mechanistic target of rapamycin (mTOR). mTOR partitions into two distinct complexes, mTORC1 and mTORC2, which despite sharing a catalytic subunit appear to have no common substrates. Strict substrate selection is enforced by direct interactions with unique adaptor protein components of the two complexes (raptor and mSin1, respectively) [67, 68]. Among these substrates, the phosphorylation site sequence appears to dictate the phosphorylation rate [69]. Interestingly, “low quality” mTORC1 phosphorylation sites were found to be sensitive to the inhibitor rapamycin (Figure 4B). By contrast, “high quality” sites were resistant to the drug, presumably because they require only a low level of kinase activity to become fully phosphorylated. These observations rationalized a long-standing mystery as to why rapamycin only blocks phosphorylation of a subset of mTORC1 substrates. Substrate quality also dictated the sensitivity of sites to nutrient withdrawal, indicating that the substrate repertoire of a kinase can be controlled by the strength of its activating signal. Such a mechanism could explain how differential cellular responses are achieved by various levels of activation of a master kinase.
Figure 4.
Substrate quality in biological context. (A) Higher quality substrates (i.e. those that are more efficiently phosphorylated by a kinase) are phosphorylated rapidly and to higher stoichiometry even at lower kinase activity. (B) Varying quality of mTOR substrates rationalizes their differential sensitivity to the mTORC1 inhibitor rapamycin. (C) Increasing CDK activity during the cell cycle contributes to the timing of substrate phosphorylation, with high quality substrates phosphorylated early (in G1/S phase) and lower quality substrates phosphorylated late (in G2/M phase).
Similar concepts may explain the timing of CDK substrate phosphorylation during the eukaryotic cell division cycle, which is important for the proper ordering of DNA replication and mitosis. Oscillating levels of various cyclin proteins generate a series of temporally distinct CDK-cyclin complexes, each responsible for phosphorylating key substrates at various phases of the cell cycle [70]. Classically cyclins act as both CDK activators and substrate adaptors, potentially explaining why different proteins are phosphorylated at different points within the cell cycle. Indeed, some cyclins directly interact with substrates through distal docking motifs and even residues close to the phosphorylation site [71–73]. Overall CDK activity rises as cells transit the cell cycle due to degradation of CDK inhibitor proteins as well as the intrinsically higher catalytic activity of late phase cyclin-CDK complexes (Figure 4C). Interestingly, the best characterized CDK substrate docking motifs are targeted by early (G1/S) phase cyclins [73]. Studies in the yeasts S. cerevisiae and S. pombe suggest that the presence of these motifs on G1/S substrates compensates for the limited CDK activity in early phases of the cell cycle, allowing for efficient phosphorylation [74, 75]. By contrast, “late” (G2/M) CDK substrates lack these motifs, are less efficient substrates, and require the higher levels of CDK activity found late in the cell cycle. Accordingly, mutation of substrate docking sites delayed phosphorylation of early substrates [74]. As seen for “high quality” mTOR substrates, early substrates were also less sensitive than late ones to CDK inhibition in S. pombe [74]. Strikingly, selectively blocking phosphorylation of late substrates by partly inhibiting CDK activity in G2 phase led to reordering of the cell cycle, supporting a model where cell cycle progression is driven by increasing CDK activity (Figure 4C). While the timing of CDK substrate phosphorylation appears to be mediated by the presence of docking sites rather than phosphorylation site quality, in S. cerevisiae early sites tend to be enriched for Ser residues. The modest preference of the yeast CDK Cdc28 may contribute to this phenomenon, but it appears to be largely driven by the high activity of the pThr-specific phosphatase Cdc55 in both interphase and mitosis [76]. In this case low CDK activity in interphase is insufficient to balance Cdc55 activity, leading to selective accumulation of phosphorylation specifically at Ser residues on early substrates.
Concluding Remarks and Future Perspectives
Our understanding of substrate targeting mechanisms utilized by kinases has grown tremendously in the past few years, but challenges still remain in relating these mechanisms to the organization of complex signaling networks. Substrate prediction based on linear kinase recognition motifs remains difficult, especially considering that multiple substrate binding modes are possible, and that indirect kinase-substrate interactions may override direct interactions in some contexts. Predicting “low quality”, yet authentic kinase substrates is a particular challenge, as both the activity of the kinase and the differential effects of levels of phosphorylation have various biological consequences. New approaches, such as NMR spectroscopy [28] and molecular dynamics simulations [10], which can address how kinase dynamics influence substrate recognition, may be useful in providing a deeper understanding of determinants of substrate quality for a given kinase. In this way, basic biochemical principles will facilitate the development of a systems-level view of protein kinases and their associated signaling networks.
Highlights.
Knowledge of kinase phosphorylation site motifs has expanded to include non-canonical binding modes and the targeting of folded, structural motifs.
Additional kinase-substrate interactions outside the catalytic cleft, such as MAPK docking sites and GPCR kinase recruitment sites, have recently been elaborated.
Understanding differences in substrate quality provides insights into the drug sensitivity and timing of phosphorylation events.
Basic principles underlying the evolution of phosphorylation networks shed light on understanding how mutations in kinases and their substrates perturb signaling networks to promote disease.
To what extent do kinase motifs show positional interdependence? That is, do preferences for specific amino acid residues at certain positions depend on the presence of particular residues at other positions? What is the structural basis for such context-dependent selectivity?
Can kinases bind substrates in both canonical and non-canonical modes? What elements of the kinase-substrate interactions determine the mode of binding?
Can kinase substrates be “primed” by post-translational modifications other than phosphorylation, such as arginine or lysine methylation? Similarly, how generally does post-translational modification of kinases change their substrate specificity?
How common are “structural consensus” motifs in substrates? Which kinases are capable of recognizing them?
Do new kinase-substrate pairs arising from cancer-associated mutations contribute to disease?
Acknowledgments
This work was supported by NIH grants R01 GM104047 and R01 GM102262. We thank Deborah Schechtman and Paulo Oliveira for sharing coordinates of the PKC-tubulin complex model, and Natalie Ahn for sharing coordinates of the ERK2-DEF substrate interaction model.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ubersax JA, Ferrell JE., Jr Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol. 2007;8:530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- 2.Good MC, et al. Scaffold proteins: hubs for controlling the flow of cellular information. Science. 2011;332:680–686. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manning G, et al. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 4.Bose R, et al. Protein tyrosine kinase–substrate interactions. Curr Opin Struct Biol. 2006;16:668–675. doi: 10.1016/j.sbi.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 5.Goldsmith EJ, et al. Substrate and docking interactions in serine/threonine protein kinases. Chem Rev. 2007;107:5065–5081. doi: 10.1021/cr068221w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bardwell L. Mechanisms of MAPK signalling specificity. Biochem Soc Trans. 2006;34:837–841. doi: 10.1042/BST0340837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen C, et al. Identification of a major determinant for serine-threonine kinase phosphoacceptor specificity. Mol Cell. 2014;53:140–147. doi: 10.1016/j.molcel.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gong J, et al. The C2 Domain and Altered ATP-Binding loop phosphorylation at Ser(359) mediate the redox-dependent increase in protein kinase C-δ activity. Mol Cell Biol. 2015;35:1727–1740. doi: 10.1128/MCB.01436-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alexander J, et al. Spatial exclusivity combined with positive and negative selection of phosphorylation motifs is the basis for context-dependent mitotic signaling. Sci Signal. 2011;4:ra42. doi: 10.1126/scisignal.2001796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shah NH, et al. An electrostatic selection mechanism controls sequential kinase signaling downstream of the T cell receptor. eLife. 2016;5:e20105. doi: 10.7554/eLife.20105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holt LJ, et al. Global Analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landry CR, et al. Weak functional constraints on phosphoproteomes. Trends Genet. 2009;25:193–197. doi: 10.1016/j.tig.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Studer RA, et al. Evolution of protein phosphorylation across 18 fungal species. Science. 2016;354:229–232. doi: 10.1126/science.aaf2144. [DOI] [PubMed] [Google Scholar]
- 14.Kettenbach AN, et al. Rapid determination of multiple linear kinase substrate motifs by mass spectrometry. Chem Biol. 2012;19:608–618. doi: 10.1016/j.chembiol.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maiolica A, et al. Modulation of the chromatin phosphoproteome by the Haspin protein kinase. Mol Cell Proteomics. 2014;13:1724–1740. doi: 10.1074/mcp.M113.034819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee S, et al. Distinct structural mechanisms determine substrate affinity and kinase activity of protein kinase Cα. J Biol Chem. 2017;292:16300–16309. doi: 10.1074/jbc.M117.804781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C, et al. Substrate recognition mechanism of atypical protein kinase Cs revealed by the structure of PKCι in complex with a substrate peptide from Par-3. Structure. 2012;20:791–801. doi: 10.1016/j.str.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 18.Pinna LA, Ruzzene M. How do protein kinases recognize their substrates? Biochim Biophys Acta. 1996;1314:191–225. doi: 10.1016/s0167-4889(96)00083-3. [DOI] [PubMed] [Google Scholar]
- 19.Duarte ML, et al. Protein folding creates structure-based, noncontiguous consensus phosphorylation motifs recognized by kinases. Sci Signal. 2014;7:ra105. doi: 10.1126/scisignal.2005412. [DOI] [PubMed] [Google Scholar]
- 20.Sheridan DL, et al. Substrate discrimination among mitogen-activated protein kinases through distinct docking sequence motifs. J Biol Chem. 2008;283:19511–19520. doi: 10.1074/jbc.M801074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee T, et al. Docking motif interactions in MAP kinases revealed by hydrogen exchange mass spectrometry. Mol Cell. 2004;14:43–55. doi: 10.1016/s1097-2765(04)00161-3. [DOI] [PubMed] [Google Scholar]
- 22.Garai Á, et al. Specificity of linear motifs that bind to a common mitogen-activated protein kinase docking groove. Sci Signal. 2012;5:ra74. doi: 10.1126/scisignal.2003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bardwell AJ, Bardwell L. Two hydrophobic residues can determine the specificity of mitogen-activated protein kinase docking interactions. J Biol Chem. 2015;290:26661–26674. doi: 10.1074/jbc.M115.691436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon EA, et al. Combining docking site and phosphosite predictions to find new substrates: Identification of Smoothelin-like-2 (SMTNL2) as a c-Jun N-terminal kinase (JNK) substrate. Cell Signal. 2013;25:2518–2529. doi: 10.1016/j.cellsig.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeke A, et al. Systematic discovery of linear binding motifs targeting an ancient protein interaction surface on MAP kinases. Mol Syst Biol. 2015;11:837. doi: 10.15252/msb.20156269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldman A, et al. The calcineurin signaling network evolves via conserved kinase-phosphatase modules that transcend substrate identity. Mol Cell. 2014;55:422–435. doi: 10.1016/j.molcel.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glatz G, et al. Structural mechanism for the specific assembly and activation of the extracellular signal regulated kinase 5 (ERK5) module. J Biol Chem. 2013;288:8596–8609. doi: 10.1074/jbc.M113.452235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Francis DM, et al. Structural basis of p38α regulation by hematopoietic tyrosine phosphatase. Nat Chem Biol. 2011;7:916–924. doi: 10.1038/nchembio.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dar AC, et al. Higher-order substrate recognition of eIF2α by the RNA-dependent protein kinase PKR. Cell. 2005;122:887–900. doi: 10.1016/j.cell.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 30.Levinson NM, et al. Structural basis for the recognition of c-Src by its inactivator Csk. Cell. 2008;134:124–134. doi: 10.1016/j.cell.2008.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamill S, et al. Structural basis for noncanonical substrate recognition of cofilin/ADF proteins by LIM kinases. Mol Cell. 2016;62:397–408. doi: 10.1016/j.molcel.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krupnick JG, Benovic JL. The role of receptor kinases and arrestins in G protein-coupled receptor regulation. Annu Rev Pharmacol Toxicol. 1998;38:289–319. doi: 10.1146/annurev.pharmtox.38.1.289. [DOI] [PubMed] [Google Scholar]
- 33.Komolov KE, et al. Structural and functional analysis of a β2-adrenergic receptor complex with GRK5. Cell. 2017;169:407–421. e416. doi: 10.1016/j.cell.2017.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Homan KT, Tesmer JJ. Structural insights into G protein-coupled receptor kinase function. Curr Opin Cell Biol. 2014;27:25–31. doi: 10.1016/j.ceb.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roach PJ. Multisite and hierarchal protein phosphorylation. J Biol Chem. 1991;266:14139–14142. [PubMed] [Google Scholar]
- 36.Valk E, et al. Multistep phosphorylation systems: tunable components of biological signaling circuits. Mol Biol Cell l. 2014;25:3456–3460. doi: 10.1091/mbc.E14-02-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.St-Denis N, et al. Systematic investigation of hierarchical phosphorylation by protein kinase CK2. J Proteomics. 2015;118:49–62. doi: 10.1016/j.jprot.2014.10.020. [DOI] [PubMed] [Google Scholar]
- 38.Frame S, et al. A common phosphate binding site explains the unique substrate specificity of GSK3 and its inactivation by phosphorylation. Mol Cell. 2001;7:1321–1327. doi: 10.1016/s1097-2765(01)00253-2. [DOI] [PubMed] [Google Scholar]
- 39.Stamos JL, et al. Structural basis of GSK-3 inhibition by N-terminal phosphorylation and by the Wnt receptor LRP6. elife. 2014;3:e01998. doi: 10.7554/eLife.01998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Donella-Deana A, et al. Phosphorylated residues as specificity determinants for an acidophilic protein tyrosine kinase. A study with src and cdc2 derived phosphopeptides. FEBS Lett. 1993;330:141–145. doi: 10.1016/0014-5793(93)80260-2. [DOI] [PubMed] [Google Scholar]
- 41.Deng Y, et al. Global analysis of human nonreceptor tyrosine kinase specificity using high-density peptide microarrays. J Proteome Res. 2014;13:4339–4346. doi: 10.1021/pr500503q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen S, et al. Tyrosine kinase BMX phosphorylates phosphotyrosine-primed motif mediating the activation of multiple receptor tyrosine kinases. Sci Signal. 2013;6:ra40. doi: 10.1126/scisignal.2003936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Begley MJ, et al. EGF-receptor specificity for phosphotyrosine-primed substrates provides signal integration with Src. Nat Struct Mol Biol. 2015;22:983–990. doi: 10.1038/nsmb.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McGrath DA, et al. Cks confers specificity to phosphorylation-dependent CDK signaling pathways. Nat Struct Mol Biol. 2013;20:1407–1414. doi: 10.1038/nsmb.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koivomagi M, et al. Multisite phosphorylation networks as signal processors for Cdk1. Nat Struct Mol Biol. 2013;20:1415–1424. doi: 10.1038/nsmb.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rock JM, et al. Activation of the yeast Hippo pathway by phosphorylation-dependent assembly of signaling complexes. Science. 2013;340:871–875. doi: 10.1126/science.1235822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gogl G, et al. The structure of an NDR/LATS kinase-Mob complex reveals a novel kinase-coactivator system and substrate docking mechanism. PLoS Biol. 2015;13:e1002146. doi: 10.1371/journal.pbio.1002146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Couzens AL, et al. MOB1 Mediated phospho-recognition in the core mammalian hippo pathway. Mol Cell Proteomics. 2017;16:1098–1110. doi: 10.1074/mcp.M116.065490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ni L, et al. Structural basis for Mob1-dependent activation of the core Mst–Lats kinase cascade in Hippo signaling. Genes Dev. 2015;29:1416–1431. doi: 10.1101/gad.264929.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Furdui CM, et al. Autophosphorylation of FGFR1 kinase is mediated by a sequential and precisely ordered reaction. Mol Cell. 2006;21:711–717. doi: 10.1016/j.molcel.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 51.Kim SY, Ferrell JE., Jr Substrate competition as a source of ultrasensitivity in the inactivation of Wee1. Cell. 2007;128:1133–1145. doi: 10.1016/j.cell.2007.01.039. [DOI] [PubMed] [Google Scholar]
- 52.Mylona A, et al. Opposing effects of Elk-1 multisite phosphorylation shape its response to ERK activation. Science. 2016;354:233–237. doi: 10.1126/science.aad1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hutti JE, et al. A rapid method for determining protein kinase phosphorylation specificity. Nat Meth. 2004;1:27–29. doi: 10.1038/nmeth708. [DOI] [PubMed] [Google Scholar]
- 54.Hennrich ML, et al. Universal quantitative kinase assay based on diagonal SCX chromatography and stable isotope dimethyl labeling provides high-definition kinase consensus motifs for PKA and human Mps1. J Proteome Res. 2013;12:2214–2224. doi: 10.1021/pr400074f. [DOI] [PubMed] [Google Scholar]
- 55.Trinh TB, et al. Profiling the substrate specificity of protein kinases by on-bead screening of peptide libraries. Biochemistry. 2013;52:5645–5655. doi: 10.1021/bi4008947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Songyang Z, et al. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr Biol. 1994;4(11):973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- 57.Miller ML, et al. Linear motif atlas for phosphorylation-dependent signaling. Sci Signal. 2008;1:ra2. doi: 10.1126/scisignal.1159433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Obenauer JC, et al. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mok J, et al. Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci Signal. 2010;3:ra12. doi: 10.1126/scisignal.2000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gwinn DM, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee YJ, et al. Reciprocal phosphorylation of yeast glycerol-3-phosphate dehydrogenases in adaptation to distinct types of stress. Mol Cell Biol. 2012;32:4705–4717. doi: 10.1128/MCB.00897-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goodwin JM, et al. An AMPK-independent signaling pathway downstream of the LKB1 tumor suppressor controls Snail1 and metastatic potential. Mol Cell. 2014;55:436–450. doi: 10.1016/j.molcel.2014.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reinhardt HC, et al. DNA damage activates a spatially distinct late cytoplasmic cell-cycle checkpoint network controlled by MK2-mediated RNA stabilization. Mol Cell. 2010;40:34–49. doi: 10.1016/j.molcel.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blethrow JD, et al. Covalent capture of kinase-specific phosphopeptides reveals Cdk1-cyclin B substrates. Proc Natl Acad Sci USA. 2008;105:1442–1447. doi: 10.1073/pnas.0708966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kanshin E, et al. Machine learning of global phosphoproteomic profiles enables discrimination of direct versus indirect kinase substrates. Mol Cell Proteomics. 2017;16:786–798. doi: 10.1074/mcp.M116.066233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Z, et al. Analysis of cellular tyrosine phosphorylation via chemical rescue of conditionally active Abl kinase. Biochemistry. 2018 doi: 10.1021/acs.biochem.7b01158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tatebe H, et al. Substrate specificity of TOR complex 2 is determined by a ubiquitin-fold domain of the Sin1 subunit. eLife. 2017;6:e19594. doi: 10.7554/eLife.19594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schalm SS, et al. TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr Biol. 2003;13:797–806. doi: 10.1016/s0960-9822(03)00329-4. [DOI] [PubMed] [Google Scholar]
- 69.Kang SA, et al. mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science. 2013;341:1236566. doi: 10.1126/science.1236566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bloom J, Cross FR. Multiple levels of cyclin specificity in cell-cycle control. Nat Rev Mol Cell Biol. 2007;8:149–160. doi: 10.1038/nrm2105. [DOI] [PubMed] [Google Scholar]
- 71.Biondi RM, Nebreda AR. Signalling specificity of Ser/Thr protein kinases through docking-site-mediated interactions. Biochem J. 2003;372:1–13. doi: 10.1042/BJ20021641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang K, et al. Structure of the Pho85-Pho80 CDK-cyclin complex of the phosphate-responsive signal transduction pathway. Mol Cell. 2007;28:614–623. doi: 10.1016/j.molcel.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bhaduri S, et al. A docking interface in the cyclin Cln2 promotes multi-site phosphorylation of substrates and timely cell-cycle entry. Curr Biol. 2015;25:316–325. doi: 10.1016/j.cub.2014.11.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Swaffer MP, et al. CDK substrate phosphorylation and ordering the cell cycle. Cell. 2016;167:1750–1761. e1716. doi: 10.1016/j.cell.2016.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Koivomagi M, et al. Dynamics of Cdk1 substrate specificity during the cell cycle. Mol Cell. 2011;42:610–623. doi: 10.1016/j.molcel.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Godfrey M, et al. PP2ACdc55 phosphatase imposes ordered cell-cycle phosphorylation by opposing threonine phosphorylation. Mol Cell. 2017;65:393–402. e393. doi: 10.1016/j.molcel.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moses AM, Landry CR. Moving from transcriptional to phospho-evolution: generalizing regulatory evolution? Trends Genet. 2010;26:462–467. doi: 10.1016/j.tig.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 78.Tan CS, et al. Comparative analysis reveals conserved protein phosphorylation networks implicated in multiple diseases. Sci Signal. 2009;2:ra39. doi: 10.1126/scisignal.2000316. [DOI] [PubMed] [Google Scholar]
- 79.Howard CJ, et al. Ancestral resurrection reveals evolutionary mechanisms of kinase plasticity. eLife. 2014;3:e04126. doi: 10.7554/eLife.04126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen C, et al. Rational redesign of a functional protein kinase-substrate interaction. ACS Chem Biol. 2017;12(5):1194–1198. doi: 10.1021/acschembio.7b00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Creixell P, et al. Unmasking determinants of specificity in the human kinome. Cell. 2015;163:187–201. doi: 10.1016/j.cell.2015.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tan CSH, et al. Positive selection of tyrosine loss in metazoan evolution. Science. 2009;325:1686–1688. doi: 10.1126/science.1174301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Heinicke S, et al. The Princeton protein orthology database (P-POD): a comparative genomics analysis tool for biologists. PLoS One. 2007;2:e766. doi: 10.1371/journal.pone.0000766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wagih O, et al. MIMP: predicting the impact of mutations on kinase-substrate phosphorylation. Nat Methods. 2015;12:531–533. doi: 10.1038/nmeth.3396. [DOI] [PubMed] [Google Scholar]
- 85.Creixell P, et al. Kinome-wide decoding of network-attacking mutations rewiring cancer signaling. Cell. 2015;163:202–217. doi: 10.1016/j.cell.2015.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lubner JM, et al. Cushing’s syndrome mutant PKAL205R exhibits altered substrate specificity. FEBS Lett. 2017;591:459–467. doi: 10.1002/1873-3468.12562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tsui T, Miller WT. Cancer-associated mutations in breast tumor kinase/PTK6 differentially affect enzyme activity and substrate recognition. Biochemistry. 2015;54:3173–3182. doi: 10.1021/acs.biochem.5b00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rennefahrt UEE, et al. Specificity profiling of Pak kinases allows identification of novel phosphorylation sites. J Biol Chem. 2007;282:15667–15678. doi: 10.1074/jbc.M700253200. [DOI] [PubMed] [Google Scholar]
- 89.Alessi DR, et al. Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 kinase. FEBS Lett. 1996;399:333–338. doi: 10.1016/s0014-5793(96)01370-1. [DOI] [PubMed] [Google Scholar]
- 90.Himpel S, et al. Specificity determinants of substrate recognition by the protein kinase DYRK1A. J Biol Chem. 2000;275:2431–2438. doi: 10.1074/jbc.275.4.2431. [DOI] [PubMed] [Google Scholar]
- 91.Tagliabracci Vincent S, et al. A single kinase generates the majority of the secreted phosphoproteome. Cell. 2015;161:1619–1632. doi: 10.1016/j.cell.2015.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Egan DF, et al. Small molecule inhibition of the autophagy kinase ULK1 and identification of ULK1 substrates. Mol Cell. 2015;59:285–297. doi: 10.1016/j.molcel.2015.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lizcano JM, et al. Molecular basis for the substrate specificity of NIMA-related kinase-6 (NEK6): Evidence that NEK6 does not phosphorylate the hydrophobic motif of ribosomal S6 protein kinase and serum- and glucocorticoid-induced protein kinase in vivo. J Biol Chem. 2002;277:27839–27849. doi: 10.1074/jbc.M202042200. [DOI] [PubMed] [Google Scholar]
- 94.Kim ST, et al. Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]