Abstract
Photoactivatable (‘caged’) pharmacological agents have revolutionized neuroscience but the palette of available compounds is limited. We describe a general method for caging tertiary amines using a stable quaternary ammonium linkage that elicits a red-shift in activation wavelength. We prepared a photoactivatable nicotine (PA-Nic), uncagable via 1- or 2-photon excitation, that is useful for optopharmacology experiments to study nicotinic acetylcholine receptors (nAChRs) in different experimental preparations and spatiotemporal scales.
Photoactivatable (‘caged’) compounds remain the primary tools for modulation of native proteins with high spatiotemporal resolution. Development of new photoactivatable compounds that are activatable with 1- or 2-photon illumination remains a critical application of chemistry to biology. Uncaging ligands for glutamate and γ-aminobutyric acid (GABA) receptors enabled seminal ‘optopharmacology’ studies on the biophysics and subcellular location of functional proteins within complex biological environments1–3, but caged ligands targeting other receptors remain rare. We developed a general strategy for preparing photoactivatable drugs through alkylation of tertiary nitrogen atoms to form photolabile quaternary linkages. Our photoactivatable nicotine (PA-Nic) exhibits ideal chemical and spectroscopic properties for interrogating endogenous nicotinic acetylcholine receptors (nAChRs) in brain tissue.
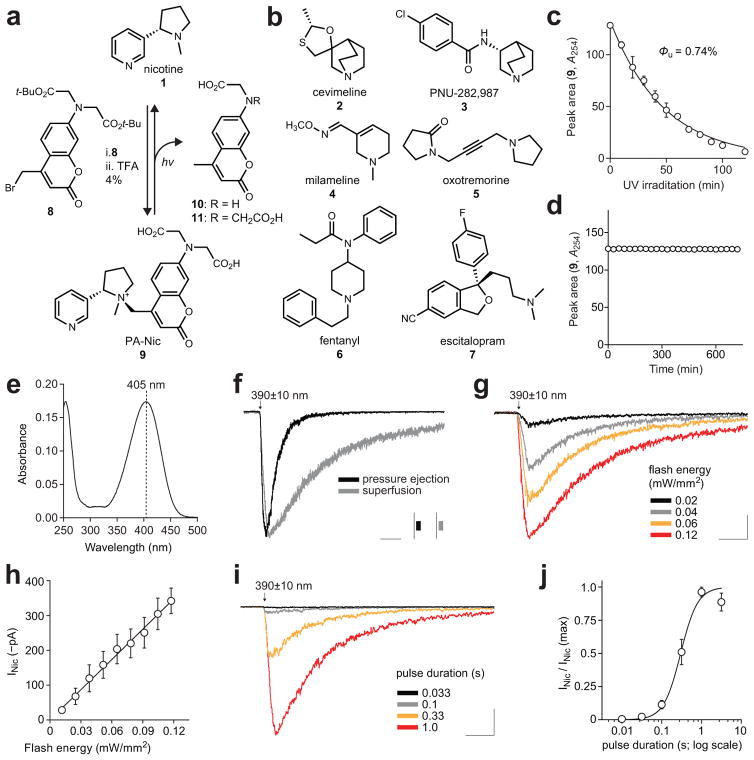
Many pharmacological agents cannot be caged using standard strategies because they lack obvious attachment sites (e.g., CO2H, OH, NH) for photolabile groups. A canonical example of an ‘uncageable’ drug is the nAChR agonist nicotine (1, Fig. 1a) but other such compounds exist, including the AChR agonists cevimeline (2), PNU-282,987 (3), milameline (4), and oxotremorine (5), as well as the opioid fentanyl (6) and the selective serotonin reuptake inhibitor escitalopram (7, Fig. 1b). A shared feature of these compounds is a tertiary nitrogen, a common motif in many pharmacological agents that is often critical for biological activity (Supplementary Note). We envisioned a general caging strategy involving covalent attachment of a coumarin4–6 cage to form a quaternary ammonium salt. Chemists have used quaternization to create photoactivatable tertiary amines such as polymer initiators7, amino acids8, mustards9, and anticancer agents10, but such compounds have not been reported in biological experiments. We applied this strategy to nicotine (1), alkylating with coumarin 8 to yield photoactivatable nicotine (PA-Nic, 9; Fig. 1a). PA-Nic releases nicotine upon UV illumination (365 nm) with a uncaging quantum yield (Φu) of 0.74%, generating coumarins 10 and 11 as byproducts, suggesting a radical-mediated photolytic mechanism (Fig. 1a, c, Supplementary Fig. 1a; Supplementary Note). The relatively low Φu value for PA-Nic is compensated, in part, by a high extinction coefficient (ε = 17,400 M−1cm−1). PA-Nic shows excellent dark stability in aqueous solution (Fig. 1d) and formation of the quaternary center at the 4-position of the coumarin elicits an unexpected ~15 nm red-shift in absorption maxima of the coumarin cage (λmax = 404 nm, Fig. 1e), making it an excellent match for ~400 nm light sources. We then applied this strategy to prepare quaternary ammonium photoactivatable derivatives of drugs 2–7, which all gave clean photoconversion with comparable Φu, excellent dark stability, red-shifted λmax, high ε, and the same coumarin byproducts (Supplementary Fig. 1b–k, Supplementary Note). We also confirmed a quaternary center is necessary for the shift in λmax (Supplementary Fig. 1 l, m).
Figure 1. Development of PA-Nic and its utility for improved pharmacological studies of native nAChRs.
(a) Chemical structure of nicotine (1) and the synthesis and photolysis of PA-Nic (9). (b) Chemical structures of ‘uncageable’ drugs 2–7. (c) Plot of mean HPLC chromatogram peak area vs. irradiation time (365 nm) provides the uncaging quantum yield (Φu); solid line is exponential fit; error bars indicate ±SD; n=2 independent samples. (d) Representative dark stability experiment of PA-Nic (9); n=3 independent samples. (e) Representative absolute absorption spectrum of PA-Nic (9, 10 μM); n=3 independent samples. (f) Light-evoked currents following PA-Nic epi-illumination photolysis. Voltage clamp traces from individual MHb neurons are shown for different methods of PA-Nic application (similar for >10 independent experiments). Photolysis: 1 s pulse, 0.12 mW/mm2. Scale: 2.5 s, 100 pA (grey), 150 pA (black) (g–j) Controllable nicotine uncaging via PA-Nic epi-illumination photolysis. (g) Representative voltage clamp traces from a MHb neuron for light pulses (1 s) of varying intensity. Scale: 2.5 s, 75 pA (h) Resulting photochemical dose-response relation for peak currents (y=2.876x+2.1, R2=0.9921). Mean (n=9/6) peak of light-activated currents plotted vs. input flash intensity. (i) Representative voltage clamp traces during light pulses (0.12 mW/mm2) of varying duration applied to a MHb neuron. Scale: 2.5 s, 250 pA (j) Graphical analysis of summary pulse duration data in i. The Hill equation was fitted to the mean data (nH (Hill slope)=2.0, duration at ½ max=0.3 s, R2=0.928) from n=5/3. All error bars indicate ±SEM.
With the generality of this caging strategy established, we focused on PA-Nic (9) first by comparing it to the previously described photoactivatable ruthenium bis(bipyridine)–nicotine complex (RuBi-Nic, 20, Supplementary Fig. 2a)11. We prepared mouse brain slices containing the medial habenula (MHb), whose neurons express high levels of nAChRs12, co-release ACh and glutamate13, and regulate affective behavior and nicotine withdrawal14 (Supplementary Fig. 2b–d). Light flashes (1 s, 390 ± 10 nm) evoked robust nicotinic currents with PA-Nic (80 μM; Fig. 1f), but RuBiNic elicited much smaller currents even with longer photolysis (15 s, 470 nm; Supplementary Fig. 2e) and proved unstable (Supplementary Fig. 2f). In addition, PA-Nic photolysis is negligible at 470 nm and 560 nm (100 ms, 0.06 mW/mm2; Supplementary Fig. 2g), enabling the use of other fluorophores or light-activated effectors. The main photochemical by-product of PA-Nic photolysis (10) was inert at nAChRs, PA-Nic itself exhibited no nAChR antagonist properties, and our preparations of PA-Nic were devoid of free nicotine (Supplementary Fig. 2h–k). PA-Nic photolysis currents were eliminated by a nAChR antagonist cocktail (Supplementary Fig. 2l) and reversed at approximately +2 mV (Supplementary Fig. 2m, n). Pressure-ejection application of nicotine and ACh provides a calibration for PA-Nic photolysis currents (Supplementary Fig. 2o, p). Epi-illumination flashes elicited responses that increased in amplitude with increasing flash energy (Fig. 1g, h) and flash duration (Fig. 1i, j). Thus, light-evoked nicotine release allowed generation of complete photochemical concentration-response relations in neurons, an advantage over existing drug-application approaches.
Next, we determined whether PA-Nic could be used during 2-photon laser scanning microscopy (2PLSM) and 2-photon photolysis. The 2-photon fluorescence action cross-section (δf; Methods) spectrum of PA-Nic was distinct from GCaMP6f (Supplementary Fig. 3a). We measured the 2-photon uncaging action cross-section (δu) to be 0.094 GM at the maximal 2P absorption (810 nm) and 0.059 GM and 0.025 GM at the commonly used 760 nm and 720 nm photolysis wavelengths, respectively (Supplementary Fig. 3b, c, Methods, Supplementary Note).4 These values are comparable to the widely used MNI-glutamate (δu = 0.06 GM at 730 nm)1. PA-Nic (100 μM) photostimulation (720 nm) evoked stable inward currents (Supplementary Fig. 3d), and no responses were evoked between 760–900 nm in the absence of PA-Nic (Supplementary Fig. 3e). At 760 nm, 2-photon photolysis amplitudes increased with longer pulse durations at a fixed laser power (Supplementary Fig. 3f), or with increasing laser power at a fixed pulse duration (Supplementary Fig. 3g).
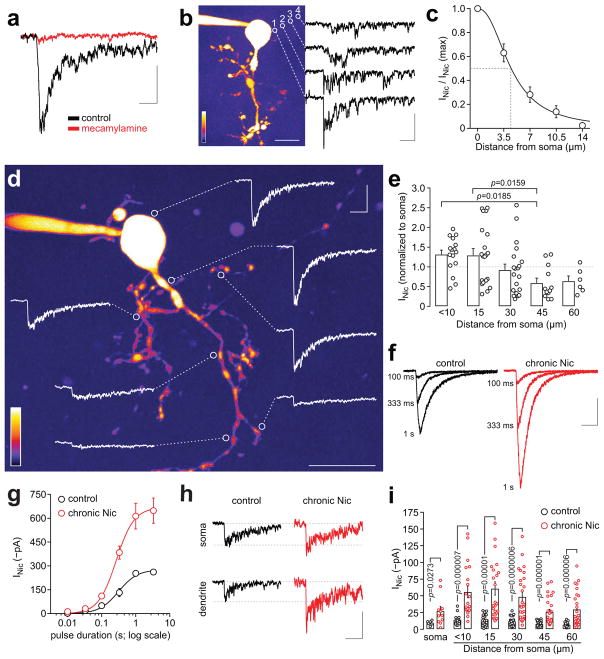
To study nAChRs in cellular compartments, MHb neuronal morphology was visualized with 2PLSM and nicotine was uncaged with peri-somatic 1-photon laser pulses (405 nm, ~1 μm spot diameter). After confirming that moderate laser powers (1–2.5 mW, 50 ms) do not elicit currents in the absence of PA-Nic (Supplementary Fig. 3h, i), we identified suitable photostimulation parameters by measuring the relationship between evoked currents versus laser power and pulse duration (Supplementary Fig. 3j–m). PA-Nic photolysis currents were antagonized by mecamylamine (Fig. 2a) and exhibited a useful spatial resolution (Fig. 2b, c). Interestingly, these currents were greater near proximal dendrites versus distal dendrites (Fig. 2d, e). These results showcase the temporal and spatial precision afforded by laser flash photolysis of PA-Nic.
Figure 2. PA-Nic enables subcellular mapping of nAChRs following chronic nicotine treatment.
(a) PA-Nic uncaging and mecamylamine antagonism. Nicotine was uncaged in a ~1 μm peri-somatic spot with a 405 nm laser pulse (10 ms, 2.9 mW). Voltage-clamp currents are shown before (black trace, similar for >10 independent experiments) and 10 min after (red trace, single experiment) mecamylamine (10 μM) superfusion. Scale: 50 ms, 100 pA (b–c) Lateral spread of uncaged nicotine, estimated electrophysiologically. (b) A representative 2PLSM image of a MHb neuron is shown. Nicotine was uncaged (white circles; 10 ms, 1.5 mW) at the surface (1; 0 μm), and at 3.5 (2), 7.0 (3), and 10.5 (4) μm from the cell surface. Representative traces from a single neuron are shown. Scales on image: (lower left = relative intensity; lower right = 20 μm). Scale (current): 500 ms, 30 pA (c) The Hill equation was fitted to the mean (±SEM) data (n (Hill slope)=2.293, R2=0.9074) (n=6/5), resulting in an estimate of 4.5 μm for the lateral distance at ½ max amplitude. (d–e) Subcellular mapping of nAChRs in MHb neurons. (d) Representative 2PLSM image of a ChAT(+) MHb neuron, marked with uncaging positions (white circles; 50 ms, 2 mW) and the evoked response at that location. Scales: (lower left = relative intensity; lower right = 20 μm, upper right = 1 s, 60 pA) (e) Summary of position-dependent uncaging data for ChAT(+) MHb neurons (n=8/5) using PA-Nic (80 μM). Nicotine uncaging responses were recorded at the soma and at dendritic locations at the indicated linear distance from the soma surface. The mean (+SEM) and individual responses are shown. P values: Tukey post-hoc after 1-way ANOVA (F(4,69)=4.3, p=0.0036). (f–i) Interrogating chronic nicotine-mediated nAChR up-regulation with PA-Nic. (f) Representative traces are shown from a control and chronic nicotine-treated ChAT(+) MHb neuron stimulated via epi-illumination photolysis (0.12 mW/mm2) for the indicated durations. Scale: 200 pA, 2 s (g) The Hill equation was fitted to photochemical dose response mean (±SEM) data from control (n=7/2) or chronic nicotine-treated neurons (n=11/3) (control: n=1.7, duration at ½ max=0.3 s, R2=0.732; chronic nicotine: n=1.6, duration at ½ max=0.3 s, R2=0.89). (h–i) Chronic nicotine up-regulates nAChRs at all cellular locations tested. (h) Representative uncaging responses are shown from a control and chronic nicotine-treated neuron stimulated at the soma and at a dendrite ~30 μm from the soma using 405 nm laser photolysis (50 ms, 2 mW) of PA-Nic. Scale: 20 pA, 2 s (i) Scatter plots (mean ±SEM) of nicotine uncaging amplitudes at the indicated cellular location are shown for ChAT(+) control (n=6/3) and chronic nicotine-treated (n=14/4) neurons. P values: two-sided Mann-Whitney test.
Next, we studied the pharmacology of nAChR up-regulation, a key feature of nicotine dependence. Chronic nicotine treatment up-regulates nAChR function in MHb neurons15, including choline acetyltransferase (ChAT)(+) neurons (Supplementary Fig. 4a–d), but it is unknown whether this reflects a numerical increase in surface receptors or a shift in nAChR sensitivity. By generating a photochemical concentration-response curve with PA-Nic, we determined that chronic nicotine increased the pharmacological efficacy of acute nicotine without affecting potency (Fig. 2f, g). This suggests that chronic nicotine treatment increases receptor number without affecting receptor sensitivity to agonist. In laser flash recordings, uncaging responses were enhanced by chronic nicotine in both somata and dendrites (Fig. 2h, i). Thus, chronic nicotine induces plastic changes in postsynaptic nAChR function that could sensitize MHb neurons to cholinergic agonists, which could modulate excitability and/or dendritic integration.
We further explored PA-Nic’s utility by studying nAChR-modulated excitability and Ca2+ mobilization. Using a restricted field stop aperture (Supplementary Fig. 2k) to constrain brief (33 ms) epi-illumination flashes (~60 μm field of view), firing was transiently enhanced when nicotine was uncaged directly over the recorded neuron, but not when nicotine was uncaged 100–200 μm away (Supplementary Fig. 5a, b). Next, we examined Ca2+ dynamics in MHb ChAT+ neurons expressing GCaMP6f16 (Supplementary Fig. 5c) and displaying spontaneous Ca2+ fluctuations and responses to exogenous nicotine (Supplementary Fig. 5d, e). PA-Nic was locally applied to the imaged neuron (Supplementary Fig. 5f) where peri-somatic flash durations of only 5 ms (405 nm, 2 mW) were sufficient to robustly enhance Ca2+ levels (Supplementary Fig. 5g). Flashes without PA-Nic were ineffective, and nAChR antagonists significantly attenuated Ca2+ signals induced by PA-Nic photolysis (Supplementary Fig. 5g, h). Together, these results show that PA-Nic can be used in electrophysiological or all-optical regimes for spatially-delimited modulation of action potential firing or activity-dependent Ca2+ increases.
Finally, we examined PA-Nic’s utility in other settings. MHb ChAT+ neurons project to interpeduncular nucleus (IPN), where neurons are densely surrounded by cholinergic, nAChR-bearing fibers (Supplementary Fig. 6a, b). PA-Nic laser flash photolysis (2 mW, 50 ms) adjacent to IPN GABAergic neurons elicited a distinct, low-amplitude but smoldering current (Supplementary Fig. 6c, d), where peak current versus net charge is monotonic (Supplementary Fig. 6e). In contrast to MHb responses, peak current and net charge did not decay with distance from the soma (Supplementary Fig. 6f, g) but were blocked by nAChR antagonists (Supplementary Fig. 6h, i). We then studied stratum radiatum interneurons in hippocampus (Supplementary Fig. 6j), which express moderate levels of postsynaptic α7 nAChRs17 (Supplementary Fig. 6k, l). Epi-illumination photolysis of PA-Nic (0.12 mW/mm2, 250 ms, 2 mM PA-Nic local perfusion) efficiently activated α7 (Supplementary Fig. 6m), confirmed by sensitivity to 10 nM MLA (Supplementary Fig. 6n). These parameters (< 1 s flash, PA-Nic local perfusion, field stop aperture restricted) may be ideal for consistent activation of desensitization-prone nAChRs such as α7 or β2-containing receptors. Together, these experiments indicate that PA-Nic is broadly useful for examining nAChRs with varying kinetics and presynaptic or postsynaptic arrangements.
In summary, we have developed a flexible strategy for preparing photoactivatable derivatives of previously uncagable drugs. Our photoactivatable nicotine (PA-Nic, 9) can be activated by relatively short wavelength 1-photon (<470 nm) or 2-photon (<900 nm) light, allowing imaging in combination with other fluorophores or sensors. It is now possible to finely tune the spatiotemporal distribution of a nAChR agonist during optopharmacology experiments, allowing modeling of different aspects of nicotine exposure. PA-Nic could prove useful for studying cholinergic volume versus point-to-point transmission18, or in nAChR functional mapping17 and imaging experiments in neurons where dendritic and/or presynaptic Ca2+ dynamics are rapidly modulated by nAChRs. Our initial mapping results (Fig. 2d, e) suggest enhanced nAChR surface expression in proximal versus distal cellular compartments, and additional studies will uncover further details of nAChR distribution in nicotine-treated animals. More generally, given the number of tertiary amine compounds in the pharmacopeia, the use of a photoscissile quaternary linkage should enable development of other photoactivatable compounds to better model drug exposure and modulate native receptor proteins in brain tissue.
Online Methods
Chemical Synthesis and Photochemistry
Experimental details, characterization for all novel compounds, and determination of photochemical byproducts of PA-Nic photolysis can be found in the Supplementary Note.
UV–Vis and Fluorescence Spectroscopy
Spectroscopy was performed using 1-cm path length, 3.5-mL quartz cuvettes from Starna Cells or 1-cm path length, 1.0-mL quartz microcuvettes from Hellma. All measurements were taken at ambient temperature (22 ± 2 °C). Absorption spectra were recorded on a Cary Model 100 spectrometer (Agilent). Fluorescence emission spectra were recorded on a Cary Eclipse (Varian). Absolute fluorescence quantum yields were recorded on a Quantaurus-QY spectrometer (model C11374, Hamamatsu). All spectroscopy measurements were performed in phosphate-buffered saline (PBS), pH 7.4 and the values of maximum absorption wavelength (λmax), extinction coefficient at λmax (ε), maximum absorption wavelength (λem), and fluorescence quantum yield (Φf) are averages (n=3; Supplementary Fig. 1k).
HPLC and LC–MS
High performance liquid chromatography (HPLC) was performed on an Agilent 1200 Analytical HPLC system equipped with autosampler and diode array detector. To measure the uncaging quantum yield (Φu; Fig. 1c), the loss of PA-Nic (9) was monitored using a 4.6 × 150 mm Kinetex C18 column (Phenomenex) with a 5–95% gradient of CH3CN in H2O containing constant 0.1% v/v TFA. To examine the release of pharmacological agents 2–7 and coumarin byproducts 10 and 11 from compounds 12–17 (Supplementary Fig. 1), samples were run on tandem liquid chromatography-mass spectrometry (LC–MS) using an Agilent 1200 LC–MS system equipped with autosampler, diode array detector, and mass spectrometry detector using a 4.6 × 150 mm Gemini NX-C18 column with a 5–95% or 5–50% gradient of CH3CN in H2O containing constant 0.1% v/v TFA. Chromatograms were measured using absorbance at 254 nm, 210 nm, or using the total ion count (TIC) depending on the optical properties of the released drug compound.
Uncaging Quantum Yield (Φu) Determination
Photochemistry was performed in 1-cm path length, 3.5-mL quartz cuvettes (Starna) in a Luzchem LZC 4V photoreactor equipped with 365 nm UV lamps, a carousel, and a timer as previously described19. Briefly, the light intensity was calibrated by potassium ferrioxalate actinometry. A solution of 60 mM K3Fe(C2O4)3 was irradiated using the photoreactor setup and released Fe2+ was determined by complexometry with 1,10 phenanthroline. Using the known quantum yield of this process (Φ = 1.21), we determined the photon flux (I) = 3.57 × 10−7 ein/min·cm2. For the conversion of PA-Nic (9) to nicotine (1), the samples were irradiated and a small aliquot (50 μL) was placed in an amber glass high recovery HPLC vial. These samples were analyzed by HPLC as described above. The uncaging quantum yield (Φu, mol/ein) was determined by fitting a plot of HPLC peak integral signal (S) versus irradiation time to a one-phase exponential decay described by equation 1:
| (1) |
where S0 = signal prior to irradiation, t = irradiation time (min), St = signal at time t, I = irradiation (ein/min·cm2), and σ = decadic extinction coefficient (in units of cm2/mol; 1000-fold higher than the ε value with units of M−1cm−1 based on cuvette geometry). For the conversion of compound 9 to compound 1, we determined Φu = 0.74% (Fig. 1c). Coumarin-based cages have found broad utility in release of small molecule modulators of biological activity4–6,20–24; we note that although the Φu value for 9 is lower than coumarin caged molecules releasing better leaving groups such as carboxylates and phosphates4–6,21,23, it is similar to the Φu value of a 7-aminocoumarin caged compound releasing an amine via a carbamate linkage6. For other caged compounds, the uncaging quantum yield was determined by illumination with 405 nm LED (LOCTITE CL20 flood array) using 9 as a standard (Supplementary Fig. 1h, Supplementary Note).
2-Photon Action Cross-Section for Fluorescence (δf) and Uncaging (δu)
Efficacy of uncaging by 2-photon excitation was quantified by determining the uncaging action cross-section (δu), which is the product of the 2-photon absorption cross-section (σ2) and the 2-photon uncaging quantum yield Φ2u. In addition, as PA-Nic (9) is moderately fluorescent (Supplementary Fig. 1k), we also characterized the 2-photon fluorescence action cross-section (δf), which is the product of the 2-photon absorption cross-section (σ2) and the 2-photon fluorescence quantum yield (Φ2), properties that are known for the reference dye fluorescein25,26. The action cross-sections δu and δf are expressed in units of Goeppert-Mayer (GM), where 1 GM is defined as 10−50 cm4·s·photon−1. To determine δf we used the experimental setup previously described27,28; 1 μM solutions of either PA-Nic (9) in phosphate-buffered saline (PBS), GCaMP6f in 30 mM MOPS buffer, pH 7.2 containing 100 mM KCl and 10 mM Ca·EGTA, or reference dye fluorescein in 50 mM sodium borate buffer, pH 9.5 were illuminated in an epi-illumination microscope (IX-81, Olympus) with light from a mode-locked femtosecond Ti:Sapphire laser (Chameleon Ultra II, Coherent). Fluorescence collected by the 1.2 NA objective (UplanSApo 60×W, Olympus) was reflected off the laser dichroic (675DCSPXR, Omega Optical), spectrally filtered (539/278, Semrock), and focused by the tube lens onto a fiber-coupled avalanche photodiode (SPCM-AQRH-14-FC, Excelitas). The system was run under computer control to set both laser wavelength (700–1080 nm) and power (1 mW at the sample plane), and to record and store the avalanche photodiode signal. The use of 1 mW of laser power focused with the 1.2 NA objective is sufficiently low laser intensity that no fluorescence is observed from the photolysis product compounds 10 or 11. At each laser wavelength, the action cross-section of PA-Nic (9) is found from equation 2:
| (2) |
where the subscripts S and F refer to the sample (PA-Nic or GCaMP6f) and fluorescein, respectively; Φ2 is the 2-photon fluorescence quantum yield (usually equal to the 1-photon fluorescence quantum yield, Φf), σ2 is the 2-photon absorption cross-section in GM, 〈F(t)〉 is the fluorescence signal recorded by the detector in counts/s, η is the detector quantum efficiency averaged over the fluorescence emission spectrum, and T is the fraction of the fluorescent light transmitted through the bandpass filter. Using this equation, together with the known fluorescein 2-photon absorption cross-section (average of values from refs. 25 and 26) and assuming Φ2F is equal to the 1-photon fluorescein fluorescence quantum yield (Φf = 0.92) we determined the 2-photon fluorescence action cross-section spectrum of PA-Nic and GCaMP6f (Supplementary Fig. 3a, c).
To determine the 2-photon uncaging action cross-section (δu) of PA-Nic (9), we adapted methods described4,29 to find the fractional amount of photolysis using HPLC (Shimadzu UFLC system with diode array detector; 4.6 × 150 mm Gemini NX-C18 column with a 5–95% a gradient of CH3CN in H2O containing constant 0.1% v/v TFA). Photolysis experiments proceeded as follows: Laser light from a Ti:Sapphire laser was focused by a 50 mm focal-length achromatic doublet lens (AC254-050-B, Thorlabs) into a sub-micro cuvette (16.10F-Q-10, Starna) containing a solution of PA-Nic (10 μM, 18 μL) in PBS buffer. The solution also contained in 50 μM of 3-(4-methoxyphenyl)propan-1-amine, a non-photolyzable internal concentration standard to correct for evaporation during the illumination period as the sealed cuvette contained ~180 μL head volume of air. Laser power of 0.75 W was used for all photolysis experiments, measured after transmission through the cuvette. As a diagnostic, a side-observing fiber-coupled spectrometer was set up to monitor the fluorescence spectrum of the PA-Nic solution in the cuvette during excitation. This allowed observation of spurious back-scattering effects from the PA-Nic solution that interrupted laser mode-locking, evidenced by abrupt disappearance of PA-Nic fluorescence. A quarter-wave plate placed between the laser and focusing lens was found sufficient to eliminate the spurious effects on laser mode-locking. To obtain the photolysis rate, PA-Nic samples were illuminated for various time intervals from 0–40 min at 810 nm, 760 nm, or 720 nm and analyzed by HPLC as described above (n=3; Supplementary Fig. 3b). To obtain the spectral dependence of photolysis, solutions of PA-Nic were illuminated for 40 mins at wavelengths between 710 nm to 930 nm then analyzed by HPLC. To determine the uncaging action cross-section (δu) of PA-Nic, we again used fluorescein as an external standard, which allows δu to be expressed according to equation 3:4,29
| (3) |
where Φ2u and σ2S are the two-photon uncaging quantum yield and absorption cross-section, respectively, of PA-Nic. Np is the rate of uncaging in molecules/s as determined by HPLC, CS is the initial concentration of PA-Nic and CF the concentration of fluorescein, 〈F(t)〉 is the time- averaged rate of fluorescence (photons/s), collected by a power detector with collection efficiency Ω, and Φ2F and σ2F are the 2-photon fluorescence quantum yield and 2-photon absorption cross-section, respectively, of fluorescein. To determine 〈F(t)〉F, fluorescein in 50 mM sodium borate buffer, pH 9.5 was placed in a cuvette (3-Q-10, Starna) and illuminated at 810 nm, 760 nm, or 720 nm with focused laser light (0.75 W) as above, and a side-observing power meter (PM100A console, S120C head, Thorlabs) was used to record the fluorescent power produced. To eliminate scattered laser light, a IR-blocking filter (720/SP, Semrock) was placed in front of the detector. The fraction of the total fluorescent power collected by the power meter is given by equation 4:4
| (4) |
where r is the radius of the power meter (4.85 mm), R is the distance from the fluorescence axis to the detector face (45 mm), y is the measured filter transmission (0.97), and n is the refractive index of water (1.33). The uncaging action cross section of PA-Nic was determined to be δu = 0.094 GM at 810 nm, δu = 0.059 at 760 nm, and δu = 0.025 GM at 720 nm. The value at 810 nm was used to set the scale for converting the spectrum of photolysis vs. excitation to a spectrum of uncaging action cross-section, the shape of which agrees with the fluorescence action cross-section (Supplementary Fig 3c), reflecting the spectral dependence of the underlying 2-photon absorption cross-section.
Reagents for Neurobiology Experiments
QX-314, CNQX, SR16584, and D-AP5 were obtained from Tocris. RuBi-nicotine was obtained from Abcam. α-conotoxin MII (α-Ctx MII) was synthesized as previously described30,31. Acetylcholine (ACh), mecamylamine (mec), atropine, picrotoxin, dihydro-β-erythroidine (DHβE), and all other chemicals without a specified supplier were obtained from Sigma.
Mice
An animal study protocol pertaining to this study (#IS00003604) was reviewed and approved by the Northwestern University Institutional Animal Care and Use Committee. Procedures also followed the guidelines for the care and use of animals provided by the National Institutes of Health Office of Laboratory Animal Welfare. Mice were housed at 22 °C on a 12-hour light/dark cycle with food and water ad libitum. Mice were weaned on postnatal day 21 and housed with same-sex littermates. Unless stated otherwise, experiments were conducted on C57BL/6J mice obtained from Jackson Laboratories (Jax #000664). ChAT-IRES-Cre32 (Jax #006410) (“ChAT-Cre”) and Ai1433 (Jax #007914) mouse strains were crossed to yield ChAT-Cre::Ai14 mice, which express tdTomato in a Cre-dependent manner (i.e., in neurons with an active ChAT promoter). Expression of tdTomato in MHb ChAT(+) neurons was confirmed in these mice via immunohistochemistry and confocal microscopy (Supplementary Fig. 2d). GAD2-Cre::Ai14 mice, which express tdTomato in GAD2(+) neurons, were created by crossing Ai14 mice to GAD2-IRES-Cre34 (Jax #010802) (“GAD2-Cre”). Pre-weanling mice (<21 days old) were used for stratum radiatum recordings. All other mice used were 8–24 wks old.
Stereotaxic Injection Surgery
AAV5.CAG.Flex.GCaMP6f.WPRE.SV40 vectors for Cre-dependent expression of GCaMP6f were obtained from University of Pennsylvania Vector Core (lot #V5532L, titer 1.74×1013 GC/mL). Male and female ChAT-Cre mice were used for surgery starting at 8 weeks of age. Mice were initially anesthetized with an intraperitoneal (i.p.) injection of a ketamine/xylazine mixture (120 mg/kg ketamine, 16 mg/kg xylazine). Mice were given additional “boost” injections of ketamine (100 mg/kg, i.p.) as needed. Alternatively, some mice were anesthetized with isoflurane: 3% (flow rate 500 mL/min) for induction and 1.5% (28 mL/min) for maintenance. Mice were secured into a stereotaxic frame and a small incision at the top of the head was made to expose the skull. Coordinates used for bilateral medial habenula injections were (relative to bregma, in mm): M/L: +/− 0.3, A/P: −1.58, D/V: −2.75 with overshoot to −3.0 before retracting to −2.75. Exact coordinates were adjusted to account for slight differences in the head size of individual mice: the bregma/lambda distance measured for each mouse was divided by the reported bregma/lambda distance for C57 mice (4.21), then multiplied by the A/P coordinate. The injection needle was slowly lowered through the drilled hole to the D/V coordinate. For AAV viruses, 300 nL of virus (per hemisphere) was infused at a rate of 50 nL/min. The injection needle was left in place for 10 min after the infusion ended before slowly retracting the needle. Sutures were used to close the incision. At the conclusion of the surgery, mice were given ketoprofen (5 mg/kg, s.c.) and placed in a recovery cage, kept warm, and observed until they were ambulatory. Mice were single-housed following virus injection surgery and were given at least 14 days to recover and for the virus to express before beginning experimental procedures.
Chronic Nicotine Treatment
Mice were treated with nicotine via drinking water as previously described35, with minor modifications. Nicotine hydrogen tartrate or L-tartaric acid (control group) were dissolved in tap water (pH 7.0) supplemented with saccharin sodium (3 mg/mL) to mask the bitter taste of nicotine. The following treatment schedule was used for nicotine (reported as nicotine free base) and tartaric acid, respectively (in μg/mL): Day 1–2 (50, 75), Day 3–4 (100, 150), Day 5 and beyond (200, 300). The latter doses were maintained by replacing drinking water solutions every 2–3 days, and mice were treated for at least 28 days prior to experimentation. We previously demonstrated up-regulation of nAChR function in MHb VI neurons using a different chronic nicotine exposure method: subcutaneous osmotic minipump implantation15. Therefore, we validated this result with the nicotine drinking water method in C57BL/6 WT mice (Supplementary Fig. 4a, b) before conducting PA-Nic uncaging experiments coupled with chronic nicotine studies on ChAT-Cre::Ai14 mice (Supplementary Fig. 4c, d and Fig. 2f–i).
Brain Slice Preparation
For epi-illumination and 405 nm laser uncaging, brain slices were prepared as previously described36. Mice were anesthetized with Euthasol (sodium pentobarbital, 100 mg/kg; sodium phenytoin, 12.82 mg/kg) before trans-cardiac perfusion with oxygenated (95% O2/5% CO2), 4 °C N-methyl-D-glucamine (NMDG)-based recovery solution that contains (in mM): 93 NMDG, 2.5 KCl, 1.2 NaH2PO4, 30 NaHCO3, 20 HEPES, 25 glucose, 5 sodium ascorbate, 2 thiourea, 3 sodium pyruvate, 10 MgSO4·7H2O, and 0.5 CaCl2·2H2O; 300–310 mOsm; pH 7.3–7.4). Brains were immediately dissected after the perfusion and held in oxygenated, 4 °C recovery solution for one minute before cutting a brain block containing the medial habenula and sectioning the brain with a vibratome (VT1200S; Leica). Coronal slices (250 μm) were sectioned through the MHb (Supplementary Fig. 2b) and transferred to oxygenated, 33 °C recovery solution for 12 min. Slices were then kept in holding solution (containing in mM: 92 NaCl, 2.5 KCl, 1.2 NaH2PO4, 30 NaHCO3, 20 HEPES, 25 glucose, 5 sodium ascorbate, 2 thiourea, 3 sodium pyruvate, 2 MgSO4·7H2O, and 2 CaCl2·2H2O; 300–310 mOsm; pH 7.3–7.4) for 60 min or more before recordings. For data in Supplementary Fig. 2f only, slices containing ventral tegmental area (~bregma −3.5 mm) were prepared.
For 2-photon uncaging, brain slices were prepared as follows. Animals were deeply anesthetized by inhalation of isoflurane, decapitated, and the brain was rapidly removed and immersed in ice-cold oxygenated artificial cerebrospinal fluid (ACSF) containing (in mM) 127 NaCl, 2.5 KCl, 25 NaHCO3, 1.25 NaH2PO4, 2.0 CaCl2, 1.0 MgCl2, and 25 glucose (osmolarity ~310 mOsm/L). Tissue was blocked and transferred to a slicing chamber containing ice-cold ACSF, supported by a small block of 4% agar. Bilateral 250 μm-thick slices containing the MHb were cut on a Leica VT1000S and transferred into a holding chamber with ACSF equilibrated with 95% O2/5% CO2. Slices were incubated at 34 °C for 15–30 minutes prior to electrophysiological recording.
UV–Vis Nicotine Uncaging
Brain slices were transferred to a recording chamber being continuously superfused at a rate of 1.5–2.0 mL/min with oxygenated 32 °C recording solution. The recording solution contained (in mM): 124 NaCl, 2.5 KCl, 1.2 NaH2PO4, 24 NaHCO3, 12.5 glucose, 2 MgSO4·7H2O, and 2 CaCl2·2H2O; 300–310 mOsm; pH 7.3–7.4). Picrotoxin (100 μM), CNQX (20 μM), and D-AP5 (50 μM) were added during MHb subcellular uncaging experiments; these three drugs plus TTX (0.5 μM) were added during IPN subcellular uncaging experiments. Holding potential for voltage clamp recordings during epi-illumination or 1P laser flash photolysis was −60 mV, except that recordings in stratum radiatum involved pooling responses from cells held at −60 mV and −70 mV. TEA (5 mM) was added to the recording solution when the holding potential was 0 mV. Patch pipettes were pulled from borosilicate glass capillary tubes (1B150F-4; World Precision Instruments) using a programmable microelectrode puller (P-97; Sutter Instrument). Tip resistance ranged from 4.5 to 8.0 MΩ when filled with internal solution. The following internal solution was used (in mM): 135 potassium gluconate, 5 EGTA, 0.5 CaCl2, 2 MgCl2, 10 HEPES, 2 MgATP, and 0.1 GTP; pH adjusted to 7.25 with Tris base; osmolarity adjusted to 290 mOsm with sucrose. For subcellular uncaging, this internal solution also contained QX-314 (2 mM) for improved voltage control. We recorded from neurons in the ventral 50–60% of the MHb, as previously described12,15.
For epi-illumination uncaging experiments, a Nikon Eclipse FN-1 upright microscope equipped with infrared and visible differential interference contrast (DIC) optics and a 40X (0.80 NA) objective was used to visualize cells within brain slices. A computer running pCLAMP 10 software (Molecular Devices) was used to acquire whole-cell recordings along with an Axopatch 200B amplifier and a 16-bit Digidata 1440 A/D converter (both from Molecular Devices). Data were sampled at 10 kHz and low-pass filtered at 1 kHz. Immediately prior to gigaseal formation, the junction potential between the patch pipette and the superfusion medium was nulled. Series resistance was uncompensated. A light emitting diode (LED) light source (XCite 110LED; Excelitas) coupled to excitation filters (400/40 nm, 470/40 nm, and 560/40 nm bandpass) was used for photostimulation. Internal LEDs in the XCite 110LED were (center wavelength/FWHM, in nm): 385/30, 470/40, 560/80, 640/40. For near-UV photostimulation, flash wavelength was therefore approximately 390 ±10 nm. Light flashes were triggered by pCLAMP via TTL pulses. Flash energy output from the LED was determined by calibration using a photodiode power sensor (Model S120C; Thor Labs).
Some experiments involved recording inward currents following pressure ejection application of drug to the recorded cell using a drug-filled pipette, which was moved to within 20–40 μm of the recorded neuron using a micromanipulator. Using a Picospritzer (Parker Hannifin), a pressure ejection dispensed drug (dissolved in the same superfusion medium used on the slice) onto the recorded neuron. Ejection volume, duration, and ejection pressure varied depending on whether a short (ms) or long (s) pulse was required. The change in amplitude between the baseline and the peak was measured. Atropine (1 μM) was present in the superfusion medium when using ACh to prevent activation of muscarinic ACh receptors. To prevent stray light from prematurely uncaging nicotine inside the drug pipette, the field stop aperture on the microscope was restricted, and the drug pipette was retracted from the cell immediately following the application.
For focal nicotine uncaging using 1-photon laser (405 nm) flash photolysis, an Olympus BX51 upright microscope with a 60× (1.0 NA) objective was used to visualize cells. Prairie View 5 (Bruker Nano) software was used for acquisition via a Multiclamp 700B patch clamp amplifier (Molecular Devices). Analog signals were sampled at 5 kHz and low-pass filtered at 1 kHz, and an A/D converter (PCI-NI6052e; National Instruments) was used for digitization. Patch clamp recordings were carried out using the internal solution mentioned above, except that Alexa 568 (50 μM) or Alexa 488 (100 μM) was also included in the recording pipette to visualize cells using 2-photon laser scanning microscopy (2PLSM). After break-in, the internal solution with the Alexa dye was allowed to equilibrate for 15–20 min before imaging was initiated. The vast majority of 2PLSM 1-photon uncaging experiments utilized Alexa 488 and a Mai Tai HP1040 (Spectra Physics) tuned to 900 nm, whereas several pilot studies utilized Alexa 568 and a Mira 900 (Coherent) infrared laser (with Verdi 10W (532 nm) pump laser) tuned to 790 nm. The laser was pulsed at 80 MHz (Mai Tai HP) or 76 MHz (Mira) (<100 fs, ~250 fs pulse duration, respectively), and a Pockels cell (ConOptics) was used for power attenuation. The dual-channel, 2-photon fluorescence was detected by two non-descanned detectors; green and red channels (dual emission filters: 525/70 nm and 595/50 nm) were detected by the following Hamamatsu photomultiplier tubes (PMTs), respectively: end-on GaAsP (7422PA-40) and side-on multi-alkali (R3896). A 405 nm continuous wave laser (100 mW OBIS FP LX; Coherent) was used for photostimulation/uncaging via a second set of x–y galvanometers incorporated into the scanhead (Cambridge Technologies). A spot diameter of ~1 μm was used for all such laser flash photolysis experiments. 405 nm laser power was measured below the sample but above the condenser using a Field Master GS (LM10 HTD sensor head). A validation study on MHb neurons was conducted with the 405 nm laser in the absence of PA-Nic to identify any laser pulse-associated artifacts (current deflections) in the voltage clamp record. Small (<10 pA) inward currents were detected in several MHb cells when stimulated with 4–5 mW pulses (50 ms) but not at <2.5 mW (Supplementary Fig. 3h, i), so laser pulses in this study did not exceed 2 mW.
PA-Nic was applied (40–80 μM in 5 mL) via superfusion to the slice using a recirculating perfusion system to conserve compound. For some MHb recordings, all MHb Ca2+ imaging, and all hippocampus recordings, local perfusion of PA-Nic (2 mM) was employed. For such studies, a glass pipette with a large diameter (20–40 μm) was back-filled with PA-Nic (2 mM) dissolved in ACSF, and was connected to a pressure ejection device. The PA-Nic-filled pipette was maneuvered to within ~50 μm of the recorded neuron and perfusion was triggered with a constant, low pressure (1–2 psi). Prairie View 5 software was used to select spots in the field of view for focal uncaging of nicotine via 405 nm laser light flashes. Typical flash durations were 5–50 ms (unless otherwise stated), which were selected following an analysis of the relationship between pulse duration and nAChR activation (Supplementary Fig. 3l, m).
2-Photon Ca2+ Imaging with UV–Vis Nicotine Uncaging
MHb neurons from ChAT-Cre mice infected with AAV5.CAG.Flex.GCaMP6f.WPRE.SV40 vectors were identified by two-photon excitation of GCaMP6f (920 nm, ~250 fs pulse duration, 2–3.5 mW) with a 60× (1.0 NA) objective. Time-lapse images (0.074-μm2 pixels, 2-μs pixel dwell time, 6× optical zoom, 0.65 s sampling rate) were acquired from GCaMP6f-expressing neurons that spontaneously oscillated between high and low Ca2+ (Supplementary Fig. 5d); neurons not showing this behavior were not selected for imaging. Spontaneously-active neurons (n=5) were tested for responsiveness to nicotine via superfusion of 100 μM nicotine (Supplementary Fig. 5e). For time-lapse images, the changes in Ca2+ were quantified by calculating the mean pixel intensity along a 2-dimensional (line scan) region of interest crossing the soma. For Ca2+ imaging experiments involving PA-Nic laser flash photolysis, PA-Nic (2 mM) was locally perfused as described above. Somatic changes in Ca2+ before and after PA-Nic photolysis were recorded with a continuous line scan. The GCaMP6f line-scan signal was acquired at 0.33 ms per line and 90 pixels per line with 0.08-μm pixels and 2 μs pixel dwell. Fluorescent intensity values were acquired from the soma and recorded as the mean of 11 pixels from the line scan. The line scan was initiated 5 s before the triggered laser flash (405nm, 5 ms, 2 mW) and continued for 15 s after the light flash. Line scan data was processed with a 21 point moving average before analysis. Ca2+ flux was expressed as the change in fluorescence from baseline (ΔF/F0), as described in equation 5:
| (5) |
where F0 is the baseline GCaMP6f signal, calculated as the mean signal from the 2 s prior to photostimulation and ΔF is the change in fluorescence as calculated as the difference between the peak response (FP, the mean signal from a 1 s window starting 4 s after photostimulation) and F0.
2-Photon Nicotine Uncaging
Slices were transferred to a recording chamber perfused with oxygenated ACSF at a flow rate of 2–3 mL/min. Whole-cell recordings were obtained from neurons of the MHb visualized under infrared Dodt contrast video microscopy using patch pipettes of ~2–5 MΩ resistance. One internal solution consisted of (in mM): 135 KMeSO3, 5 KCl, 0.5 CaCl2, 5 HEPES, 5 EGTA, 10 phosphocreatine disodium salt, 2 ATP, 0.5 GTP. A second internal solution, used in mecamylamine blockade experiments, contained (in mM): 120 CsMeSO4, 15 CsCl, 8 NaCl, 10 TEA-Cl, 10 HEPES, 2 QX-314, 4 ATP, 0.3 GTP, 0.2 EGTA. Alexa Fluor 488 (10–20 μM) was added to the internal solution to visualize cell morphology and confirm cell identity and location. Recordings were made using a Multiclamp 700B amplifier (Molecular Devices). Data were sampled at 10 kHz and filtered at 3 kHz, acquired in MATLAB (MathWorks). Series resistance, measured with a 5 mV hyperpolarizing pulse in voltage clamp, was under 30 MΩ and was left uncompensated. All 2P uncaging voltage clamp recordings were made at a holding potential of −70 mV. ACh sensitivity, observed in 100% of recorded neurons in the experiments validating PA-Nic uncaging efficacy, was tested using pressure ejection application of 1 mM ACh in ACSF.
Two-photon laser-scanning microscopy and two-photon laser photoactivation were accomplished on a modified Scientifica microscope with a 60× (1.0 NA) objective. Two mode-locked Ti:Sapphire lasers (Mai-Tai eHP Deep See and Mai-Tai eHP; Spectra Physics) were separately tuned, with beam power controlled by independent Pockels cells (ConOptics). The beams were separately shuttered, recombined using a polarization-sensitive beam-splitting cube (Thorlabs), and guided into the same galvanometer scanhead (Cambridge). The Mai Tai eHP Deep See was tuned to 910 nm for excitation of Alexa 488, and the Mai Tai eHP variably tuned between 690 and 1000 nm to uncage PA-Nic. A modified version of ScanImage was used for data acquisition37. PA-Nic was added by superfusion (100 μM), or via pressure ejection. For PA-Nic pressure ejection, 300 ms pulses of 200 μM solution in ACSF were delivered at 5–10 psi through a patch pipette placed 20–60 μm away from the recorded cell. Successful photoactivation of PA-Nic, confirmed by blockade with mecamylamine (pre-mec: 11.06 ± 0.9 pA; post-mec: 8.56 ± 1.2 pA; n=11; p=0.008, paired t-test), was observed at the following parameter ranges: 3–20 ms pulse widths, 680–880 nm uncaging laser tuning, and 10–80 mW power measured at the sample plane. A spot diameter of ≤0.8 μm, based on measurements of 0.5 μm beads (17152-10; Polysciences Inc.) imaged with the uncaging laser, was used for all 2-photon laser flash photolysis experiments. Two GaAsP photosensors (Hamamatsu, H7422) with 520/28 nm band pass filters (Semrock), mounted above and below the sample, were used for imaging Alexa 488 fluorescence signals.
Immunohistochemistry and Confocal Microscopy
Mice were anesthetized with sodium pentobarbital (100 mg/kg, i.p.) and transcardially perfused with 10 mL of heparin-containing phosphate buffered saline (PBS) followed by 30 mL of 4% paraformaldehyde. Brains were dissected, postfixed in 4% paraformaldehyde overnight at 4 °C, and dehydrated in 30% sucrose. Coronal brain slices (50 μm) were cut on a freezing/sliding microtome (SM2010R; Leica). MHb-containing slices were stained using the following procedure. Slices were first permeabilized for 2 min via incubation in PBST (0.3% Triton X-100 in PBS), followed by a 60 min incubation in blocking solution (0.1% Triton X-100, 5% horse serum in Tris-buffered saline (TBS)). Primary antibodies used in this study were as follows: goat anti-ChAT (Millipore; cat# AB144P), rabbit anti-DsRed (Clontech; cat# 632496). Primary antibodies were diluted in blocking solution (anti-ChAT at 1:100, anti-DsRed at 1:500). Slices were incubated in primary antibodies overnight at 4 °C. Three 5 min washes in TBST (0.1% Triton X-100 in TBS) were done before transferring slices to secondary antibodies for a 60-min incubation at room temperature. Secondary antibodies, diluted 1:500 in blocking solution, were as follows: anti-goat Alexa 488 (Invitrogen; cat# A11055), anti-rabbit Alexa 647 (Invitrogen; cat# A31573). Slices were washed as before, mounted on slides, and coverslipped with Vectashield. Staining in the MHb or IPN was imaged as previously described12 with a Nikon A1 laser-scanning confocal microscope.
Statistics and Data Analysis
For spectroscopy and photolysis experiments, n denotes the number of independent samples tested, typically prepared from a singular stock solution in DMSO or water. For biology experiments, two-sided statistical tests employed GraphPad Prism 6 software. Flash photolysis responses in IPN were analyzed in MATLAB; for these recordings only, peak inward current was defined as the maximum change in current responses within 100 ms of laser flash onset. For all other laser flash photolysis recordings, the peak current occurred at ± 200 ms of laser flash offset. For pharmacological experiments involving animal models, sample sizes were selected based on prior similar studies12,15, and mice of the appropriate age (2–24 wks) were randomly (informal randomization) chosen for assignment to specific experimental groups. For some studies involving recordings from neurons derived from mice treated with nicotine or control drinking water, the experimenter was blind to the treatment during data collection. Typically, multiple slices were cut from each brain (each animal), and often multiple samples (neurons) were analyzed within each slice depending on the experimental design requirements. During superfusion of brain slices with pharmacological agents such as receptor antagonists, slices were discarded at the conclusion of each recording. For experiments involving variations in flash strength, pulse duration, etc., it was possible to record multiple data points from the same neuron. All experiments involved a minimum of 2 independent replicates. Where samples were taken from animals, every reasonable effort was made to replicate findings in more than one animal. Outlier data points were removed from data sets via the ROUT method (Q=1%) as previously described38. P values for Supplementary Figures 3f and 5b were corrected for multiple comparisons. For presentation purposes in figures, most electrophysiology traces were downsampled to a final sampling interval of 1–2.5 ms, baseline adjusted such that pre-stimulation current/voltage levels were approximately zero, and filtered with a moving average smoothing filter. Group sizes involving recordings from neurons in mouse brain slices are presented as number of cells/number of mice. Unless otherwise stated and where applicable, summary data show mean and standard error of the mean (SEM).
Data Availability
The data that support the findings of this study, if not explicitly contained within the paper, Supplementary Information, or Supplementary Note, are available from the corresponding authors upon reasonable request.
Supplementary Material
Acknowledgments
We thank members of the Drenan and Lavis laboratories for helpful advice and discussion and T. Lerner for contributing viral reagents. This work was supported by the Howard Hughes Medical Institute (to S.B. J.J.M, and L.D.L.), National Institutes of Health (NIH) grants (DA035942 and DA040626 to R.M.D., MH099114 to A.C., DA037161 to H.A.L., NS054850 to D.J. Surmeier, GM103801 and GM48677 to J.M.M.), PhRMA Foundation Fellowship (to M.C.A.), Beckman Young Investigator Award (to Y.K.), Bernice E. Bumpus Foundation Early Career Innovation Award (to. Y.K.), Rita Allen Foundation Award (to Y.K), Searle Scholar Award (to Y.K.), Sloan Research Fellowship (to Y.K.), NINDS F32 NS103243 (to N.M.B.), funding from the JPB Foundation, and funds from Northwestern University.
Footnotes
Author Contributions
R.M.D., M.C.A., H.A.L., S.B., K.R.G., and L.D.L. conceived the project. M.C.A., N.M.B., D.L.W., X.J., J.J.M., Y.W., C.P., V.J.K., J.J.M., A.C., Y.K., R.M.D., S.B., and L.D.L. planned and/or executed experiments. D.L.W., Y.K., J.M.M., and K.R.G. contributed essential reagents and expertise. R.M.D., M.C.A., S.B., and L.D.L. wrote the paper with input from all authors. R.M.D. and L.D.L. supervised all aspects of the work.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Matsuzaki M, Ellis-Davies GC, Nemoto T, et al. Nat Neurosci. 2001;4:1086–1092. doi: 10.1038/nn736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellis-Davies GC. Nat Methods. 2007;4:619–628. doi: 10.1038/nmeth1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsuzaki M, Hayama T, Kasai H, et al. Nat Chem Biol. 2010;6:255–257. doi: 10.1038/nchembio.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Furuta T, Wang SS, Dantzker JL, et al. Proc Natl Acad Sci USA. 1999;96:1193–1200. doi: 10.1073/pnas.96.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hagen V, Dekowski B, Nache V, et al. Angew Chem Int Ed. 2005;44:7887–7891. doi: 10.1002/anie.200502411. [DOI] [PubMed] [Google Scholar]
- 6.Hagen V, Dekowski B, Kotzur N, et al. Chem Eur J. 2008;14:1621–1627. doi: 10.1002/chem.200701142. [DOI] [PubMed] [Google Scholar]
- 7.Sarker AM, Kaneko Y, Neckers DC. J Photochem Photobiol A. 1998;117:67–74. [Google Scholar]
- 8.Petersson EJ, Choi A, Dahan DS, et al. J Am Chem Soc. 2002;124:12662–12663. doi: 10.1021/ja028206i. [DOI] [PubMed] [Google Scholar]
- 9.Mccarron ST, Feliciano M, Johnson JN, et al. Bioorg Med Chem Lett. 2013;23:2395–2398. doi: 10.1016/j.bmcl.2013.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asad N, Deodato D, Lan X, et al. J Am Chem Soc. 2017;139:12591–12600. doi: 10.1021/jacs.7b06363. [DOI] [PubMed] [Google Scholar]
- 11.Filevich O, Salierno M, Etchenique R. J Inorg Biochem. 2010;104:1248–1251. doi: 10.1016/j.jinorgbio.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Shih PY, Engle SE, Oh G, et al. J Neurosci. 2014;34:9789–9802. doi: 10.1523/JNEUROSCI.0476-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren J, Qin C, Hu F, et al. Neuron. 2011;69:445–452. doi: 10.1016/j.neuron.2010.12.038. [DOI] [PubMed] [Google Scholar]
- 14.Salas R, Sturm R, Boulter J, et al. J Neurosci. 2009;29:3014–3018. doi: 10.1523/JNEUROSCI.4934-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shih PY, Mcintosh JM, Drenan RM. Mol Pharmacol. 2015 doi: 10.1124/mol.115.101444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen TW, Wardill TJ, Sun Y, et al. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khiroug L, Giniatullin R, Klein RC, et al. J Neurosci. 2003;23:9024–9031. doi: 10.1523/JNEUROSCI.23-27-09024.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parikh V, Kozak R, Martinez V, et al. Neuron. 2007;56:141–154. doi: 10.1016/j.neuron.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grimm JB, English BP, Choi H, et al. Nat Methods. 2016;13:985–988. doi: 10.1038/nmeth.4034. [DOI] [PubMed] [Google Scholar]
- 20.Schoenleber RO, Giese B. Synlett. 2003:501–504. [Google Scholar]
- 21.Shembekar VR, Chen Y, Carpenter BK, et al. Biochemistry. 2007;46:5479–5484. doi: 10.1021/bi700280e. [DOI] [PubMed] [Google Scholar]
- 22.Schaal J, Dekowski B, Wiesner B, et al. Chembiochem. 2012;13:1458–1464. doi: 10.1002/cbic.201200110. [DOI] [PubMed] [Google Scholar]
- 23.Herbivo C, Omran Z, Revol J, et al. Chembiochem. 2013;14:2277–2283. doi: 10.1002/cbic.201300425. [DOI] [PubMed] [Google Scholar]
- 24.Nadler A, Yushchenko DA, Muller R, et al. Nat Commun. 2015;6:10056. doi: 10.1038/ncomms10056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu C, Webb WW. J Op Soc Am B. 1996;13:481–491. [Google Scholar]
- 26.Makarov NS, Drobizhev M, Rebane A. Opt Express. 2008;16:4029–4047. doi: 10.1364/oe.16.004029. [DOI] [PubMed] [Google Scholar]
- 27.Mutze J, Iyer V, Macklin JJ, et al. Biophys J. 2012;102:934–944. doi: 10.1016/j.bpj.2011.12.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grimm JB, English BP, Chen J, et al. Nat Methods. 2015;12:244–250. doi: 10.1038/nmeth.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis MJ, Kragor CH, Reddie KG, et al. J Org Chem. 2009;74:1721–1729. doi: 10.1021/jo802658a. [DOI] [PubMed] [Google Scholar]
- 30.Azam L, Dowell C, Watkins M, et al. J Biol Chem. 2005;280:80–87. doi: 10.1074/jbc.M406281200. [DOI] [PubMed] [Google Scholar]
- 31.Azam L, Maskos U, Changeux JP, et al. FASEB J. 2010;24:5113–5123. doi: 10.1096/fj.10-166272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossi J, Balthasar N, Olson D, et al. Cell Metab. 2011;13:195–204. doi: 10.1016/j.cmet.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madisen L, Zwingman TA, Sunkin SM, et al. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taniguchi H, He M, Wu P, et al. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao-Shea R, Degroot SR, Liu L, et al. Nat Commun. 2015;6:6770. doi: 10.1038/ncomms7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Engle SE, Broderick HJ, Drenan RM. J Vis Exp. 2012:e50034. doi: 10.3791/50034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pologruto TA, Sabatini BL, Svoboda K. Biomed Eng Online. 2003;2:13. doi: 10.1186/1475-925X-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Motulsky HJ, Brown RE. BMC Bioinformatics. 2006;7:123. doi: 10.1186/1471-2105-7-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study, if not explicitly contained within the paper, Supplementary Information, or Supplementary Note, are available from the corresponding authors upon reasonable request.