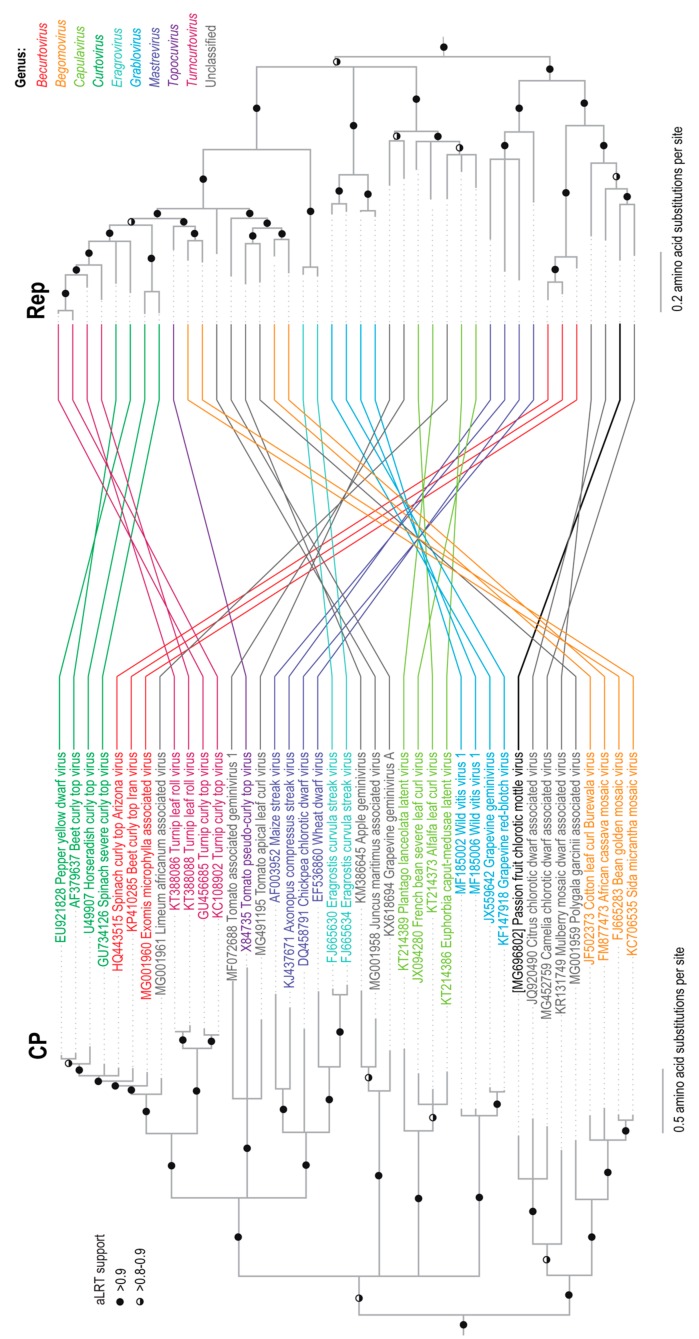
Figure 2.
Maximum likelihood phylogenetic tree inferred using the substitution models rtREV+G+I+F and rtREV+G+I for capsid protein (CP) and Rep amino acid sequences, respectively, with an approximate likelihood ratio test (aLRT) for branch support of representative geminiviruses and PCMoV identified in this study. Both trees are rooted with the CP and Rep amino acid sequences of the genomovirus, Sclerotinia sclerotiorum hypovirulence associated DNA virus 1 (KM598384) and MSSI2.225 virus (LK931485). Branches with <0.8 aLRT branch support have been collapsed.