Abstract
Recently, a Salmonella Typhi isolate producing CTX-M-15 extended spectrum β-lactamase (ESBL) and with decreased ciprofloxacin susceptibility was isolated in the Democratic Republic of the Congo. We have selected bacteriophages that show strong lytic activity against this isolate and have potential for phage-based treatment of S. Typhi, and Salmonella in general.
Keywords: typhoid fever, Salmonella Typhi, extended-spectrum beta lactamases (ESBL), Democratic Republic of the Congo, bacteriophages
Although effective oral antibiotics used to be readily available, today antimicrobial resistance in Salmonella Typhi is becoming an increasingly serious public health concern, especially in low- and middle-income countries (LMICs). Resistance to all first-line antimicrobials used in the treatment of S. Typhi infections emerged sequentially, leading to multidrug resistance (MDR) in the 1990s, and more recently, high levels of fluoroquinolone resistance in South Asia [1]. Recent data from the Typhoid Fever Surveillance in Africa Program (TSAP) indicates that the incidence rate for typhoid fever in Africa has been underestimated and is equal to, or even greater than, incidences reported in Asia [2]. Therefore, extended spectrum β-lactamase (ESBL) producing Enterobacteriaceae and fluoroquinolone resistant Salmonellae were included in the high-priority tier of the recent WHO priority list of antibiotic-resistant bacteria [3].
In general, it is acknowledged that global antimicrobial resistance (AMR) poses a fundamental long-term threat to human health, the production of food, and sustainable development. Based on scenarios of rising drug resistance for only six pathogens, experts estimated that by 2050, up to 10 million people could die every year from the effects of AMR and it could also impose an economic burden of US$100 trillion [4]. Recently, the UN committed to supporting the development of new antimicrobial agents and therapies [5].
Phage therapy is one of the promising “new” treatments that has been increasingly featured [6]. Bacteriophages (phages in short) are naturally occurring viruses of bacteria. Since the early phases of evolution, phages have controlled bacteria on our planet. In the early twentieth century, humans discovered them and immediately applied them to medicine. This was especially true in the former Soviet Union, where the use of phages continued after the advent of commercial antibiotics [7]. They can be selected to kill only certain bacteria of concern (e.g., bacteria causing infectious diseases) while leaving non-pathogenic bacteria and mammalian cells unharmed. As such, they can be effective against antibiotic-resistant bacteria and, in contrast to broad-spectrum antibiotics, spare the gut microbiota, which could particularly benefit malnourished and immunocompromised patients. In addition, phages can be easily isolated from environmental sources such as river or sewage water, using basic tools available in LMICs [8].
In 2017, a case of typhoid fever in a six-year-old boy in the Democratic Republic of the Congo (DRC) caused by an S. Typhi isolate producing CTX-M-15 extended spectrum β-lactamase (ESBL) and showing decreased ciprofloxacin susceptibility, was reported [9]. CTX-M-15 is part of the M1 group that includes six plasmid-mediated enzymes [10]. This isolate, named Typhi 10040_15, was sent to the Eliava Institute of Bacteriophage, Microbiology & Virology (EIBMV) in Tbilisi (Georgia) to determine phage susceptibility. The 14 Salmonella phage clones of the Eliava R&D collection and five batches of the commercial phage cocktail “INTESTI phage” were tested. Phage screening against this S. Typhi strain was performed at the EIBMV’s BSL-2 Plus laboratory, meeting the required safety requirements. The fourteen phage clones were isolated from the river Mtkvari in Tbilisi, from the Black Sea (Batumi) and from the Tbilisi sewage water supply system in the period 2013–2017 (Table 1). The five tested INTESTI phage batches were #M 067 (produced in July 2017), #M2 901 (November 2017), #84 of (February 2017), #82 (January 2017), and #78 (December 2016).
Table 1.
Characteristics of 14 Salmonella phages from the Eliava collection tested on the Typhi 10040_15_DRC_2015 isolate from the Democratic Republic of the Congo (DRC).
| Nr | Name | GenBank Accession Numbers | Source | Isolation Year | Host Strain | Family | Homology to Other Phages | Host Range (%) *All Serovars/S. Enteritidis/S. Typhimurium/S. Dublin (Total Number of Tested Isolates) | Lytic Activity on the Typhi 10040_15_DRC_2015 Isolate | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Streak Method | Spot Test | Gratia’s Method | Appelmans’ Method | |||||||||||||
| Titer (pfu/mL) | Dilution | 6 h | 18 h | 24 h | ||||||||||||
| 001 | GE_vB_N3 | ND | Mtkvari river water | 2013 | S. Enteritidis 3 | Siphoviridae | ND | 78/98/73/87 (239) | + | + | + | 109 | −1 | − | − | + |
| 108 | −2 | − | − | + | ||||||||||||
| 107 | −3 | − | + | + | ||||||||||||
| 002 | GE_vB_N5 | MG969412 | Mtkvari river water | 2013 | S. Enteritidis 3 | Siphoviridae | E. coli T5 strain ATCC 11303-B5 | 45/80/25/74 (239) | + | + | + | 108 | −1 | − | + | + |
| 107 | −2 | − | + | + | ||||||||||||
| 106 | −3 | − | + | + | ||||||||||||
| 003 | GE_vB_N8 | MG969413 | Mtkvari river water | 2013 | S. Enteritidis 3 | Siphoviridae | phage SPC35 | 65/84/67/78 (239) | + | + | − | 109 | −1 | − | − | + |
| 108 | −2 | − | − | + | ||||||||||||
| 107 | −3 | − | + | + | ||||||||||||
| 004 | GE_vB_MG | MG969411 | Tbilisi sewage water | 2013 | S. Enteritidis 3 | Myoviridae | S. phage PVP-SE1 | 47/49/59/43 (239) | − | − | + | 109 | −1 | − | − | − |
| 108 | −2 | − | − | − | ||||||||||||
| 107 | −3 | − | − | − | ||||||||||||
| 005 | GE_vB_BS | MG969407 | Black Sea water | 2013 | S. Typhimurium 4 | Myoviridae | S SPT-1, partial genome | 81/96/93/83 (239) | + | + | + | 1010 | −1 | + | − | − |
| 109 | −2 | + | − | − | ||||||||||||
| 108 | −3 | + | − | − | ||||||||||||
| 006 | GE_vB_B1 | MG969405 | Mtkvari river water | 2013 | S. Typhimurium 6 | Myoviridae | S. phage Mushroom | 80/93/83/87 (239) | + | + | + | 109 | −1 | + | − | − |
| 108 | −2 | + | − | − | ||||||||||||
| 107 | −3 | + | − | − | ||||||||||||
| 007 | GE_vB_B3 | MG969406 | Mtkvari river water | 2013 | S. Typhimurium 6 | Myoviridae | S. phage Mushroom | 81/98/73/87 (239) | + | + | + | 109 | −1 | + | − | − |
| 108 | −2 | + | − | − | ||||||||||||
| 107 | −3 | + | − | − | ||||||||||||
| 008 | GE_vB_NS7 | MG969414 | Raw cow milk | 2015 | S. Typhimurium 6 | Myoviridae | S. phage Mushroom | 75/91/81/78 (239) | + | + | + | 109 | −1 | + | − | − |
| 108 | −2 | − | + | − | ||||||||||||
| 107 | −3 | − | + | + | ||||||||||||
| 009 | GE_vB_M4 | MG969409 | Black Sea water | 2016 | S. Enteritidis 232 | Siphoviridae | S. phage vB_SenS-Ent3 | 23/64/18/22 (218) | + | + | + | 1010 | −1 | + | − | − |
| 109 | −2 | + | − | − | ||||||||||||
| 108 | −3 | − | − | − | ||||||||||||
| 010 | GE_vB_M5 | MG969410 | Black Sea water | 2016 | S. Enteritidis 407 | Siphoviridae | S. phage vB_SenS-Ent3 | 33/66/26/61 (218) | + | + | + | 108 | −1 | + | − | − |
| 107 | −2 | + | − | − | ||||||||||||
| 106 | −3 | + | − | − | ||||||||||||
| 011 | GE_vB_TR | MG969415 | Mtkvari river water | 2017 | S. Typhimurium 641 | Podoviridae | S. phage BTP1 | 40/90/28/59 (141) | − | − | + | 109 | −1 | − | − | − |
| 108 | −2 | − | − | − | ||||||||||||
| 107 | −3 | − | − | − | ||||||||||||
| 012 | GE_vB_HIL | MG969408 | Mtkvari river water | 2017 | S. Enteritidis 765 | Siphoviridae | S. phage vB_SenS-Ent3 | 58/81/75/77 (141) | + | + | − | 1010 | −1 | + | − | − |
| 109 | −2 | − | − | − | ||||||||||||
| 108 | −3 | − | + | + | ||||||||||||
| 013 | GE_vB_7A | MG969404 | Mtkvari river water | 2017 | S. Typhimurium 1328 | Myoviridae | S. phage BPS15Q2 | 37/62/28/23 (141) | + | + | + | 108 | −1 | − | − | − |
| 107 | −2 | − | − | − | ||||||||||||
| 106 | −3 | − | − | − | ||||||||||||
| 014 | GE_vB_M1 | ND | Black Sea water | 2016 | S. Enteritidis 104 | Podoviridae | ND | 12/20/19/0 (77) | + | + | − | 109 | −1 | − | − | − |
| 108 | −2 | − | − | − | ||||||||||||
| 107 | −3 | − | − | − | ||||||||||||
* The host range of the phages was determined for the total Salmonella strain collection (all serovars), with the total number of strains indicated between brackets, and for the three main serovars (S. Enteritidis/S. Typhimurium/S. Dublin) separately. ND, not done; “+”, phage lytic activity; “−”, no phage lytic activity; S. phage: Salmonella phage.
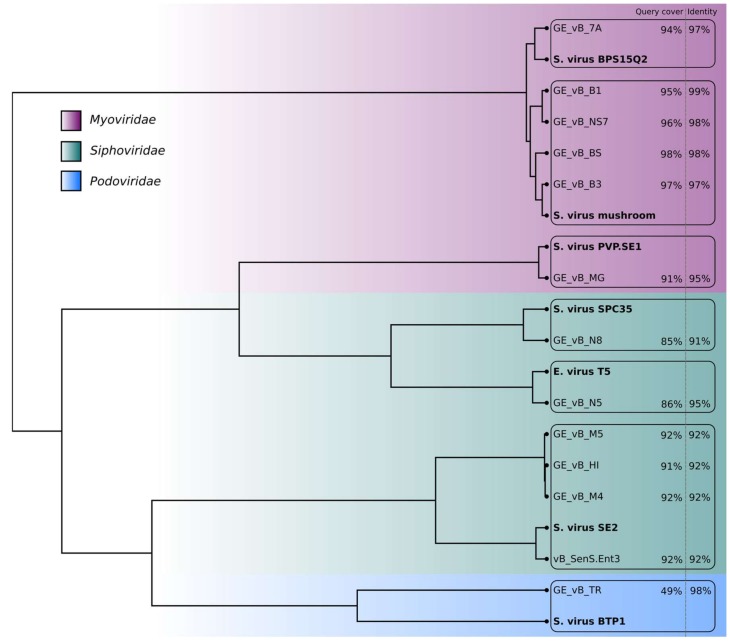
As in many other pathogenic bacteria, temperate phages contribute to virulence in Salmonella enterica through the acquisition and exchange of virulence factors [11]. To assert the strictly lytic nature of these phages, high-resolution genome maps of 12 of the 14 individual phages were obtained using nanopore sequencing [12]. A pooled library consisting of barcoded genomic DNA of the phages was prepared using native barcodes and the 1D ligation kit from Oxford Nanopore Technology (ONT). The result was then sequenced on a MinION device, equipped with an R9.4 flowcell. For the data analysis, Albacore v2.1 (ONT, Oxford, UK) was used for base-calling the reads, followed by porechop v0.2.1 (https://github.com/rrwick/Porechop) in order to remove barcode sequences. Genome map assembly was performed with Canu v1.6 (https://github.com/marbl/canu) [13]. All the assembled genomes were subsequently processed with Racon v0.5 for better consensus sequences [14], and nanopolish v0.8.3 (https://github.com/jts/nanopolish) for higher accuracy of base-called nucleotides in the sequences. Considering the intrinsic properties of nanopore sequencing, together with the run coverage (30× to 60×), we define these assemblies as high-resolution phage maps, rather than fully accurate genome sequences. Known homologous phage isolates were first located using the blastn tool on the NCBI nucleotide database [15]. For each of our new isolates, the closest match (highlighted in bold) in terms of query coverage and identity was identified and the corresponding genome downloaded. The genomic distance between all the pairs of phages was calculated using Mash [16] and the resulting distance matrix was used to build the clustering tree (Figure 1) with the hclust function found in the R stats package [17]. No known toxin genes were present and the genomes did not contain recognizable integrase genes, corroborating the lytic nature of these bacteriophages. Sequences were submitted to GenBank (Accession numbers: MG969404-15).
Figure 1.
Clustering tree based on the genomic distance matrix generated for the Salmonella phages from the Eliava R&D collection and their closest matches (in bold) in the NCBI database. No genome maps were obtained for phages GE_vB_N3 (Siphovirus) and GE_vB_M1 (Podovirus).
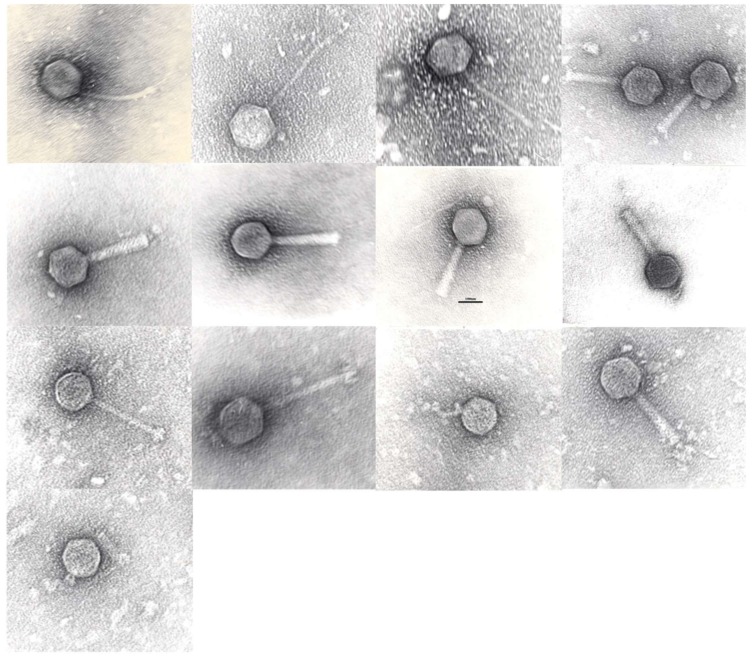
Morphological analysis using Transmission Electron Microscopy (TEM) of the phage clones confirmed their general classification (Table 1). Purification and staining of the samples was performed according to Hans-W. Ackermann [18] and preparations were examined with JEOL-JEM-1400 TEM (Figure 2). Phages belonged to the families of the Siphoviridae (n = 6), Myoviridae (n = 6) and Podoviridae (n = 2). The genome map-based grouping allowed us to further assign these phage clones to individual phage species/genera, as indicated in Table 1.
Figure 2.
Tansmission electron micrographs of phages in left->right, top->bottom order: GE_vB_N3, GE_vB_N5, GE_vB_N8, GE_vB_MG, GE_vB_BS, GE_vB_B1, GE_vB_B3, GE_vB_NS7, GE_vB_M4, GE_vB_M5, GE_vB_TR, GE_vB_7A, GE_vB_M1. Scale bar, 100 nm.
The host range of the phages was assessed by screening their lytic activity and using the spot test against 118 clinical and 121 veterinary Salmonella. spp. isolates from Georgia (20), Armenia (71), Germany (7), and Ireland (141). These isolates belonged to the following serotypes: S. Typhimurium (95), S. Enteritidis (45), S. Dublin (23), S. Anatum (11), S. Infantis (9), S. Newport (8), S. Derbey (8), S. Bredney (5), S. Branderburg (3), S. Germinara (2), S. Uganda (2), S. Senftenberg (2), S. Kentucky (2), S. Reading (2), S. Parat. B (2), S. Java (1), S. Bareilly (1), S. Virchow (1), S. Goldcost (1), S. Kottbus (1), S. Agona (1), and S. Poona (1). Thirteen isolates were not attributed to any known serotypes. Two hundred microlitres of S. enterica mid-log phase cultures were mixed with 5 mL of lukewarm 0.6% Lysogeny Broth (LB, Merck, Darmstadt, Germany) agar and overlaid on LB agar plates. The LB broth consisted of 10 g peptone from casein, 5 g yeast extract and 10 g NaCl in 1 L of deionized water. After the plates had cooled, 5 µL of the phage clones, with a titer of 107 plaque forming units (pfu)/mL was spotted on the lawn. Drops were air-dried and plates were incubated for 18 h at 37 °C. After the incubation, the plates were checked for zones of clearance resulting from phage activity [19]. The host range of the phages varied from 12 to 81% of the S. enterica strains (Table 1). It should be noted that the spot test is usually performed to determine bacterial susceptibility and host range using as many bacterial strains as possible because this method is simple, quick and inexpensive. A significant part of bacterial cell killing can be due to “lysis from without”, i.e., the destruction of bacterial cells by the adherence of a sufficiently high number of phages to the bacterial cell, and the destruction of an essential cell wall structure by an extracellular lytic enzyme with subsequent lysis, but without phage replication.
The lytic activity of the 14 phage clones and of the five INTESTI phage batches against the Typhi 10040_15 isolate from the DRC was evaluated using the spot test and the streak method. For the streak method, mid-log phase S. Typhi was plated as a single line on an agar plate, air dried and 5 µL of single phage clones (titer 107 pfu/mL) was dropped on the bacterial line. Drops were air-dried and plates were incubated for 18 h at 37 °C. After incubation, plates were checked for zones of clearance resulting from phage activity [20]. Twelve out of 14 phage clones and three out of five batches of the commercial preparation INTESTI phage formed clear lysis zones on the CTX-M-15, producing an S. Typhi strain from the DRC. This occurred in the streak method, as well as in the spot test. The batches #M 067 and #M2 901 showed the strongest lytic activity with clear confluent zones, while batch #84 only developed weakly distinguishable discrete lytic zones. Only two phage clones, phage GE_vB_MG (Myoviridae, Vequintavirinae, Se1virus, Salmonella virus SE1) and phage GE_vB_TR, a potential lysogenic phage (Podoviridae, P22virus), showed no activity against the Typhi 10040_15 isolate (Table 1).
To assess the ability of phages to multiply inside the bacterial cells (creating phage plaques), the activity of the 14 phage clones (but not the INTESTI phage batches) was tested using the method of Gratia. A 200 µL mid-log phase culture of S. Typhi and 1 mL of a ten-fold diluted phage suspension, ranging from 106 to 1010 pfu/mL, were mixed with 5 mL of lukewarm 0.6% LB agar and overlaid on LB agar. Plates were incubated at 37 °C for 18 h and checked for plaque formation [21]. All phage clones, with the exception of three (GE_vB_N8, GE_vB_HIL and GE_vB_M1), were found to form plaques on the bacterial lawn (Table 1).
Finally, to confirm the ability of the phages to lyse the DRC S. Typhi strain in aqueous solutions, the lytic activity of the 14 phage clones was determined using Appelman’s method. The 14 phage clones were diluted ten-fold to range from 106 to 1010 pfu/mL in 5 mL of LB broth and inoculated with 150 µL (109 cfu/mL) overnight S. Typhi culture. Mixtures were incubated at 37 °C without shaking and the turbidity of the samples was checked visually after 6, 18 and 24 h. As reference and control samples, phage-free bacterial culture and diluted phages without bacteria were tested under the same conditions [22]. Ten phage clones showed activity at different time points and concentrations, five of which showed the ability to lyse the CTX-M-15 producing S. Typhi isolate without forming phage-resistant mutants after 24 h of incubation, which is indicative of their inherent ability to limit the growth of phage-resistant mutants during phage therapy: GE_vB_N3, GE_vB_N5, GE_vB_N8, GE_vB_NS7 and GE_vB_HIL (Table 1). It should be noted that phages GE_vB_MG and GE_vB_TR are not active according to the spot and the streak tests, while still forming plaques according to the method of Gratia. The difference between these two methods is not that uncommon and could, for example, be caused by differences between phages’ “lysis from without” (accentuated in the spot test) and phage infection, or “lysis from within” (accentuated in the Gratia test) capabilities. In other words, some phages could be less efficient in adhering to, and entering bacterial cells, while being very efficient once the normal lytic cycle is initiated. The spot test and the streak method are rapid ways to check whether a phage can infect a bacterium by trickling small droplets of the phage suspensions to be tested on a plate prepared with a bacterial isolate. Limitations of these tests compared with the Gratia and Appelmans methods are that clear zones on the bacterial lawn (a positive test), may be the result of abortive infection or lysis from without, both forming clear zones without new phages being produced.
The evolution of bacterial resistance to phages is often observed in vitro. Phages have evolved multiple strategies to overcome the antiviral mechanisms they encounter when infecting bacterial cells [23]. In experimental settings, phage-resistant bacteria emerged rapidly, but often at significant fitness costs, shown by the reduced growth rate in the absence of phages [24]. The in vivo evolution of bacterial resistance to phages in human clinical practice, however, was poorly documented until now. It has been suggested that phages could be combined with antibiotics to improve phage activity (synergy) [25].
In conclusion, in a short time frame (two days), at least five phage clones from the Eliava collection were found to exhibit excellent in vitro lytic activity against the ESBL producing S. Typhi isolate from the DRC. Phages can be considered a potential additional tool for the treatment of MDR Salmonella infections and a (food) decontamination agent. Antimicrobials that address foodborne diseases are particularly important for LMICs as many of them lack reliable cold chain infrastructure and adequate hygiene practices [8]. In Western countries, several phage products are currently approved for the control of food pathogens, including Salmonella. In addition, phage preparations can be developed and produced faster and cheaper than conventional drugs. They can also be (freeze-)dried [26] so that they require no refrigeration [8].
Author Contributions
All authors contributed to the conception and writing of this paper.
Conflicts of Interest
The authors declare no conflict of interest.
Funding
This work was supported by the International Science and Technology Center (grant number ISTC-A 2140), the Science Foundation Ireland (12/R1/2335), the KU Leuven Orchestrated Research Action (Phage Biosystems to RL), the Fonds Wetenschappelijk Onderzoek Vlaanderen (1S64718N to CL), the Belgian Directorate of Development Cooperation (DGD) (Project 2.01 of the 3rd Framework Agreement between the Belgian DGD and the ITM Antwerp, Belgium), the Baillet-Latour Fund (The Bacterial Infections in the Tropics (BIT) research cluster at ITM Antwerp, Belgium) and the Department of Economy, Science and Innovation in Flanders (EWI funding ITM Antwerp).
References
- 1.Karkey A., Thwaites G.E., Baker S. The evolution of antimicrobial resistance in Salmonella Typhi. Curr. Opin. Gastroenterol. 2018;34:25–30. doi: 10.1097/MOG.0000000000000406. [DOI] [PubMed] [Google Scholar]
- 2.Marks F., von Kalckreuth V., Aaby P., Adu-Sarkodie Y., El Tayeb M.A., Ali M., Aseffa A., Baker S., Biggs H.M., Bjerregaard-Andersen M., et al. Incidence of invasive Salmonella disease in sub-Saharan Africa: A multicentre population-based surveillance study. Lancet Glob. Health. 2017;5:e310–e323. doi: 10.1016/S2214-109X(17)30022-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D.L., Pulcini C., Kahlmeter G., Kluytmans J., Carmeli Y., et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2017;18:234–236. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 4.Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. The Review on Antimicrobial Resistance. [(accessed on 27 January 2018)];2016 Available online: https://amr-review.org/sites/default/files/160525_Final paper_with cover.pdf.
- 5.United Nations Draft Political Declaration of the High-Level Meeting of the General Assembly on Antimicrobial Resistance (16-16108 (E)) [(accessed on 17 December 2017)]; Available online: http://www.un.org/pga/71/wp-content/uploads/sites/40/2016/09/DGACM_GAEAD_ESCAB-AMR-Draft-Political-Declaration-1616108E.pdf.
- 6.Watts G. Phage therapy: Revival of the bygone antimicrobial. Lancet. 2017;390:2539–2540. doi: 10.1016/S0140-6736(17)33249-X. [DOI] [PubMed] [Google Scholar]
- 7.Chanishvili N. Bacteriophages as Therapeutic and Prophylactic Means: Summary of the Soviet and Post Soviet Experiences. Curr. Drug Deliv. 2016;13:309–323. doi: 10.2174/156720181303160520193946. [DOI] [PubMed] [Google Scholar]
- 8.Nagel T.E., Chan B.K., de Vos D., El-Shibiny A., Kang’ethe E.K., Makumi A., Pirnay J.P. The Developing World Urgently Needs Phages to Combat Pathogenic Bacteria. Front. Microbiol. 2016;7:882. doi: 10.3389/fmicb.2016.00882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phoba M.F., Barbé B., Lunguya O., Masendu L., Lulengwa D., Dougan G., Wong V.K., Bertrand S., Ceyssens P.J., Jacobs J., et al. Salmonella enterica serovar Typhi Producing CTX-M-15 Extended Spectrum β-Lactamase in the Democratic Republic of the Congo. Clin. Infect. Dis. 2017;65:1229–1231. doi: 10.1093/cid/cix342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonnet R. Growing group of extended-spectrum β-lactamases: The CTX-M enzymes. Antimicrob. Agents Chemother. 2004;48:1–14. doi: 10.1128/AAC.48.1.1-14.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho T.D., Figueroa-Bossi N., Wang M., Uzzau S., Bossi L., Slauch J.M. Identification of GtgE, a novel virulence factor encoded on the Gifsy-2 bacteriophage of Salmonella enterica serovar Typhimurium. J. Bacteriol. 2002;184:5234–5239. doi: 10.1128/JB.184.19.5234-5239.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deamer D.W., Akeson M. Nanopores and nucleic acids: Prospects for ultrarapid sequencing. Trends Biotechnol. 2000;18:147–150. doi: 10.1016/S0167-7799(00)01426-8. [DOI] [PubMed] [Google Scholar]
- 13.Koren S., Walenz B.P., Berlin K., Miller J.R., Bergman N.H., Phillippy A.M. Canu: Scalable and accurate long-read assembly via adaptive K-mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaser R., Sović I., Nagarajan N., Šikić M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 2017;27:737–746. doi: 10.1101/gr.214270.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coordinators N.R. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2016;44:D7. doi: 10.1093/nar/gkv1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ondov B.D., Treangen T.J., Melsted P., Mallonee A.B., Bergman N.H., Koren S., Phillippy A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016;17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2015. [(accessed on 24 March 2018)]. Available online: https://www.R-project.org/ [Google Scholar]
- 18.Ackermann H.W. Basic phage electron microscopy. In: Clokie M.R.J., Kropinski A., editors. Bacteriophages: Methods and Protocols. 1st ed. Vol. 1. Humana Press; New York, NY, USA: 2009. pp. 113–126. [DOI] [PubMed] [Google Scholar]
- 19.Kutter E. Phage host range and efficiency of plating. In: Clokie M.R.J., Kropinski A., editors. Bacteriophages: Methods and Protocols. 1st ed. Vol. 1. Humana Press; New York, NY, USA: 2009. pp. 141–149. [DOI] [PubMed] [Google Scholar]
- 20.Matsuzaki S., Uchiyama J., Takemura-Uchiyama I., Ujihara T., Daibata M. Isolation of Bacteriophages for Fastidious Bacteria. In: Azeredo J., Sillankorva S., editors. Bacteriophage Therapy. 1st ed. Humana Press; New York, NY, USA: 2018. pp. 3–10. [DOI] [PubMed] [Google Scholar]
- 21.Gratia A. Des relations numeriques entre bacteries lysogenes et particules de bacteriophage. Ann. Inst. Pasteur. 1936;57:652–676. [Google Scholar]
- 22.Appelmans R. Le dosage du bactériophage. C. R. Seances Soc. Biol. 1921;85:1098. [Google Scholar]
- 23.Labrie S.J., Samson J.E., Moineau S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010;8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 24.Hall A.R., de Vos D., Friman V.P., Pirnay J.P., Buckling A. Effects of sequential and simultaneous applications of bacteriophages on populations of Pseudomonas aeruginosa in vitro and in wax moth larvae. Appl. Environ. Microbiol. 2012;78:5646–5652. doi: 10.1128/AEM.00757-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamal F., Dennis J.J. Burkholderia cepacia complex Phage-Antibiotic Synergy (PAS): Antibiotics stimulate lytic phage activity. Appl. Environ. Microbiol. 2015;81:1132–1138. doi: 10.1128/AEM.02850-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merabishvili M., Vervaet C., Pirnay J.P., De Vos D., Verbeken G., Mast J., Chanishvili N., Vaneechoutte M. Stability of Staphylococcus aureus phage ISP after freeze-drying (lyophilization) PLoS ONE. 2013;8:e68797. doi: 10.1371/journal.pone.0068797. [DOI] [PMC free article] [PubMed] [Google Scholar]