Abstract
The thiol tripeptides glutathione (GSH) and homoglutathione (hGSH) are very abundant in legume root nodules and their synthesis is catalyzed by the enzymes γ-glutamylcysteine synthetase (γECS), GSH synthetase (GSHS), and hGSH synthetase (hGSHS). As an essential step to elucidate the role of thiols in N2 fixation we have isolated cDNAs encoding the three enzymes and have quantified the transcripts in nodules. Assay of enzyme activities in highly purified nodule organelles revealed that γECS is localized in the plastids, hGSHS in the cytosol, and GSHS in the cytosol and mitochondria. These results are consistent with sequence analyses. Subcellular fractionation of nodules also showed that bacteroids contain high thiol concentrations and high specific γECS and GSHS activities. Results emphasize the role of nodule plastids in antioxidant protection and in control of thiol synthesis, and suggest that plastids may be important in the stress response of nodules. Overall, our results provide further evidence that thiol synthesis is critical for nodule functioning.
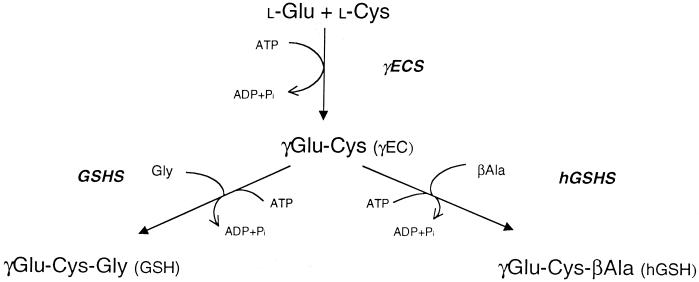
The thiol tripeptide glutathione (GSH; γGlu-Cys-Gly) is very abundant in plants where it performs a multiplicity of important functions ranging from scavenging of reactive oxygen species to heavy metal detoxification (Hausladen and Alscher, 1993; Rennenberg, 1997; May et al., 1998a). The synthesis of GSH involves two reactions, catalyzed by γ-glutamylcysteine synthetase (γECS; EC 6.3.2.2) and GSH synthetase (GSHS; EC 6.3.2.3), which are strictly dependent on ATP and Mg2+ (Fig. 1). In the leaves the synthesis of GSH is thought to take place in the chloroplasts and cytosol (Hausladen and Alscher, 1993; Rennenberg, 1997; Noctor and Foyer, 1998).
Figure 1.
Pathway for GSH and hGSH synthesis in legumes. The two ATP-dependent reactions leading to GSH synthesis are, respectively, the condensation of Glu and Cys to form γEC (catalyzed by γECS) and the addition of Gly to the C terminus of γEC (catalyzed by GSHS). For hGSH synthesis, βAla replaces Gly in the second reaction (catalyzed by hGSHS).
Legumes may contain another thiol tripeptide, homoglutathione (hGSH; γGlu-Cys-βAla), partially or fully replacing GSH (Fig. 1). The synthesis of hGSH from γEC and βAla is catalyzed by a specific hGSH synthetase (hGSHS) with high affinity for βAla and low affinity for Gly (Macnicol, 1987; Klapheck et al., 1988). Based on analyses of thiol metabolites and thiol synthetase activities in different organs and nodule tissues of eight legumes of agronomic relevance, we proposed the hypothesis that GSH plays a critical role in N2 fixation (Matamoros et al., 1999b). As an essential step to elucidate this role, we have initiated the molecular study of γECS, GSHS, and hGSHS of legume nodules. Using a strategy combining PCR screening of nodule cDNA libraries, 5′-RACE, and reverse-transcription (RT) PCR of nodule and leaf RNA, we have obtained the complete cDNA sequences encoding the three enzymes from the nodule host cells. Results showed that the synthesis of GSH and hGSH in nodules involves the participation of several cell compartments. Subcellular fractionation studies also showed that bacteroids contain high thiol concentrations and the highest specific activities of γECS and GSHS for any nodule fraction, providing further evidence that GSH is critical for nodule functioning.
RESULTS
Isolation and Sequence Analyses of Thiol Synthetase cDNAs
We have previously reported the isolation of cDNAs encoding γECS of pea (Pisum sativum) and bean (Phaseolus vulgaris) nodules (Matamoros et al., 1999b). To complete the molecular study of nodule thiol synthetases, the first part of this work was devoted to isolate cDNA clones encoding the enzymes GSHS and hGSHS, which catalyze the second step of GSH and hGSH synthesis in legumes (Fig. 1). Screening of a pea nodule library by PCR with primers based on conserved sequences of GSHS from other higher plants produced a number of positive clones. The cDNA inserts were sequenced and shown to correspond to two different genes. The complete sequences of the cDNAs, designated GSHS1 and GSHS2, were obtained and 5′-RACE analysis was used to confirm the starting ATG codons. Pea GSHS1 and GSHS2 shared 74% identity and both were approximately 65% identical with the homologous complete cDNAs of Arabidopsis, Indian mustard, and tomato. The pea sequences were also 74% to 88% identical with two partial sequences obtained from a Medicago truncatula cDNA library made from 4-d-old nodules (Frendo et al., 1999) and with a full-length sequence of soybean recently deposited in the databases (accession no. AJ272035).
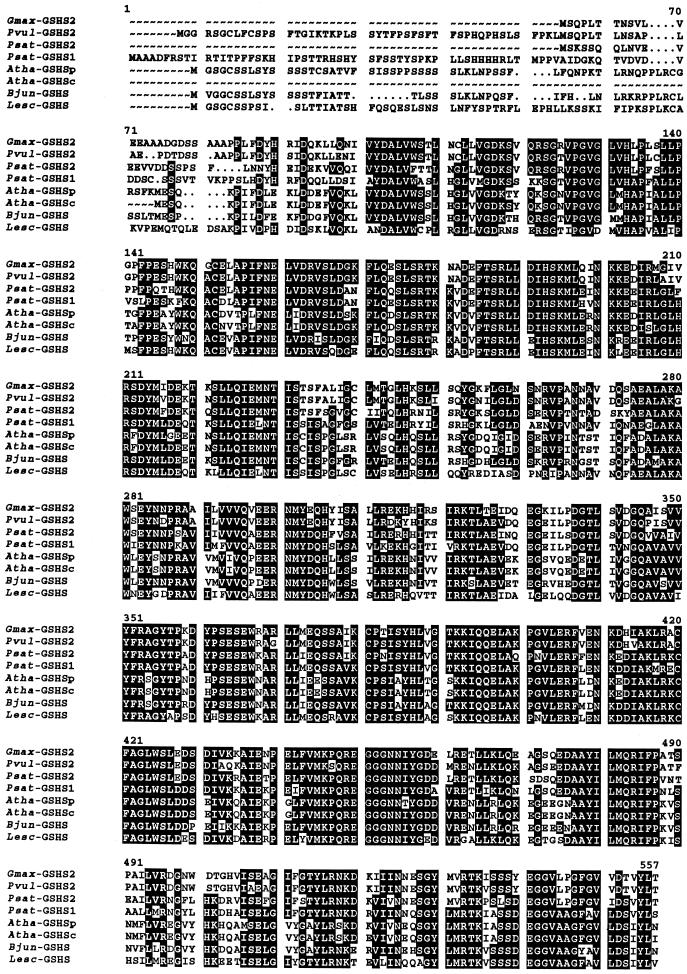
The same primers were used to screen a bean nodule library, but in this case only cDNA clones corresponding to a single gene could be isolated. All bean nodule cDNA clones examined were truncated at the 5′ end. The sequence was completed by 5′-RACE, which provided 19 bp extra in the open reading frame (including the starting ATG) and 9 bp in the 5′-untranslated region (UTR). The complete bean sequence showed approximately 63% identity with the GSHS cDNAs of Arabidopsis, Indian mustard, and tomato, 73% identity with pea GSHS1 and GSHS2, and 72% to 87% identity with the sequences of M. truncatula and soybean. At the protein level (Fig. 2), the bean sequence shows higher homology with pea GSHS2 (73% identity) than with pea GSHS1 (66% identity), and the bean nodule cDNA was therefore designated GSHS2. In a similar manner, the soybean enzyme was designated as GSHS2 because of its higher homology at the protein level with pea GSHS2 (76.2% identity) and bean GSHS2 (91.1% identity) than with pea GSHS1 (71.3%).
Figure 2.
Alignment of complete deduced amino acid sequences of GSHS and hGSHS from higher plants. Abbreviations and GenBank accession numbers are as follows: soybean GSHS (Gmax-GSHS2, AJ272035), bean GSHS2 (Pvul-GSHS2, AF258320), pea GSHS2 (Psat-GSHS2, AF258319), pea GSHS1 (Psat-GSHS1, AF231137), Arabidopsis plastidic GSHS (Atha-GSHSp, AJ243813), Arabidopsis cytosolic GSHS (Atha-GSHSc, U22359), Indian mustard GSHS (Bjun-GSHS, Y10984), and tomato GSHS (Lesc-GSHS, AF017984). Dots denote gaps to maximize alignment. Residues in white lettering on a black background are identical in at least five sequences.
Predicted Properties and Phylogenetic Analysis of Thiol Synthetases
The predicted properties of thiol synthetases from nodules are indicated in Table I. The M. truncatula enzymes were not included because the corresponding cDNAs lack the 5′ regions and therefore no predictions can be made with respect to their subcellular localization. The γECS proteins contained a putative cleavage site motif, Ile-X-Ala↓Ala, for plastid targeting. We have now confirmed these findings with the isolation of a soybean γECS cDNA bearing the complete 5′ end (accession no. AF128453). The deduced amino acid sequence of soybean γECS also contained, at the N terminus, a cleavage site motif (Ile-Val-Ala↓Ala) and a plastid transit peptide (56 amino acids). In contrast, no such motif was found for any of the GSHS proteins from nodules (Fig. 2). Prediction programs indicated that pea GSHS2 has no signal peptide and is localized in the cytosol, and that pea GSHS1 has a signal peptide for mitochondrial, rather than plastidic, targeting. However, the putative subcellular localization of bean GSHS2 is more ambiguous. The PSORT program gave rather similar probabilities for localization in the plastids and cytosol, whereas ChloroP indicated a plastidic localization with a putative cleavage site between residues 59 and 60. The sequence alignment shown in Figure 2 shows that the length of bean GSHS2 is similar to those of Arabidopsis GSHSp, Indian mustard GSHS, and tomato GSHS, all of which are predicted to be localized in organelles. As expected, the four sequences are in turn considerably longer than those of the cytosolic enzymes, namely, pea GSHS2 and Arabidopsis GSHSc. All these comparisons strongly suggest that bean GSHS2 bears a signal peptide.
Table I.
Predicted properties of thiol synthetases from pea and bean nodules
| Enzyme | Lengtha | Signalb | Mrc | pId | Cleavage Site | Subcellular Localization |
|---|---|---|---|---|---|---|
| Pea γECS | 499 | 51 | 56.6 | 6.22 | RLIVA↓ASPP | Plastids |
| Bean γECS | 508 | 60 | 57.6 | 6.12 | RVIVA↓ASPP | Plastids |
| Pea GSHS1 | 552 | 20 | 61.5 | 6.50 | FFSKH↓IPST | Mitochondria |
| Pea GSHS2 | 495 | – | 55.9 | 5.54 | – | Cytosol |
| Bean GSHS2 | 543 | 59? | 60.2 | 5.64 | NSAPL↓AEPD? | Plastids and/or cytosol |
No. of amino acid residues of precursor protein.
No. of amino acid residues of putative signal peptide.
Molecular mass of precursor protein.
Isoelectric point of precursor protein.
The programs PSORT, ChloroP, and MitoProtII were also used, as a control, to predict the subcellular localization of the other plant GSHS proteins shown in Figure 2. The programs correctly localized Arabidopsis GSHSp and tomato GSHS in the chloroplasts; however, they predicted that Indian mustard GSHS, assumed to be a mitochondrial enzyme (Schäfer et al., 1998), is targeted to the plastids and that soybean GSHS2, assumed to be a plastidic enzyme (accession no. AJ272035), is cytosolic. The latter case is also evidenced by an almost identical length of soybean GSHS2 and pea GSHS2 (Fig. 2).
In addition, the criteria of von Heijne et al. (1989), based on the Ser-to-Arg ratio to discriminate between plastidic and mitochondrial transit peptides, identified Arabidopsis GSHSp and Indian mustard GSHS as plastidic enzymes and pea GSHS1 as a mitochondrial enzyme. Figure 2 also shows that there is considerable homology among the complete GSHS sequences of all higher plants examined from residue 92 onwards (numbering is based on the pea GSHS1 sequence) and little homology in the region before residue 92, which includes the purported signal peptides. There was also relatively low homology in short stretches interspersed in the proteins, particularly between residues 233 and 273, 329 and 343, 470 and 474, and 488 and 505.
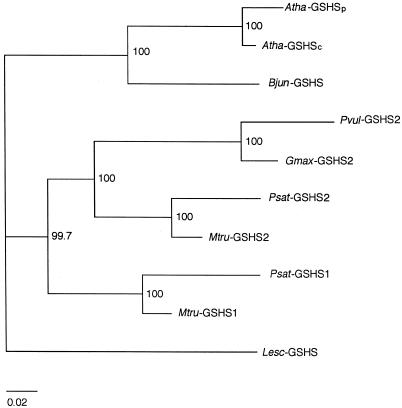
The deduced GSHS sequences of plants, including a complete sequence of soybean GSHS2 and the two partial sequences of M. truncatula, were used to construct an unrooted phylogenetic tree (Fig. 3). Sequences were aligned using PileUp and analyzed using CLUSTAL W. The tree reveals that legume GSHS proteins cluster together with respect to the non-legume proteins, which in turn cluster in two groups. However, the most interesting results for the purposes of this paper are that, within legumes, the GSHS1 and GSHS2 proteins cluster separately, and that, within GSHS2 proteins, those from legumes with determinate nodules (Phaseoleae) group separately from those with indeterminate nodulation (Vicieae, Trifolieae).
Figure 3.
Unrooted phylogenetic tree of GSHS proteins from higher plants. The tree was calculated using the neighbor-joining method of the CLUSTAL W suite of programs. The numbers correspond to percentages of 1,000 “bootstraps.” The bar represents 0.02 substitutions per site.
Sequence Assignment and Expression of Thiol Synthetases
Previous work showed that legumes contain exclusively GSH or hGSH in their leaves and that the distribution of thiols in nodules is determined by the respective synthetases (Matamoros et al., 1999a, 1999b). We then reasoned that the leaves also express a single thiol synthetase and this was demonstrated by measuring enzyme activities in pea, bean, and cowpea (Vigna unguiculata) leaves (Table II). Pea leaves express only GSHS and bean leaves only hGSHS. This finding was very useful to assign the cDNA sequences of pea and bean nodules to the GSHS or hGSHS groups of enzymes. Cowpea was introduced at this stage of the study because we needed, for localization studies, an additional legume species producing exclusively GSH and amenable for subcellular fractionation of nodules.
Table II.
Thiol tripeptide synthetase activities in legume leaves and nodules
| Enzyme | Pea
|
Bean
|
Cowpea
|
|||
|---|---|---|---|---|---|---|
| Leaves | Nodulesa | Leaves | Nodulesa | Leaves | Nodulesa | |
| nmol min−1 g−1 fresh wt | ||||||
| GSHS | 5.0 ± 0.2 | 11.4 ± 1.2 | 0 | 0.7 ± 0.2 | 10.5 ± 3.5 | 15.8 ± 1.8 |
| hGSHS | 0 | 6.9 ± 0.4 | 2.6 ± 0.1 | 7.0 ± 1.0 | 0 | 0 |
Values are means ± se of three to six samples obtained from two series of independently grown plants.
Data taken from previous work (Matamoros et al., 1999b).
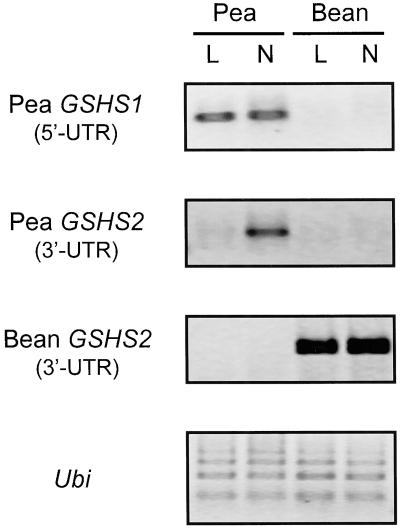
RT-PCR analysis using gene-specific primers based on the 5′-UTR of pea GSHS1 and 3′-UTR of pea GSHS2 revealed that GSHS1 is equally expressed in leaves and nodules, but GSHS2 is expressed only in nodules (Fig. 4). Therefore, pea GSHS1 encodes a GSHS, whereas the product of GSHS2 can be tentatively identified as a hGSHS because pea plants express hGSHS in nodules, but not in leaves (Table II). The same analysis using primers based on the 3′-UTR sequence of bean GSHS2 indicated that this gene is expressed at the same level in the leaves and nodules (Fig. 4). Therefore, bean GSHS2 encodes a hGSHS. The assignments of pea GSHS1 as GSHS and of pea and bean GSHS2 as hGSHS were confirmed by the cluster analysis described above. Thus, M. truncatula GSHS2 (Frendo et al., 1999) and soybean GSHS2 (M. Skipsey, C.J. Andrews, J.K. Townson, I. Jepson, and R. Edwards, unpublished data) have been characterized as hGSHS enzymes. The two proteins cluster together with pea and bean GSHS2 and separately from GSHS1 (Fig. 3), indicating, together with our expression data and those of Frendo et al. (1999), that GSHS2 cDNAs encode hGSHS enzymes, whereas GSHS1 cDNAs encode GSHS enzymes.
Figure 4.
Expression of GSHS in leaves and nodules of pea and bean plants. RT-PCR analysis was performed using primers designed to 5′-UTRs or 3′-UTRs of pea (GSHS1 and GSHS2) and bean (GSHS2) cDNAs, as shown in the left. Ubiquitin (Ubi) was used as a control for uniform loading.
The same gene-specific primers along with primers designed to the 3′-UTR of γECS cDNA were used to quantify expression of γECS, GSHS1, and GSHS2 in pea nodules by RT-PCR. However, there were no major variations in the abundance of any of the three transcripts during natural (aging) or stress-induced nodule senescence (data not shown). This contrasts with the decline in the corresponding enzyme activities observed in aging pea nodules or the increase in γECS activity observed in dark-stressed pea nodules (Matamoros et al., 1999b), and suggests that thiol synthetase activities may be regulated at the post-transcriptional level, as shown by May et al. (1998b) for γECS in Arabidopsis cell cultures.
Localization of Thiol Synthetases
The tentative localization of thiol synthetases in the various nodule compartments, based on the presence or absence of recognizable cleavage site motifs, required verification by purification of organelles on density gradients. Marker enzymes and leghemoglobin were used to assess cross-contamination among nodule organelles. Specific protocols had to be employed to purify plastids and bacteroids, whereas a single method served to purify mitochondria, peroxisomes, and cytosol. Mitochondria were <10% contaminated with peroxisomes and plastids, and showed no detectable contamination with the cytosol or bacteroids. Plastids showed <20% contamination with bacteroids, <10% with mitochondria and peroxisomes, and negligible contamination with cytosol. Chloroplasts, mitochondria, and cytosol were also purified from the leaves of the same plants to verify results. Similar protocols were followed for organelle purification, and chlorophyll and marker enzymes were used to assess purity (Corpas et al., 1991; Jiménez et al., 1997). Chloroplasts were essentially free of contamination with cytosol or mitochondria. However, leaf mitochondria had still substantial contamination (20%–30%) with thylakoids. Crude extracts of nodules and leaves, in which the enzymes were released from organelles by prolonged sonication, were also analyzed as parallel controls.
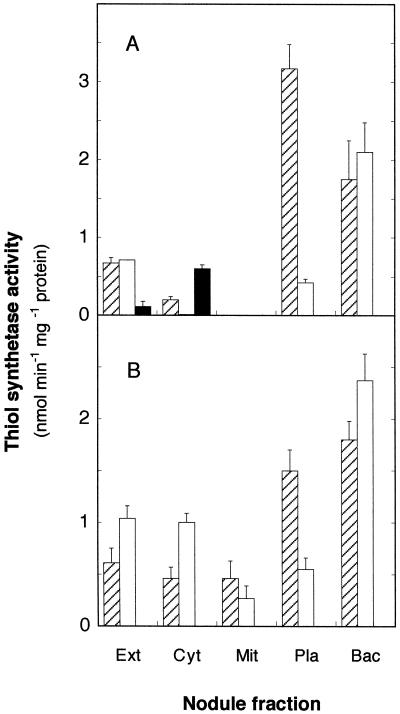
Bean nodule extracts showed γECS, GSHS, and hGSHS activities (Fig. 5A). However, when the extracts were not sonicated, only γECS and hGSHS activities could be detected, suggesting that GSHS activity originated in the bacteroids. Bean and cowpea bacteroids have very high GSHS activities (Fig. 5, A and B) and the small GSHS activity detected in the plastids of the two legumes (15% of the specific activity of bacteroids) was due to cross-contamination. Thus, when the plastid fractions were made up to 0.01% (v/v) Triton X-100 and centrifuged, the GSHS activity remained in the sediment (contaminating bacteroids) and not in the supernatant (broken plastids).
Figure 5.
Subcellular localization of γECS (▨), GSHS (□), and hGSHS (▪) in bean (A) and cowpea (B) nodules. Values are means ± se of three or four independent experiments. Ext, Crude extract; Cyt, cytosol; Mit, mitochondria; Pla, plastids; Bac, bacteroids.
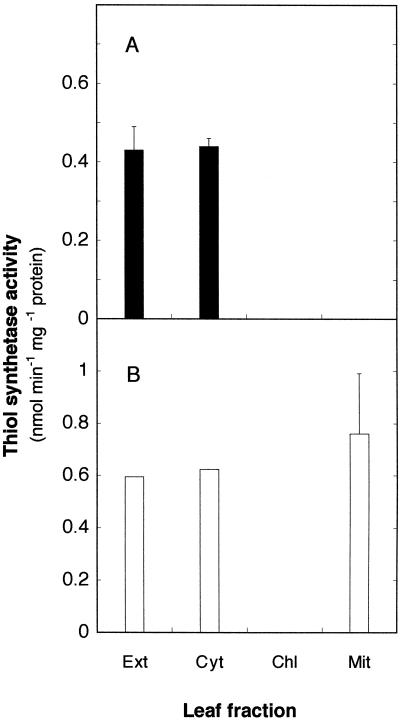
Most γECS activity of bean nodules was localized in the plastids and bacteroids, with somewhat less activity being present in the cytosol; in contrast, the hGSHS activity was localized in the cytosol, with no measurable activity in the mitochondria, plastids, peroxisomes, or bacteroids (Fig. 5A). Assay of enzymes in nodules of soybean, another hGSH-producing legume (Matamoros et al., 1999b), confirmed that hGSHS activity was localized in the cytosol (data not shown). The same location was found for the hGSHS of bean leaves, with no measurable activity in chloroplasts or mitochondria (Fig. 6A). Furthermore, when large amounts of bean leaves were processed to partially purify hGSHS, we were unable to detect GSHS activity in the extracts and the hGSHS activity invariably remained in the soluble fraction (cytosol) after sedimentation of organelles in isosmotic conditions. In consequence, the subcellular localization data demonstrate that hGSHS is the only thiol tripeptide synthetase present in bean leaves and nodule host cells, and that there is at least a hGSHS isoenzyme in the cytosol.
Figure 6.
Subcellular localization of GSHS (□) and hGSHS (▪) in bean (A) and cowpea (B) leaves. Values are means ± se of two or three independent experiments. Ext, Crude extract; Cyt, cytosol; Mit, mitochondria; Chl, chloroplasts.
As could be anticipated for a GSH-producing legume (Matamoros et al., 1999b), crude extracts of cowpea nodules exhibited γECS and GSHS activities, but not hGSHS activity (Fig. 5B). The majority of γECS activity was localized in the plastids and bacteroids, and the majority of GSHS activity in the cytosol and bacteroids. Unlike bean nodules, however, we found γECS and GSHS activities in the mitochondria of cowpea nodules (Fig. 5B). Mitochondria preparations showed no detectable contamination with bacteroids and negligible contamination with plastids, and therefore we decided to purify leaf organelles to verify results. Subcellular fractionation of cowpea leaves revealed that the mitochondria, but not the chloroplasts, contain GSHS (Fig. 6B). This study also confirmed the presence of low levels of γECS in cowpea mitochondria (data not shown).
Thiols and Thiol Synthetases of Bacteroids
Bacteroids purified on Percoll gradients were essentially free of contamination with host cell organelles or cytosol, as judged by the assay of marker proteins. Additional controls, consisting of bacteroids that had been washed up to four times or repurified on two sequential Percoll gradients prior to sonication, yielded identical results. Bacteroids showed high specific γECS and GSHS activities, but no hGSHS activity, regardless of the main thiol tripeptide synthetase present in the nodules (Fig. 5, A and B).
The high capacity of bacteroids for GSH synthesis and the lack of any previous information on their contribution as a source of thiols within the nodules prompted us to measure the thiol content of bacteroids. Bean and cowpea nodule bacteroids contained 0.29 nmol Cys and approximately 10 nmol GSH per mg of protein (Table III). As expected, cowpea bacteroids had no hGSH, but bean bacteroids contained 1.5 nmol hGSH per mg of protein. This small but significant hGSH concentration was not due to thiol adsorbed to the bacteroid surface since it remained constant after repeated washings of bacteroids. Soybean bacteroids (strain USDA110) had significantly higher concentrations—4.5 nmol hGSH per mg of protein. Because bacteroids do not express hGSHS (Fig. 5, A and B), we conclude that the hGSH found in the bacteroids was synthesized by the host plant and taken up through the symbiosome membrane.
Table III.
Thiol contents of bacteroids from bean and cowpea nodules
| Thiola | Bean (Strain 3622) | Cowpea (Strain 32H1) |
|---|---|---|
| nmol mg−1 protein | ||
| Cys | 0.29 ± 0.05 | 0.29 ± 0.03 |
| GSH | 9.40 ± 0.68 | 12.70 ± 0.87 |
| hGSH | 1.53 ± 0.32 | 0 |
Data are means ± se of four to six samples of bacteroids isolated from at least two series of independently grown plants.
The content of γEC was <0.06 nmol mg−1 protein in both legumes.
DISCUSSION
Legumes are the only plants known so far to contain hGSH in addition to or in place of GSH (Klapheck, 1988). The first enzyme committed to the synthesis of the thiol tripeptides, γECS, is ubiquitous in leaves and nodules, whereas expression of the second enzymes, GSHS and hGSHS, determines the relative abundance of GSH and hGSH in the different legume species and plant tissues (Matamoros et al., 1999a, 1999b). To ascertain the specific roles of thiols in legume nodules and particularly in N2 fixation, it is essential to characterize the genes and localize the three enzymes involved in their synthesis.
In this paper we demonstrate by subcellular fractionation that the γECS of nodule host cells is localized in the plastids (Fig. 5). Previous studies showed that the enzyme is present in the chloroplasts and cytosol of leaves (Hell and Bergmann, 1990) and in root plastids (Rüegsegger and Brunold, 1993). However, we found only very low γECS activity in the nodule cytosol and this is probably attributable to contamination with the enzyme of plastids, which are extremely fragile organelles (Atkins et al., 1997). There are some significant differences between the γECS from legume nodules and that from tobacco suspension cells. The nodule enzymes have a predicted molecular mass of approximately 51 kD, significantly smaller than the 60 kD found for the enzyme purified from tobacco (Hell and Bergmann, 1990). Likewise, the γECS activities of tobacco cells and of pea and spinach leaves, but not of Arabidopsis or maize (May and Leaver, 1994), were inhibited by reductants. In tobacco, the inhibition was due to dissociation of the protein into subunits (Hell and Bergmann, 1990). In contrast, we have observed that 5 mm dithioerythritol enhanced the assayable γECS activity of nodules from 1.2- to 3.5-fold (depending on species), which suggests that the nodule enzymes are active as monomers.
Knowledge on non-photosynthetic plastids lags well behind that on chloroplasts partly due to difficulties encountered in their isolation (for review, see Emes and Neuhaus, 1997). The same holds true for nodule plastids. As more biochemical and molecular information on nodule metabolism becomes available, the picture is emerging that plastids perform multiple functions essential for N2 fixation. The best known functions of nodule plastids are related to their participation in ammonia assimilation (Temple et al., 1998) and purine synthesis (Atkins et al., 1997). We report here that another function, so far overlooked, is protection against reactive oxygen. Thus, the specific localization in plastids of γECS (Fig. 7), along with glutathione reductase (Tang and Webb, 1994), ferritin (Matamoros et al., 1999a), and Fe-superoxide dismutase (M.C. Rubio and M. Becana, unpublished data), emphasize that these organelles are a primary line of antioxidative defense in nodules. The three antioxidant proteins are responsive to stress and may be directly induced by reactive oxygen species (Lobréaux et al., 1995; May et al., 1998a; Matamoros et al., 1999a, 1999b). In particular, γECS is the regulatory step of GSH synthesis and is post-transcriptionally activated in response to stressful conditions (May et al., 1998a, 1998b). Therefore, plastids from nodules and probably from other non-green tissues have an important complement of antioxidant proteins that enable them to sense and respond to conditions generating oxidative stress in the plant.
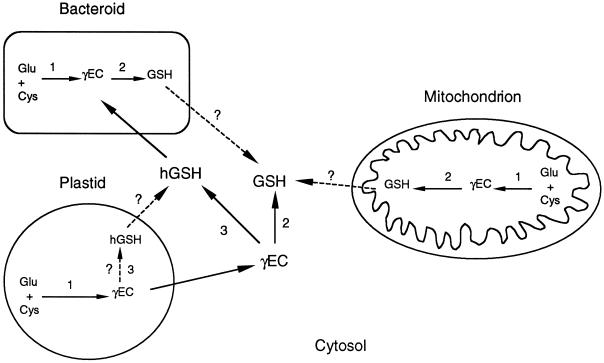
Figure 7.
Diagram depicting the subcellular localization of GSH and hGSH synthesis in nodules. Enzymes are γECS (1), GSHS (2), and hGSHS (3). In GSH-producing legume nodules, reaction 1 occurs in the bacteroids, plastids, and mitochondria, and reaction 2 in the bacteroids, mitochondria, and cytosol. In hGSH-producing legume nodules, reaction 1 occurs in the bacteroids and plastids, reaction 2 in the bacteroids, and reaction 3 in the cytosol and probably also in the plastids. Arrows in discontinuous lines indicate possible contributions of bacteroids and mitochondria to the cytosolic GSH pool and of the plastids to the cytosolic hGSH pool.
Our results also indicate that the second step of GSH and hGSH synthesis in nodule host cells takes place predominantly in the cytosol (Fig. 7). This is only in partial agreement with earlier studies of subcellular localization in leaves. In pea and spinach leaves, between 47% and 69% of total GSHS activity is localized in the chloroplasts and the rest in the cytosol (Klapheck et al., 1987; Hell and Bergmann, 1990). We have found that the majority of GSHS in nodules and leaves of cowpea was localized in the cytosol. Indeed, we have also isolated a cDNA clone from cowpea nodules that encodes a cytosolic GSHS (J.F. Moran and M. Becana, unpublished data). A novel observation is, however, that there is also GSHS in the mitochondria (but not in the plastids or chloroplasts) of cowpea leaves and nodules (Fig. 7), implying a further protective role of GSH against oxidants generated during respiration. In the leaves the specific activity of GSHS in mitochondria was, in fact, slightly higher than that in the crude extract and cytosol (Fig. 6B). The GSHS activity of nodule and leaf mitochondria is genuinely restricted to these organelles because they were not contaminated with bacteroids or nodule plastids and because, although leaf mitochondria were contaminated with chloroplasts, these do not contain GSHS. The finding of GSHS activity in mitochondria is consistent with the sequence analysis of pea nodule GSHS1, which encodes a protein bearing a putative mitochondrial transit peptide (Fig. 2).
We have found hGSHS activity only in the cytosol from nodules and leaves of bean (Figs. 5A and 6A) and soybean (data not shown). However, we cannot exclude the possibility that an additional hGSHS isozyme exists in the chloroplasts and nodule plastids in view of the cDNA sequence, bean GSHS2, that has been isolated in the course of this work. Although this sequence is consistent with both a plastidic and a cytosolic localization of the enzyme, it might be that the hGSHS activity in the chloroplasts and plastids is very low or unusually labile, escaping detection. This explanation would be more in agreement with, to our knowledge, the only previous report addressing hGSHS localization in plants. Klapheck et al. (1988) estimated that 17% of the hGSHS activity of Phaseolus coccineus leaves is in the chloroplasts, whereas the rest was assumed to be in the cytosol.
We have also reported that the majority of hGSHS activity of bean nodules is in the cortex (Matamoros et al., 1999b) and have now expanded this observation to soybean nodules. These have distinctly different thiol synthetase activities (nmol min−1 g−1 fresh weight) in the cortex (GSHS = 0, hGSHS = 9.9 ± 0.3) and in the infected zone (GSHS = 1.5 ± 0.8, hGSHS = 3.3 ± 1.4). Therefore, the predominant localization of hGSHS at the subcellular (cytosol) and tissue (nodule cortex) levels appears to be widespread in hGSHS-producing nodules.
Results of this work also reveal that bacteroids have very high γECS and GSHS activities and thiol concentrations (Fig. 5; Table III), and hence actively synthesize GSH (Fig. 7). These and previous findings (Matamoros et al., 1999b) suggest that bacteroids are a major source of GSH and would explain, at least in part, why this thiol is so abundant in the infected zone of indeterminate and determinate nodules. In bacteroids, as in other prokaryotes, GSH may have multiple functions. One such function has been demonstrated very recently. A Rhizobium tropici mutant strain containing 3% of the GSH present in the wild type was more sensitive to osmotic and acid stress and was less competitive in co-inoculation experiments, suggesting an important role of GSH in stress tolerance (Riccillo et al., 2000). Bacteroids cannot synthesize hGSH, but this thiol, produced by the plant, can apparently cross the symbiosomal membrane and reach the bacteroids (Fig. 7). Whether these are energy-intensive or simple diffusive processes awaits further investigation.
MATERIALS AND METHODS
Plant Growth
Nodulated pea (Pisum sativum L. cv Lincoln × Rhizobium leguminosarum biovar viciae NLV8), bean (Phaseolus vulgaris L. cv Contender × Rhizobium leguminosarum biovar phaseoli 3622), soybean (Glycine max Merr. cv Williams × Bradyrhizobium japonicum USDA110), and cowpea (Vigna unguiculata Walp. cv California no. 5 × Bradyrhizobium sp. [Vigna] 32H1) plants were grown under controlled environment conditions as described (Gogorcena et al., 1997). All legumes were at the vigorous vegetative growth stage (30–35 d) when leaves and nodules were harvested. Leaves and nodules to be used for RT-PCR experiments were collected with gloves and immediately frozen in liquid N2. All plant material was stored at −80°C except nodules to be used for dissection and subcellular fractionation studies, which were processed immediately after harvest.
Thiol Synthetase Assays
Thiol compounds were derivatized with monobromobimane and quantified by HPLC with fluorometric detection (Fahey and Newton, 1987) with the modifications described in detail elsewhere (Matamoros et al., 1999b). Thiol synthetase activities in nodule extracts and organelles were determined using the same HPLC method based on the γEC synthesized from Cys and Glu (γECS), GSH synthesized from γEC and Gly (GSHS), and hGSH synthesized from γEC and βAla (hGSHS). The optimized extraction and assay media for the enzymes were identical to those previously reported (Matamoros et al., 1999b), with the only exception that the dithioerythritol concentration for the assay of γECS activity was increased from 0.5 to 5 mm. Enzyme activities in all nodule fractions and organelles were expressed on a protein basis. Protein was quantified by the dye-binding microassay (Bio-Rad Laboratories, Hercules, CA), using bovine serum albumin as a standard.
Isolation of cDNA Clones Encoding Thiol Synthetase cDNAs
The cDNA clones encoding γECS were isolated by PCR screening of nodule libraries using oligonucleotide primers designed to conserved sequences (GenBank accession nos. in parentheses) of Arabidopsis (Y09944), tomato (AF017983), and Indian mustard (Y10848). Pea and bean λZAP cDNA libraries were provided by Dr. Carroll Vance (U.S. Department of Agriculture-University of Minnesota, St. Paul), and the soybean λgt11 cDNA library was provided by Dr. Robert Klucas (University of Nebraska, Lincoln).
The same libraries were PCR-screened to isolate cDNA clones encoding GSHS and hGSHS of legume nodules. Degenerate primers were designed based on the complete GSHS cDNA sequences of Arabidopsis (U22359 and AJ243813), tomato (AF017984), and Indian mustard (Y10984), and on the partial GSHS cDNA sequences of Medicago truncatula (AF075699 and AF075700). Primers used to isolate internal cDNA sequences were: sense, 5′-CG[A/C]AACATGTA[C/T]GA[C/T]CA[A/G]CATT-3′; and antisense, 5′-CCTTCTCT[C/T]TG[A/G]GG[C/T]TTCAT-3′. The 5′ and 3′ ends of all nodule clones, except those of pea nodule GSHS2, were amplified using the above primers in combination with T3 and T7 primers. The PCR mixture contained 0.5 μm of primers, 0.2 mm dNTPs, 2.5 mm MgCl2, 0.05% (v/v) W-1 detergent, and 1.5 units of Taq polymerase (Life Technologies, Paisley, UK), in a final volume of 25 μL of PCR buffer (20 mm Tris-HCl, pH 8.4, and 50 mm KCl). PCR conditions were exactly as previously described (Matamoros et al., 1999b).
Primers used for PCR amplification of the 5′ and 3′ ends of pea nodule GSHS2 were: sense, 5′-GCAGTCGCAATCGTTTACTTCC-3′; and antisense, 5′-CCCACCTTCATCAAATAATGATGG-3′. These were used in combination with T3 and T7 primers. The PCR mixture contained 0.2 μm of both primers, 0.24 mm dNTPs, 1.5 mm MgCl2, 0.05% (v/v) W-1 detergent, and 1.25 units of Taq polymerase, in a final volume of 25 μL of PCR buffer. The PCR cycling protocol consisted of an initial denaturation step at 95°C for 3 min, 40 cycles (95°C for 45 s, 62°C for 45 s, and 72°C for 90 s), and a final elongation step at 72°C for 10 min.
RACE-PCR and RT-PCR
For 5′-RACE the manufacturer's instructions (Life Technologies) were followed using the primer 5′-CCTTCTCT[C/T]TG[A/G]GG[C/T]TTCAT-3′ to generate specific cDNA. For the subsequent PCR of pea nodule GSHS1, pea nodule GSHS2, and bean nodule GSHS2, the antisense primers 5′-CGGAAGAAGAACAAGAATCGTCG-3′, 5′-TGGTGTATAGCCAGCTCGGAAG-3′, and 5′-CCAAACTCACACGATCAACAAGC-3′ were used, respectively.
Total RNA was extracted from nodules using the hot phenol method followed by LiCl precipitation (de Vries et al., 1982). For the RT-PCR analysis of leaves and nodules, total RNA (5 μg) was treated with 2 units of DNase I at 37°C for 10 min to remove traces of contaminating DNA. After addition of 2.5 mm EDTA, samples were incubated at 65°C for 15 min to inactivate DNase. For RT, RNA samples were annealed to the primer 5′-CTCGAGGATCCGCGGCCGC-(T)20-3′at 70°C for 10 min, and then the cDNAs were synthesized using 200 units of reverse transcriptase (Superscript, Life Technologies) in a buffer containing 10 mm dithiothreitol and 1.25 mm dNTPs. The reaction proceeded at 42°C for 55 min and was stopped at 70°C for 15 min. The remaining RNA present in the samples was removed by incubation with 1 unit of RNase H at 37°C for 20 min. The reaction mix was diluted to 120 μL, and 5 μL was used as template for PCR amplification.
For the PCRs, gene-specific primers were designed based on the UTR sequences. Oligonucleotides used were as follows: For pea GSHS1, sense, 5′-CCCCTTTCTTCTCCAAACACATTC-3′, and antisense, 5′-CGGAAGAA GAACAAGAATCGTCG-3′; for pea GSHS2, sense, 5′-GTTGTTGATTGATGGCTTGCATG-3′, and antisense, 5′-GCGCCAAAATCCATTGTGAAC-3′; for bean GSHS2, sense, 5′-GAAAGTGGCTATATGGTGCG-3′, and antisense, 5′-GACACCATTCAGTAGGAAAAGC-3′. The reaction mixture contained 5 μL of first-strand cDNA, 0.25 mm dNTPs, 1.5 mm MgCl2, 0.2 μm of primers, and 1.25 units of Taq polymerase (Perkin-Elmer Applied Biosystems, Foster City, CA) in a total volume of 25 μL. The PCR cycling conditions comprised an initial denaturation step at 94°C for 2 min, 30 to 35 cycles (94°C for 30 s, 60°C for 30 s, and 72°C for 45 s), and a final elongation step at 72°C for 10 min. As an internal control, PCR was performed simultaneously using ubiquitin primers (Horvath et al., 1993). In all cases, preliminary runs were used to verify that the number of amplification cycles was well below that required for signal saturation.
Cloning and Sequencing
The cDNA bands of the expected sizes were gel purified (Concert, Life Technologies, or QIAquick gel extraction kit, Qiagen, Santa Clarita, CA) and subcloned into pCRII or pCR2.1 (Invitrogen, Carlsbad, CA). All sequencing was conducted on both strands of cDNA from at least two clones with an ABI Prism 377 sequencer (Applied Biosystems) using AmpliTaq DNA polymerase, FS dye-terminator cycle sequencing chemistry. Homology searches were done with the BLAST algorithm (Altschul et al., 1997). Sequence alignments and homology analyses were performed using the PileUp and Gap programs, respectively, of the Genetics Computer Group (Madison, WI). Phylogenetic analysis was performed with the CLUSTAL W (1.75) suite of programs (Thompson et al., 1994). Signal peptide analyses and predictions of subcellular localization were performed using the programs MitoProtII (Claros, 1995), PSORT (Nakai and Kanehisa, 1992), and ChloroP and TransitP (Center for Biological Sequence Analysis, Department of Biotechnology, Technical University of Denmark, Denmark).
Organelle Purification for Assay of Thiol Synthetases
Nodule host-cell organelles and bacteroids were purified from nodules at 0°C to 4°C using Percoll gradients. For the purification of the mitochondria and peroxisomes, nodules (10 g) were gently ground in a mortar with 30 mL of a medium containing 0.3 m mannitol, 50 mm Tris-HCl (pH 8.0), 2 mm EDTA, 20 mm MgCl2, and 2% (w/v) polyvinylpolypyrrolidone. The homogenate was filtered through four layers of cheesecloth (moistened with extraction medium). An aliquot (1 mL) of the filtrate was sonicated in an ice bath (4 × 30 s with 30-s breaks; Branson sonifier) and centrifuged at 13,000g for 5 min. The cleared supernatant (“crude extract”) was saved for enzyme analysis. The rest of the homogenate was centrifuged twice at 4,000g for 5 min and then at 12,000g for 15 min. An aliquot of the supernatant (“cytosol”) was also saved for subsequent enzyme analysis. The pellet was washed with 25 mL of washing medium (extraction medium omitting polyvinylpolypyrrolidone) and resuspended in 1.8 mL of washing medium. The whole volume was loaded on a first Percoll gradient essentially as described by Sandalio et al. (1987). Peroxisomes, which banded between the 35% and 50% (v/v) Percoll layers, were freed from Percoll with two washes with washing medium. Mitochondria, which banded between the 15% and 35% (v/v) Percoll layers, were freed from Percoll, as was done for peroxisomes, and loaded on a second Percoll gradient following the method of Struglics et al. (1993). Mitochondria and peroxisomes were broken by resuspension in 0.5 mL of hypotonic medium (50 mm Tris-HCl [pH 8.0], 0.2 mm EDTA, and 20 mm MgCl2) and overnight incubation at 0°C. Broken organelles were then centrifuged and immediately used for enzyme analyses. Freezing and thawing of organelles did not significantly affect the yield and activity of enzymes.
For plastid purification, nodules (10 g) were carefully ground in a mortar with 30 mL of a medium containing 0.3 m Suc, 30 mm Tris-HCl (pH 8.0), 1 mm EDTA, and 20 mm MgCl2. The homogenate was filtered through one layer of cheesecloth and centrifuged at 3,000g for 5 min. The pellet was resuspended in 25 mL of extraction medium. After a new centrifugation at 200g for 5 min, the pellet was discarded and the supernantant was centrifuged at 3,000g for 5 min. The plastid-enriched pellet was resuspended in 2 mL of extraction medium and the plastids were purified by using sequentially two 35% (v/v) Percoll gradients as described by Atkins et al. (1997). Plastids were broken as indicated for mitochondria and peroxisomes.
For bacteroid purification, nodules (1 g) were carefully ground in a mortar with 1 mL of a medium containing 50 mm KH2PO4 and 150 mm NaCl (pH 8.0). The residue was further ground with 1 mL of the same medium and the pooled extract was filtered through four layers of cheesecloth. The extract (1.5 mL) was loaded on 70% (v/v) Percoll made in extraction buffer, and bacteroids were purified as described by Reibach et al. (1981). Bacteroids were broken by sonication (4 × 30 s with 30-s breaks) in an ice bath. For the thiol analysis of bacteroids, the same extraction buffer was used except that the pH was adjusted to 6.5. The purified bacteroids were broken by resuspension in 200 mm methanesulfonic acid (containing 0.5 mm diethylenetriaminepentaacetic acid) and by subsequent sonication.
Organelle Purification for Assay of Marker Proteins
Similar procedures, but at pH 7.2, were used to monitor the purification process of organelles with the assistance of the following marker proteins: β-hydroxybutyrate dehydrogenase and Ala dehydrogenase (bacteroids; Reibach et al., 1981), Cyt c oxidase (mitochondria; Schnarrenberger et al., 1971), uricase and catalase (peroxisomes; Hanks et al., 1981), NADH-Glu synthase (plastids; Atkins et al., 1997), and leghemoglobin (cytosol; LaRue and Child, 1979). Nodule crude extracts, cytosol, mitochondria, and peroxisomes were purified using an extraction medium comprising 0.35 m mannitol, 30 mm MOPS [3-(N-morpholino)-propanesulfonic acid, pH 7.2], 2 mm EDTA, 10 mm KH2PO4, and 2% (w/v) polyvinylpolypyrrolidone. The purified organelles were washed with the same medium omitting polyvinylpolypyrrolidone. Plastids were purified using an extraction and washing medium containing 0.3 m Suc, 30 mm MOPS (pH 7.2), 1 mm EDTA, and 20 mm MgCl2. Bacteroids were purified using an extraction and washing medium containing 50 mm KH2PO4 and 150 mm NaCl (pH 7.2).
To assay β-hydroxybutyrate dehydrogenase and Ala dehydrogenase, nodule crude extracts and purified fractions were sonicated 4 × 30 s (with 30-s breaks) in an ice bath. To assay Cyt c oxidase, uricase, catalase, and leghemoglobin, nodule extracts and fractions were made to 0.05% (v/v) Triton X-100. To assay NADH-Glu synthase, nodule extracts and fractions were made (immediately after isolation) to 94 mm MES [2-(N-morpholino)-ethanesulfonic acid, final pH 6.5], 0.05% (v/v) Triton X-100, and 270 mm β-mercaptoethanol. This enzyme was found to be labile at pH 8.0 and in the absence of β-mercaptoethanol, as previously reported by others (Groat and Vance, 1981).
ACKNOWLEDGMENTS
The authors are most grateful to Carroll Vance and Robert Klucas for the gift of cDNA libraries. We also thank Gloria Rodríguez for growing the plants.
Footnotes
This work was supported by the Comisión Interministerial de Ciencia y Tecnología and the European Commission (grant nos. 2FD97–1101 and PB98–0522 to M.B.), by the Dirección General de Enseñanza Superior e Investigación Científica and the British Council (Acción Hispano-Británica HB98–163 to M.B. and N.J.B.), by a Marie Curie grant from the European Commission (to I.I.-O.), and by the Biotechnology and Biological Sciences Research Council (to N.J.B.). J.F.M., M.A.M., M.C.R., and M.R.C. were the recipients, respectively, of a postdoctoral contract from the Ministry of Education and Culture (Spain), a predoctoral fellowship from the Gobierno Vasco, and two predoctoral fellowships from the Ministry of Education and Culture.
LITERATURE CITED
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins CA, Smith PMC, Storer PJ. Reexamination of the intracellular localization of the novo purine synthesis in cowpea nodules. Plant Physiol. 1997;113:127–135. doi: 10.1104/pp.113.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros MG. MitoProt, a Macintosh application for studying mitochondrial proteins. Comput Appl Biosci. 1995;11:441–447. doi: 10.1093/bioinformatics/11.4.441. [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Sandalio LM, Palma JM, Leidi EO, Hernández JA, Sevilla F, del Río LA. Subcellular distribution of superoxide dismutase in leaves of ureide-producing leguminous plants. Physiol Plant. 1991;82:285–291. [Google Scholar]
- de Vries SC, Springer J, Wessels JHG. Diversity of abundant messenger-RNA sequences and patterns of protein synthesis in etiolated and greened pea seedlings. Planta. 1982;156:129–135. doi: 10.1007/BF00395427. [DOI] [PubMed] [Google Scholar]
- Emes MJ, Neuhaus HE. Metabolism and transport in non-photosynthetic plastids. J Exp Bot. 1997;48:1995–2005. [Google Scholar]
- Fahey RC, Newton GL. Determination of low-molecular-weight thiols using monobromobimane fluorescent labeling and high-performance liquid chromatography. Methods Enzymol. 1987;143:85–96. doi: 10.1016/0076-6879(87)43016-4. [DOI] [PubMed] [Google Scholar]
- Frendo P, Gallesi D, Turnbull R, Van de Sype G, Hérouart D, Puppo A. Localization of glutathione and homoglutathione in Medicago truncatula is correlated to a differential expression of genes involved in their synthesis. Plant J. 1999;17:215–219. [Google Scholar]
- Gogorcena Y, Gordon AJ, Escuredo PR, Minchin FR, Witty JF, Moran JF, Becana M. N2 fixation, carbon metabolism, and oxidative damage in nodules of dark-stressed common bean plants. Plant Physiol. 1997;113:1193–1201. doi: 10.1104/pp.113.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groat RG, Vance CP. Root nodule enzymes of ammonia assimilation in alfalfa (Medicago sativa L.) Plant Physiol. 1981;67:1198–1203. doi: 10.1104/pp.67.6.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks JF, Tolbert NE, Schubert KR. Localization of enzymes of ureide biosynthesis in peroxisomes and microsomes of nodules. Plant Physiol. 1981;68:65–69. doi: 10.1104/pp.68.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausladen A, Alscher RG. Glutathione. In: Alscher RG, Hess JL, editors. Antioxidants in Higher Plants. Boca Raton, FL: CRC Press; 1993. pp. 1–30. [Google Scholar]
- Hell R, Bergmann L. γ-Glutamylcysteine synthetase in higher plants: catalytic properties and subcellular localization. Planta. 1990;180:603–612. doi: 10.1007/BF02411460. [DOI] [PubMed] [Google Scholar]
- Horvath B, Heidstra R, Lados M, Moerman M, Spaink HP, Promé JC, van Kammen A, Bisseling T. Lipo-oligosaccharides of Rhizobium induce infection-related early nodulin gene expression in pea root hairs. Plant J. 1993;4:727–733. doi: 10.1046/j.1365-313x.1993.04040727.x. [DOI] [PubMed] [Google Scholar]
- Jiménez A, Hernández JA, del Río LA, Sevilla F. Evidence for the presence of the ascorbate-glutathione cycle in mitochondria and peroxisomes of pea leaves. Plant Physiol. 1997;114:275–284. doi: 10.1104/pp.114.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapheck S. Homoglutathione: isolation, quantification, and occurrence in legumes. Physiol Plant. 1988;74:727–732. [Google Scholar]
- Klapheck S, Latus C, Bergmann L. Localization of glutathione synthetase and distribution of glutathione in leaf cells of Pisum sativum L. J Plant Physiol. 1987;131:123–131. [Google Scholar]
- Klapheck S, Zopes H, Levels HG, Bergmann L. Properties and localization of the homoglutathione synthetase from Phaseolus coccineus leaves. Physiol Plant. 1988;74:733–739. [Google Scholar]
- LaRue TA, Child JJ. Sensitive fluorometric assay for leghemoglobin. Anal Biochem. 1979;92:11–15. doi: 10.1016/0003-2697(79)90618-3. [DOI] [PubMed] [Google Scholar]
- Lobréaux S, Thoiron S, Briat JF. Induction of ferritin synthesis in maize leaves by an iron-mediated oxidative stress. Plant J. 1995;8:443–449. [Google Scholar]
- Macnicol PK. Homoglutathione and glutathione synthetases of legume seedlings: partial purification and substrate specificity. Plant Sci. 1987;53:229–235. [Google Scholar]
- Matamoros MA, Baird LM, Escuredo PR, Dalton DA, Minchin FR, Iturbe-Ormaetxe I, Rubio MC, Moran JF, Gordon AJ, Becana M. Stress-induced legume root nodule senescence: physiological, biochemical, and structural alterations. Plant Physiol. 1999a;121:97–111. doi: 10.1104/pp.121.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matamoros MA, Moran JF, Iturbe-Ormaetxe I, Rubio MC, Becana M. Glutathione and homoglutathione synthesis in legume root nodules. Plant Physiol. 1999b;121:879–888. doi: 10.1104/pp.121.3.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May MJ, Leaver CJ. Arabidopsis thaliana γ-glutamylcysteine synthetase is structurally unrelated to mammalian, yeast, and Escherichia coli homologs. Proc Natl Acad Sci USA. 1994;91:10059–10063. doi: 10.1073/pnas.91.21.10059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May MJ, Vernoux T, Leaver C, Van Montagu M, Inzé D. Glutathione homeostasis in plants: implications for environmental sensing and plant development. J Exp Bot. 1998a;49:649–667. [Google Scholar]
- May MJ, Vernoux T, Sánchez-Fernández R, Van Montagu M, Inzé D. Evidence for posttranscriptional activation of γ-glutamylcysteine synthetase during plant stress responses. Proc Natl Acad Sci USA. 1998b;95:12049–12054. doi: 10.1073/pnas.95.20.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Foyer CH. Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- Reibach PH, Mask PL, Streeter JG. A rapid one-step method for the isolation of bacteroids from root nodules of soybean plants, utilizing self-generating Percoll gradients. Can J Microbiol. 1981;27:491–495. doi: 10.1139/m81-072. [DOI] [PubMed] [Google Scholar]
- Rennenberg H. Molecular approaches to glutathione biosynthesis. In: Cram WJ, DeKok LJ, Stulem I, Brunold C, Rennenberg H, editors. Sulphur Metabolism in Higher Plants. Leiden, The Netherlands: Backhuys Publishers; 1997. pp. 59–70. [Google Scholar]
- Riccillo PM, Muglia CI, de Bruijn FJ, Roe AJ, Booth IR, Aguilar OM. Glutathione is involved in environmental stress responses in Rhizobium tropici, including acid tolerance. J Bacteriol. 2000;182:1748–1753. doi: 10.1128/jb.182.6.1748-1753.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüegsegger A, Brunold C. Localization of γ-glutamylcysteine synthetase and glutathione synthetase activity in maize seedlings. Plant Physiol. 1993;101:561–566. doi: 10.1104/pp.101.2.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandalio LM, Palma JM, Del Río LA. Localization of manganese superoxide dismutase in peroxisomes isolated from Pisum sativum L. Plant Sci. 1987;51:1–8. [Google Scholar]
- Schäfer HJ, Haag-Kerwer A, Rausch T. cDNA cloning and expression analysis of genes encoding GSH synthesis in roots of the heavy-metal accumulator Brassica juncea L.: evidence for Cd-induction of a putative mitochondrial γ-glutamylcysteine synthetase isoform. Plant Mol Biol. 1998;37:87–97. doi: 10.1023/a:1005929022061. [DOI] [PubMed] [Google Scholar]
- Schnarrenberger CA, Oeser A, Tolbert NE. Development of microbodies in sunflower cotyledons and castor bean endosperm during germination. Plant Physiol. 1971;48:566–574. doi: 10.1104/pp.48.5.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struglics A, Fredlund KM, Rasmusson AG, Møller IM. The presence of a short redox chain in the membrane of intact potato tuber peroxisomes and the association of malate dehydrogenase with the peroxisomal membrane. Physiol Plant. 1993;88:19–28. [Google Scholar]
- Tang X, Webb MA. Soybean root nodule cDNA encoding glutathione reductase. Plant Physiol. 1994;104:1081–1082. doi: 10.1104/pp.104.3.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple SJ, Vance CP, Gantt JS. Glutamate synthase and nitrogen assimilation. Trends Plant Sci. 1998;3:51–56. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G, Steppuhn J, Herrmann RG. Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem. 1989;180:535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]