Abstract
Background
There is an urgent need for the development of global analytic frameworks that can perform analyses in a privacy-preserving federated environment across multiple institutions without privacy leakage. A few studies on the topic of federated medical analysis have been conducted recently with the focus on several algorithms. However, none of them have solved similar patient matching, which is useful for applications such as cohort construction for cross-institution observational studies, disease surveillance, and clinical trials recruitment.
Objective
The aim of this study was to present a privacy-preserving platform in a federated setting for patient similarity learning across institutions. Without sharing patient-level information, our model can find similar patients from one hospital to another.
Methods
We proposed a federated patient hashing framework and developed a novel algorithm to learn context-specific hash codes to represent patients across institutions. The similarities between patients can be efficiently computed using the resulting hash codes of corresponding patients. To avoid security attack from reverse engineering on the model, we applied homomorphic encryption to patient similarity search in a federated setting.
Results
We used sequential medical events extracted from the Multiparameter Intelligent Monitoring in Intensive Care-III database to evaluate the proposed algorithm in predicting the incidence of five diseases independently. Our algorithm achieved averaged area under the curves of 0.9154 and 0.8012 with balanced and imbalanced data, respectively, in κ-nearest neighbor with κ=3. We also confirmed privacy preservation in similarity search by using homomorphic encryption.
Conclusions
The proposed algorithm can help search similar patients across institutions effectively to support federated data analysis in a privacy-preserving manner.
Keywords: privacy, federated environment, similarity learning, hashing, homomorphic encryption
Introduction
Data-Driven Decision Making in Medical Fields
Electronic health records (EHRs) are becoming ubiquitous across almost all medical institutions. They provide insight into diagnoses [1-6], as well as prognoses [7-10] and can assist in the development of cost-effective treatment and management programs [8,11-14]. All kinds of data across institutions are being collected in EHRs, including diagnosis, medication, lab results, procedures, and clinical notes. In the recently announced precision medicine initiative, many more other types of data including omics data such as genomic and proteomic data and behavior data such as activity sensor data are being generated and collected by doctors and patients. As such rich and heterogeneous health data become available, the entire medical research and practice are shifting from the knowledge or guideline-driven approaches to the data or evidence-driven paradigm, where effective and efficient algorithms become the key for clinical research and practice.
Limitations of Single-Institutional Studies
Previously, many biomedical studies were conducted within a single institution having limited EHR data because of the lack of federated data analysis framework and the institutional privacy concerns on data sharing. However, such an approach has many limitations. For example, it has been demonstrated that genome-wide association studies on EHR data often failed to discover known biomarkers from a single institution because of limited sample size [15,16]. To enable cross-institutional studies, many collaborative networks have been proposed, such as mini-sentinel [17], Observational Health Data Sciences and Informatics [18], National Patient-Centered Clinical Research Network [19], and i2b2 Shared Health Research Informatics Network [20]. These frameworks enable certain analyses (such as database queries with very specific inclusion or exclusion criteria) to be conducted efficiently in a federated manner. However, more sophisticated analyses such as predictive models [21] and context-specific patient similarity search [22] are still a challenge for most existing frameworks, as cross-institutional EHR data exchange is required to build such models, which is usually infeasible because of the institutional privacy and security concerns. There is an urgent need for the development of novel frameworks that can perform analyses in a privacy-preserving federated environment across multiple institutions. In this way, global analytic models can be built collectively without sharing raw EHR data. A few studies on the topic of federated clinical analysis [23-26] have been conducted recently with the focus on different algorithms. However, none of them have solved the problem of similar patient matching, which is important for many biomedical studies. Therefore, we plan to develop a privacy-preserving analytic platform that focuses on a suite of algorithmic challenges on patient similarity learning.
Patient Similarity Learning
Patient similarity learning aims to develop computational algorithms for defining and locating clinically similar patients to a query patient under a specific clinical context [7,27-30]. The patient similarity search is very challenging because the raw EHR data is sparse, high-dimensional, and noisy, which makes finding an exact match among patients using EHR data almost impossible. Besides, patient similarity learning is often context-specific. For example, patient similarity measure for heart disease management can be very different from cancer management. The fundamental challenge is how we can perform effective context-specific patient similarity learning in a federated setting, which enables many different applications:
Cohort construction: cross-institution observational studies are challenging but necessary as many studies require a large and specific patient cohort that does not exist within a single institution. To conduct such a study, an efficient similarity search needs to be conducted across institutions to identify the focused patient cohort.
Disease surveillance: The Centers for Disease Control and Prevention monitors thousands of hospitals for potential epidemics. When a suspicious case is reported, there is a need to find similar cases across geographies.
Clinical trial recruitment: pharmaceutical companies often need to spend significant amount of time and resources to identify targeted patients through many different clinical institutions. Ideally, they would like to be able to perform patient similarity search across all clinical institutions to identify where those relevant patients are. Then they can quickly focus on recruiting patients from the right clinical institutions.
Patient similarity learning involves two computational phases: (1) patient representation learning is to learn the context-specific representation of patients based on their EHR data. For example, patients may be given different representations in heart disease management versus cancer management and (2) patient similarity search is to find similar patients based on their corresponding representations. In a federated environment where multiple institutions exist, patient similarity learning has many unique challenges: (1) how to design an efficient but flexible patient representation that enables fast similarity search? (2) how to learn patient representation from heterogeneous data sources? and (3) how to preserve privacy while still allowing the computation of the patient representation and the search of similar patients across institutions?
Research Objective
The main objective of this paper was to develop a privacy-preserving analytic platform for patient similarity learning in a distributed manner. We propose to learn context-specific binary hash codes to represent patients across institutions. The similarities between patients can be efficiently computed as the hamming distance using the resulting hash codes of corresponding patients; the hamming distance is defined to be the number of places where two binary codes differ. As patient data are heterogeneous from multiple sources such as diagnosis, medication, and lab results, we propose a multi-hash approach that learns a hash function for each data source. Then, the patient similarity is calculated by hash codes from data sources. To avoid the potential security risk because of the attack from malicious users, we also adopt homomorphic encryption [31] to support secure patient similarity search in a federated setting. Finally, the proposed algorithm is applied and validated on real data.
Methods
Feature Construction
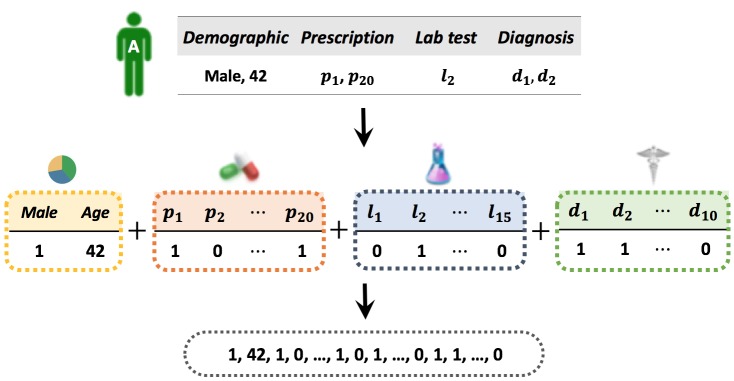
For K feature domains, we assume a vector-based representation for patients in every feature domain (1≤k≤K). There are different ways to construct the feature vectors: (1) for nominal features with standard dictionaries, such as diagnosis and procedure codes, we can use either binary value for presence, or code frequency within the observation period (where the features are extracted from); (2) for continuous features such as age or lab test values, we can use them as they are or we can first quantize them and treat each quantized region as a nominal feature. For example, the values of a specific lab test can be quantized as critical low, low, normal, high, and critical high; and (3) for time-evolving features, if we want to consider the temporal trends in the feature construction process, we can first construct a temporal pattern dictionary with either data-driven method or expertise knowledge, and then treat each pattern as a nominal feature. For example, if there are four types of features including two demographics, 20 prescriptions, 15 lab tests, and 10 diagnoses, we can construct a vector-based representation for patient A as shown in Figure 1. We represent gender as a binary value and age as it is. For diagnosis, prescription, and lab test, we add a one-hot representation of each event (ie, {0,1}|C| with the number of codes | C |).
Figure 1.
Example of feature construction. Prescription, lab test, and diagnosis are denoted by p, l, and d, respectively.
Hashing
In general, hashing is an approach of transforming the data item to a low-dimensional representation, or equivalently a short code consisting of a sequence of bits (Figure 2).
Figure 2.
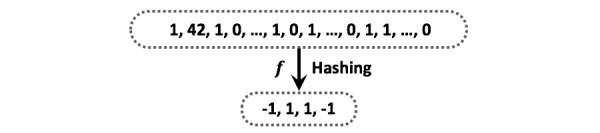
Example of hashing.
Hashing technologies can be applied in many applications such as Bloom filter [32] and cryptography [33]. Similarity-based hashing [34] is one specific type of hashing that aims to preserve the data similarities in their original space with hash codes. On the basis of the availability of supervision information, a similarity-based hashing method can be categorized as unsupervised [35-40], semi-supervised [41-43], or supervised hashing [44,45]. Unsupervised methods learn hash functions purely based on data distributions. Supervised methods exploit the labeled pairwise relationship between entities to capture the high-level data semantics. Semi-supervised methods lie in between them, that is, they explore both data distribution characteristics and labeled pairwise data relationships to learn the hash functions. Most of these existing methods assume a single vector-based representation for every data object.
However, one challenge in our scenario is that the patient features are highly heterogeneous, that is, the features for characterizing the patients are of different types. In this case, it may not be effective to represent each patient as a single vector (simple concatenation will not work as different features are of different types and have different value range). There are some existing multi-modal hashing methods [46-52] that aim to derive a unified single-hash table for encoding the data objects with heterogeneous features. The problem with single-hash (or uni-hash) table is that it is difficult to discover the latent similarity components [53] derived from different feature types, which is crucial in our scenario. For example, it is important to know how similar two patients are, but also why (eg, patients A and B are similar to each other mainly because of their similar demographics and patients B and C are similar because of their similar diagnosis history and lab test values).
Federated Patient Hashing Framework
Symbols used in this paper are listed in Textbox 1.
List of symbols.
M: the number of local sites
K: the number of feature types (domains)
Si: i-th local site
Pi: patient population in Si
Ni: patient population size of Si
Pik: patient population for k-th type of feature in Si
pij: j-th column of Pi, j-th patient in Pi
pijk: j-th column of Pik, k-th type of feature vector for pij
fk: k-th hash function
dk: dimensionality of the k-th feature type
bk: the number of bits of the learned hash codes for the k-th feature type
Wk: function coefficients of the hash function for the k-th feature type
wik: i-th column of Wk
Qik: numerical data transformed from Pik
sign(Qik): signed Qik
Hik: hash codes for Pik(=sign(Qik))
hijk: j-th column of Hik, the hash codes of pijk
Ψ({Wk}Kk=1): reconfiguration error term for {Wk}Kk=1
S ({Wk}Kk=1): supervised loss term for {Wk}Kk=1
U ({Wk}Kk=1): unsupervised loss term for {Wk}Kk=1
Ω ({Wk}Kk=1): term related to {Wk}Kk=1 itself
L (x, y): loss function between x and y
λS: regularizer of S ({Wk}Kk=1)
λU: regularizer of U ({Wk}Kk=1)
λW: regularizer of Ω ({Wk}Kk=1)
λ: regularizer of a supervised loss term
η: regularizer of a Frobenius norm for Q
σikuv: similarity between piuk and pivk
Ri: pairwise relationship of Ri for labeled information
Sik: pairwise similarity of Pik
riuv: relationship between piuk and pivk for labeled information
sikuv: similarity between piuk and pivk
SL(Qik): approximated sign function for Qik
Figure 3 illustrates the overall federated patient matching framework. Suppose there are M sites with the i-th site Si which owns a patient population Pi. We use pij to represent the j-th patient in Pi. Then, our problem is, given a query patient, how to retrieve similar patients from those M sites without explicitly accessing the patient feature vectors. Our plan is to resolve this problem using similarity based hashing, which transforms the patient’s raw features into a binary vector representing patient characteristics (patient representation learning). The pairwise patient similarities will be evaluated as the pairwise distance based on those signatures (patient similarity search). In this paper, we will focus on feature-based hashing, that is, those binary patient signatures are obtained by proper transformation from patient features. Therefore, to perform hashing, we need to first construct feature-based representation for patients.
Figure 3.
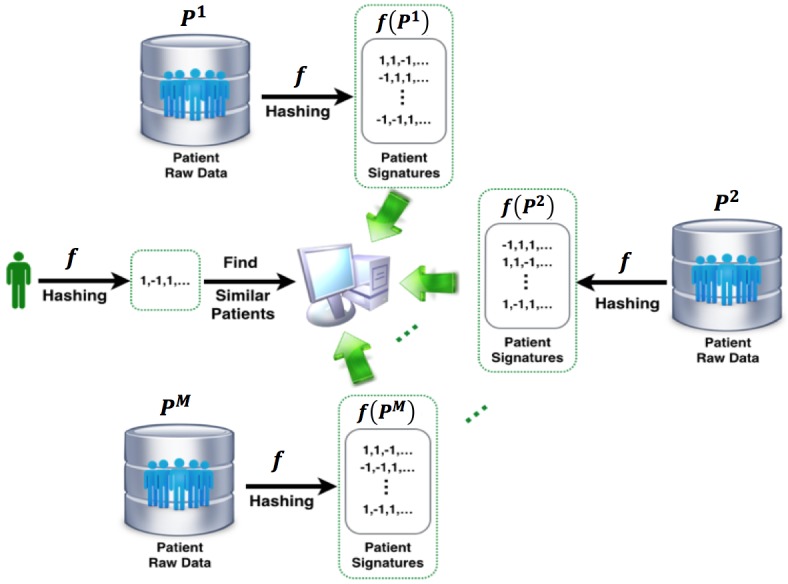
The whole process of patient matching in a federated environment. The user sends a patient matching request to the service center, which is delegated to patient data resources from several clinical sites. Due to the privacy concerns, the center does not have access to the raw patient data. All patients within different sites need to be first hashed, and the center only has the patient’s signatures after hashing. The hash functions are shared across different sites.
Without the loss of generality, we assume there are K different feature types to characterize every pij, and we use pijk (k=1,2,…, K) to represent the k-th type of feature vector of pij. The goal is to derive an effective computational framework for patient matching in a federated environment, and the key idea is to learn a good hash function that can transform the patient features into binary hash codes. A uni-hash table approach shown in Figures 2 and 3 is to learn only one hash function for the feature vector f: Rd→{-1,+1}b, where d is the dimensionality of the whole feature vector, and b is the number of bits of the hash codes learned d by f. In this paper, we propose a multi-hash approach for patient hashing that aims to learn a hash function fk: Rdk→{-1,+1}bk for every patient feature type k (k=1,2,…, K); dk is the dimensionality of the k-th feature type, and bk is the number of bits of the learned hash codes for the k-th feature type. Each fk (k=1,2,…, K) is shared across all the M sites. We use the sign function to construct the hash codes, that is, sign (Qik)∈{-1,+1}bkⅹNi , where Qik is transformed numerical data from original data of i-th site for k-th type of feature Pik∈ RdkⅹNi by a hash function fk that incorporates function coefficients for the k-th feature type Wk∈dkⅹbk; Ni is the population size of i-th site. How these components are formulated is described in the next paragraph in detail. We use Hki=sign(Qik) to denote the hash codes of k-th feature type for the patients at i-th site. Figure 4 shows the process of patient similarity calculation with a multi-hash approach.
Figure 4.
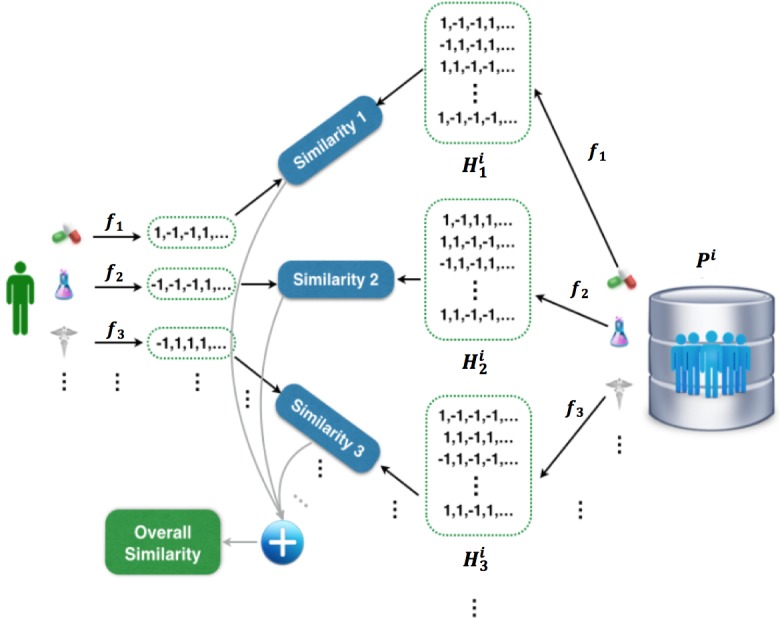
The process of calculating patient similarity with a multi-hash approach.
The u-th column of Hki, hiuk∈{-1,+1}bk is the hash codes of piuk. Then, the similarity between piuk and piuk can be evaluated as the inner product of hiuk and hiuk as shown in equation 1:
| (1) s i kuv = 1/ b k(hi uk) T(h i vk) |
Thus, the overall similarity can be computed as the average of K similarities, as shown in equation 2, which is bounded on the interval of (−1,1).
| (2) s i uv = 1/ K∑k(h i uk) T(h i vk) |
Here, we suggest a general framework for learning {Wk}Kk=1, which is the most important component. The framework basically constructs an objective function in terms of {Wk}Kk=1 such as shown in equation 3, where λS, λU, and λW are regularizers of S ({Wk}Kk=1), U ({Wk}Kk=1), and Ω({Wk}Kk=1), respectively, and then minimizes (or maximizes) it:
| (3) J ({W k}K k=1) = Ψ ({W k}K k=1) + λ S S ({W k}K k = 1) + λ U U ({W k}K k = 1) + λ WΩ ({W k}K k = 1) |
Ψ ({Wk}Kk=1) is a reconfiguration error term between the low-dimensional representation of the original data and hash codes, which is the main term of the objective function, and generates the hash codes from the original data, as shown in equation 4, where ||·||F is a Frobenius norm [54]. On the basis of this term, the hash function in our framework is formed as fk(Pki)=WkTPki, and this transformation results in Hki=sign(Qik).
| (4) Ψ ({W k}K k=1) = ∑i∑k||Wk T P k i - H k i||2 F |
The objective function can incorporate regularizers, as well as the main term to obtain better solutions of {Wk}Kk=1 by (1) introducing additional information to improve either unsupervised or supervised learning if desired, (2) solving an ill-posed problem, and 3) preventing overfitting. Possible regularizers are listed as follows:
S({Wk}Kk=1) is a supervised loss term that measures the quantization loss during the hashing process when supervision information is available for the patients. Here, the supervision information could be the labels of the patients, such as the disease the patients have. For example, if both piu and piv have the same disease, then their relationship riuv=1, otherwise riuv=-1. Then, we can set S({Wk}Kk=1) as shown in equation 5:
| (5) S ({W k}K k=1) = ∑i∑k∑u,v - s i kuv r i uv |
The possible choices of supervised loss term could be any loss function L (x,y), and examples include L(x,y)=-xy and well-known binary loss functions such as (1) logistic loss, L (x,y)=log(1+exp(- xy)) and (2) hinge loss, L (x,y)=max(0,1-xy).
Note that U({Wk}Kk=1) is an unsupervised term that exploits the intrinsic data distribution and enforces the resultant hash codes to comply with the distribution. For example, we can request similar patients to have similar hash codes on each feature type. This can be achieved by minimizing the below regularizer, as shown in equation 6, where σikuv is a similarity between piuk and pivk based on, for example, a Gaussian function for continuous valued features or a cosine function after Term Frequency-Inverse Document Frequency normalization on bag-of-code (eg, diagnosis code or procedure code):
| (6) U ({W k}K k=1) = ∑i∑k∑u,v σ i kuv||h i uk- h i uk||2 F |
Ω({Wk}Kk=1) is a term related to {Wk}Kk=1 themselves, which is independent of the patient features. Examples of Ω({Wk}Kk=1) include (1) Frobenius norm regularizer ∑Kk=1||Wk||2F, which can be used for improving the numerical stability of the solution process and (2) orthogonality regularizer ∑Kk=1∑i≠j||wTikwjk||2, where wik is the i-th column of Wk, which can encourage the diversity of the learned hash codes and thus improve their representation effectiveness.
Figure 5 shows a running example of the proposed hashing methodology. Such optimization problems can be solved with Block Coordinate Descent technologies [55], with {Wk}Kk=1 as variable blocks that alternatively update Wk (1≤k≤K) one by one. Moreover, as different sites are continuously receiving new patients (or new patient features), we will need to continuously update the hash functions as well. Fortunately, as can be observed from equations 4, 5, and 6, those terms are fully decomposable with respect to different sites. Therefore, we can update the hash functions in an asynchronous manner, that is, we can update the current {Wk}Kk=1 as soon as new patient data is received on site i.
Figure 5.
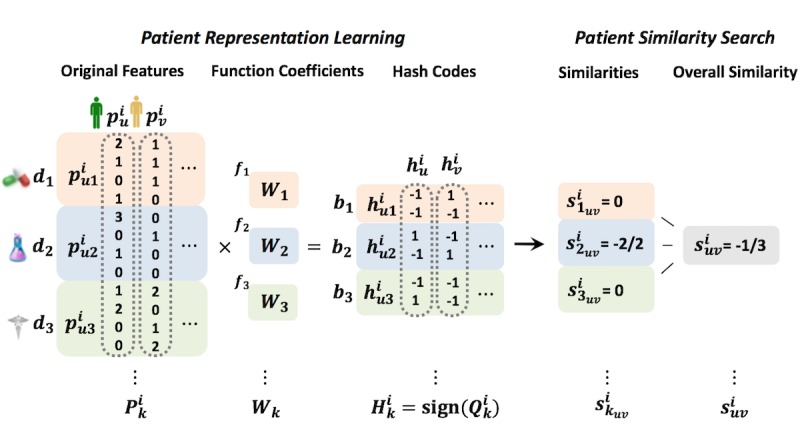
Example of transformation of patient vectors into hash codes and computation of similarity between hash codes.
Privacy-Preserving Patient Representation Learning in a Federated Setting
Without loss of generality, let us instantiate the objective function with the regularizer λ of empirical error on the labeled data for a family of hash codes; this choice might be the most basic approach to similar patient learning based on the fact that supervised learning is more commonly used than unsupervised learning because data generated in the medical field usually have label information.
When solving the initiated objective function, two possible problems because of the sign function for Q arise. First, Q may not be a unique solution, and thus, the objective function is difficult to converge without considering any regularizer about Q. We add a Frobenius norm regularizer η to solve this problem. In addition, the objective function f(W,Q) is nondifferentiable in terms of Q. We can approximate the sign function with the surrogate function. Then, we have the final objective function, as shown in equation 7, where Ri∈ RNiⅹNi is the pairwise relationship of Pi for labeled information:
| (7) f (W, Q) = min∑i∑k||W k T P i k -S L( Q i k)||2 F + λ ∑i∑k tr(-S L( Q i k) R i S L( Q i k)T ) + η ∑ i∑ k|| Q i k||2 F |
If both piu and piv have the same disease, then their relationship riuv=1, otherwise riuv=-1, and SL(·) is the surrogate function, as shown in equation 8, where ∘ is the hadamard (elementwise) product:
| (8) S L (Q i k) = (Q i k∘ Q i k + ξ) -1/2∘Q i k |
The detailed process to derive the final objective function is given in Multimedia Appendix 1 (Note ⊗ is the Kronecker product [56]). The objective function for {Wk}Kk=1 and {Qik}K,Mk,i=1 can be solved one by one iteratively as variable blocks [55] by using the Newton-Raphson method [57] until the estimates converge. To be specific, this approach first allows us to update Wk for each of k (k=1,2,…, K) with other Wl for all l (1≤l≠k≤K) and Q being fixed:
| (9) W new k = W k - (∂ 2 f /∂ W 2 k) -1∂ f /∂ W k |
Then, similarly, we update Qik for each combination of (i, k) (1≤i≤M,1≤k≤K) with other combinations of (j, l) (1≤j≠i≤M,1≤l≠k≤K) and W being fixed:
| (10) Q i,new k = Q i k - (∂ 2 f /∂ Q i k 2) -1∂ f /∂ Q i k |
The derivation process for the first and second derivatives of W and Q is described in Multimedia Appendix 1. As derivatives are linearly decomposable by sites i, the objective function defined in equation 7 can be computed in a distributed manner. This means the optimization only requires locally computed statistics to be delivered to estimate the {Wk}Kk=1 iteratively until convergence.
The time complexity at each iteration depends on feature type k and site i. When updating Wk for each of k (1≤k≤K) with other Wl for all l (1≤l≠k≤K) and Q being fixed, the time complexity is O (dk3) because each site has to inverse the dkⅹ dk Hessian matrix. When updating Qik for each combination of (i, k) (1≤i≤M, 1≤k≤K) with all other combinations of (j, l) (1≤j≠i≤M, 1≤l≠k≤K) and W being fixed, the time complexity is O (bk3Ni3) because Si has to inverse the bkNiⅹbkNi Hessian matrix. Therefore, parameters that have a significant effect on time complexity include original and projection dimensions by feature type and population size by site. Other parameters such as the number of sites M and the number of feature types K along with the number of iterations are excluded in the big O notation because they are just constants. That is unless the number of site or the number of feature type goes to infinity, it only has a small impact on the complexity.
Privacy-Preserving Patient Similarity Search in a Federated Setting
To find similar patients across sites, hash codes for each site Hi (ie, {Hik}Kk=1 have to be exchanged across institutions originally. However, when all other sites expect for i-th site receive Hi for similarity search, the patient-level information of i-th site can be leaked by equation 4; other sites and a server can be united for reverse engineering to extract Pibecause they have both {Wk}Kk=1 and Hi, as well as their information in equation 4. Figure 6 illustrates the situation mentioned.
Figure 6.
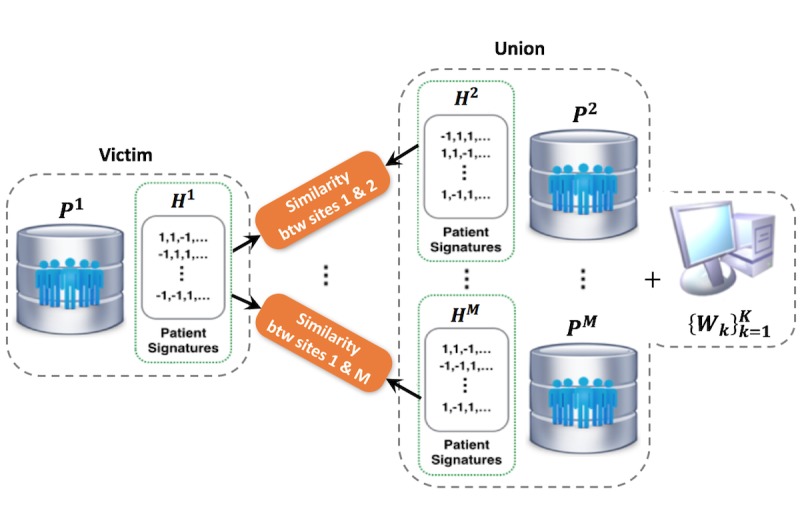
Example of potential privacy leakage in patient similarity search across sites.
Therefore, we suggest the way to search similarity among different sites by avoiding revealing Hik but able to compute similarities based on Hik. We introduce homomorphic encryption specifically that is a form of encryption where a specific algebraic operation performed on the plaintext is equivalent to another algebraic operation performed on the cipher-text, and an encrypted result, when decrypted, matches the result of the same operation performed on the plaintext. Unlike traditional encryption schemes that do not allow any computations to be performed on the cipher-text without first decrypting it, homomorphic encryption allows computations to be performed without decrypting the data. The results of the computations remain encrypted and can only be read and interpreted by someone with access to the decryption key. Therefore, it is appropriate to use homomorphic encryption in our case that other sites and a server can attack maliciously. It enables cross-site comparison of health care statistics with protecting privacy for each site. The procedure of homomorphic encryption in this paper is summarized as follows: first, i-th site encrypts hash codes for its query data and delivers encrypted codes to j-th site. Next, j-th site performs the computation between delivered encrypted codes of i-th site and encrypted codes of j-th site without a decryption key and sends the computed value to i-th site. Finally, i-th site decrypts the value to get the hamming distance of hash codes between query data and data of j-th site. Each site is restricted to only answer the hamming distance to avoid the risk of privacy leakage. This process is depicted in Figure 7.
Figure 7.
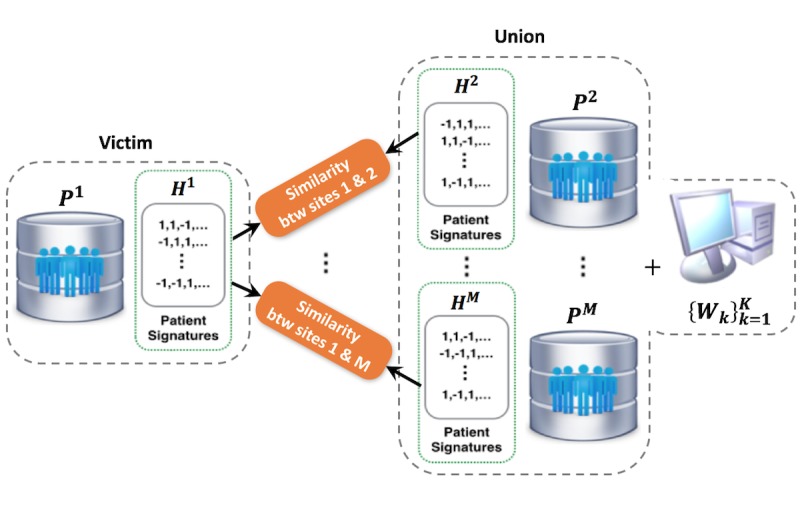
Privacy-preserving patient similarity search by homomorphic encryption; green key: encryption (public) key, blue key: decryption (private) key.
We note that homomorphic encryption provides an extra layer of privacy protection especially during patient similarity search.
Security
There are several participants in our framework.
Data custodians (DCs) represent institutions or hospitals who have access to patient data and would like to collaborate in learning about similar patients.
Crypto service provider (CSP) generates public and private keys. The public key is provided to the data custodians to safeguard the intermediary statistics.
Cloud server (CS) computes over summary statistics from individual data custodians to obtain a global patient similarity model.
Our goal is that a DC does not learn patient-level information from other DCs during the process. We also want to ensure CS cannot infer patient-level information from the data. We assume a CSP is trustworthy and provides encryption keys (public and private). In the threat model, we assume the CS to be semi-honest, that is, it is honest to follow the protocol but curious about patient’s private information while executing the protocol. We make the following basic assumptions: (1) DC and CS do not collude, (2) CS and CSP also do not collude, and (3) DC always receives correct keys from the CSP. To evaluate the security of our system, it is assumed that the security of the system is compromised if patient-level data or intermediary statistics that can infer patient-level data are leaked. CSP is only involved in generating public and private keys and transferring those keys to DCs, and no access to unintended fine-grained local information is involved in this process.
The leakage is related to computation of {Wk}Kk=1, and possible scenarios according to the participants are as follows:
Leakage to CSP in each computation: CSP does not participate in computation at all. Therefore, there is no leakage.
Leakage to DC in each computation: each DC cannot indirectly learn patient data from other DCs only with {Wk}Kk=1 and its local information {Wk}Kk=1 and {Qik}Kk=1. If all DCs except for one collude, it is infeasible for the other DCs to reconstruct Pik of that one DC because the first and second derivatives of Wk have a nonlinear relationship for Pik. Specifically, it is not possible to specify a certain matrix only given information of covariance matrix because of insufficient equations. They also do not have information (first and second derivatives) about Qik.
Leakage to CS in each computation: CS cannot infer patient data from {Wk}Kk=1. Even though CS receives local information for the first and second derivatives of {Wk}Kk=1, it is infeasible for CS to recover {Pik}Kk=1 for the same reason as the collusion among DCs. In finding similar patients, hash codes for each site {Hik}Kk=1 have to be exchanged across institutions originally, but the use of homomorphic encryption prevents direct exchange of hash codes {Hik}Kk=1 between DCs, and thus, there is no leakage.
Results
Experimental Setting
We conducted experiments to validate our proposed method on real data. For comparison, we assumed two different systems against our system according to connection among M sites: open and closed system. In the open system, M sites can exchange their patient’s information without any restrictions; in the closed system, each site can only utilize patient's information in each site. Ours is in the middle of two systems. For better understanding of these systems, let us assume that there are three sites A, B, and C with the same number of patients N. In this situation, an open system means that every site can access the complete information of the entire patient cohort (3ⅹ N), including information from other sites as well, and thus, three sites work like one site without any concerns on privacy. On the other hand, closed system indicates that each site can only access its patient-level information (N) exclusively. Open system and closed system are derived based on an idealistic situation and a realistic situation, respectively, and our system is in between these two systems, which cannot access patient’s information from other sites but can utilize it through { Wk}Kk=1. Then, we predicted the incidence of a certain disease and compared the standard κ-nearest neighbor (κ-NN) classification results based on hamming distance of multi-hash codes from our system with those based on hamming distance of multi-hash codes from open and closed systems, as well as uni-hash codes from open and closed systems. We also provided baseline results based on four similarity distances of raw data without using hashing for open and closed systems: Euclidean, cityblock, cosine, and correlation. We utilized five-fold cross validation (CV) that randomly splits patients into five folds with the equal size; we used four folds for training, and one fold for testing. As an evaluation measure, we used the area under the curve (AUC) where the true positive rate (TPR; ie, the number of true positives divided by the sum of true positives and false negatives) is plotted against the false positive rate (ie, the number of false positives divided by the sum of false positives and true positives) at various thresholds. AUC as a summarized single value for the curve has desirable properties that are independent to the threshold and invariant to a priori class probability distributions. An area of 1 represents a perfect model, and an area of 0.5 represents a worthless model. As we repeated CV ten times, we obtained ten vectors consisting of probabilities based on κ nearest neighbors’ voting. The program was implemented by MATLAB 2015b (MathWorks).
Temporal Sequence Construction
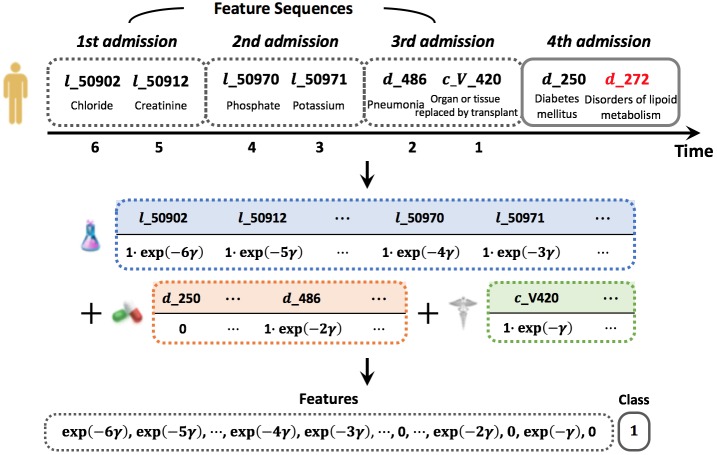
A sequence is composed of lab tests, prescriptions, diagnoses, conditions, and symptoms that were given to a patient in multiple hospital admissions. We only extracted common lab tests, prescriptions, diagnoses, conditions, and symptoms (prefixed with “ l_,” “ p_,” “ d_,” “ c_,” and “ s_,” respectively). We used the International Classification of Diseases, 9th revision (ICD-9) level 3 codes instead of level 4 or 5 to avoid extreme sparsity of diagnoses. We assumed space in time between all events to be same. Then, we constructed data for incidence of a target disease as follows: for patients in which a target disease occurs, we sliced the very admission that includes the diagnosis event of a target disease out of the sequence, and used only events before that admission as a feature sequence. For other patients, we used all events. We utilized temporal information of a sequence to make a time-decayed vector representation; when we add a one-hot representation for each event, it is multiplied by the time decaying function (ie, exp(-γt) with the decay constant γ) that enables to weaken the effect of old event but to strengthen the effect of recent event. A graphical illustration of this sequence and its vector representation is presented in Figure 8.
Figure 8.
Example of constructing temporal sequence with target disease in red and its vector representation.
Multiparameter Intelligent Monitoring in Intensive Care-III Database
We used Multiparameter Intelligent Monitoring in Intensive Care-III (MIMIC-III) database that contains health-related data associated with 46,520 patients and 58,976 admissions to the intensive care unit of Beth Israel Deaconess Medical Center from 2001 to 2012. The database consists of detailed information about patients, including demographics such as gender, age, and race; admissions; lab test results; prescription records; procedures; and discharge ICD diagnoses. On the basis of this database, we randomly selected several common diseases (ie, diseases with relatively large number of positives) as a target disease to verify that our method can perform well in general not only for a specific disease. Then, we extracted temporal sequences and constructed following six feature vectors (K=6) for patients in i-th site: demographic information Pi1∈Rd1ⅹNi, lab results Pi2∈ Rd2ⅹNi, diagnoses Pi3∈Rd3ⅹNi, prescriptions Pi4∈Rd4ⅹNi, conditions Pi5∈Rd5ⅹNi, and symptoms Pi6∈ Rd6ⅹNi. Time decay constant γ was set to 0.01. We note that the feature vector of diagnoses in each dataset does not include its outcome of interest. Information of original datasets is described in Table 1.
Table 1.
Description of five datasets from Multiparameter Intelligent Monitoring in Intensive Care-III (MIMIC-III) database.
| Disease | Data size (negative or positive) | Dimension (dk, k=1,…,6) |
| Disorders of lipoid metabolism | 4546/2990 | (12,204,814,1338,262,170) |
| Hypertensive chronic kidney disease | 5652/1884 | (12,204,822,1338,266,169) |
| Cardiac dysrhythmias | 3878/3658 | (12,204,817,1338,263,169) |
| Heart failure | 4167/3369 | (12,204,819,1338,265,169) |
| Acute renal failure | 4182/3354 | (12,204,809,1338,268,170) |
To test three-site scenario, we made datasets balanced and horizontally partitioned the dataset into three, assuming data are evenly partitioned among sites (M=3), P1k∈Rdkⅹ125, P2k∈Rdkⅹ125, and P3k∈Rdkⅹ125 for every k=1,…6; federated system is needed when each institution has a limited sample size that is not enough for an analysis. In addition, from the complexity analysis, time to implement the algorithm exponentially increases in proportion to the number of patients. On the basis of these, we randomly selected and placed 125 patients in each site. Then, we predicted the incidence of five diseases independently. We set parameters for regularizers λ=0.5 and η=10-3 in common. In addition, for multi-hash approach, we reduced the original dimensions for each feature to ten (ie, bk=10 for k=2,…,6) except for the demographic feature that was reduced to two (ie, b1=2), and for uni-hash approach we reduced the total dimensionality to the sum of projection dimensions in multi-hash approach (ie, b=52). We note that the results would be robust to the projection dimensionality unless we have too many or too few of it. Table 2 shows the results of κ-NN with κ=3 based on hamming distance for the following configurations: our system, open and closed systems with multi-hash, as well as open and closed systems with uni-hash.
Table 2.
Averaged area under the curve (AUC) with SD of κ-NN (κ=3) based on hamming distance from our, open and closed systems with multi-hash approach and from open and closed systems with uni-hash approach and based on cosine distance from open and closed systems.
| Disease | Multi-hash | Uni-hash | Baseline | ||||
|
|
Our system, Averaged AUC (SD) | Open system, Averaged AUC (SD) | Closed system, Averaged AUC (SD) | Open system, Averaged AUC (SD) | Closed system, Averaged AUC (SD) | Open system, Averaged AUC (SD) | Closed system, Averaged AUC (SD) |
| Disorders of lipoid metabolism | 0.9330 (0.0086) | 0.9343 (0.0125) | 0.9002 (0.0285) | 0.9159 (0.0255) | 0.8486 (0.0271) | 0.8079 (0.0222) | 0.7945 (0.0308) |
| Hypertensive chronic kidney disease | 0.9078 (0.0346) | 0.9283 (0.0432) | 0.8538 (0.0421) | 0.9270 (0.0350) | 0.8501 (0.0305) | 0.7823 (0.0261) | 0.7762 (0.0262) |
| Cardiac dysrhythmias | 0.9135 (0.0287) | 0.9368 (0.0492) | 0.8833 (0.0397) | 0.9072 (0.0414) | 0.8236 (0.0328) | 0.7695 (0.0151) | 0.7340 (0.0343) |
| Heart failure | 0.9058 (0.0282) | 0.9351 (0.0326) | 0.8798 (0.0414) | 0.9089 (0.0376) | 0.8471 (0.0248) | 0.7986 (0.0292) | 0.7733 (0.0421) |
| Acute renal failure | 0.9169 (0.0397) | 0.9477 (0.0374) | 0.8637 (0.0320) | 0.8821 (0.0403) | 0.7929 (0.0378) | 0.7434 (0.0380) | 0.7289 (0.0341) |
Additionally, Table 2 presents a baseline result based on cosine distance obtained from open and closed systems, which has the highest AUC among baseline results. We note that the results for closed systems are the average of three sites.
Figure 9 shows the comparison for heart failure of our open and closed systelabelms with multi-hash approach as an example. The prediction performance of our system is moderate between those of open and closed systems. It is encouraging that our system approaches open system without sharing local data. Figure 10 also shows the comparison result for heart failure of our system with multi-hash approach and open and closed system with uni-hash approach. We can see the superior performance of our system over closed system as before. However, in this case, our system is comparable with open system and even outperformed it for three diseases; this may come from multi-hash approach is more effective than uni-hash approach to construct context-specific hash codes. Figure 11 shows the results of our system with different κ. The detailed results with different κ are presented in Multimedia Appendix 2. AUC generally increases as κ increases.
Figure 9.
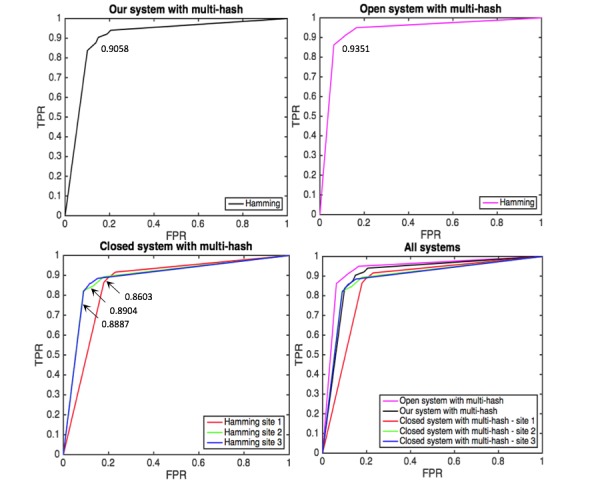
Averaged area under the curve (AUC) of κ-NN (κ=3) for heart failure based on hamming distance from our, open and closed systems with multi-hash approach.
Figure 10.
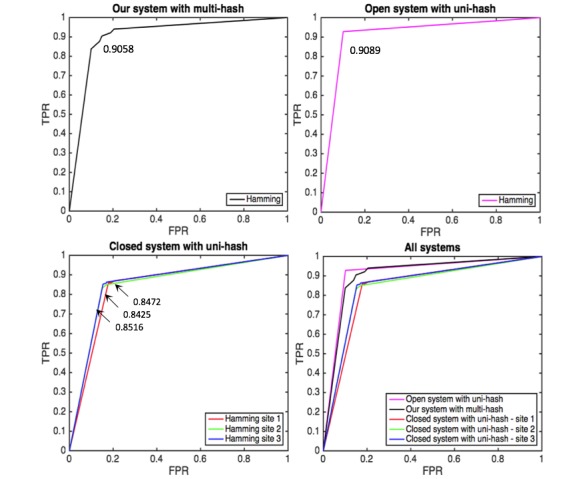
Averaged area under the curve (AUC) of κ-NN (κ=3) for heart failure based on hamming distance from our system with multi-hash approach and open and closed systems with uni-hash approach.
Figure 11.
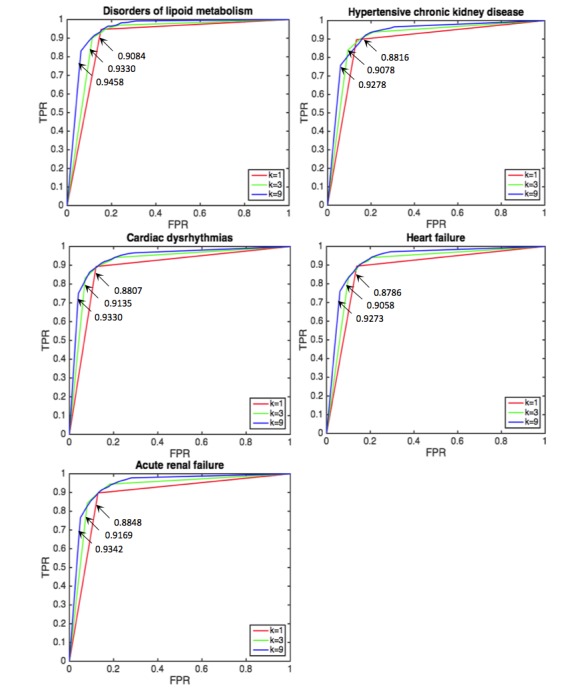
Averaged area under the curve (AUC) of κ-NN with different κ (κ=1,3,9) for five diseases from our system.
However, in real life, different sites have a different specialty and have a different distribution in patient data. To see how our platform works in random and skewed distribution, we differentiated the ratio of samples having negative and positive classes by site. We assumed that three sites, respectively, have 10%, 30%, and 50% of positive class for five diseases. Note that all other settings including the number of sites and patients for each site, projection dimensions, and parameters were set the same as before to test only the change originated from the class imbalance and for experimental convenience; we omitted the uni-hash approach, which is expected to have the similar trend about multi-hash approach to that shown in Table 2. Table 3 shows the averaged AUC results from κ-NN with κ=3 based on hamming distance for our system, open and closed systems with multi-hash, and based on cosine distance for open and closed systems with raw data. For more elaborate comparison, F1, sensitivity (ie, TPR), and specificity (ie, the number of true negatives divided by the sum of true negatives and false positives) [58] were also measured along with AUC (Multimedia Appendix 3); F1 is the harmonic mean of precision and recall where it reaches its best value at 1 and worst at 0. It can be interpreted as weighted average of the precision (ie, the number of true positives divided by the sum of true positives and false positives) and recall (ie, TPR, sensitivity) with their equal contribution.
Table 3.
Averaged area under the curve (AUC) with SD of κ-NN (κ=3) based on hamming distance from our, open, and closed systems with multi-hash approach and based on cosine distance from open and closed systems.
| Disease | Multi-hash | Baseline | |||
|
|
Our system, AUC (SD) | Open system, AUC (SD) | Closed system, AUC (SD) | Open system, AUC (SD) | Closed system, AUC (SD) |
| Disorders of lipoid metabolism | 0.8056 (0.0386) | 0.8309 (0.0412) | 0.7629 (0.0295) | 0.7525 (0.0212) | 0.7104 (0.0187) |
| Hypertensive chronic kidney disease | 0.7637 (0.0367) | 0.7924 (0.0209) | 0.7275 (0.0266) | 0.7296 (0.0215) | 0.7141 (0.0207) |
| Cardiac dysrhythmias | 0.7840 (0.0301) | 0.7937 (0.0228) | 0.7659 (0.0223) | 0.7638 (0.0198) | 0.7385 (0.0188) |
| Heart failure | 0.8287 (0.0283) | 0.8832 (0.0278) | 0.7459 (0.0331) | 0.7735 (0.0206) | 0.6778 (0.0213) |
| Acute renal failure | 0.8239 (0.0326) | 0.8704 (0.0335) | 0.7558 (0.0263) | 0.7304 (0.0218) | 0.7415 (0.0225) |
Most of the results can be interpreted in the same context as Table 2, but it should be noted that the degree of performance degradation in our system (~13%) is greater than that at baseline (~5%). Given these results from open and closed systems, as well as our system with multi-hash approach, accuracy might be lost because of the instability caused by updating weights {Wk}Kk=1 with information from skewed distributions. However, it is encouraging that sensitivity is obtained stably in multi-hash approach rather than baseline. Sensitivity is an important measure in medical analysis because it is much more dangerous to diagnose that the disease has not occurred even though it has already developed than the opposite case. The fact that F1 is significantly larger is consistent with this. Therefore, considering all the results, we believe that our system is a useful alternative.
Next, we performed secure data aggregation and data comparison among different sites in a federated setting by which each site is able to retrieve its hamming distance under certain criteria in a privacy-preserving manner. In our experiments with balanced data, each row has 52 bits (hash code), and a 128-bit encryption key is used for homomorphic encryption. We measured the execution time of some key cryptographic operations in a workstation with an Intel 2.5 GHz CPU, where all the results are averaged over five-fold CV of total time for six cases (three test sets by two training sets). The execution time of each basic cryptographic operation has been profiled and shown in Table 4.
Table 4.
Averaged execution time of each basic cryptographic operation for five diseases.
| Operation | Time (seconds) | ||||
|
|
Disorders of lipoid metabolism | Hypertensive chronic kidney disease | Cardiac dysrhythmias | Heart failure | Acute renal failure |
| Homomorphic encryption | 1.9 | 2.2 | 2.2 | 2.3 | 2.2 |
| Initialization | 5.2 | 6.3 | 5.8 | 6.5 | 6.0 |
| Comparison | 994.2 | 1243.9 | 1067.1 | 1131.7 | 1066.5 |
| Homomorphic decryption | 0.4 | 0.4 | 0.4 | 0.4 | 0.4 |
We confirmed that the calculated similarities across sites are the same when exchanging raw {Hik}Kk=1 directly with each other (ie, without homomorphic encryption) or exchanging encrypted {Hik}Kk=1 (ie, with homomorphic encryption) with each other. Therefore, the results after homomorphic encryption were obtained exactly the same as the results in Tables 2 and 3 and Figures 9 to 11 without any privacy leakage.
Discussion
Principal Findings
There are several limitations in the proposed framework. When learning hash functions, the assumption is that each site has common feature events that should be needed. However, different sites, for example, hospitals, may have different event types, and additionally, the notation system for each event type cannot be standardized except for diagnoses, symptoms, and conditions that are based on ICD-9. Even though we have the limitation of common feature events, we believe that our methodology can be still useful for cooperating hospitals eager to find similar patients across sites at the point of care. We are planning to develop a new and more practical approach to relax this assumption.
Basically, our system works better when all the participants have similar distributions. However, we have confirmed through the imbalance class experiment that our system still works well with different distributions, as well at the cost of some performance degradation. We will address more generalized imbalance data problem in future work.
Next, even if we have computational benefits by adopting a multi-hash approach compared with a uni-hash approach, and the computational complexity is not prohibitive in practice, a technical challenge still remains in scalable hash function learning when the sample size and the feature dimensionality are large. This is because the complexity for inverting Hessian matrices in our algorithm is affected by the sample size and the feature dimensionality. This is an expensive operation of time complexity and requires a lot of memory. We can solve this problem by using parallelization or graphics processing units or utilizing a gradient descent method that replaces the inversion of Hessian matrix with a constant or a variable varying with the iteration number.
We demonstrated the feasibility of privacy-preserving similarity search, and the experiments were conducted on a single machine (with different processes) to serve as a proof of concept. In practice, we need to deploy the algorithm in multiple computers, and that is a trivial task. We will execute this algorithm using secure multiparty computation such as in the Secure Multi-pArty Computation Grid LOgistic REgression [59] in future work.
We have also listed several limitations to consider for more elaborate future work. When constructing temporal sequences, it assumes the sequence events are sampled at the same frequency for simplicity, which means the temporal effect has not been represented in this work. We roughly determined parameters of projection dimension and decay factor, which might not be optimal. In our experiment, we used 3-digit ICD to show a proof of concept, but the granularity of the ICD code will affect the performance in real applications, especially if the interest is related to the rare ones.
Conclusions
We proposed a federated patient hashing framework and developed a privacy-preserving patient similarity learning algorithm. This technique allows to learn hash codes for each patient reflecting information of different sites without sharing patient-level data. Using MIMIC-III database, we conducted experiments to demonstrate the accuracy and usability of the proposed algorithm. By utilizing the multi-hash approach, our algorithm obtained more usable and practical results than the uni-hash approach. To avoid privacy leakage in patient similarity search, we also applied homomorphic encryption able to calculate the hamming distance without transmitting hash codes. As a result, we confirmed the same results without any privacy leakage.
Acknowledgments
This work was supported by NHGRI grants R00HG008175, R01HG008802, and R01HG007078, NIGMS R01GM114612, NLM grants R00LM011392, R21LM012060, and NHLBI grant U54HL108460. The work of FW is supported by NSF IIS-1650723 and IIS-1716432.
Abbreviations
- AUC
area under the curve
- CS
cloud server
- CSP
crypto service provider
- CV
cross validation
- DC
data custodian
- EHR
electronic health record
- ICD-9
International Classification of Diseases, 9th revision
- MIMIC-III
Multiparameter Intelligent Monitoring in Intensive Care-III
- TPR
true positive rate
Privacy-preserving patient representation learning in a federated setting.
Prediction performance of balanced class datasets.
Prediction performance of imbalanced class datasets.
Prediction performance of balanced class datasets.
Footnotes
Conflicts of Interest: None declared.
References
- 1.Kononenko I. Machine learning for medical diagnosis: history, state of the art and perspective. Artif Intell Med. 2001 Aug;23(1):89–109. doi: 10.1016/s0933-3657(01)00077-x.S0933-3657(01)00077-X [DOI] [PubMed] [Google Scholar]
- 2.Roque FS, Jensen PB, Schmock H, Dalgaard M, Andreatta M, Hansen T, Søeby K, Bredkjær S, Juul A, Werge T, Jensen LJ, Brunak S. Using electronic patient records to discover disease correlations and stratify patient cohorts. PLoS Comput Biol. 2011 Aug;7(8):e1002141. doi: 10.1371/journal.pcbi.1002141. http://dx.plos.org/10.1371/journal.pcbi.1002141 .PCOMPBIOL-D-11-00196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Savage N. Better medicine through machine learning. Commun ACM. 2012 Jan 01;55(1):17. doi: 10.1145/2063176.2063182. http://paperpile.com/b/4F2MYV/dbIe . [DOI] [Google Scholar]
- 4.Sun J, Hu J, Luo D, Markatou M, Wang F, Edabollahi S, Steinhubl SE, Daar Z, Stewart WF. Combining knowledge and data driven insights for identifying risk factors using electronic health records. AMIA Annu Symp Proc. 2012;2012:901–10. http://europepmc.org/abstract/MED/23304365 . [PMC free article] [PubMed] [Google Scholar]
- 5.Visweswaran S, Angus DC, Hsieh M, Weissfeld L, Yealy D, Cooper GF. Learning patient-specific predictive models from clinical data. J Biomed Inform. 2010 Oct;43(5):669–85. doi: 10.1016/j.jbi.2010.04.009. http://linkinghub.elsevier.com/retrieve/pii/S1532-0464(10)00056-0 .S1532-0464(10)00056-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu J, Roy J, Stewart WF. Prediction modeling using EHR data: challenges, strategies, and a comparison of machine learning approaches. Med Care. 2010 Jun;48(6 Suppl):S106–13. doi: 10.1097/MLR.0b013e3181de9e17. [DOI] [PubMed] [Google Scholar]
- 7.Ebadollahi S, Sun J, Gotz D, Hu J, Sow D, Neti C. Predicting patient's trajectory of physiological data using temporal trends in similar patients: a system for near-term prognostics. AMIA Annu Symp Proc. 2010 Nov 13;2010:192–6. http://europepmc.org/abstract/MED/21346967 . [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet. 2012 May 02;13(6):395–405. doi: 10.1038/nrg3208.nrg3208 [DOI] [PubMed] [Google Scholar]
- 9.Saria S, Rajani AK, Gould J, Koller D, Penn AA. Integration of early physiological responses predicts later illness severity in preterm infants. Sci Transl Med. 2010 Sep 08;2(48):48ra65. doi: 10.1126/scitranslmed.3001304. http://stm.sciencemag.org/cgi/pmidlookup?view=long&pmid=20826840 .2/48/48ra65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun J, Sow D, Hu J, Ebadollahi S. A system for mining temporal physiological data streams for advanced prognostic decision support. IEEE 10th International Conference on Data Mining (ICDM); December 13-17, 2010; Sydney, NSW, Australia. http://ieeexplore.ieee.org/document/5694085/ [Google Scholar]
- 11.Bennett C, Doub T, Selove R. EHRs connect research and practice: Where predictive modeling, artificial intelligence, and clinical decision support intersect. Health Policy Technol. 2012 Jun;1(2):105–114. doi: 10.1016/j.hlpt.2012.03.001. http://paperpile.com/b/4F2MYV/3Szd . [DOI] [Google Scholar]
- 12.Greengard S. A new model for healthcare. Commun ACM. 2013 Feb 01;56(2):17–19. doi: 10.1145/2408776.2408783. http://paperpile.com/b/4F2MYV/49Kc . [DOI] [Google Scholar]
- 13.Ramakrishnan N, Hanauer D, Keller B. Mining electronic health records. Computer. 2010 Oct;43(10):77–81. doi: 10.1109/mc.2010.292. http://paperpile.com/b/4F2MYV/f42V . [DOI] [Google Scholar]
- 14.Romano MJ, Stafford RS. Electronic health records and clinical decision support systems: impact on national ambulatory care quality. Arch Intern Med. 2011 May 23;171(10):897–903. doi: 10.1001/archinternmed.2010.527. http://europepmc.org/abstract/MED/21263077 .archinternmed.2010.527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newton KM, Peissig PL, Kho AN, Bielinski SJ, Berg RL, Choudhary V, Basford M, Chute CG, Kullo IJ, Li R, Pacheco JA, Rasmussen LV, Spangler L, Denny JC. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc. 2013 Jun;20(e1):e147–54. doi: 10.1136/amiajnl-2012-000896. http://europepmc.org/abstract/MED/23531748 .amiajnl-2012-000896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denny JC, Bastarache L, Ritchie MD, Carroll RJ, Zink R, Mosley JD, Field JR, Pulley JM, Ramirez AH, Bowton E, Basford MA, Carrell DS, Peissig PL, Kho AN, Pacheco JA, Rasmussen LV, Crosslin DR, Crane PK, Pathak J, Bielinski SJ, Pendergrass SA, Xu H, Hindorff LA, Li R, Manolio TA, Chute CG, Chisholm RL, Larson EB, Jarvik GP, Brilliant MH, McCarty CA, Kullo IJ, Haines JL, Crawford DC, Masys DR, Roden DM. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013 Dec;31(12):1102–10. doi: 10.1038/nbt.2749. http://europepmc.org/abstract/MED/24270849 .nbt.2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Platt R, Carnahan RM, Brown JS, Chrischilles E, Curtis LH, Hennessy S, Nelson JC, Racoosin JA, Robb M, Schneeweiss S, Toh S, Weiner MG. The U.S. Food and Drug Administration's Mini-Sentinel program: status and direction. Pharmacoepidemiol Drug Saf. 2012;21 Suppl 1:1–8. doi: 10.1002/pds.2343. http://paperpile.com/b/4F2MYV/s6o0 . [DOI] [PubMed] [Google Scholar]
- 18.Hripcsak G, Duke JD, Shah NH, Reich CG, Huser V, Schuemie MJ, Suchard MA, Park RW, Wong IC, Rijnbeek PR, van der Lei J, Pratt N, Norén GN, Li YC, Stang PE, Madigan D, Ryan PB. Observational Health Data Sciences and Informatics (OHDSI): opportunities for observational researchers. Stud Health Technol Inform. 2015;216:574–8. http://europepmc.org/abstract/MED/26262116 . [PMC free article] [PubMed] [Google Scholar]
- 19.Fleurence RL, Curtis LH, Califf RM, Platt R, Selby JV, Brown JS. Launching PCORnet, a national patient-centered clinical research network. J Am Med Inform Assoc. 2014;21(4):578–82. doi: 10.1136/amiajnl-2014-002747. http://europepmc.org/abstract/MED/24821743 .amiajnl-2014-002747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber GM, Murphy SN, McMurry AJ, Macfadden D, Nigrin DJ, Churchill S, Kohane IS. The Shared Health Research Information Network (SHRINE): a prototype federated query tool for clinical data repositories. J Am Med Inform Assoc. 2009;16(5):624–30. doi: 10.1197/jamia.M3191.M3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ng K, Ghoting A, Steinhubl SR, Stewart WF, Malin B, Sun J. PARAMO: a PARAllel predictive MOdeling platform for healthcare analytic research using electronic health records. J Biomed Inform. 2014 Apr;48:160–70. doi: 10.1016/j.jbi.2013.12.012. http://linkinghub.elsevier.com/retrieve/pii/S1532-0464(13)00203-7 .S1532-0464(13)00203-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallego B, Walter SR, Day RO, Dunn AG, Sivaraman V, Shah N, Longhurst CA, Coiera E. Bringing cohort studies to the bedside: framework for a 'green button' to support clinical decision-making. J Comp Eff Res. 2015 May 11;:1–7. doi: 10.2217/cer.15.12. https://www.futuremedicine.com/doi/10.2217/cer.15.12 . [DOI] [PubMed] [Google Scholar]
- 23.Wu Y, Jiang X, Kim J, Ohno-Machado L. Grid Binary LOgistic REgression (GLORE): building shared models without sharing data. J Am Med Inform Assoc. 2012;19(5):758–64. doi: 10.1136/amiajnl-2012-000862. http://europepmc.org/abstract/MED/22511014 .amiajnl-2012-000862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu H, Jiang X, Jaideep V. Privacy-preserving SVM using nonlinear kernels on horizontally partitioned data. The ACM symposium on Applied computing; April 23-27, 2006; Dijon, France. [Google Scholar]
- 25.Yu H, Vaidya J, Jiang X. Privacy-preserving SVM classification on vertically partitioned data. In: Ng WK, Kitsuregawa M, Li J, Chang K, editors. Advances in Knowledge Discovery and Data Mining. PAKDD 2006. Lecture Notes in Computer Science, vol 3918. Berlin, Heidelberg: Springer; 2006. pp. 647–656. [Google Scholar]
- 26.Lu CL, Wang S, Ji Z, Wu Y, Xiong L, Jiang X, Ohno-Machado L. WebDISCO: a web service for distributed cox model learning without patient-level data sharing. J Am Med Inform Assoc. 2015 Nov;22(6):1212–9. doi: 10.1093/jamia/ocv083. http://europepmc.org/abstract/MED/26159465 .ocv083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang F, Sun J, Ebadollahi S. Integrating Distance Metrics Learned from Multiple Experts and its Application in Patient Similarity Assessment. The SIAM International Conference on Data Mining; April 28, 2011; Mesa, Arizona. [DOI] [Google Scholar]
- 28.Wang F, Hu J, Sun J. Medical prognosis based on patient similarity and expert feedback. 21st International Conference on Pattern Recognition (ICPR) 2012; November 11-15, 2012; Tsukuba, Japan. [Google Scholar]
- 29.Sun J, Wang F, Hu J, Edabollahi S. Supervised patient similarity measure of heterogeneous patient records. SIGKDD Explor Newsl. 2012 Dec 10;14(1):16–24. doi: 10.1145/2408736.2408740. [DOI] [Google Scholar]
- 30.Wang F, Sun J, Ebadollahi S. Composite distance metric integration by leveraging multiple experts' inputs and its application in patient similarity assessment. Stat Anal Data Min. 2012 Feb 17;5(1):54–69. doi: 10.1002/sam.11135. [DOI] [Google Scholar]
- 31.Gentry C. Fully homomorphic encryption using ideal lattices. Proceedings of the 41st Annual ACM Symposium on Theory of computing; May 31-June 2, 2009; Bethesda, MD. [DOI] [Google Scholar]
- 32.Bloom BH. Space/time trade-offs in hash coding with allowable errors. Commun ACM. 1970 Jul;13(7):422–426. doi: 10.1145/362686.362692. http://dmod.eu/deca/ft_gateway.cfm.pdf . [DOI] [Google Scholar]
- 33.Schneier B. 2004. Aug, [2018-03-02]. Schneier on Security: Cryptanalysis of MD5 and SHA: Time for a New Standard https://www.schneier.com/essays/archives/2004/08/cryptanalysis_of_md5.html .
- 34.Wang J, Shen HT, Song J, Ji J. 2014. Aug, [2018-03-03]. Hashing for Similarity Search: A Survey http://arxiv.org/abs/1408.2927 .
- 35.Indyk P, Motwani R. Approximate nearest neighbors: towards removing the curse of dimensionality. Proceedings of the Thirtieth Annual ACM Symposium on Theory of computing; May 24-26, 1998; Dallas, Texas. [DOI] [Google Scholar]
- 36.Broder AZ, Charikar M, Frieze AM, Mitzenmacher M. Min-Wise Independent Permutations. J Comput Syst Sci. 2000 Jun;60(3):630–659. doi: 10.1006/jcss.1999.1690. [DOI] [Google Scholar]
- 37.Liu W, Wang J, Chang SF, Kumar S. Hashing with graphs. Proceedings of the 28th International Conference on Machine Learning; June 28, 2011; Bellevue, Washington. http://www.icml-2011.org/papers/6_icmlpaper.pdf . [Google Scholar]
- 38.Gong Y, Lazebnik S. Iterative quantization: a procrustean approach to learning binary codes. IEEE Conference on Computer Vision and Pattern Recognition (CVPR); June 20-25, 2011; Colorado Springs, CO. [DOI] [Google Scholar]
- 39.Kong W, Li WJ. Isotropic hashing. In: Bartlett P, Weinberger KQ, Burges CJ, Bottou L, Pereira FC, editors. Advances in Neural Information Processing Systems 25. Red Hook, NY, USA: Curran Associates, Inc; 2012. [Google Scholar]
- 40.Gong Y, Kumar S, Verma V, Lazebnik S. Angular quantization-based binary codes for fast similarity search. Proceedings of the 25th International Conference on Neural Information Processing Systems (NIPS) - Volume 1; December 3-6, 2012; Lake Tahoe, Nevada. http://paperpile.com/b/4F2MYV/noQb . [Google Scholar]
- 41.Wang J, Kumar S, Chang SF. Semi-supervised hashing for scalable image retrieval. IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2010; June 13-18, 2010; San Francisco, CA. [DOI] [Google Scholar]
- 42.Wang J, Kumar S, Chang SF. Semi-supervised hashing for large-scale search. IEEE Trans Pattern Anal Mach Intell. 2012 Dec;34(12):2393–406. doi: 10.1109/TPAMI.2012.48. http://www.ee.columbia.edu/ln/dvmm/publications/12/PAMI_SSHASH.pdf . [DOI] [PubMed] [Google Scholar]
- 43.Yang X, Fu H, Zha H, Barlow J. Semi-supervised Nonlinear Dimensionality Reduction. The 23rd International Conference on Machine Learning; June 25-29, 2006; Pittsburgh, Pennsylvania. 2006. [DOI] [Google Scholar]
- 44.Liu W, Wang J, Ji R, Jiang YG, Chang SF. Supervised Hashing with Kernels. IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2012; June 16-21, 2012; Providence, RI. [Google Scholar]
- 45.Wang J, Kumar S, Chang SF. Sequential projection learning for hashing with compact codes. The 27th International Conference on Machine Learning (ICML) 2010; June 21-24, 2010; Haifa, Israel. [Google Scholar]
- 46.Bronstein MM, Bronstein AM, Michel F, Paragios N. Data fusion through cross-modality metric learning using similarity-sensitive hashing. IEEE Computer Society Conference on Computer Vision and Pattern Recognition; June 13-18, 2010; San Francisco. [Google Scholar]
- 47.Kumar S, Udupa R. Learning hash functions for cross-view similarity search. Proceedings of the 22nd International Joint Conference on Artificial Intelligence (IJCAI) 2011; July 16-22, 2011; Barcelona, Spain. p. 1360. [Google Scholar]
- 48.Song J, Yang Y, Huang Z, Shen HT, Hong R. Multiple Feature Hashing for Real-time Large Scale Near-duplicate Video Retrieval. Proceedings of the 19th ACM International Conference on Multimedia; November 28-December 1, 2011; Scottsdale, AZ. [Google Scholar]
- 49.Zhen Y, Yeung DY. A probabilistic model for multimodal hash function learning. Proceedings of tThe 18th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining - KDD 2012; August 12-16, 2012; Beijing, China. [Google Scholar]
- 50.Zhen Y, Yeung DY. Co-Regularized Hashing for Multimodal Data. Proceedings of the 25th International Conference on Neural Information Processing Systems 2012 (NIPS 2012); December 3-6, 2012; Lake Tahoe, Nevada. pp. 1376–1384. http://paperpile.com/b/4F2MYV/6Qrp . [Google Scholar]
- 51.Song J, Yang Y, Yang Y, Huang Z, Shen HT. Inter-media hashing for large-scale retrieval from heterogeneous data sources. Proceedings of the 2013 International Conference on Management of Data - SIGMOD '13; June 22-27, 2013; New York, NY, USA. [DOI] [Google Scholar]
- 52.Zhu X, Huang Z, Shen HT, Zhao X. Linear cross-modal hashing for efficient multimedia search. Proceedings of the 21st ACM International Conference on Multimedia; October 21-25, 2013; Barcelona, Spain. pp. 143–152. http://paperpile.com/b/4F2MYV/LBef . [DOI] [Google Scholar]
- 53.Changpinyo S, Liu K, Sha F. Similarity component analysis. Neural Information Processing Systems 2013 (NIPS 2013); October 2013; Lake Tahoe, Nevada. pp. 1511–1519. http://www-scf.usc.edu/~kuanl/papers/nips13_sca.pdf . [Google Scholar]
- 54.Horn RA, Johnson CR. Norms for vectors and matrices. In: Horn RA, editor. Matrix Analysis. Cambridge, England: Cambridge University Press; 1990. pp. 313–386. [Google Scholar]
- 55.Ortega JM, Rheinboldt WC. Iterative Solution of Nonlinear Equations in Several Variables. Philadelphia: SIAM; 1970. [Google Scholar]
- 56.Zehfuss G. Ueber eine gewisse determinante? J Appl Math Phys. 1858;3:298–301. [Google Scholar]
- 57.Goldberger AL, Amaral LA, Glass L, Hausdorff JM, Ivanov PC, Mark RG, Mietus JE, Moody GB, Peng C, Stanley HE. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation. 2000 Jun 13;101(23):e215–e220. doi: 10.1161/01.CIR.101.23.e215. http://paperpile.com/b/4F2MYV/MWQ1 . [DOI] [PubMed] [Google Scholar]
- 58.Han H, Wang WY, Mao BH. Borderline-SMOTE: A New Over-Sampling Method in Imbalanced Data Sets Learning. Proceedings of the 2005 International Conference on Advances in Intelligent Computing (ICIC); August 23-26, 2005; Hefei, China. pp. 878–887. [Google Scholar]
- 59.Shi H, Jiang C, Dai W, Jiang X, Tang Y, Ohno-Machado L, Wang S. Secure Multi-pArty Computation Grid LOgistic REgression (SMAC-GLORE) BMC Med Inform Decis Mak. 2016 Dec 25;16 Suppl 3:89. doi: 10.1186/s12911-016-0316-1. https://bmcmedinformdecismak.biomedcentral.com/articles/10.1186/s12911-016-0316-1 .10.1186/s12911-016-0316-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Privacy-preserving patient representation learning in a federated setting.
Prediction performance of balanced class datasets.
Prediction performance of imbalanced class datasets.
Prediction performance of balanced class datasets.