Abstract
Fetal cytotrophoblast invasion of maternal decidual vasculature is necessary to normal pregnancy. In preeclampsia, there is shallow invasion and abnormal remodeling of the uterine vasculature that lead to significant maternal and perinatal morbidity and mortality. The placental basement membrane (BM) proteins (e.g. laminin, collagen) has been implicated in the development of placenta while the level of laminin is significantly lower in preeclampsia. However, there are very limited studies, if any, on the effect of extracellular matrix (ECM) microenvironment on the invasion of cytotrophoblast. In this study, we hypothesized that placental BM proteins are required for effective cytotrophoblast invasion. Using proteomics, we found that more than 80% of ECM proteins in placental basal plate (pECM) were BM proteins. In addition to upregulating expressions of MMP2 (1.5 fold) and MMP9 (6.3 fold), pECM significantly increased the motility rates of cytotrophoblasts by 13 fold (from 5.60±0.95 to 75.5±21.8 μm/day) to achieve an effective invasion rate that was comparable to in vivo results. Treatments with PI3K inhibitors completely removed the pECM-enhanced invasive phenotypes and genotypes of cytotrophoblasts, suggesting its dominant role in cytotrophoblast-ECM interactions. Our results described, for the first time, the substantial effects of the ECM microenvironment on regulating cytotrophoblast invasion, an area that is less investigated but appear to be critical in the pathogenesis of preeclampsia. Moreover, the approach presented in this work that fabricates organ models with organ-specific ECM can be an attractive option to screen and develop novel therapeutics and biomarkers not only in preeclampsia but also other diseases such as cancer metastasis.
Keywords: bioprinting, tissue-specific ECM, preeclampsia, basement membrane protein, placenta
Introduction
In normal pregnancy, embryo implantation and placentation occur by cytotrophoblast invasion of the maternal spiral arteries to create a low-resistance, high-flow maternal uteroplacental circulation1–4. Impaired cytotrophoblast invasion of the maternal spiral arteries results in abnormal placentation leading to reduced placental perfusion and oxidative stress1–5. One of the major clinical complications of impaired cytotrophoblast invasion is preeclampsia (PE)6. PE is a leading cause of maternal and perinatal morbidity and mortality affecting 3 to 8% of all pregnancies1–3,7,8 and resulting in 29,000 deaths yearly in US9. The only treatment option for PE available is the premature delivery of baby and placenta because the mechanisms of cytotrophoblast invasion are poorly defined1,3,10.
In early placental development, cytotrophoblasts differentiate and lose their organized epithelial phenotype and transitioning to a more migratory and invasive mesenchymal phenotype which allows them to infiltrate the maternal decidua and spiral arteries. This transition has been recently suggested as a type of epithelial-mesenchymal transition (EMT) that involves upregulating and secreting matrix metalloproteinase (MMP) and gaining the ability to migrate and invade11–15. EMT has been reported to be mediated by tyrosine kinase receptors (TKR) and transforming growth factor beta (TGF-β) family14–16. Among these receptors, TKR appears to have a direct and specific impact on endometrial susceptibility to implantation in vivo based on a murine TKR-ablated uterus model (epidermal growth factor receptor, EGFR, knock-out)17. Moreover, EMT is mediated through the action of phosphoinositide 3-kinase (PI3K) signaling pathway14,16, a major signaling pathway located downstream of TKR that regulates cellular processes including motility, proliferation, survival and growth– which are critical for cytotrophoblast invasion5,18,19. However, the effect of extracellular matrix (ECM) microenvironment on EMT and PI3K signaling on cytotrophoblast invasion remains poorly understood.
Cell-ECM interactions play a fundamental role in the growth, differentiation and invasion of cytotrophoblasts20. Prior to implantation of blastocysts, the maternal endometrium undergoes substantial remodeling and differentiation to become decidua, a process known as decidualization17,21. When decidualization occurs, the maternal decidual stromal cells (those in direct contact with cytotrophoblasts) produce pericellular basement membrane (BM) proteins that are critical to placental development and successful embryogenesis. For example, knocking out laminin genes in murine models cause embryonic lethal outcomes (Lama1; Lama5; Lamb1; Lamc1) and considerable abnormalities in vascular and cytotrophoblast differentiation during placental development (Lama5)22,23. These placental abnormalities are potentially caused by the lack of stable adhesion between cytotrophoblasts and Lama5. Moreover, human term placenta from preeclamptic pregnancies have lower expression levels of laminin compare to those of normal pregnancies24–26. The expressions of laminin alpha 2 appears to also be downregulated in the basal plate of preeclamptic term placenta27. Even though these evidences implicate the vital role of BM proteins in placentation and cytotrophoblast invasion, the majority of published literature focuses on the effect of soluble factors20. The intricate and highly ordered nature of ECM makes it difficult to reproduce using synthetic or purified components and these BM-proteins are often tissue-specific and work in concert instead of individually20,23,28. These differences in ECM compositions between native tissue and in vitro culture techniques affect cellular genotypes and phenotypes20. Therefore, studies on cell invasion utilizing single ECM components, while still valuable, may not represent the in vivo environment.
Our hypothesis is that placental BM proteins isolated from basal plate of human placenta are required for effective cytotrophoblast invasion. To test this hypothesis, we isolated and characterized ECM from the basal plate of term human placenta, which is defined as pECM for the rest of the work. Through proteomics, we determined that more than 80% of pECM consists of BM proteins. Our results showed that the addition of isolated placental BM proteins substantially increased the invasion rates by 13 fold while significantly upregulated the gene expressions of MMP2 and MMP9 (surrogate markers for invasion and EMT19,29). The addition of LY294002, a well-established PI3K inhibitor5,18, significantly reduced the enhanced invasive rates and expressions of MMP2 and MMP9. These results demonstrated that the placental BM proteins stimulated cytotrophoblast invasion predominantly through PI3K signaling - first direct evidence indicating that the cytotrophoblast differentiation and invasion are critically modulated by their surrounding ECM microenvironment.
Materials and Methods
Cell Culture
BeWo cells were purchased from American Type Culture Collection (ATCC) and cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, ATCC), 15% (vol/vol) Fetal Bovine Serum (FBS; Thermo Fisher Scientific), and 1% penicillin/streptomycin (Pen/Strep; Thermo Fisher Scientific). Cells are cultured in standard cell culture incubator (Thermo Fisher Scientific) at 37°C and 5% CO2 with humidity control.
Tissue Collection and Isolation
Five ex vivo placenta from normal pregnancies were collected from MedStar Washington Hospital Center (maternal age=29.1±1.2 years; gestational age=37±0.84 weeks) according to protocol approved by the MedStar Research Institute Institutional Review Board (IRB# 2015-131). The placentas are frozen at −80°C immediately after delivery until tissue isolation. Surgical tools (e.g. scissors, scalpels, forceps) were utilized to carefully harvest the top slice of the placenta from the maternal side (no more than 3 mm). The isolated tissue was minced and washed using until the effluent become clear.
Decellularization, Digestion and Characterization of Placental Basal Plate
The decellularization and digestion protocols were developed based on previous studies28,30. The minced tissues were treated with 1% (v/v) antibiotic/antifungal (Thermo Fisher Scientific) and placed on a shaker overnight at room temperature. The tissue was then treated in 0.1% Sodium Dodecyl Sulfate (SDS) in distilled water for 48 hours to decellularize the tissue without any buffer change. Following SDS treatment, the tissue is washed in distilled water for another 48 hours and lyophilized. The lyophilized, decellularized tissue was crushed using a mortar and pestle set and digested in pepsin (1 g of pepsin / 10 g of dry tissue) in 0.5M acetic acid in 4°C for 48 hours. The pH of the tissue digest was adjusted to 7.4 using an aqueous base solution (e.g. NaOH) and then centrifuged at 3000 g for 5 mins. The supernatant was filtered using a cell strainer (100 μm, Thermo Fisher Scientific) and stored at −80°C until further use. The resulting supernatant was defined as pECM for the current work. The DNA content of pECM was tested using a Quant-iT PicoGreen dsDNA Assay kit (Life Technologies) following the manufacturer’s protocol. Collagen content of pECM was tested using a hydroxyproline assay kit (Sigma-Aldrich).
Gelatin Methacrylate Synthesis
Gelatin methacrylate (GelMA) was synthesized according to previously published methods31–33. Briefly, Type A porcine skin gelatin (Sigma-Aldrich; 300 bloom) was mixed at 10% (w/v) in phosphate buffered saline (PBS; Thermo Fisher Scientific) at 50°C for 20 minutes. Methacrylic anhydride (Sigma-Aldrich) was then added at 0.6 g of Methacrylic anhydride / g gelatin under rigorous stirring for one hour. The reactants were then diluted 2x with PBS to stop the reaction. The mixture was centrifuged and the pellet was discarded. The supernatant was dialyzed against deionized water at 50°C using dialysis cassettes (10kDa MWCO, Thermo Fisher Scientific) for at least 3 days. The deionized water was changed twice a day to remove salts and excess methacrylic acid. The dialyzed GelMA was then lyophilized for at least 3 days and store at −80°C until further use.
Proteomics
The digested ECM sample was dissolved in SDS-loading buffer and separated by size using a 10% acrylamide gel by SDS-PAGE. The gel was then fixed and cut into 10 pieces of similar sizes for proteomics analysis using clean razors using the ladder as a guide. Briefly, the proteins in gel bands were digested with sequencing grade Trypsin (Promega) and the resulting peptides were extracted with acetonitrile-formic acid buffer. The peptide mixtures from each band were sequentially analyzed by liquid chromatography-mass spectrometry (LC-MS) using nano-LC system (Easy nLC 1000, Thermo Fisher Scientific) connected to Q Exactive mass spectrometer (Thermo Fisher Scientific). The peptides were eluted at a flow rate of 300 nL/min using linear gradients of 2–25 % Acetonitrile for 35 min, followed by 25–40% for 10 min, 40–60% for 1 min, and static flow at 60% for 14 min. The MS raw data were searched against UniProt database that also included common contaminants using MaxQuant software (version 1.5.5.1).
Size Exclusion Chromatography (SEC)
250μL of pECM (prepared as specified above) were loaded onto a ENrich™ SEC 650 column (Bio-Rad) using the NGC Quest™ 10 chromatography system (Bio-Rad) which was equilibrated with 1x PBS. Proteins were eluted from the column using 1x PBS as mobile phase at a flow of 1mL min−1 while monitoring the absorbance at 280nm. Samples were collected in 0.7mL fractions using a fraction collector (BioFrac™, Bio-Rad) and frozen at −20°C until further use.
Cell Adhesion Study
Adhesion of cytotrophoblasts to pECM was evaluated using the washing assay. This assay relies on seeding cells onto substrates of interest and washing off ‘nonadherent’ cells with physiological buffers and then counting the remaining cells34–37. Briefly, a 24 well plate (not tissue culture treated, Corning) was coated with desired ECM overnight, both at 4°C. The following ECMs were used for coating: GelMA (0.1 g/mL), pECM (0.1 g/mL) and fibronectin (0.1 g/mL). Cytotrophoblasts were stained with Calcein AM (Thermo Fisher Scientific) following manufacturer’s recommendations and seeded on the treated cell culture wells in culture media (30,000 cells/cm2). Cytotrophoblasts were allowed to adhere for 2 hours at 37°C and then an initial fluorescent readings (F1) was taken using a fluorescent plate reader taken at 530 nm. Then the wells were washed twice with PBS to remove non-adherent cells and the fluorescence was read again (F2). Fraction of adhesion (FA) was calculated using the formula: FA= 1 − (F1−F2)/F1.
Bioprinting of Placenta Model with pECM
All bioprinting work was completed using a commercial bioprinter (3D-Bioplotter; EnvisionTEC). To prepare the prepolymer solution, lyophilized GelMA was dissolved in complete growth media at 50°C (10% w/v) for 20 minutes. Photoinitiator (2-hydroxy-1-(4-(hydroxyethoxy)phenyl)-2-methyl-1-propanone, also known as Irgacure 2959; BASF) was then added into the GelMA solution at 0.1% (w/v) at 50°C for 15 minutes under vigorous stirring. The prepolymer solution was loaded into the low-temperature printer head and allowed to equilibrate for 30 minutes at 37°C. Additional biomaterials such as pECM, cells, and/or growth factors of interest were added at this time, and the functionalized prepolymer solution was equilibrate to printing temperature (e.g. ~23°C) for another 30 minutes prior to printing. The following settings were utilized for all bioprinting work: needle diameter=0.4 mm; printing temperature = 23°C; pressure = 0.5–1 bar; layer thickness = 0.35 mm; speed = 10 mm/s. The placenta model was a cylindrical construct (height = 2 mm, diameter = 10 mm) with a central source of chemoattractant (height = 2mm, diameter = 3 mm; epidermal growth factor, EGF, loading level = 10 uM) at the center and an outer layer of cytotrophoblasts in the periphery. Printed constructs were UV-cured for 30 seconds (0.09 mW/cm2) using a UV box (VWR). The Live/Dead assay (Thermo Fisher Scientific) was used to assess cell viability within printed constructs according to the manufacturer’s instructions. An inverted fluorescent microscope (Olympus CKX41) with mercury lamp (Olympus U-RFLT50) were used to image the stained cells.
Mechanical Analysis
Unconfined compression testing was performed on the bioprinted 3D cylindrical GelMA discs (diameter= 10 mm; h = 1mm) containing different concentrations of pECM (0, 0.02, 0.2 mg/mL) (Fig 1f). Hydrated samples were placed between two compression plates and were loaded at a strain rate of 15% per minute using a mechanical tester (Instron). Young’s modulus were defined as the slope of the linear portion of the stress-strain curve comprised between 20–40% strain.
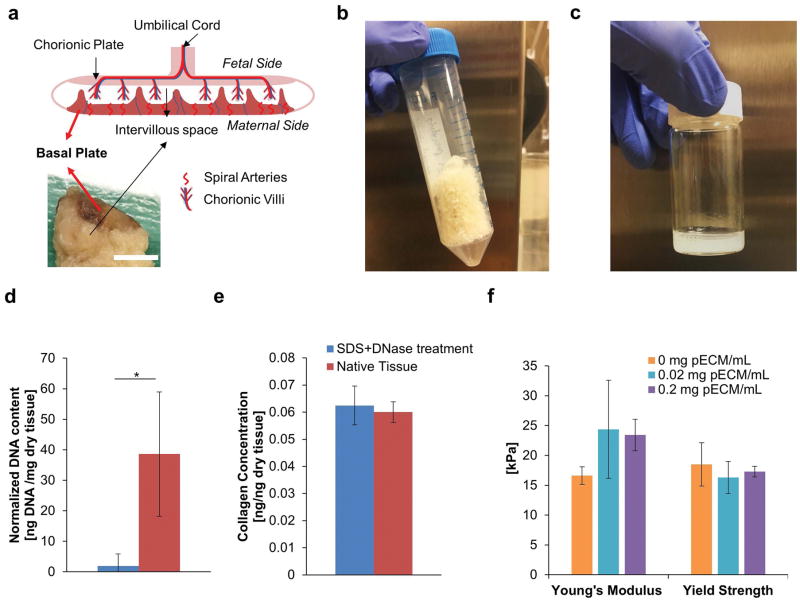
Figure 1. Development of Decellularized Placental ECM (pECM).
(a) Anatomy of Ex Vivo Placenta. Top image illustrates the major structures of human placenta and bottom image demonstrated a cross section of human placenta sample. Red arrows indicated the location of basal plate. Scale bar= 10 mm. (b) Representative Image of Lyophilized pECM. The tissue of basal plate was lyophilized for long term storage after it has been decellularized. The lyophilized pECM was a porous scaffold with white-yellow appearance. (c) Representative Image of Solubilized pECM. The lyophilized pECM was solubilized in pepsin under acidic condition at a desired concentration to be bioprinted later. (d) DNA Content of Solubilized ECM. After treatments of SDS and DNase (blue), 95% of the DNA from the native tissue was removed (red, n=3). (e) Collagen Content of Solubilized ECM. There was no significant difference between the collagen contents of solubilized decellularized and native tissue, which suggested the integrity of the ECM was well-preserved after decellurlarization treatment (n=3). (f) Effect of pECM on the Mechanical Properties of Bioprinted Constructs (n=6). Our results showed that the addition of pECM into bioprinted GelMA constructs did not alter the Young’s modulus and Yield Strength significantly.
Invasion Study
For the invasion experiments, cells were bioprinted (2 million cells/mL) along the periphery of the cylindrical placenta model with a chemotactic source (epidermal growth factor, 10 μM) at the center as previously described38. pECM were added at various concentration to determine its effect on invasion rates (0, 0.02, 0.2 mg/mL pECM). Cytotrophoblasts were cultured in growth media with reduced serum concentration (5%) and phase-contrast images were taken on day 2 and day 7 using an inverted microscope (Olympus CKX41). The distance between the edge of the placenta model and the invasion fronts were measured and averaged over time to obtain invasion rates. For samples with reduced PI3K signaling, samples were treated with LY294002 (25 μM) on days 4 and 6 for 24 hours.
DNA Isolation and Quantification
Sample discs (10 mm diameter and 2 mm height, 2 million cells/mL) were bioprinted for DNA quantification study. Bioprinted discs were cultured in standard cell culture incubator (37°C, 5% CO2) with humidity control for seven days. On day 7, bioprinted discs were treated with papain (4 mg/mL) for 30 mins at 37°C to dissolve the gel and isolate the cells. The isolated cells were lysed with RIPA Lysis and Extraction Buffer (Thermo Fisher Scientific). The DNA content of the cell lysates were directly tested using a Quant-iT PicoGreen dsDNA Assay kit (Life Technologies) following the manufacturer’s protocol.
RNA Isolation and qRT-PCR
Sample discs (10 mm diameter and 2 mm height, 2 million cells/mL) were bioprinted for gene expression studies. Bioprinted discs were cultured in standard cell culture incubator (37°C, 5% CO2) with humidity control for seven days. On day 7, bioprinted discs were dissolved by treating them with papain (4 mg/mL) for 20 mins at 37°C, and dissolved in Trizol (Thermo Fisher Scientific). Total RNA was isolated from the cytotrophoblasts using the RNeasy Plus Mini Kit (Qiagen) and then reverse transcribed to complementary DNA (cDNA) using a High Capacity cDNA Archive Kit (Thermo Fisher Scientific). Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) was performed by combining the cDNA solution with a Universal Master Mix (Thermo Fisher Scientific), as well as oligonucleotide primers and Taqman probes for MMP2, MMP9 and the endogenous gene control glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Life Technologies). The reaction was performed using a 7900HT real-time PCR System (Applied Biosystems) at thermal conditions of 2 min at 50°C, 10 min at 95°C, 40 cycles of 15 s at 95°C, and 1 min at 60°C. The relative gene expression level of each target gene was normalized to the mean of GAPDH in each group then the fold change was determined relative to the Bioprinted Placenta Model without pECM. Fold change was calculated using the ΔΔCT relative comparative method as described previously39.
Measurement of MMP Activity
Sample discs (10 mm diameter and 2 mm height, 2 million cells/mL) were bioprinted for MMP activity studies. Bioprinted discs were cultured in standard cell culture incubator (37°C, 5% CO2) with humidity control for 7 days. On day 7, supernatant from cultures of bioprinted placenta model with different dosage of pECM were collected after 7 days of culture. The MMP activity was measured using the MMP Activity Assay Kit (Abcam) according to manufacturer’s instructions.
Statistical Analysis
Error bars indicate standard deviation and * indicates statistically significant differences between groups. Data from all the studies were analyzed using the Multiple Comparison Test function in MATLAB. A confidence level of 95% was chosen, and p values below 0.05 were considered significant. All experiments were done in triplicates (biological replicates) unless stated otherwise.
Results
Isolation and Characterization of Placental Basement Membrane Proteins
To harvest relevant ECM material to study cytotrophoblast invasion, we isolated the basal plate of the fresh human term placentae by removing a thin layer (less than 3 mm) of tissue from the maternal side (Figure 1a, red arrows). Following decellularization and solubilization, the amount of DNA present in it the processed tissue was 95% lower than that of solubilized native tissue (Figure 1d) and was below the level indicated to induce immune response40. The decellularized and solubilized ECM isolated from basal plate of term placenta is referred to as pECM for the rest of the work (Figure 1c). Next, we compared the amounts of collagen in pECM to solubilized native tissue and showed no significant differences (Figure 1e). These data showed that the decellularization treatment was effective in removing the DNA contents and kept the ECM components intact which was demonstrated by the similar collagen levels. Since the mechanical properties of the substrate have a profound impact on cellular behavior41, we used compression testing and determined that the pECM has no significant impact (p>0.05) on the Young’s modulus and yield strength of bioprinted GelMA constructs (Figure 1f). Therefore, any biological effects induced by pECM on cytotrophoblasts were due to differences in ECM composition rather than bulk mechanical properties.
Effect of pECM on Proliferation and Adhesion of Cytotrophoblast
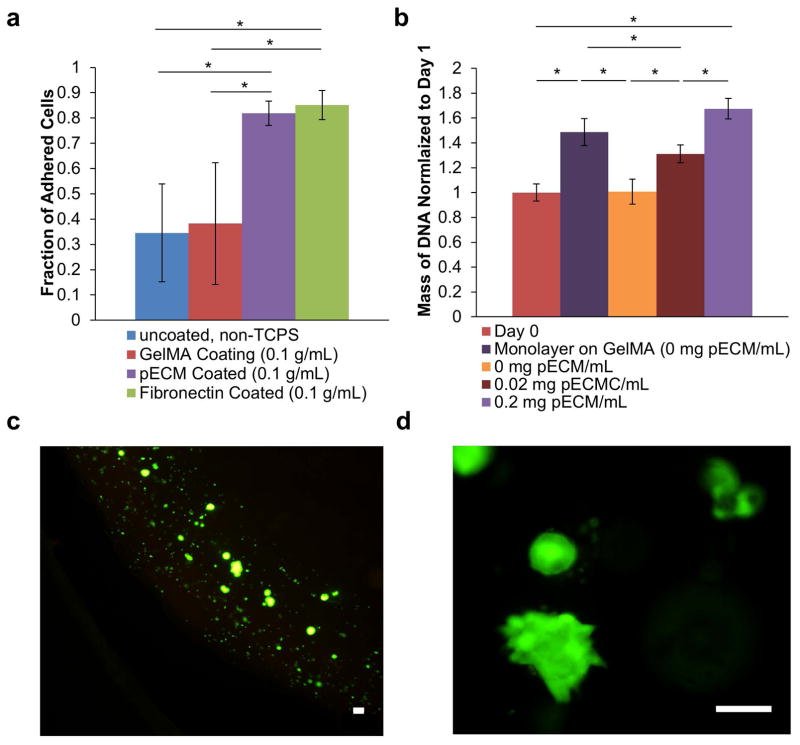
Since cytotrophoblasts are anchorage-dependent and their binding to ECM is required to invade the decidual matrix, we tested the effect of pECM on proliferation and adhesion of cytotrophoblast. As shown in Figure 2a, the fraction of adhered cells for pECM-coated group (0.82±0.05) was significantly higher than those of untreated plastic (0.35±0.19, p<0.05) and GelMA coated plastic (0.38±0.24, p<0.05). GelMA was used as a control because it was the basis material for our bioprinting work. In addition, the fraction of adhered cells on pECM coated wells was comparable to that of the fibronectin coated wells (0.85±0.06), a widely-used ECM matrix that promotes cell adhesion21,38. In addition, we observed that the addition of pECM enhances the proliferation of bioprinted cytotrophoblasts in a dosage-dependent manner, expressed as fold change from day 1 after 7 days of culture (Figure 2b). Monolayer of cytotrophoblasts were grown on top of GelMA in 2D as a control to ensure cytotrophoblasts were proliferating on GelMA alone (dark purple bar in Figure 2b). Consistent with published literature, we observed a higher proliferation rates in 2D compare to cells encapsulated in 3D (dark purple vs. orange bars in Figure 2b)42.
Figure 2. Effect of pECM on Growth of Cytotrophoblasts.
(a) Effect of pECM on Relative Adhesion. A washing adhesion assay, which is qualitative, was performed to test the relative adhesion of cytotrophoblasts on different substrate coating.34,36,37,57 Our results show that pECM coating significantly increased the adhesion of cytotrophoblasts compared to non-tissue culture treated polystyrene and GelMA coating. In addition, the fraction of adhesion was not significantly different from fibronectin, the most extensively utilized ECM to promote cell adhesion in biomedical research. (b) Effect of pECM on Proliferation of Cytotrophoblasts. The mass of DNA of bioprinted cytotrophoblasts encapsulated in different concentrations of pECM were quantified after 7 days of culture (normalized to day 0). Our results demonstrated a dosage-dependent response of DNA mass and concentration of pECM. Monolayer of cytotrophoblasts grown on GelMA in 2D was used as control to ensure GelMA alone supports cytotrophoblast growth. (c) Representative Live/dead Images of Bioprinted Cytotrophoblasts. The cells are stained with calcein-AM (green) for live cells and propidium iodide (red) for dead cells. The majority of the bioprinted cytotrophoblasts were stained live (green) with very little dead (red). Scale bar = 100 μm. (d) Representative Live/Dead Images of Cytotrophoblasts Demonstrating Aggregate Formation. The cells are stained with calcein-AM (green) for live cells and propidium iodide (red) for dead cells. Scale Bars = 100μm.
Qualitative images using live/dead viability staining kit demonstrated that the additional pECM did not affect the viability of bioprinted cytotrophoblasts with mostly viable cells (green) and very few dead cells (red, Figure 2c). Encapsulated cytotrophoblasts form a sub-population of aggregates and appeared differently than 2D culture in bioprinted GelMA constructs, consistent with published literature with cells encapsulated in hydrogels (Figure 2d)42. These results demonstrated that pECM supported the adhesion and proliferation of cytotrophoblast.
Characterization of Complete and Fractionated pECM
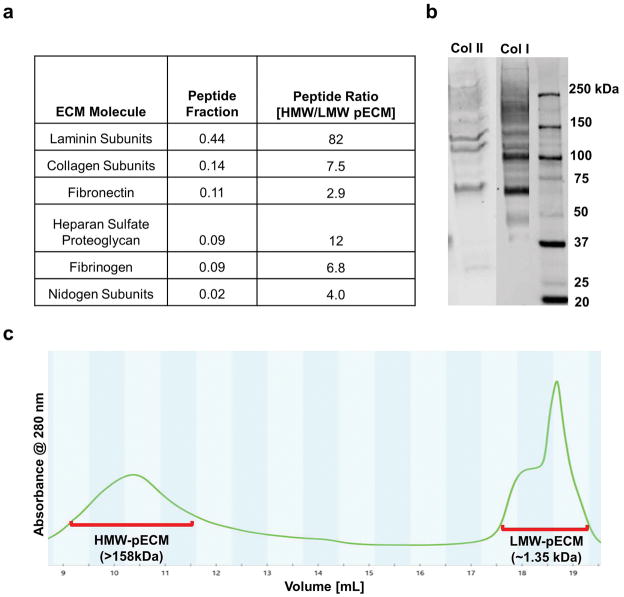
Since pECM is a mixture of ECM proteins, we used proteomics to determine its composition to elucidate the mechanism behind the interactions between pECM and cytotrophoblast. Using proteomics, we found the pECM was composed of mostly BM proteins (80% by peptide counts), including laminin, collagen, fibronectin, Heparan sulfate proteoglycan (Figure 3a), and these results were validated by Western blot analyses (Figure 3b). To evaluate if the effect of pECM was due to the placental BM proteins, we used size-exclusion based chromatography (SEC) to separate pECM components into high molecular weight (>158 kDa, referred to as HMW-pECM) and low molecular weight (<3.5 kDa, referred to as LMW-pECM) as indicated by the two peaks in Figure 3c. Relative amount of ECM proteins in these two fractions were determined using proteomics (Figure 3a). Our results indicate that key placental BM proteins such as laminin, collagen, fibronectin, heparan sulfate proteoglycan, and nidogen were depleted in LMW-pECM as their amounts are lower than that of HMW-pECM by at least 3 fold.
Figure 3. Characterization of Fractionated and Complete pECM.
(a) Compositions of Fractionated and Complete pECM. We found that there is ample basement membrane (BM) proteins including laminin, collagen (VI, XII, XIV and XV), fibronectin, BM-specific heparan sulfate proteoglycan in pECM which consist of more than 80% peptide fractions. Moreover, we quantified the peptides of ECM molecules and calculated the ratio of HMW over LMW pECM. We found that the amounts of placental BM proteins are at least two orders of magnitude higher in HMW pECM than that of LMW pECM. (b) Western Blot Results. SDS-page and positive immunostaining of collagen I, II serves as confirmation of the proteomics results. (c) Chromatogram of Solubilized pECM After Eluting Through a SEC Column. Samples were taken from the high molecular weight fractions (defined as HMW pECM, MW>158 kDa) and low molecular weight fractions (defined as LMW pECM, MW~1.35 kDa).
Effects of Fractionated pECM on Cytotrophoblast Invasion
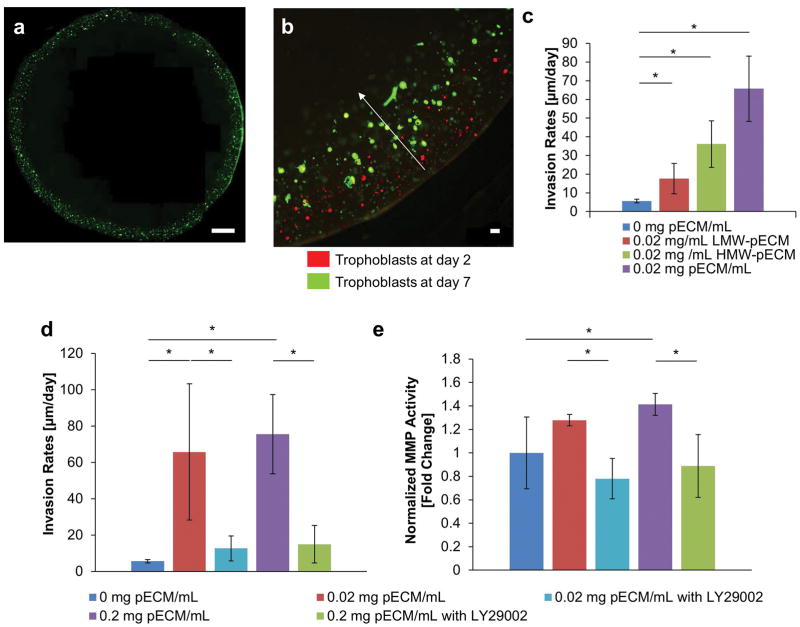
Once the compositions of the fractionated and complete pECM were determined, we tested their effects on cytotrophoblast invasion rates of BeWo cells, an epithelial trophoblastic cell line that is extensively used to study placental development10,43, in a bioprinted placenta model (BPM)38. BeWo cells was used instead of HTR8 because we wanted to study epithelial-mesenchymal transition of cytotrophoblasts and HTR8 is considered mesenchymal-like based their positive expression for vimentin44. The concentrations of ECM protein were kept at 0.02 mg/mL, a level that was comparable to current in vitro studies38 across all groups in Figure 4c. The cytotrophoblast bioprinted in LMW-pECM had a lower invasion rate compared to those of HMW-pECM (17.6±8.19 versus 36.1±12.5 μm/day, respectively; Figure 4c). Interestingly, however, both invasion rates were lower than that of complete pECM (65.8±37.5 μm/day; Figure 4c). These results indicated the effect of complete pECM on cytotrophoblast invasion cannot be reproduced using individual fractions alone, suggesting a combination mechanism of action of both placental BM proteins and peptide components. To confirm that invasion required the expression of MMP, a known group of enzymes that are responsible for the degradation of ECM proteins and are upregulated during EMT and invasion of cytotrophoblasts29,45, we tested the effect of fractionated and complete pECM on the gene expressions of MMP2 and MMP9 (Figure 5)18,46. Using the groups without any pECM as reference points, HMW-pECM upregulated the expressions of MMP2 and MMP9 significantly (p<0.05) compared to LMW-pECM and the control group without any pECM (Figure 5a,b). LMW-pECM, on the other hand, did not have a significant impact (p<0.05) on MMP2 or MMP9 expression. Even though HMW-pECM upregulated MMP2 and MMP9 expressions, the magnitudes remained lower than that of complete pECM (1.40 versus 1.55 and 2.39 versus 5.99 fold change, respectively), again suggesting there may be synergistic effects between the different components of pECM which was consistent with the invasion rate results.
Figure 4. Effect of pECM on Cytotrophoblast Invasion.
(a) Overview of Bioprinted Placenta Model. The bioprinted placenta model (BPM) was based on previously published work that has a layer of cytotrophoblasts along the periphery of a cylindrical disc (green) with a chemoattractant at the center. Placental BM proteins were incorporated with GelMA throughout BPM. (b) Representative Images of Cytotrophoblast Invasion. By taking images of cytotrophoblasts at day 2 (red) and day 7 (green), we observed the shift in the invasion fronts. We calculated the invasion rates by taking the differences between the invasion fronts and average over time. (c) Effect of Fractionated pECM on Invasion Rates of Cytotrophoblasts. The cytotrophoblasts bioprinted in complete pECM has the highest invasion rates, followed by HMW-pECM and then LMW-pECM (n=3). (d) Trophoblast Invasion Rates as a Function of pECM and PI3K Inhibitor. The pECM at both dosages (0.02 and 0.2 mg pECM/mL) significantly increased invasion rates of cytotrophoblasts compared to the control BPM without any pECM. The addition of LY294002, a PI3K inhibitor, significantly reduced the effect of pECM on the invasion rates when compared to their respective dosage of pECM. Moreover, there’s no statistically significant differences between the invasion rates of control BPM with no pECM and groups treated with LY294002 (n=3), which indicated that the effect of pECM was completely removed by LY294002. (e) Effect of pECM and PI3K Inhibitor on MMP Activities. A similar pattern was observed in the amount of MMP activity where pECM increased MMP activities significantly at both dosages (0.2 and 0.02 mg pECM/mL) and the addition of PI3K inhibitor significantly reduced the effect of pECM. (n=3).
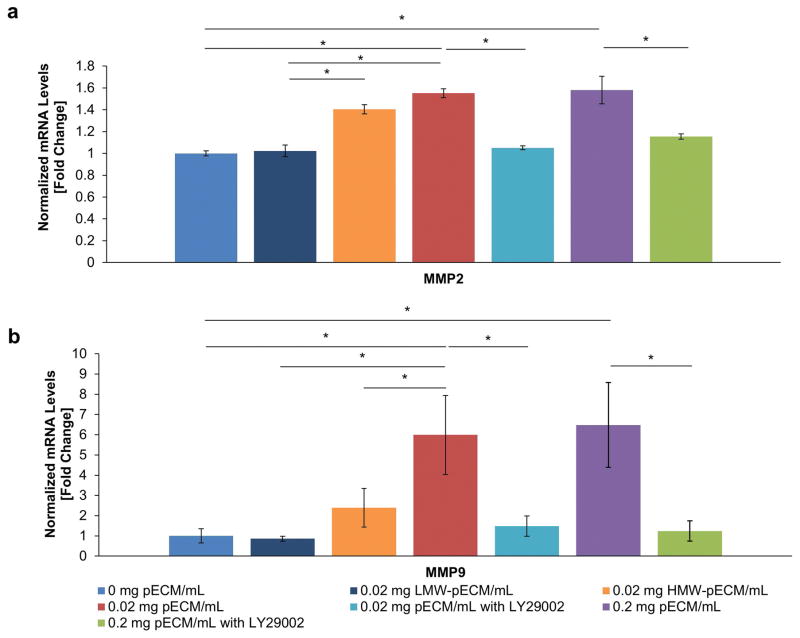
Figure 5. Mechanisms of pECM-enhanced Cytotrophoblast Invasion.
(a) Effect of pECM on the Expressions of MMP2 of Cytotrophoblasts. The HMW-pECM increased the expressions of MMP2 significantly compared to LMW-pECM and control with no pECM. The MMP2 expressions for HMW-pECM was lower than that of complete pECM with no significant differences. LMW-pECM did not have a significant impact on the MMP2 expression (n=3). The addition of pECM significantly increased the MMP2 gene expression compared to control. However, the effect of pECM was completely removed when PI3K was inhibited with no significant differences with the control (n=3). (b) Effect of pECM on the Expressions of MMP9 of Cytotrophoblasts. The HMW-pECM increased the expressions of MMP9 significantly compared to LMW-pECM. However, the magnitude of HMW-pECM’s impact on MMP9 was substantially lower than that of complete pECM with significant differences. On the other hand, LMW-pECM does not change MMP9 expressions significantly compared to control (n=3). Moreover, the treatment of pECM increased the MMP9 gene expression compared to control (0 mg pECM/mL) but again the effect was completely removed when PI3K inhibitor added with no significant differences with the control (n=3).
Mechanism of pECM-Induced Cytotrophoblast Invasion
Finally, we determined cytotrophoblast invasion rates as a function of pECM concentrations to test our hypothesis that placental BM proteins are required for effective cytotrophoblast invasion. Our results showed that pECM, at both dosages (0.02 and 0.2 mg pECM/mL), significantly increased invasion rates of cytotrophoblasts (65.8±37.5, and 75.5±21.8 μm/day, respectively; p<0.05; Figure 4d) compared to control BPM with no pECM (5.6±1.0 μm/day). Similarly, the addition of pECM increased MMP activities (normalized to control group without any pECM) but only significantly at a higher pECM concentration (0.2 mg pECM/mL; p<0.05; Figure 4e). Since PI3K signaling is upstream of secretion of MMP and has been previously implicated in embryonic implantation, we tested its role cytotrophoblast invasion17,18,47. The addition of a PI3K inhibitor (LY294002; 25 μM) in the presence of pECM significantly reduced the invasion rates (12.6±10.4 and 15.0±6.90 μm/day; p<0.05) compared to their respective dosages of pECM (65.8±37.5 and 75.5±21.8, respectively). Moreover, there were no statistically significant differences (p>0.05) between the invasion rates of control BPM and pECM groups treated with PI3K inhibitor, indicating that inhibiting PI3K signaling completely removed the effects of pECM. A similar pattern was observed in the amount of MMP activities (normalized to control group without any pECM) where pECM increased MMP activities (1.28±0.049 and 1.41±0.10; figure 4e) and the addition of PI3K inhibitor significantly reduced them (0.78±0.17 and 0.89±.27, respectively; p<0.05).
At the genetic level, the addition of complete pECM (0.2 and 0.02 mg pECM/mL) significantly upregulated MMP2 gene expression, expressed as fold change from the control group without any pECM, (1.55±0.04 and 1.97±0.08, respectively; p<0.05) and were reduced when PI3K inhibitor was added (1.15±0.02 and 1.05±0.02, respectively; Figure 5a; p<0.05). Similarly, the addition of pECM significantly upregulated MMP9 (6.48±2.1 and 5.99±1.96, respectively; p<0.05) and were equally reduced when the PI3K was inhibited (1.24±1.1 and 1.48±0.50, respectively; p<0.05; Figure 5b). Additionally, there were no statistically significant differences (p<0.05) between control BPM without pECM and PI3K inhibited groups in terms of MMP2 and MMP9 expressions, indicating that the effect of pECM on the gene expressions was completely removed by PI3K inhibitors. These results indicated that the pECM-enhanced invasion rates were correlated directly with upregulated of MMP2 and MMP9, and PI3K signaling appeared to play a dominant role in pECM-enhanced cytotrophoblast invasion.
Discussion
Cytotrophoblast-ECM interactions play a fundamental role in placentation and are critical to placentation and overall pregnancy success20. However, there are very limited studies, if any, that investigate the effect of ECM microenvironment on cytotrophoblast invasion and differentiation. In this research, for the first time, the necessary and critical roles of placental BM proteins on cytotrophoblast invasion were demonstrated. The addition of pECM, which was rich in BM proteins, significantly increased the invasion rates by 13 fold (Figure 4c) and upregulated the gene expressions of MMP2 (1.5 fold) and MMP9 (6.3 fold) of cytotrophoblasts, all compared to samples without pECM (Figure 5). These pECM-enhanced invasive responses were completely removed upon the addition of PI3K inhibitor, suggesting placental BM protein stimulated cytotrophoblast invasion predominantly through PI3K signaling (Figure 4,5).
In preparation for embryonic implantation, the maternal endometrium undergoes substantial remodeling and differentiation to become the decidua17,21 and causes the maternal decidual stromal cells (those in direct contact with cytotrophoblasts) produce pericellular basement membrane (BM)26. Published studies have established the significance of BM proteins in placentation and embryogenesis in murine BM-proteins ablated models23, human placental tissue25,48,49, and cytotrophoblast invasion in vitro25,50,51. However, none of the studies to date have directly determined the impact of extracellular placental BM proteins on cytotrophoblast invasion. We found the placental BM proteins significantly increased cytotrophoblast invasion rates and MMP expressions compare to samples without pECM (Figure 4,5). If we used the invasion rates obtained through the placenta model loaded with pECM and assumed a decidual thickness of 5 mm52, the maternal decidua would be fully invaded by cytotrophoblasts in 7 weeks, consistent with in vivo results from human pregnancies52. Without the presence of placental BM proteins, the cytotrophoblast invasion was critically impaired, a condition that is associated with preeclampsia1–5. These findings, for the first time, exemplifies the necessary and critical role of placental BM proteins in normal cytotrophoblast invasion. Since placental BM proteins are secreted from decidualization, our findings validate the importance of endometrial remodeling prior to the arrival of blastocyst. Future studies will involve incorporating decidual stromal cells into our tissue-specific placental model to assess their roles in cytotrophoblast invasion.
Our results showed that the placental BM proteins promoted cytotrophoblast invasion predominantly through PI3K signaling, a critical signaling pathway downstream of TKR that regulates cell motility, proliferation, survival and growth16,18. We demonstrated that the improved invasive phenotype and genotypes of cytotrophoblasts in the presence of were completely removed when PI3K signaling was inhibited (Figure 4,5). This result was not surprising because other study demonstrated laminin binding activates the PI3K pathway in mammalian cells53. However, our results suggest that the reason for the failed implantation in TKR-ablated uterus in a murine model17 may be due to impaired cytotrophoblast invasion and differentiation. Since PI3K signaling appeared to be an important signaling pathway that regulates the interactions between cytotrophoblast and placental BM proteins, it could be an attractive potential therapeutic target for preeclampsia. However, further testing for other molecular pathways and inhibitors are necessary before preclinical testing.
The ECM microenvironment plays a fundamental role in the maintenance of cellular phenotypes such as invasion20,28,54. There is strong evidence supporting that the interaction between ECM microenvironment and cells is mediated through receptor-ligand interactions and biomechanical stimuli, which in turn control cellular function28,54. Recent studies have validated tissue-specific ECM improves cell function and intricate tissue formation28,55. These ECM scaffolds were shown to have the capacity to direct tissue-specific cell lineage commitment and maintaining the phenotype of mature cell populations.28 The cell-ECM interactions are extremely complex in nature and justify the need for a tissue-specific approach to recreate the native cytotrophoblast niche to encourage and maintain their invasive phenotypes28,56. As such, we isolated placental BM proteins from the basal plate of fresh term placenta because it is a rich source of invasive extravillous cytotrophoblasts56. This concept was confirmed when we found that we could not reproduce the biological effects of pECM using its fractionated components alone (pECM vs. LMW-pECM vs. HMW-pECM), which indicated that all components of pECM were required, as demonstrated in Figure 4b and Figure 5. Therefore, current literature on cytotrophoblast biology that utilizes soluble, single ECM components and/or tissue culture polystyrene, while still valuable, may not represent the in vivo environment. Utilizing additive manufacturing technologies, we combined the unparalleled features of pECM with bioprinting to construct a cytotrophoblast-laden placenta model with micro-patterned biomaterials (Figure 4a). The ability to pattern cells and chemoattractant was crucial because it enabled the quantifications of cell invasion rates (demonstrated in Figure 4b) rather than the binary read-outs in current conventional, 2-dimensional (2D) invasion assays (e.g. transwell). In vitro culture using pECM supports adhesion, proliferation and invasion of cytotrophoblasts for at least 7 days (Figure 2). Altogether, we demonstrated that bioprinting with tissue-specific ECM is an attractive option for constructing in vitro placental tissue model to screen for novel therapeutics for placental diseases and their pathology. Our tissue specific, bioprinted placental model can potentially predict in vivo drug administration outcome for PE. Furthermore, other tissue models such as cancer can be developed based on the method presented in this work by recreating the key microenvironmental characteristics (complex ECM composition, mechanical properties, cellularity) that are representative to the in vivo environment.
Conclusion
We successfully incorporated placental BM protein isolated from basal plate of human placenta in our established bioprinting platform. Our results showed that pECM significantly increased adhesion, proliferation, and invasion of cytotrophoblasts and these effects could not be reproduced using fractions of pECM. Particularly, the addition of pECM, which is rich in basement membrane proteins, was required for cytotrophoblasts to achieve effective invasion rates that is comparable normal pregnancies and PI3K appear to play a dominant role in this process. Our results illustrated, for the first time, the significant effects of the ECM microenvironment in cytotrophoblast invasion, a parameter that is less investigated but critical in the development of preeclampsia. Moreover, the approach of bioprinting organ models with organ-specific ECM can be used to screen for novel therapeutics for placental and other diseases such as cancer invasion.
Acknowledgments
This research was funded by the Sheikh Zayed Institute for Pediatric Surgical Innovation, a part of Children’s National Health System, and by NIH Center for Engineering Complex Tissues (CECT). The size-exclusion based chromatography work was supported by the Board of Visitor’s Grant of Children’s National Health System. In addition, we would like to thank the Cellular Neuroscience Imaging core of Children’s Research Institute (CRI) for providing microscopy instruments. The resources associated with proteomics were supported by Award Number 1U54HD090257-01 from the NIH, District of Columbia Intellectual and Developmental Disabilities Research Center Award (DC-IDDRC) program. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the DC-IDDRC or the NIH. Lastly, the authors would like to thank Drs. Stephanie Val and Aswini Panigrahi for their help with proteomics.
References
- 1.Christopher W, Redman ILS. Latest Advances in Understanding Preeclampsia. SCIENCE. 2005;308:1592–1594. doi: 10.1126/science.1111726. [DOI] [PubMed] [Google Scholar]
- 2.Tinnakorn Chaiworapongsa PC, Yeo Lami, Romero Roberto. Pre-eclampsia part 1: current understanding of its paothophsiology. Nature Nephrology. 2014;10 doi: 10.1038/nrneph.2014.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young BC, Levine RJ, Karumanchi SA. Pathogenesis of preeclampsia. Annu Rev Pathol. 2010;5:173–192. doi: 10.1146/annurev-pathol-121808-102149. [DOI] [PubMed] [Google Scholar]
- 4.Mohd Nordin Noraihan PSaABEJ. Report of 50 cases of eclampsia. 2005;31:302–309. doi: 10.1111/j.1447-0756.2005.00292.x. [DOI] [PubMed] [Google Scholar]
- 5.Luo J, Manning BD, Cantley LC. Targeting the PI3K-Akt pathway in human cancer: Rationale and promise. Cancer Cell. 2003;4:257–262. doi: 10.1016/s1535-6108(03)00248-4. [DOI] [PubMed] [Google Scholar]
- 6.McMaster MT, Zhou Y, Fisher SJ. Abnormal placentation and the syndrome of preeclampsia. Seminars in Nephrology. 2004;24:540–547. doi: 10.1016/s0270-9295(04)00124-x. [DOI] [PubMed] [Google Scholar]
- 7.Eric AP, Steegers PvD, Duvekot Johannes J, Pijnenborg Robert. Pre-eclampsia. The Lancet. 2010;376:631–644. doi: 10.1016/s01406736(10)60279-6. [DOI] [PubMed] [Google Scholar]
- 8.Tinnakorn Chaiworapongsa PC, Lami Yeo, Romero Roberto. Pre-eclampsia part 2: prediction prevention and management. Nature Reviews Nephrology. 2014;10:531–540. doi: 10.1038/nrneph.2014.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collaborators GMaCoD. Global, regional, and national age–sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. The Lancet. 2015;385:117–171. doi: 10.1016/s0140-6736(14)61682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orendi K, et al. Placental and trophoblastic in vitro models to study preventive and therapeutic agents for preeclampsia. Placenta. 2011;32(Suppl):S49–54. doi: 10.1016/j.placenta.2010.11.023. [DOI] [PubMed] [Google Scholar]
- 11.Davies JE, et al. Epithelial-mesenchymal transition during extravillous trophoblast differentiation. Cell Adhesion & Migration. 2016;10:310–321. doi: 10.1080/19336918.2016.1170258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vicovac L, Aplin JD. Epithelial-mesenchymal transition during trophoblast differentiation. Acta Anatomica. 1996;156:202–216. doi: 10.1159/000147847. [DOI] [PubMed] [Google Scholar]
- 13.Ackland ML, et al. Epidermal growth factor-induced epithelio-mesenchymal transition in human breast carcinoma cells. Laboratory Investigation. 2003;83:435–448. doi: 10.1097/01.lab.0000059927.97515.fd. [DOI] [PubMed] [Google Scholar]
- 14.Ahmed N, et al. Molecular pathways regulating EGF-induced epithelio-mesenchymal transition in human ovarian surface epithelium. American Journal of Physiology-Cell Physiology. 2006;290:C1532–C1542. doi: 10.1152/ajpcell.00478.2005. [DOI] [PubMed] [Google Scholar]
- 15.Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. Journal of Clinical Investigation. 2009;119:1429–1437. doi: 10.1172/jci36183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nature Reviews Molecular Cell Biology. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Large MJ, et al. The Epidermal Growth Factor Receptor Critically Regulates Endometrial Function during Early Pregnancy. Plos Genetics. 2014;10 doi: 10.1371/journal.pgen.1004451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen JS, et al. Involvement of PI3K/PTEN/AKT/mTOR pathway in invasion and metastasis in hepatocellular carcinoma: Association with MMP-9. Hepatology Research. 2009;39:177–186. doi: 10.1111/j.1872-034X.2008.00449.x. [DOI] [PubMed] [Google Scholar]
- 19.Qiu Q, Yang M, Tsang BK, Gruslin A. EGF-induced trophoblast secretion of MMP-9 and TIMP-1 involves activation of both PI3K and MAPK signalling pathways. Reproduction. 2004;128:355–363. doi: 10.1530/rep.1.00234. [DOI] [PubMed] [Google Scholar]
- 20.Zhang YY, et al. Tissue-specific extracellular matrix coatings for the promotion of cell proliferation and maintenance of cell phenotype. Biomaterials. 2009;30:4021–4028. doi: 10.1016/j.biomaterials.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Babiarz B, Romagnano L, Afonso S, Kurilla G. Localization and expression of fibronectin during mouse decidualization in vitro: Mechanisms of cell:matrix interactions. Developmental Dynamics. 1996;206:330–342. doi: 10.1002/(sici)1097-0177(199607)206:3<330::aid-aja10>3.3.co;2-7. [DOI] [PubMed] [Google Scholar]
- 22.Miner JH, Cunningham J, Sanes JR. Roles for laminin in embryogenesis: Exencephaly, syndactyly, and placentopathy in mice lacking the laminin alpha 5 chain. Journal of Cell Biology. 1998;143:1713–1723. doi: 10.1083/jcb.143.6.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiradjaja F, DiTommaso T, Smyth I. Basement Membranes in Development and Disease. Birth Defects Research Part C-Embryo Today-Reviews. 2010;90:8–31. doi: 10.1002/bdrc.20172. [DOI] [PubMed] [Google Scholar]
- 24.Ma KD, et al. A Proteomic Analysis of Placental Trophoblastic Cells in Preeclampsia-Eclampsia. Cell Biochemistry and Biophysics. 2014;69:247–258. doi: 10.1007/s12013-013-9792-4. [DOI] [PubMed] [Google Scholar]
- 25.Kurdoglu M, et al. Expression of laminin receptor 1 in human placentas from normal and preeclamptic pregnancies and its relationship with the severity of preeclampsia. Journal of Perinatal Medicine. 2011;39:411–416. doi: 10.1515/jpm.2011.024. [DOI] [PubMed] [Google Scholar]
- 26.Risteli J, Foidart JM, Risteli L, Boniver J, Goffinet G. THE BASEMENT-MEMBRANE PROTEINS LAMININ AND TYPE-IV COLLAGEN IN ISOLATED VILLI IN PRE-ECLAMPSIA. Placenta. 1984;5:541–550. doi: 10.1016/s0143-4004(84)80008-9. [DOI] [PubMed] [Google Scholar]
- 27.Winn VD, Gormley M, Fisher SJ. The impact of preeclampsia on gene expression at the maternal-fetal interface. Pregnancy Hypertension-an International Journal of Womens Cardiovascular Health. 2011;1:100–108. doi: 10.1016/j.preghy.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pati F, et al. Printing three-dimensional tissue analogues with decellularized extracellular matrix bioink. Nature communications. 2014;5:3935. doi: 10.1038/ncomms4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radisky ES, Radisky DC. Matrix Metalloproteinase-Induced Epithelial-Mesenchymal Transition in Breast Cancer. Journal of Mammary Gland Biology and Neoplasia. 2010;15:201–212. doi: 10.1007/s10911-010-9177-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flynn L, Semple JL, Woodhouse KA. Decellularized placental matrices for adipose tissue engineering. Journal of Biomedical Materials Research Part A. 2006;79A:359–369. doi: 10.1002/jbm.a.30762. [DOI] [PubMed] [Google Scholar]
- 31.Billiet T, Gevaert E, De Schryver T, Cornelissen M, Dubruel P. The 3D printing of gelatin methacrylamide cell-laden tissue-engineered constructs with high cell viability. Biomaterials. 2014;35:49–62. doi: 10.1016/j.biomaterials.2013.09.078. [DOI] [PubMed] [Google Scholar]
- 32.De Cock LJ, et al. Engineered 3D microporous gelatin scaffolds to study cell migration. Chemical communications. 2012;48:3512–3514. doi: 10.1039/c2cc17006j. [DOI] [PubMed] [Google Scholar]
- 33.An I, Van Den Bulke BB, DeRooze Nadine, Schacht Etienne H, Cornelissen Maria, Berghmans Hugo. Structural and Rheological Properties of Methacrylamide Modified Gelatin Hydrogels. Biomacromolecules. 2000;1:31–38. doi: 10.1021/bm990017d. [DOI] [PubMed] [Google Scholar]
- 34.Friedrichs J, Helenius J, Muller DJ. Quantifying cellular adhesion to extracellular matrix components by single-cell force spectroscopy. Nature Protocols. 2010;5:1353–1361. doi: 10.1038/nprot.2010.89. [DOI] [PubMed] [Google Scholar]
- 35.Klebe RJ. ISOLATION OF A COLLAGEN-DEPENDENT CELL ATTACHMENT FACTOR. Nature. 1974;250:248–251. doi: 10.1038/250248a0. [DOI] [PubMed] [Google Scholar]
- 36.Christ KV, Turner KT. Methods to Measure the Strength of Cell Adhesion to Substrates. Journal of Adhesion Science and Technology. 2010;24:2027–2058. doi: 10.1163/016942410x507911. [DOI] [Google Scholar]
- 37.Khalili AA, Ahmad MR. A Review of Cell Adhesion Studies for Biomedical and Biological Applications. International Journal of Molecular Sciences. 2015;16:18149–18184. doi: 10.3390/ijms160818149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuo CY, et al. Development of a 3D Printed, Bioengineered Placenta Model to Evaluate the Role of Trophoblast Migration in Preeclampsia. Acs Biomaterials Science & Engineering. 2016;2:1817–1826. doi: 10.1021/acsbiomaterials.6b00031. [DOI] [PubMed] [Google Scholar]
- 39.Nguyen BNB, Ko H, Moriarty RA, Etheridge JM, Fisher JP. Dynamic Bioreactor Culture of High Volume Engineered Bone Tissue. Tissue Engineering Part A. 2016;22:263–271. doi: 10.1089/ten.tea.2015.0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crapo PM, Gilbert TW, Badylak SF. An overview of tissue and whole organ decellularization processes. Biomaterials. 2011;32:3233–3243. doi: 10.1016/j.biomaterials.2011.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 42.Edmondson R, Broglie JJ, Adcock AF, Yang LJ. Three-Dimensional Cell Culture Systems and Their Applications in Drug Discovery and Cell-Based Biosensors. Assay and Drug Development Technologies. 2014;12:207–218. doi: 10.1089/adt.2014.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakatsuji Y, et al. Epidermal growth factor enhances invasive activity of BeWo choriocarcinoma cells by inducing alpha 2 integrin expression. Endocrine Journal. 2003;50:703–714. doi: 10.1507/endocrj.50.703. [DOI] [PubMed] [Google Scholar]
- 44.Daoud G, Barrak J, Abou-Kheir W. Assessment Of Different Trophoblast Cell Lines As In Vitro Models For Placental Development. Faseb Journal. 2016;30 [Google Scholar]
- 45.DaSilva-Arnold S, James JL, Al-Khan A, Zamudio S, Illsley NP. Differentiation of first trimester cytotrophoblast to extravillous trophoblast involves an epithelial-mesenchymal transition. Placenta. 2015;36:1412–1418. doi: 10.1016/j.placenta.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 46.Ren QA, Guan S, Fu JL, Wang AG. Spatio-Temporal Expression of Matrix Metalloproteinases-2 and-9 in Porcine Endometrium During Implantation. Journal of Animal and Veterinary Advances. 2010;9:2074–2081. [Google Scholar]
- 47.Karar J, Maity A. PI3K/AKT/mTOR pathway in angiogenesis. Frontiers in Molecular Neuroscience. 2011;4 doi: 10.3389/fnmol.2011.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kilic F, et al. Shear wave elastography of placenta: in vivo quantitation of placental elasticity in preeclampsia. Diagnostic and Interventional Radiology. 2015;21:202–207. doi: 10.5152/dir.2014.14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pijnenborg R, Dhooghe T, Vercruysse L, Bambra C. Evaluation of trophoblast invasion in placental bed biopsies of the baboon, with immunohistochemical localisation of cytokeratin, fibronectin, and laminin. Journal of Medical Primatology. 1996;25:272–281. doi: 10.1111/j.1600-0684.1996.tb00210.x. [DOI] [PubMed] [Google Scholar]
- 50.Shan N, et al. Laminin alpha 4 (LAMA4) expression promotes trophoblast cell invasion, migration, and angiogenesis, and is lowered in preeclamptic placentas. Placenta. 2015;36:809–820. doi: 10.1016/j.placenta.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 51.Wang LL, Yu Y, Guan HB, Liu T, Qiao C. 67-kDa Laminin receptor contributes to hypoxia-induced migration and invasion of trophoblast-like cells by mediating matrix metalloproteinase-9. Clinical and Experimental Pharmacology and Physiology. 2015;42:549–558. doi: 10.1111/1440-1681.12389. [DOI] [PubMed] [Google Scholar]
- 52.Wong HS, Cheung YK. Sonographic study of the decidua basalis in early pregnancy loss. Ultrasound in Obstetrics & Gynecology. 2010;36:362–367. doi: 10.1002/uog.7736. [DOI] [PubMed] [Google Scholar]
- 53.Langenbach KJ, Rando TA. Inhibition of dystroglycan binding to laminin disrupts the PI3K/AKT pathway and survival signaling in muscle cells. Muscle & Nerve. 2002;26:644–653. doi: 10.1002/mus.10258. [DOI] [PubMed] [Google Scholar]
- 54.Guilak F, et al. Control of Stem Cell Fate by Physical Interactions with the Extracellular Matrix. Cell Stem Cell. 2009;5:17–26. doi: 10.1016/j.stem.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sellaro TL, et al. Maintenance of Human Hepatocyte Function In Vitro by Liver-Derived Extracellular Matrix Gels. Tissue Engineering Part A. 2010;16:1075–1082. doi: 10.1089/ten.tea.2008.0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Borbely AU, et al. The term basal plate of the human placenta as a source of functional extravillous trophoblast cells. Reproductive Biology and Endocrinology. 2014;12 doi: 10.1186/1477-7827-12-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ridley AJ, Hall A. THE SMALL GTP-BINDING PROTEIN RHO REGULATES THE ASSEMBLY OF FOCAL ADHESIONS AND ACTIN STRESS FIBERS IN RESPONSE TO GROWTH-FACTORS. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]