Abstract
Magnolias are widely cultivated for their beautiful flowers, but despite their popularity, the molecular mechanisms regulating flower bud differentiation have not been elucidated. Here, we used paraffin sections and RNA-seq to study the process of flower bud differentiation in Magnolia sinostellata. Flower bud development occurred between 28 April and 30 May 2017 and was divided into five stages: undifferentiated, early flower bud differentiation, petal primordium differentiation, stamen primordium differentiation, and pistil primordium differentiation. A total of 52,441 expressed genes were identified, of which 11,592 were significantly differentially expressed in the five bud development stages. Of these, 82 genes were involved in the flowering. In addition, MADS-box and AP2 family genes play critical roles in the formation of flower organs and 20 differentially expressed genes associated with flower bud differentiation were identified in M. sinostellata. A qRT-PCR analysis verified that the MADS-box and AP2 family genes were expressed at high levels during flower bud differentiation. Consequently, this study provides a theoretical basis for the genetic regulation of flowering in M. sinostellata, which lays a foundation for further research into flowering genes and may facilitate the development of new cultivars.
Keywords: flower bud differentiation, flowering gene, RNA-seq, qRT-PCR, Magnolia sp.
1. Introduction
Flower bud differentiation is an important stage in angiosperm development. The vegetative meristem is transformed into the floral meristem, which forms the base of the floral organ before developing into the floral tissues. The complex process of floral development arises in response to the integration of signals from the external environment and internal factors [1]. In Arabidopsis thaliana, flower development was found to involve six pathways; the photoperiod, gibberellin, vernalization, autonomous, senescence, and ambient-temperature pathways [2]. These pathways co-regulate the flower-specific genes, causing the vegetative meristem to physiologically transition into the floral meristem. Many genes have been found to integrate the signals received via these pathways, such as FT (FLOWER LOCUS T) and SOC1 (SUPPRESSOR OF OVEREXPRESSION OF CO 1), both of which promote floral development and lead to early flowering in plants [3,4]. LFY (LEAFY) integrates the signals of multiple floral induction pathways and activates floral organ development [5,6]. These integrative genes are highly conserved in plants. The FT genes of apple (Malus × domestica) and longan (Dimocarpus longan) influenced flowering when heterologously expressed in Arabidopsis [7,8], while the Arabidopsis FT and LFY genes regulated flowering when heterologously expressed in the satsuma mandarin (Citrus unshiu) [9] and poplar (Populus sp.) [10].
Flower development is controlled by floral organ trait genes. The classic ABC model of floral organ development, developed by Meyerowitz and Coen [11,12], proposes that class A genes control the formation of the sepals and petals, class B genes control the formation of the petals and stamens, and class C genes control the formation of the stamens and carpels. Members of these gene categories have been cloned from a variety of plants [13]. Later studies found that FBP11 (FLORAL BINDING PROTEIN11) controls ovule development in Petunia sp. [14], while other positive regulators of many aspects of floral development, such as SEP (SEPALLATA), were identified in Arabidopsis sp. and other model plants [15]. Thus, the ABC model was replaced by the ABCDE model, which states that A- and E-class genes determine the first whorl, the calyx; A-, B- and E-class genes interact to form the petals in the second whorl; B-, C-, and E-class genes interact to regulate the third whorl, the stamens; C- and E-class genes control carpel development in the fourth whorl; and D- and E-class genes are involved in the formation of the ovary [16,17]. The floral development mode is composed of the activities of MADS-box and AP2 family transcription factors [13,18]. AP2 gene plays a role as A-class genes in a flower development model and MADS-box transcription genes participate in various types [13].
Magnolias (Magnolia sp.) are widely distributed shrubs and trees that produce beautiful flowers in the spring. The Magnoliaceae have long been considered an ancient angiosperm family and much of the current research on members of this family focuses on their systematic evolution [19,20]. The studies of Magnoliaceae flower bud differentiation mostly involve morphological observation [21], leaving the molecular mechanisms underpinning their flower development largely unclear. Homologs of AP3 (APETALA3) and AG (AGAMOUS) have been cloned from the flower buds of Magnolia wufengensis and their expression patterns have been reported [22,23], while other studies investigated the transcriptomic regulation of petal color in Magnolia sprengeri [24] and the biosynthetic pathways regulating floral volatile organic compounds in Magnolia champaca [25].
Magnolia sinostellata is an endangered magnolia shrub species with high ornamental value [26]. Its morphological features are similar to Magnolia stellata [27]; however, the two species can be differentiated by their twig color, by staining during meiosis [26], and because M. sinostellata blooms earlier than M. stellata. The flowers of M. sinostellata wither in late March and the leaves begin to grow nearly a month later. During the process of flower bud differentiation, the outer bud sequentially forms two to three layers of shallow, brown, spathe-like bracts. Flowers develop once a year, beginning at the end of April and ending around the beginning of June, for a total differentiation period of almost 40 days.
We have previously investigated flower bud differentiation in Magnolia biondii, Magnolia denudata and Magnolia × soulangeana ‘Red lucky’ [28]; however, the molecular regulation of flower bud differentiation has not been previously reported for Magnolia sp. Here, we explored the morphological characteristics of flower bud differentiation in M. sinostellata and performed a transcriptomic analysis to identify genes that were differentially transcribed at the five floral bud stages. The results of this study provide a foundation for future studies and breeding efforts focusing on flowering in Magnolia sp.
2. Materials and Methods
2.1. Plant Materials
The M. sinostellata individuals (approximately 10 years old) used in this study are maintained outdoor of the nursery in Zhejiang Agriculture and Forestry University (30°15′14″ N and 119°43′39″ E). The subtropical monsoon climate is warm and humid, with sufficient light and an annual average precipitation of approximately 1600 mm. All mature M. sinostellata trees were managed according to ordinary culture practices.
2.2. Morphological Observation
A total of 15 flower buds were collected from the middle and upper regions of the tree crown every month from June 2016 to June 2017 and were stored in FAA fixative (8:1:1 ratio of 50% ethanol: formaldehyde: glacial acetic acid). The initiation of a new leaf, and the flower buds were collected every seven days and were observed after the flower bud began to differentiate. Additionally, the sampling time was shortened to every three days until the end of differentiation. The experiment had three biological replications, and each replication had 10 buds. The development process was observed using an anatomical microscope [29]. Samples were sliced to a 10-μm thickness and subjected to safranin fast green staining for 30 s. The samples were sealed with neutral balata and then observed and photographed using an Axio Imager A2 positive fluorescence microscope (Carl Zeiss, Oberkochen, Germany).
2.3. RNA Extraction and cDNA Library Construction
Total RNA was extracted from flower buds at the same stages as those observed above using an RNAprep Pure Plant Kit (TaKaRa, Dalian, China), with three biological replicates at each stage. The quality and purity of the RNA samples were assessed using an RNA 6000 Nano LabChip Kit and a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA), using an RNA integrity number (RIN) of >7.0. Poly-(A)-containing mRNA was purified using oligo (dT) magnetic beads and an Oligotex mRNA Kit (Qiagen, Hilden, Germany). Fragmentation buffer was added to disrupt the mRNA strands into short fragments, which were used as templates to synthesize the first-strand cDNA using reverse transcriptase and random hexamer primers. The second-strand cDNA was synthesized using buffer, dNTPs, RNase H, and DNA polymerase I. The double-stranded cDNA fragments were subjected to end repair and adapter ligation. Adapter-modified fragments were selected using gel purification and PCR amplified to create the final cDNA library.
2.4. Illumina Sequencing, Assembly and Annotation
The cDNA library was sequenced on an Illumina HiSeq 4000 sequencing platform (Illumina, San Diego, CA, USA) by LC Sciences (Hanhzhou, China) to yield 2 × 150-bp paired-end raw reads. The sequenced raw reads were subjected to a quality check using FastQC [30]. The adapter sequences were removed from the raw reads. Reads with a ratio of ambiguous N nucleotides greater than 5% and those with low-quality sequences (quality score of less than 20) were removed. Sequencing reads were de novo assembled using Trinity software under default parameters and with a k-mer size of 25 [31]. The transcriptomes were assembled using pooled reads from all replications and stages. Assembly quality was critically assessed by LC Sciences Company (http://www.lc-bio.com/) before subsequent analyses. The assembled transcriptome sequences were named ‘unigenes’.
All unigenes were queried against six commonly used databases using BLASTx search to identify homologs (E-value < 10–10). The databases used were Swiss-prot [32], Nr [33], KEGG [34], KOG [35], Pfam [36], and GO [37].).
2.5. Differential Gene Expression Analysis
To analyze the differentially expressed genes (DEGs) during flower bud differentiation, the number of reads of each of the contigs was converted to reads per kilobase per million (RPKM) using RSEM 1.2.31 [38]. DESeq was used to determine the FDR (false discovery rate) threshold. If the FDR was less than 0.05 in the multi-group comparison, it was considered to be a significantly different expression level. The DEGs were further annotated with GO functional and KEGG pathway analyses using the software TBtools [39]. p-values generated from the enrichment analyses were subjected to multiple hypotheses testing by PerlScript [40], and a p-value of <0.05 considering statistically significant. Furthermore, the DEGs were annotated by a BLAST search against known flowering genes in Arabidopsis sp. [16,41] and then screened for functions known to be involved in floral development.
2.6. qRT-PCR Analysis
Total RNA was extracted from flower buds at the five developmental stages. After treatment with DNase, the total RNA was reverse transcribed to produce cDNA using a reverse transcription system with PrimeScript™ RT Master Mix (Perfect Real Time, TaKaRa, Dalian, China). qRT-PCR was conducted using a Light Cycler 480II (Roche Applied Science, Penzberg, Germany). Each 20-µL qRT-PCR reaction contained 10 µL SYBR Premix Ex TaqTMII (TaKaRa, Dalian, China), 2 µL cDNA (80 ng/µL), 0.8 µL forward primer (10 μM), 0.8 µL reverse primer (10 μM) and 6.4 µL H2O. The primers were designed using Primer 5.0 (Table S1). The amplification program was as follows: 30 s at 95 °C, followed by 40 cycles of 5 s at 95 °C and 30 s at 60 °C. All qRT-PCR experiments were conducted in triplicate with three biological replicates. EF-1α was used as the reference gene (Table S2, Figure S1) and the relative transcript abundances were calculated using the 2−∆∆Ct method [42].
3. Results
3.1. Morphological Changes during the Different Stages of Flower Bud Differentiation
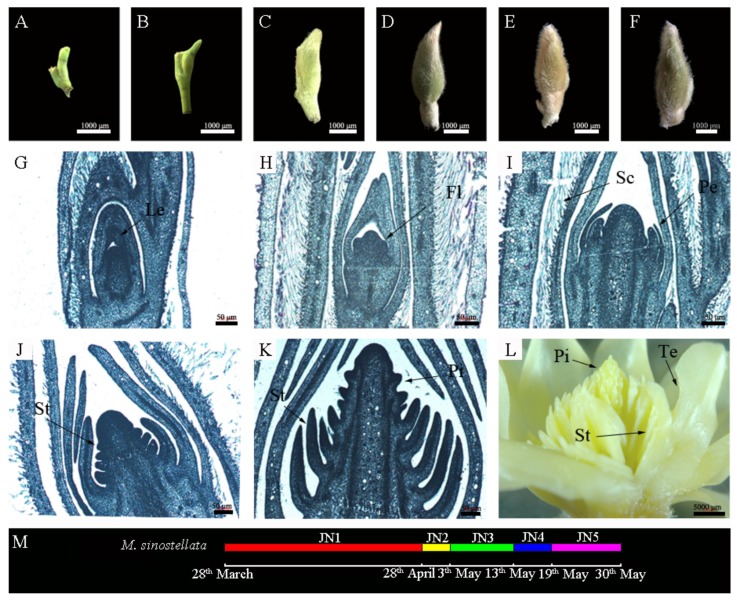
Based on the morphological features of floral bud differentiation in M. sinostellata, we divided the process into the following five stages (Figure 1): undifferentiated (JN1), early flower bud differentiation (JN2), petal primordium differentiation (JN3), stamen primordium differentiation (JN4), and pistil primordium differentiation (JN5).
Figure 1.
Morphological characteristics of the flower bud differentiation process in Magnolia sinostellata. (A,G) Undifferentiated stage (JN1); (B,H) Early flower bud differentiation stage (JN2); (C,I) Petal primordium differentiation stage (JN3); (D,J) Stamen primordium differentiation stage (JN4); (E,K) Pistil primordium differentiation stage (JN5); (F) The fully developed flower bud; (L) A dissected flower bud; (M) A timeline of flower bud differentiation. Le: leaf; Fl: Flower primordium; Sc: Scales; Pe: Petal primordium; St: Stamen primordium; Pi: Pistil base; Te: Tepal.
In the undifferentiated stage, the buds were yellowish green, smooth on the outside, and lacked scale hairs. The tip of the bud was pointed and clung to the petiole of the neighboring leaf (Figure 1A). The growth point of the flower bud was small and the differentiating primordium cells were small and closely arranged (Figure 1G). At the early flower bud differentiation stage, the basal region of the bud began to expand and the outer surface grew yellowish brown hairs. Inside the growing flower bud, the spathe-like bracts began to be stratify (Figure 1B). The floral primordium became larger and uplift deformation was observed, although the cells were still closely arranged (Figure 1H). At the petal primordium differentiation stage, the bud grew longer and wider and became distinct from the leaf primordia. Inside the bud were a few layers of spathe-like bracts with scale hairs (Figure 1C). The tip of the developing floral meristem had an undulating surface, indicating the initiation of petal primordium differentiation (Figure 1I). During the stamen primordium differentiation stage, the buds were enlarged and the outer yellowish-brown hairs gradually became denser (Figure 1D). The differentiation region of the inner bud became wider and elongated, forming a hump shade with a smooth tip. The outer cells of the bud meristem were small and compact, while the inner cells were separated from each other. Inside the petal primordia, many rows of small protruding spots were formed around the bottom of the meristem, which were determined to be the differentiating stamen primordia (Figure 1J). At the pistil primordium differentiation stage, the bud volume further increased and the external scale hairs became fluffy (Figure 1E,F). The inner developing bud tip elongated and its base became thick, while its top formed a smooth conical shape. At this stage, the upper region of the meristem formed multiple round bulges, indicating that the pistil primordia had begun to differentiate (Figure 1K). The bulges were retained until the differentiation was complete (Figure 1L).
3.2. Differentiating Flower Bud Transcriptome Sequencing, Assembly, and Functional Annotation
Separate transcriptomes were obtained from the five stages of M. sinostellata flower bud differentiation, with three biological replicates performed for each stage. A total of 110.56 Gb raw data was obtained during transcriptomic sequencing. The raw data were uploaded to NCBI under the accession numbers SRP129819 and SRR6475480–SRR6475509. After removing the unqualified reads from the raw data, the Q20 was above 97.53% and the GC number was 49.14–51.65% (Table S3). The mapping rate for the assembled transcriptomes was more than 86.08% (Table S4). The N50 length was 1126 bp and the size of the unigenes was generally between 200 bp and 2000 bp, with a mean length of 648 bp (Table S5). After assembly, a total of 52,441 Unigenes were obtained from 15 samples at different developmental stages, which were compared with Swiss-prot [32], Nr [33], KEGG [34], KOG [35], Pfam [36], and GO [37] for annotation and analysis (Table 1, Table S6). In the distribution of species, 59.8% of unigenes could be annotated based on their sequence similarity to unigenes from other species; the species providing the highest number of significant hits were grapevine (Vitis vinifera) and date palm (Phoenix dactylifera), which were used to annotate 23.2% and 19.2% of the transcripts, respectively (Figure S2).
Table 1.
Functional unigene annotations
| Gene Number | Unigenes Annotated Using These Databases | ||||||
|---|---|---|---|---|---|---|---|
| Nr | Pfam | KEGG | KOG | Swiss-Prot | GO | ||
| Unigenes | 52,441 | 25,531 | 18,736 | 8612 | 20,977 | 15,850 | 14,055 |
| Percentage | 100% | 48.69% | 35.73% | 16.42% | 40.00% | 30.22% | 26.80% |
3.3. Genes Expression in Each Stage of Flower Bud Differentiation
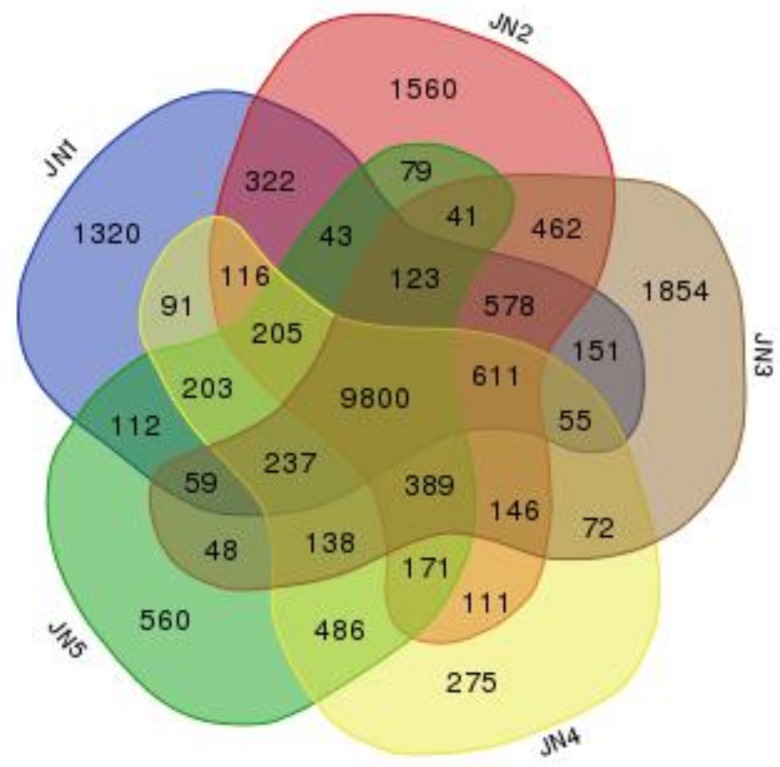
The genes number was analyzed between the five stages of flower bud differentiation (Figure 2, Figure S3). The number of genes expressed in the five stages (from undifferentiated to pistil primordium differentiation) was 14,026; 14,757; 14,764; 13,106; and 12,794, respectively. The number of genes specifically expressed in each stage was 1320; 1560; 1854; 275; and 560, respectively (Figure 2). According to the criteria FDR < 0.05, the differentially expressed genes of the three replicates for each floral differentiation stage showed in Table S6, indicating that gene expression changed significantly during M. sinostellata flower bud development and that fewer genes were specifically expressed in the late stages of flower bud differentiation.
Figure 2.
Venn diagram of gene number in the five developmental stages.
3.4. GO and KEGG Enrichment Analysis of Differentially Expressed Genes
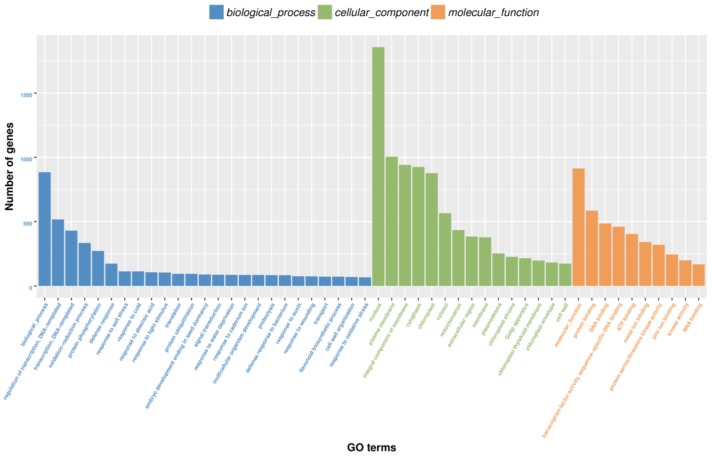
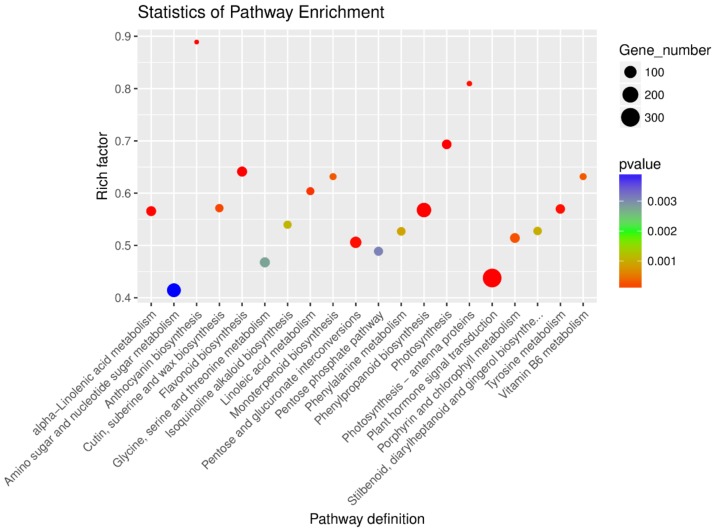
The RNA-seq analysis revealed a total of 52,441 genes, of which 11,592 genes were significantly differentially expressed in the five stages (FDR < 0.05). A gene ontology (GO) enrichment analysis of the DEGs revealed a significant enrichment of 371 GO terms throughout the differentiation process (p < 0.05) (Table S7). The GO terms were divided into three major categories—i.e., molecular function, cell component, and biological process—as well as 50 minor classes (Figure 3). The DEGs were successfully annotated as members of 135 pathways and the number of significantly enriched KEGG pathways was 36 (p < 0.05) (Table S8). Relatively high numbers of genes were annotated as plant hormone signal transduction (map04075; 303 genes), phenylpropanoid biosynthesis (map00940; 155 genes), amino sugar and nucleotide sugar metabolism (map00520; 138 genes), pentose and glucuronate interconversions (map00040; 83 genes), and flavonoid biosynthesis (map00941; 59 genes) pathways, with Rich factors of 0.44, 0.57, 0.41, 0.51, and 0.64, respectively (Figure 4). A Rich factor is the ratio of the number of DEGs annotated with a pathway term relative to the total number of genes annotated with this pathway term. The larger the Rich factor, the greater the enrichment of this KEGG pathway.
Figure 3.
GO function classification in flower bud differentiation differentially expressed genes (DEGs).
Figure 4.
The top 20 of KEGG pathway enrichments in flower bud differentiation DEGs.
3.5. DEGs and Transcription Factors Associated with the Five Stages of Flower Bud Differentiation
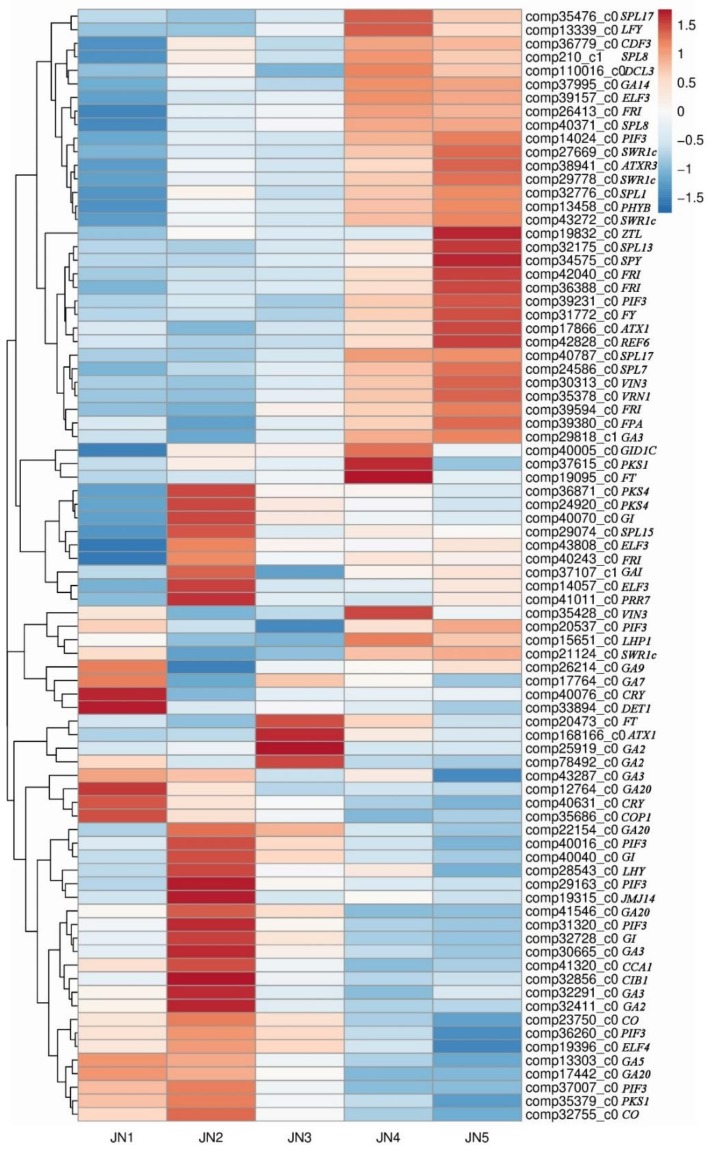
We identified 82 significantly differentially expressed M. sinostellata genes that were putative homologs of the flower bud differentiation genes in A. thaliana (Figure 5). In the circadian rhythm pathway, the expression levels of CRY (CRYPTOCHROME) decreased and ELF3 (EARLY FLOWERING 3) increased, while the expression of GI (GIGANTEA) and CO (CONSTANS) increased during the early stages of M. sinostellata floral bud differentiation and decreased in the later stages (Table S9). The expression levels of FY, FPA, and SPL (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE) were lower than those of flower bud differentiation. The expression of FRI (FRIGIDA) increased during flower bud differentiation. A total of 15 DEGs were associated with the gibberellin pathway; however, no consistent changes in the expression of these genes were observed during floral development. The expression of FT, an integrator of flowering regulatory signals [3], increased and then decreased as floral differentiation progressed.
Figure 5.
The expression of DEGs putatively related to flowering-pathway genes in Arabidopsis thaliana. Comp indicates the prefix of the gene ID. Red and Blue represents up- and down-regulated DEGs, respectively.
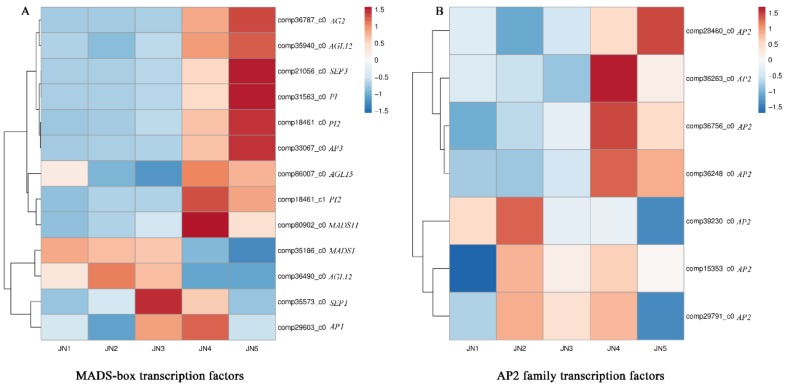
The floral development model is composed of the activities of MADS-box and AP2 family transcription factors [13,18]. The expression levels of MADS-box and AP2 family transcription factor-related genes differed between the five stages of floral development (Figure 6). Other genes encoding putative transcription factors were also differentially expressed throughout flower bud differentiation; for example, 36 genes encoding putative MYB transcription factors were differentially expressed during flower bud differentiation (Figure S4), as were 18 NAC family genes (Figure S5), 27 WRKY genes (Figure S6) and 13 GATA genes (Figure S7).
Figure 6.
The expression of DEGs putatively encoding MADS-box and AP2 family transcription factors. (A) is the expression of DEGs putatively encoding MADS-box; (B) is the expression of DEGs putatively encoding AP2 family transcription factors. Comp indicates the prefix of the gene ID. Red and Blue represents up- and down-regulated DEGs, respectively.
3.6. Verification of Relative Gene Expression in Different Stages of Flower Bud Differentiation
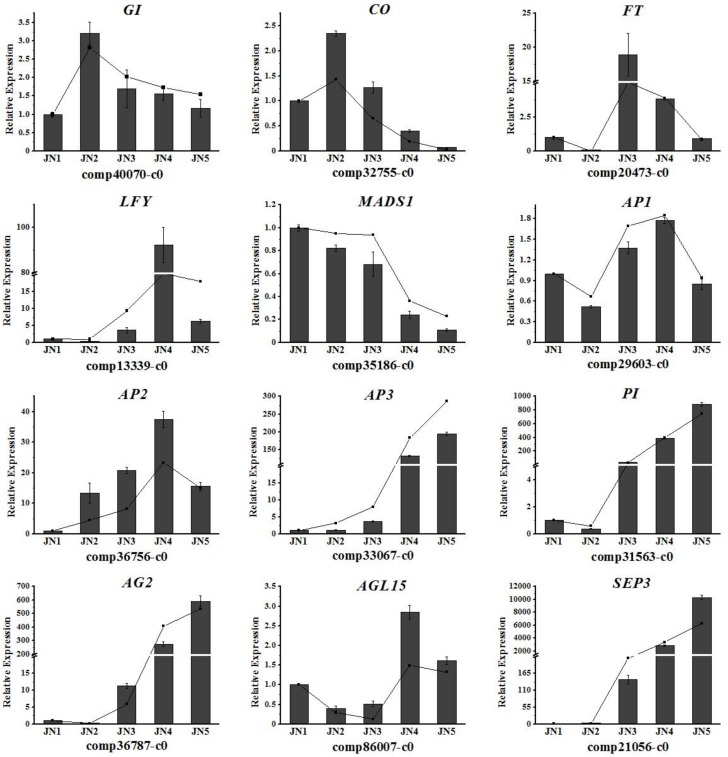
Twelve putative MADS-box family, AP2 family, and circadian rhythm pathway genes were selected from the DEGs: GI, CO, FT, LFY, AP1 (APETALA1), AP2, AP3, PI (PISTILLATA), MADS1, AG2 (AGAMOUS2), AGL15 (AGAMOUS-LIKE-15), and SEP3. The expression patterns of these 12 genes during flower bud differentiation were verified using qRT-PCR (Figure 7) and their expression trends were found to be similar to those obtained by RNA-seq, suggesting that the RNA-seq data reliably reflect the gene expression trends. According to the RNA-seq and qRT-PCR results, GI, CO, FT, and LFY expression increased before the onset of flower bud differentiation. AP1 and AP2 belong to the A-class genes and their expression levels increased during petal primordium differentiation and decreased gradually at the later stages. The expression levels of AP3, PI, AG2, AGL15, and SEP3 were significantly increased after petal primordium differentiation, which was consistent with the expression levels of B-, C-, and E-class genes. MADS1 expression decreased during flower bud differentiation, suggesting that low levels of MADS1 expression induce flower formation from the buds in the early stages of floral development.
Figure 7.
qRT-PCR analysis of the expression 12 DEGs. These 12 putative MADS-box family, AP2 family and circadian rhythm pathway genes were related to the formation of flower. The undifferentiated stage was used as a control group when calculating the relative expression quantity. Line were from the results of transcriptome data; column charts were from the results of qRT-PCR.
4. Discussion
Flower bud differentiation is the turning point from vegetative to reproductive growth, which occurs after the plant has had time to accumulate the required nutrients [43,44]. M. sinostellata buds take almost a month to develop and the paraffin section staining performed here showed that the flower bud primordia began to elongate and broaden as they differentiated. The floral tissues developed gradually and sequentially, from the outer sepals and petals to the inner stamens and pistils. The observed morphological changes were similar to those of Liriodendron tulipifera, Magnolia championii, Magnolia delavayi, Magnolia grandiflora, and Magnolia paenetalaum [20]. The flower bud differentiation of M. sinostellata could be divided into five stages; the undifferentiated stage, early flower bud differentiation, petal primordium differentiation, stamen primordium differentiation, and pistil primordium differentiation. These five stages are consistent with the flower bud differentiation stages observed in Ziziphus jujube [45], Prunus armeniaca [46], and Cerasus tianschanica [47].
In this study, transcriptome information was obtained by the high-throughput sequencing of flower buds at the five stages of flower bud differentiation. We identified 11,592 genes that were significantly differentially expressed during differentiation (FDR < 0.05). The numbers of genes specifically expressed in the undifferentiated, early flower bud differentiation and petal primordial differentiation stages were 1320, 1560, and 1854, respectively, while the numbers of genes specifically expressed in the stamen primordium differentiation and pistil primordium differentiation stages were only 275 and 560, respectively. This indicated many genes regulate flower bud differentiation and that the majority were involved in the early stages of floral development [48]. The GO annotations in the DEGs focused mainly on the regulation of biological processes, nucleus, and molecular function. The KEGG annotations showed that the DEGs were highly enriched in plant hormone signal transduction, indicated that many endogenous hormones were involved in the flowering of M. sinostellata, not just gibberellins at this work [49]. Many DEGs were related to pentose, glucuronate interconversions, amino sugar, and nucleotide sugar metabolism suggesting that sugar metabolism may be another important pathway to regulate the flowering of M. sinostellata [50]. In addition, DEGs were also enriched in circadian rhythm and flavonoid biosynthesis. These indicated that the plant received light signals which induced the transcriptional regulation of a variety of pathways and led to the accumulation of flavonoids in floral tissues [51].
The putative MYB, NAC, WRKY, and GATA transcription factors were differentially regulated during floral differentiation in M. sinostellata. These transcription factors are known to be involved in plant growth and development. The MYBs participate in plant secondary metabolism, responding to phenylalanine metabolism, signals from the external environment and phytohormones and play major roles in the flavonoid and pigment biosynthesis metabolic pathways [52,53]. The MYB transcription factors are also involved in the signal transduction of gibberellin, which regulates LFY expression to control flowering [54,55]. The NAC transcription factors are also involved in a variety of developmental and physiological processes and play important roles in hormone signal transduction, bud apical meristem formation, and the maintenance of flower development and morphology [56]. The WRKYs also play a role in the transduction of hormone signals during the growth and development of plants [57]. The GATA transcription factors have been shown to affect the regulation of floral development in response to the transmission of light signals [58,59]. The effects of these transcription factors are consistent with the changes in their expression observed during flower bud differentiation.
Some of the genes involved in the six major pathways regulating Arabidopsis flower formation were also found to be differentially expressed in flower bud differentiation in M. sinostellata. The red-light and blue-light receptor genes, PHY and CRY, were differentially expressed; CRY expression was decreased, while the expression of GI was increased, with later increases observed in the expression of CO, which promotes M. sinostellata flowering. LFY, which is expressed in the meristem, is considered to be the switch that determines flower development [60,61]. The expression of LFY was significantly altered during flower bud differentiation in M. sinostellata, indicating that LFY also plays a key role in the development of magnolia flowers. Our transcriptome sequencing results indicated that many different genes and pathways are involved in flower bud differentiation. FLC is a negative regulator of flowering involved in the vernalization and autonomous pathways [62]. FY and FPA act in the autonomous pathway to promote the expression of FT by inhibiting FLC, while in the vernalization pathway, FRI promotes FLC expression to inhibit flowering [41,63]. FLC was not found amongst the expressed genes of flower bud differentiation in M. sinostellata, but the expression levels of FY, FPA, FRI, and FT were all elevated, indicating that FY, FPA, and FRI may have other ways of promoting flowering. The expression of SPL, associated with the senescence pathway, increased during flower development, suggesting that this gene might also promote AP1 and LFY expression to induce flowering in M. sinostellata [64,65]. Studies have shown that gibberellin plays an important role in flower bud differentiation [65,66]. A total of 15 DEGs were found to be related to gibberellin-pathway genes; however, their expression did not follow any obvious patterns and their mechanism of action requires further study. These results suggest that flower bud differentiation in M. sinostellata is not the result of a single category of gene function, but rather the result of many genetic interactions.
The various pathways involved in flower induction eventually converge on the floral meristem, where the MADS-box and AP2 family transcription factors play a role in flower development [67] (Figure S8). According to our RNA-seq and qRT-PCR results, the expression levels of AP1, a flowering marker gene [68], began to increase when the petal primordium differentiated. The related gene AP2 is also an A-class gene and has a more pronounced flowering-development expression pattern. AP3 and PI are B-class genes and were found to have lower levels of expression prior to stamen differentiation, after which their expression increased dramatically [69]. The expression patterns of the C-class genes were found to contrast with those of the A-class genes, indicating the possible existence of negative feedback regulation between the two groups, which was previously reported in the classic ABC model [70].
5. Conclusions
In this study, we characterized the morphological changes of flower bud differentiation in M. sinostellata and divided the differentiation period into five distinct stages. The expression patterns of genes during these different stages were determined using the transcriptome sequencing of flower buds during their differentiation and those specifically involved in flower bud development were identified. The results of a qRT-PCR analysis enabled us to identify the transcriptional changes of genes involved in the differentiation of the different floral organs throughout flower development. These results represent a first step towards illuminating the molecular mechanisms of flower development in Magnolia sp. and provide abundant genomic resources and new candidate genes for the study of flowering regulation, as well as highlighting possible molecular breeding targets in this plant species.
Acknowledgments
This study was funded by the 13th Five-Year-Plan for Floriculture Special Breeding of Zhejiang Province, China (2016C02056-12), the National Natural Science Foundation of China (31400599), and the Public Welfare Forestry Industry Project of State Forestry Administration, China (no. 201504322).
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4425/9/4/212/s1, Table S1: Primers used to analyze the expression of the flower bud differentiation-related genes and the reference gene; Table S2: Analysis of internal reference genes expression stability and ranking by BestKeeper software. Table S3: Sequencing data quality preprocessing results; Table S4: Mapping rates of sequencing data assembly; Table S5: Sequencing data assembly results; Table S6: All genes in five stages; Table S7: GO enrichment of the DEGs; Table S8: KEGG pathway enrichment of the DEGs; Table S9: The 82 DEGs related to flowering-pathway genes in Arabidopsis sp.; Figure S1:Analysis of internal reference genes expression stability by geNorm software. The smaller the M value, the better the stability. Figure S2: The distribution of species used to annotate the transcripts; Figure S3: Pearson correlation between samples; Figure S4: MYB transcription factor expression levels during flower bud differentiation.; Figure S5: NAC transcription factor expression levels during flower bud differentiation.; Figure S6: WRKY transcription factor expression levels during flower bud differentiation.; Figure S7: GATA transcription factor expression levels during flower bud differentiation. Figure S8: The pathway of flowering in M. sinostellata. Se: Scales; Pe: Petal primordium; St: Stamen primordium; Pi: Pistil base; Yellow is B-class genes, Purple is A-class gene, and Red is C- and E-class genes.
Author Contributions
Lijie Fan contributed to the bioinformatics analysis and the writing of the manuscript. Yamei Shen conceived the study and its design. Bin Dong, Yaling Wang, and Shouzhou Zhang contributed to the revisions of the manuscript. Mengqian Chen and Ninghang Wang participated in the preparation of experimental materials. Qin Yu, Xingli Wang, and Lingjuan Xuan helped with the paraffin sectioning and the qRT-PCR. All the authors have read and approved the final manuscript.
Conflicts of Interest
The authors declare that they have no competing interests.
References
- 1.Zhang Y., Zhou Y., Chen Q., Huang X., Tian C. Molecular basis of flowering time regulation in Arabidopsis. Chin. Bull. Bot. 2014;49:469–482. [Google Scholar]
- 2.Mouradov A., Cremer F., Coupland G. Control of flowering time: Interacting pathways as a basis for diversity. Plant Cell. 2002;14:S111–S130. doi: 10.1105/tpc.001362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fornara F., de Montaigu A., Coupland G. Snapshot: Control of flowering in Arabidopsis. Cell. 2010;141:550. doi: 10.1016/j.cell.2010.04.024. [DOI] [PubMed] [Google Scholar]
- 4.Lee J., Lee I. Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 2010;61:2247–2254. doi: 10.1093/jxb/erq098. [DOI] [PubMed] [Google Scholar]
- 5.Nilsson O., Lee I., Blázquez M.A., Weigel D. Flowering-time genes modulate the response to leafy activity. Genetics. 1998;150:403–410. doi: 10.1093/genetics/150.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wagner D., Sablowski R.W., Meyerowitz E.M. Transcriptional activation of APETALA1 by LEAFY. Science. 1999;285:582–584. doi: 10.1126/science.285.5427.582. [DOI] [PubMed] [Google Scholar]
- 7.Mimida N., Kidou S.-I., Iwanami H., Moriya S., Abe K., Voogd C., Varkonyi-Gasic E., Kotoda N. Apple FLOWERING LOCUS T proteins interact with transcription factors implicated in cell growth and organ development. Tree Physiol. 2011;31:555–566. doi: 10.1093/treephys/tpr028. [DOI] [PubMed] [Google Scholar]
- 8.Winterhagen P., Tiyayon P., Samach A., Hegele M., Wünsche J.N. Isolation and characterization of fLOWERING LOCUS T subforms and APETALA1 of the subtropical fruit tree Dimocarpus longan. Plant Physiol. Biochem. 2013;71:184–190. doi: 10.1016/j.plaphy.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 9.Nishikawa F., Endo T., Shimada T., Fujii H., Shimizu T., Omura M., Ikoma Y. Increased CiFT abundance in the stem correlates with floral induction by low temperature in Satsuma mandarin (Citrus unshiu Marc.) J. Exp. Bot. 2007;58:3915–3927. doi: 10.1093/jxb/erm246. [DOI] [PubMed] [Google Scholar]
- 10.Hsu C.-Y., Adams J.P., Kim H., No K., Ma C., Strauss S.H., Drnevich J., Vandervelde L., Ellis J.D., Rice B.M. FLOWERING LOCUS T duplication coordinates reproductive and vegetative growth in perennial poplar. Proc. Natl. Acad. Sci. USA. 2011;108:10756–10761. doi: 10.1073/pnas.1104713108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coen E. Goethe and the ABC model of flower development. Comptes Rendus de l’Académie des Sciences-Series III-Sciences de la Vie. 2001;324:523–530. doi: 10.1016/S0764-4469(01)01321-X. [DOI] [PubMed] [Google Scholar]
- 12.Coen E.S., Meyerowitz E.M. The war of the whorls: Genetic interactions controlling flower development. Nature. 1991;353:31. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- 13.Litt A., Kramer E.M. The abc Model and the Diversification of Floral Organ Identity. Elsevier; New York, NY, USA: 2010. Seminars in cell & developmental biology; pp. 129–137. [DOI] [PubMed] [Google Scholar]
- 14.Angenent G.C., Franken J., Busscher M., van Dijken A., van Went J.L., Dons H., van Tunen A.J. A novel class of MADS box genes is involved in ovule development in petunia. Plant Cell. 1995;7:1569–1582. doi: 10.1105/tpc.7.10.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pelaz S., Ditta G.S., Baumann E., Wisman E., Yanofsky M.F. B and c floral organ identity functions require sepallata MADS-box genes. Nature. 2000;405:200. doi: 10.1038/35012103. [DOI] [PubMed] [Google Scholar]
- 16.Bowman J.L., Smyth D.R., Meyerowitz E.M. The ABC model of flower development: Then and now. Development. 2012;139:4095–4098. doi: 10.1242/dev.083972. [DOI] [PubMed] [Google Scholar]
- 17.Theißen G. Development of floral organ identity: Stories from the MADS house. Curr. Opin. Plant Biol. 2001;4:75–85. doi: 10.1016/S1369-5266(00)00139-4. [DOI] [PubMed] [Google Scholar]
- 18.Kim S., Koh J., Yoo M.J., Kong H., Hu Y., Ma H., Soltis P.S., Soltis D.E. Expression of floral MADS-box genes in basal angiosperms: Implications for the evolution of floral regulators. Plant J. 2005;43:724–744. doi: 10.1111/j.1365-313X.2005.02487.x. [DOI] [PubMed] [Google Scholar]
- 19.Liu W.-Z., Hilu K., Wang Y.-L. From leaf and branch into a flower: Magnolia tells the story. Bot. Stud. 2014;55:28. doi: 10.1186/1999-3110-55-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu F., Rudall P. Comparative floral anatomy and ontogeny in magnoliaceae. Plant Syst. Evol. 2006;258:1–15. doi: 10.1007/s00606-005-0361-1. [DOI] [Google Scholar]
- 21.Wu Z., Hu J., Si H., Tang J., Sun J., Hu Y. An observation on differentiation of Magnolia denudata Desr. flower buds. China J. Chin. Mater. Med. 1991;2:4. [PubMed] [Google Scholar]
- 22.Jing D., Liu Z., Zhang B., Ma J., Han Y., Chen F. Two ancestral APETALA3 homologs from the basal angiosperm Magnolia wufengensis (Magnoliaceae) can affect flower development of Arabidopsis. Gene. 2014;537:100–107. doi: 10.1016/j.gene.2013.11.076. [DOI] [PubMed] [Google Scholar]
- 23.Wu W., Chen F., Jing D., Liu Z., Ma L. Isolation and characterization of an AGAMOUS-like gene from Magnolia wufengensis (Magnoliaceae) Plant Mol. Biol. Rep. 2012;30:690–698. doi: 10.1007/s11105-011-0385-3. [DOI] [Google Scholar]
- 24.Shi S.-G., Yang M., Zhang M., Wang P., Kang Y.-X., Liu J.-J. Genome-wide transcriptome analysis of genes involved in flavonoid biosynthesis between red and white strains of Magnolia sprengeri pamp. BMC Genom. 2014;15:706. doi: 10.1186/1471-2164-15-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dhandapani S., Jin J., Sridhar V., Sarojam R., Chua N.-H., Jang I.-C. Integrated metabolome and transcriptome analysis of Magnolia champaca identifies biosynthetic pathways for floral volatile organic compounds. BMC Genom. 2017;18:463. doi: 10.1186/s12864-017-3846-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang S.-Z. Magnolia sinostellata and relatives (Magnoliaceae) Phytotaxa. 2013;154:47–58. [Google Scholar]
- 27.Xia N.H., Liu Y.H., Nooteboom H.P. Flora of China. Science Press; Beijing, China: Missouri Botanical Garden Press; St. Louis, MO, USA: 2008. Magnoliaceae; pp. 47–91. [Google Scholar]
- 28.Fan L., Chen M., Wang N., Shen Y., Zhang S. Flower bud differention of three species of magnolia. J. Northeast For. Univ. 2018;46:27–30. [Google Scholar]
- 29.Xu Y.C., Zhou L.H., Hu S.Q., Hao R.M., Huang C.j., Zhao H.-B. The differentiation and development of pistils of hermaphrodites and pistillodes of males in androdioecious Osmanthus fragrans L. and implications for the evolution to androdioecy. Plant Syst. Evol. 2014;300:843–849. doi: 10.1007/s00606-013-0923-6. [DOI] [Google Scholar]
- 30.Andrews S. Fastqc: A Quality Control Tool for High Throughput Sequence Data. Babraham Institute; Cambridge, UK: 2010. [Google Scholar]
- 31.Haas B.J., Papanicolaou A., Yassour M., Grabherr M., Blood P.D., Bowden J., Couger M.B., Eccles D., Li B., Lieber M. De novo transcript sequence reconstruction from rna-seq using the trinity platform for reference generation and analysis. Nat. Protocols. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 33.Srikanth A., Schmid M. Regulation of flowering time: All roads lead to Rome. Cell. Mol. Life Sci. 2011;68:2013–2037. doi: 10.1007/s00018-011-0673-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔct method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Boeckmann B., Bairoch A., Apweiler R., Blatter M.C., Estreicher A., Gasteiger E., Martin M.J., Michoud K., O’donovan C., Phan I. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li W., Godzik A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 37.Kanehisa M., Goto S. Kegg: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krylov D.M., Wolf Y.I., Rogozin I.B., Koonin E.V. Gene loss, protein sequence divergence, gene dispensability, expression level and interactivity are correlated in eukaryotic evolution. Genome Res. 2003;13:2229–2235. doi: 10.1101/gr.1589103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen C., Zhang Y., Xu Z., Luan A., Mao Q., Feng J., Xie T., Gong X., Wang X., Chen H. Transcriptome profiling of the pineapple under low temperature to facilitate its breeding for cold tolerance. PLoS ONE. 2016;11:e0163315. doi: 10.1371/journal.pone.0163315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y., Xiu Y., Bi K., Ou J., Gu W., Wang W., Meng Q. Integrated analysis of mrna-seq in the haemocytes of eriocheir sinensis in response to spiroplasma eriocheiris infection. Fish Shellfish Immunol. 2017;68:289–298. doi: 10.1016/j.fsi.2017.07.036. [DOI] [PubMed] [Google Scholar]
- 41.Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L. The pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Consortium G.O. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peng F., Tian M., Wang C. Morphological and anatomical characteristics along with nutrient content changes during flower bud differentiation in oncidium. J. Zhejiang Agric. Univ. 2012;29:7–11. [Google Scholar]
- 44.Pang F., Zhao M., Wang Y., Yu H., Xia J. Studies on floral bud differentiation and biochemical changes of strawberry ‘ningyu’. J. Fruit Sci. 2014;31:1117–1122. [Google Scholar]
- 45.Niu H., Zhang H., Yuan B. Flower formation and endogenous hormones dynamic in chinese jujube. Acta Hortic. Sin. 2015;42:655–664. [Google Scholar]
- 46.Liu L.Q., Qin W., Liao K., He F.-J., Zhang D.-H., Xu L., Fan W.-M. Study on the floral morphodifferentiation of some apricot cultivars in Xinjiang. Xinjiang Agric. Sci. 2007;5:004. [Google Scholar]
- 47.Chao W., Long Z., Ni P., Xin Z. Morphological differentiation of flower buds of Cerasus tianschanica Pojark. Acta Agric. Boreali-Occident. Sin. 2015;5:20. [Google Scholar]
- 48.Liu H., Sun M., Du D., Pan H., Cheng T., Wang J., Zhang Q. Whole-transcriptome analysis of differentially expressed genes in the vegetative buds, floral buds and buds of Chrysanthemum morifolium. PLoS ONE. 2015;10:e0128009. doi: 10.1371/journal.pone.0128009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamaguchi N., Wu M.F., Winter C.M., Berns M.C., Nole-Wilson S., Yamaguchi A., Coupland G., Krizek B.A., Wagner D. A molecular framework for auxin-mediated initiation of flower primordia. Dev. Cell. 2013;24:271. doi: 10.1016/j.devcel.2012.12.017. [DOI] [PubMed] [Google Scholar]
- 50.Matsoukas I.G. Interplay between sugar and hormone signaling pathways modulate floral signal transduction. Front. Genet. 2014;5:218. doi: 10.3389/fgene.2014.00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Apel L., Kammerer D.R., Stintzing F.C., Spring O. Comparative metabolite profiling of triterpenoid saponins and flavonoids in flower color mutations of Primula veris L. Int. J. Mol. Sci. 2017;18:153. doi: 10.3390/ijms18010153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Q., Tang H., Dong X., Hou Y., Luo Y., Jiang Y., Huang Q. Progress in the study of plant MYB transcription factors. Genom. Appl. Biol. 2009;28:365–372. [Google Scholar]
- 53.Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15:573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 54.Palatnik J.F., Allen E., Wu X., Schommer C., Schwab R., Carrington J.C., Weigel D. Control of leaf morphogenesis by microRNAs. Nature. 2003;425:257. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- 55.Park W., Li J., Song R., Messing J., Chen X. CARPEL factory, a dicer homolog and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 2002;12:1484–1495. doi: 10.1016/S0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen N., Jiang J., Cao B., Lei J., Chen C. The latest progresses on plant NAC transcription factors function. Mol. Plant Breed. 2015;13:1407–1414. [Google Scholar]
- 57.Rushton P.J., Somssich I.E., Ringler P., Shen Q.J. WRKY transcription factors. Trends Plant Sci. 2010;15:247–258. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 58.Hayama R., Coupland G. Shedding light on the circadian clock and the photoperiodic control of flowering. Curr. Opin. Plant Biol. 2003;6:13–19. doi: 10.1016/S1369-5266(02)00011-0. [DOI] [PubMed] [Google Scholar]
- 59.Reyes J.C., Muro-Pastor M.I., Florencio F.J. The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol. 2004;134:1718–1732. doi: 10.1104/pp.103.037788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wagner D., Meyerowitz E.M. Switching on flowers: Transient LEAFY induction reveals novel aspects of the regulation of reproductive development in Arabidopsis. Front. Plant Sci. 2011;2:60. doi: 10.3389/fpls.2011.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y., Ballard H.E., Sternberger A.L., Wyatt S.E., Stockinger E.J., Nadella V. The potential role of two LEAFY orthologs in the chasmogamous/cleistogamous mixed breeding system of Viola pubescens (Violaceae) J. Torrey Bot. Soc. 2017;144:206–217. doi: 10.3159/TORREY-D-16-00023R2. [DOI] [Google Scholar]
- 62.Caicedo A.L., Stinchcombe J.R., Olsen K.M., Schmitt J., Purugganan M.D. Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc. Natl. Acad. Sci. USA. 2004;101:15670–15675. doi: 10.1073/pnas.0406232101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He Y. Control of the transition to flowering by chromatin modifications. Mol. Plant. 2009;2:554–564. doi: 10.1093/mp/ssp005. [DOI] [PubMed] [Google Scholar]
- 64.Wang J.W., Czech B., Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138:738–749. doi: 10.1016/j.cell.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 65.Yamaguchi A., Wu M.F., Yang L., Wu G., Poethig R.S., Wagner D. The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL and APETALA1. Dev. Cell. 2009;17:268–278. doi: 10.1016/j.devcel.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Galvão V.C., Horrer D., Küttner F., Schmid M. Spatial control of flowering by DELLA proteins in Arabidopsis thaliana. Development. 2012;139:4072–4082. doi: 10.1242/dev.080879. [DOI] [PubMed] [Google Scholar]
- 67.Pařenicová L., de Folter S., Kieffer M., Horner D.S., Favalli C., Busscher J., Cook H.E., Ingram R.M., Kater M.M., Davies B. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis. Plant Cell Online. 2003;15:1538–1551. doi: 10.1105/tpc.011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jaya E.S., Clemens J., Song J., Zhang H., Jameson P.E. Quantitative expression analysis of meristem identity genes in Eucalyptus occidentalis: AP1 is an expression marker for flowering. Tree Physiol. 2009;30:304–312. doi: 10.1093/treephys/tpp117. [DOI] [PubMed] [Google Scholar]
- 69.Wu Y.Q., Li Z.Y., Zhao D.Q., Tao J. Comparative analysis of flower-meristem-identity gene APETALA2 (AP2) codon in different plant species. J. Integr. Agric. 2017;6:60345–60347. doi: 10.1016/S2095-3119(17)61732-5. [DOI] [Google Scholar]
- 70.Kim S., Yoo M.-J., Albert V.A., Farris J.S., Soltis P.S., Soltis D.E. Phylogeny and diversification of b-function MADS-box genes in angiosperms: Evolutionary and functional implications of a 260-million-year-old duplication. Am. J. Bot. 2004;91:2102–2118. doi: 10.3732/ajb.91.12.2102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.