Inherited retinal diseases (IRDs) are genetically and clinically heterogeneous disorders. Together, they have an estimated incidence of 1:2000 and thereby are the leading cause of vision loss in persons between 15 and 45 years of age [1,2,3]. IRDs can be clinically classified based on disease progression and the retinal cell types that are primarily involved in disease pathogenesis. They can be stationary, as for instance observed in most cases with congenital stationary night blindness (CSNB) and achromatopsia (ACHM), or progressive, such as in retinitis pigmentosa (RP), which is basically a rod-cone dystrophy, but also in cone-rod dystrophy (CRD) and Stargardt disease (STGD1). A second classification is based on the primary dysfunction or degeneration of the rod or cone photoreceptor cells. We can distinguish CSNB, which represents a dysfunction of retinal signaling from rods and cones to bipolar cells [4], and ACHM, or color blindness [5], in which one or more of the three types of cone cells are dysfunctional. In persons with CRD and STGD1, cones are affected first, followed by the degeneration of rods. This means that affected individuals initially experience central vision defects, which expand towards the mid-periphery. In persons with RP, this is the converse: initial clinical symptoms are night blindness and tunnel vision due to rod degeneration; central vision can also become impaired with progressive disease when cones also degenerate, eventually leading to legal blindness. Late in the disease process, both cones and rods are affected in CRD, RP, and STGD1, which make it difficult to come to a clear diagnosis. The most severe form of IRD is Leber congenital amaurosis (LCA), in which not only cones and rods of the neural retina can be affected simultaneously, but in which the retinal pigment epithelium (RPE) can also be primarily involved [6,7].
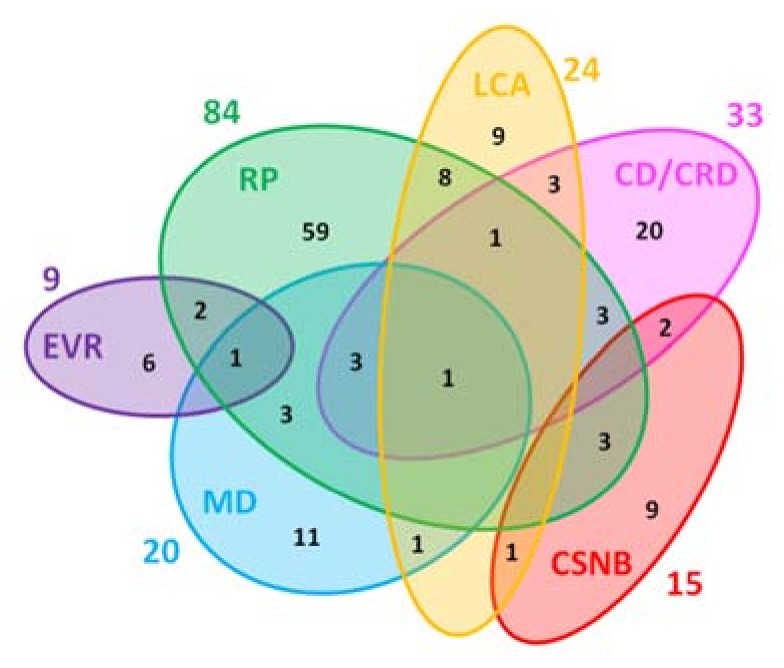
A few phenotypes, such as choroideremia (CHM) and STGD1, are caused by mutations in single genes, CHM and ABCA4, respectively [8,9]. In most of the IRDs, mutations in many different genes can cause very similar phenotypes. For example, mutations in 84 different genes underlie RP [10], 33 genes are implicated in cone dystrophy (CD)/CRD, 20 genes are involved in macular dystrophies (MD), 15 genes are involved in CSNB, and 9 genes are mutated in familial exudative vitreoretinopathy (FEVR) (Figure 1) [11].
Figure 1.
Genetic heterogeneity among the six major non-syndromic inherited retinal diseases (IRDs). Numbers outside of the ellipses correspond to the number of non-syndromic IRD genes responsible for the specific disease, while numbers within the ellipses correspond either to disease-specific genes or to genes mutated in two or more diseases. The non-redundant total of genes associated with these non-syndromic IRDs is 146. RP: retinitis pigmentosa; LCA: Leber congenital amaurosis; CD/CRD: cone dystrophy/cone-rod dystrophy; CSNB: congenital stationary night blindness; MD: macular dystrophy; EVR: exudative vitreoretinopathy.
Different variants in one gene can cause autosomal recessive (ar) or dominant (ad) retinal dystrophies (RDs), as exemplified by GUCY2D in which ad variants result in CRD and ar variants cause LCA [12,13]. Similarly, variants in rhodopsin (RHO) and RP1 can be involved inadRP and arRP [14,15,16,17]. Although many arRDs are caused by mutations that result in the absence of functional protein, there are also several examples in which there is residual protein activity. Different combinations of mutations in some genes thereby can be associated with IRDs with different severity. For example, two null alleles in ABCA4 result in early-onset CRD, whereas combinations of severe and mild variants result in intermediate or late-onset STGD1 [9,18,19,20,21]. Different combinations of mutations in the same gene can also cause syndromic and non-syndromic forms of arRDs. Some USH2A mutations either cause non-syndromic arRP or Usher syndrome type 2 [22,23]. Mutations in Bardet–Biedl syndrome (BBS)-associated genes, such as BBS1, can also be found in non-syndromic arRP [24,25,26], and CEP290 variants underlie LCA, Senior–Løken syndrome, Joubert syndrome, or Meckel–Gruber syndrome [27,28,29,30]. Finally, bi-allelic null mutations in some IRD-associated genes may be lethal. The far majority of LCA cases carrying NMNAT1 variants carry one hypomorphic variant and one null allele [31,32,33,34,35], and it was hypothesized that two NMNAT1 null alleles could be lethal [35] or are associated with syndromic IRD.
The first retinal disease-associated gene identified was the ornithine aminotransferase (OAT) gene involved in gyrate atrophy. Reduced ornithine aminotransferase activity was measured in a patient’s cells in 1977 [36], and in 1988, the OAT gene was cloned, and the first mutation was identified [37]. Two years later, the second and third IRD-associated genes were identified. Mutations in the RHO gene, encoding the rod-specific light-sensitive chromophore, were identified in patients with adRP using a candidate gene approach [14] after linkage analysis in a large Irish adRP family had pointed towards a genomic region encompassing this gene [38]. In the same year, the CHM gene was identified using a positional cloning approach by mapping deletions in patients with syndromic and non-syndromic choroideremia [8].
The candidate gene approach (i.e., the search for IRD-associated variants in genes encoding proteins with known crucial functions in the retina) has been very successful. Similarly, comparing phenotypes of existing animal models with a known gene defect and subsequent screening of the respective candidate gene has identified many genes underlying IRD [4]. The identification of IRD-associated genes through their genomic position (i.e., positional cloning) as determined by linkage analysis has been used effectively, though this generally requires the availability of large families or a large set of families in which the same locus is involved. Linkage studies can be performed using microarrays that test thousands of single nucleotide polymorphisms (SNPs) spread across the genome. SNP microarrays have also proven very valuable for homozygosity and identity-by-descent (IBD) mapping of recessive disease genes, not only in consanguineous families [27,39], but also in small families and single patients of non-consanguineous marriages [40,41]. We are witnessing a new era in disease gene identification with the introduction of next-generation sequencing, allowing the analysis of all genes implicated in IRD [42] in a defined linkage interval, all exons in the genome (whole exome sequencing (WES)) [43,44,45,46], or even the entire genomic sequence (whole genome sequencing (WGS)) [47]. This also brings new challenges, such as data analysis and interpretation of genomic variants. Given the huge number of variants present in a patient’s genome, positional information on where the causative gene may be localized (e.g., by linkage analysis and/or homozygosity mapping) remains very helpful to pinpoint the genetic defect. Employing WGS, thousands of rare single nucleotide variants (SNVs) and structural variations (SVs) are found in every individual, and it remains very challenging to identify the causal variant(s). A functional read-out is required to identify the culprit variant(s). Gene-specific mRNA analysis or genome-wide mRNA analysis (transcriptome analysis) may identify quantitative or structural defects in mRNAs.
Whole exome sequencing and gene-panel sequencing analysis genetically solve 55–60% of these cases [42,48,49,50]. The hidden genetic variations may be unrecognized SVs and deep-intronic variations, which can be identified by WGS or gene-specific locus sequencing. Copy number variations (CNV)s can explain up to 18% of previously unsolved cases [51,52].
Receiving a molecular diagnosis becomes increasingly important with the development of (gene) therapy for IRDs. Up to 10 years ago, it was not possible to slow down, stabilize, or treat the vision impairment in patients with IRDs. This changed for a small group of patients with RPE65 mutations, as gene augmentation was successfully and safely applied through subretinal injections of recombinant adeno-associated viruses (rAAVs) in Phase 1/2 trials [53,54,55]. Recombinant adeno-associated viruses transduce the RPE cells, upon which the viruses are shuttled to the nucleus, and the rAAV vector remains a stable extrachromosomal element. In the meantime, many more patients have been treated in two centers in Philadelphia and one in London. Vision improvement was variable and, in general, modest and appears to be more effective in younger patients. A Phase 3 trial was conducted in one center using an improved rAAV vector, which resulted in increased subjective and objective vision in the treated eye versus the untreated eye [56]. Gene therapy for RPE65—associated with LCA or RP—in the form of Luxturna is now an approved treatment in the United States (U.S. Food and Drug Administration STN: 125610). Gene augmentation targeting photoreceptors and the RPE was also successfully performed in a Phase 1/2 trial in choroideremia patients [57,58]. In addition, an oral 9-cis retinoid supplementation therapy seems effective in patients with RPE65 and LRAT mutations [59,60]. Several therapies that will be developed in the next years will be gene-, or even mutation-specific, emphasizing the importance for patients to receive a molecular diagnosis. An overview of all ongoing gene therapy trials can be found on the internet [61].
Proving the involvement of a gene defect in an IRD and often a definite genetic diagnosis are dependent on modeling of the identified mutation(s) in animals or in ex vivo assays. Defects in several genes were previously found as naturally occurring or were modeled in rodents, zebrafish, and Drosophila [62] (and references therein). Human mutations have also been modeled by over-expression of wild-type and mutant proteins with a presumed dominant effect in cell culture [63]. More sophisticated approaches have been developed to study potential splicing alterations or hypomorphic alleles [64,65]. Some assays can be performed directly in cells available from patients [27]. However, in the absence of patients’ somatic cells that express the gene of interest, robust in vitro RNA splice assays can be set up for every human gene. In case retina-specific splice defects could play a role, photoreceptor precursor cells can be derived from induced pluripotent stem cells generated from blood cells or fibroblasts [66,67,68,69,70,71]. In addition to the new functional assays, the recent entrance of CRISPR/Cas opens great opportunities for IRD research, as it enables a more efficient introduction of specific mutations into animal models or cells [72]. This technology also offers a new therapeutic potential [73].
Only a few examples of digenic inheritance and modifier genes for IRDs have been reported [74,75,76,77,78]. Nevertheless, there are many examples of significant differences between phenotypes (e.g., age at onset) in IRD cases that carry the same mutation(s), both within and between families. Reduced penetrance of variants might explain several autosomal dominant conditions, but as yet we have few clues regarding the genetic and possibly non-genetic modifiers. To study the mechanism of variable expression and non-penetrance, large case/control cohorts and genome-wide analysis techniques, such as WES and WGS, are required.
What are the future challenges in IRD research and diagnostics?
How can we determine causality for ultra-rare mutations in novel candidate genes? Apart from in silico tools that predict the potential causality of rare variants, we need to share our findings in a global manner. Tools for this are GeneMatcher [79,80] and intense collaborations, such as the European Retinal Disease Consortium (ERDC) [81].
How can we identify and functionally test non-coding variants?
To fully understand genotype–phenotype correlations, what are the effects of coding variants on RNA splicing and protein function?
Can we begin to understand phenotypic differences (and non-penetrance) of persons with the same mutations or the same types of mutations due to cis and trans modifiers or digenic inheritance?
With this special issue of the journal Genes, we address all the challenges mentioned above, except for the identification and functional testing of non-coding variants. In the paper by Astuti et al. [82] several probands carrying ultra-rare defects are presented in 11 novel candidate IRD genes through a European collaboration. The high aggregate carrier frequency of autosomal recessive variants associated with retinal dystrophies (up to 15%) [83] in some families with multiple affected individuals can result in the identification of independently acting defects in different genes, as shown by Gustafson et al. [84]. Llavona et al. [85] report on allelic mRNA imbalances for selected IRD-associated genes, which is very relevant to understanding phenotypic differences between individuals carrying the same genetic defects. El Shamieh et al. [86] report on additional RP-associated mutations in KIZ, encoding a ciliary protein, and the need to establish retina organoids from patient-derived iPS cells to understand the effect of these mutations on ciliary structure. The remaining nine manuscripts deal with genotype–phenotype correlations. They range from very large genotyping studies (e.g., the Target5000 study by Dockery et al. [87]) to targeted genotyping studies in pericentral RP (Comander et al. [88]) and early-onset RP and LCA families (Di Iorio et al. [89] and Porto et al. [90]). Brandl et al. [91] studied two genes encoding homologous proteins (IMPG1 and IMPG2) that are mutated in vitelliform macular dystrophies. Gene-specific studies were reported by McGuigan et al. (EYS—arRP) [92], Roosing et al. (CEP290—oligocone trichromacy) [93], Tracewska-Siemiątkowska et al. (YARS—RP, deafness, agenesis of the corpus callosum, and liver disease) [94], and Littink et al. (NRL—enhanced S-cone syndrome) [95].
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Rattner A., Sun H., Nathans J. Molecular genetics of human retinal disease. Annu. Rev. Genet. 1999;33:89–131. doi: 10.1146/annurev.genet.33.1.89. [DOI] [PubMed] [Google Scholar]
- 2.Hamel C. Retinitis pigmentosa. Orphanet J. Rare Dis. 2006;1:40. doi: 10.1186/1750-1172-1-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krumpaszky H.G., Ludtke R., Mickler A., Klauss V., Selbmann H.K. Blindness incidence in Germany. A population-based study from Württemberg-Hohenzollern. Ophthalmologica. 1999;213:176–182. doi: 10.1159/000027415. [DOI] [PubMed] [Google Scholar]
- 4.Zeitz C., Robson A.G., Audo I. Congenital stationary night blindness: An analysis and update of genotype-phenotype correlations and pathogenic mechanisms. Prog. Retin. Eye Res. 2015;45:58–110. doi: 10.1016/j.preteyeres.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Hofmann L., Palczewski K. Advances in understanding the molecular basis of the first steps in color vision. Prog. Retin. Eye Res. 2015;49:46–66. doi: 10.1016/j.preteyeres.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Den Hollander A.I., Black A., Bennett J., Cremers F.P. Lighting a candle in the dark: Advances in genetics and gene therapy of recessive retinal dystrophies. J. Clin. Investig. 2010;120:3042–3053. doi: 10.1172/JCI42258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roosing S., Thiadens A.A., Hoyng C.B., Klaver C.C., den Hollander A.I., Cremers F.P. Causes and consequences of inherited cone disorders. Prog. Retin. Eye Res. 2014;42:1–26. doi: 10.1016/j.preteyeres.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Cremers F.P., van de Pol D.J., van Kerkhoff L.P., Wieringa B., Ropers H.H. Cloning of a gene that is rearranged in patients with choroideraemia. Nature. 1990;347:674–677. doi: 10.1038/347674a0. [DOI] [PubMed] [Google Scholar]
- 9.Allikmets R., Singh N., Sun H., Shroyer N.F., Hutchinson A., Chidambaram A., Gerrard B., Baird L., Stauffer D., Peiffer A., et al. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive stargardt macular dystrophy. Nat. Genet. 1997;15:236–246. doi: 10.1038/ng0397-236. [DOI] [PubMed] [Google Scholar]
- 10.Verbakel S.K., van Huet R.A.C., Boon C.J.F., den Hollander A.I., Collin R.W.J., Klaver C.C.W., Hoyng C.B., Roepman R., Klevering B.J. Non-syndromic retinitis pigmentosa. Prog. Retin. Eye Res. 2018 doi: 10.1016/j.preteyeres.2018.03.005. in press. [DOI] [PubMed] [Google Scholar]
- 11.RetNet. [(accessed on 17 March 2018)]; Available online: http://www.sph.uth.tmc.edu/RetNet/
- 12.Kelsell R.E., Gregory-Evans K., Payne A.M., Perrault I., Kaplan J., Yang R.B., Garbers D.L., Bird A.C., Moore A.T., Hunt D.M. Mutations in the retinal guanylate cyclase (RETGC-1) gene in dominant cone-rod dystrophy. Hum. Mol. Genet. 1998;7:1179–1184. doi: 10.1093/hmg/7.7.1179. [DOI] [PubMed] [Google Scholar]
- 13.Perrault I., Rozet J.M., Calvas P., Gerber S., Camuzat A., Dollfus H., Chatelin S., Souied E., Ghazi I., Leowski C., et al. Retinal-specific guanylate cyclase gene mutations in Leber’s congenital amaurosis. Nat. Genet. 1996;14:461–464. doi: 10.1038/ng1296-461. [DOI] [PubMed] [Google Scholar]
- 14.Dryja T.P., McGee T.L., Reichel E., Hahn L.B., Cowley G.S., Yandell D.W., Sandberg M.A., Berson E.L. A point mutation of the rhodopsin gene in one form of retinitis pigmentosa. Nature. 1990;343:364–366. doi: 10.1038/343364a0. [DOI] [PubMed] [Google Scholar]
- 15.Rosenfeld P.J., Cowley G.S., McGee T.L., Sandberg M.A., Berson E.L., Dryja T.P. A null mutation in the rhodopsin gene causes rod photoreceptor dysfunction and autosomal recessive retinitis pigmentosa. Nat. Genet. 1992;1:209–213. doi: 10.1038/ng0692-209. [DOI] [PubMed] [Google Scholar]
- 16.Sullivan L.S., Heckenlively J.R., Bowne S.J., Zuo J., Hide W.A., Gal A., Denton M., Inglehearn C.F., Blanton S.H., Daiger S.P. Mutations in a novel retina-specific gene cause autosomal dominant retinitis pigmentosa. Nat. Genet. 1999;22:255–259. doi: 10.1038/10314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khaliq S., Abid A., Ismail M., Hameed A., Mohyuddin A., Lall P., Aziz A., Anwar K., Mehdi S.Q. Novel association of RP1 gene mutations with autosomal recessive retinitis pigmentosa. J. Med. Genet. 2005;42:436–438. doi: 10.1136/jmg.2004.024281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cremers F.P., van de Pol D.J., van Driel M., den Hollander A.I., van Haren F.J., Knoers N.V., Tijmes N., Bergen A.A., Rohrschneider K., Blankenagel A., et al. Autosomal recessive retinitis pigmentosa and cone-rod dystrophy caused by splice site mutations in the Stargardt’s disease gene ABCR. Hum. Mol. Genet. 1998;7:355–362. doi: 10.1093/hmg/7.3.355. [DOI] [PubMed] [Google Scholar]
- 19.Maugeri A., Klevering B.J., Rohrschneider K., Blankenagel A., Brunner H.G., Deutman A.F., Hoyng C.B., Cremers F.P.M. Mutations in the ABCA4 (ABCR) gene are the major cause of autosomal recessive cone-rod dystrophy. Am. J. Hum. Genet. 2000;67:960–966. doi: 10.1086/303079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Westeneng-van Haaften S.C., Boon C.J., Cremers F.P., Hoefsloot L.H., den Hollander A.I., Hoyng C.B. Clinical and genetic characteristics of late-onset Stargardt’s disease. Ophthalmology. 2012;119:1199–1210. doi: 10.1016/j.ophtha.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 21.Zernant J., Lee W., Collison F.T., Fishman G.A., Sergeev Y.V., Schuerch K., Sparrow J.R., Tsang S.H., Allikmets R. Frequent hypomorphic alleles account for a significant fraction of ABCA4 disease and distinguish it from age-related macular degeneration. J. Med. Genet. 2017;54:404–412. doi: 10.1136/jmedgenet-2017-104540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eudy J.D., Weston M.D., Yao S., Hoover D.M., Rehm H.L., Ma-Edmonds M., Yan D., Ahmad I., Cheng J.J., Ayuso C., et al. Mutation of a gene encoding a protein with extracellular matrix motifs in usher syndrome type IIa. Science (N. Y.) 1998;280:1753–1757. doi: 10.1126/science.280.5370.1753. [DOI] [PubMed] [Google Scholar]
- 23.Rivolta C., Sweklo E.A., Berson E.L., Dryja T.P. Missense mutation in the USH2A gene: Association with recessive retinitis pigmentosa without hearing loss. Am. J. Hum. Genet. 2000;66:1975–1978. doi: 10.1086/302926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Estrada-Cuzcano A.I., Koenekoop R.K., Senechal A., De Baere E.B., de Ravel T., Banfi S., Kohl S., Ayuso C., Sharon D., Hoyng C.B., et al. BBS1 mutations in a wide spectrum of phenotypes ranging from nonsyndromic retinitis pigmentosa to bardet-biedl syndrome. Arch. Ophthalmol. (Chic. IL 1960) 2012;130:1425–1432. doi: 10.1001/archophthalmol.2012.2434. [DOI] [PubMed] [Google Scholar]
- 25.Mykytyn K., Nishimura D.Y., Searby C.C., Shastri M., Yen H.J., Beck J.S., Braun T., Streb L.M., Cornier A.S., Cox G.F., et al. Identification of the gene (BBS1) most commonly involved in bardet-biedl syndrome, a complex human obesity syndrome. Nat. Genet. 2002;31:435–438. doi: 10.1038/ng935. [DOI] [PubMed] [Google Scholar]
- 26.Katsanis N., Ansley S.J., Badano J.L., Eichers E.R., Lewis R.A., Hoskins B.E., Scambler P.J., Davidson W.S., Beales P.L., Lupski J.R. Triallelic inheritance in bardet-biedl syndrome, a mendelian recessive disorder. Science ( N. Y.) 2001;293:2256–2259. doi: 10.1126/science.1063525. [DOI] [PubMed] [Google Scholar]
- 27.Den Hollander A.I., Koenekoop R.K., Yzer S., Lopez I., Arends M.L., Voesenek K.E., Zonneveld M.N., Strom T.M., Meitinger T., Brunner H.G., et al. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am. J. Hum. Genet. 2006;79:556–561. doi: 10.1086/507318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sayer J.A., Otto E.A., O’Toole J.F., Nurnberg G., Kennedy M.A., Becker C., Hennies H.C., Helou J., Attanasio M., Fausett B.V., et al. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat. Genet. 2006;38:674–681. doi: 10.1038/ng1786. [DOI] [PubMed] [Google Scholar]
- 29.Valente E.M., Silhavy J.L., Brancati F., Barrano G., Krishnaswami S.R., Castori M., Lancaster M.A., Boltshauser E., Boccone L., Al-Gazali L., et al. Mutations in CEP290, which encodes a centrosomal protein, cause pleiotropic forms of Joubert syndrome. Nat. Genet. 2006;38:623–625. doi: 10.1038/ng1805. [DOI] [PubMed] [Google Scholar]
- 30.Frank V., den Hollander A.I., Bruchle N.O., Zonneveld M.N., Nurnberg G., Becker C., Du Bois G., Kendziorra H., Roosing S., Senderek J., et al. Mutations of the CEP290 gene encoding a centrosomal protein cause Meckel-Gruber syndrome. Hum. Mutat. 2008;29:45–52. doi: 10.1002/humu.20614. [DOI] [PubMed] [Google Scholar]
- 31.Chiang P.W., Wang J., Chen Y., Fu Q., Zhong J., Chen Y., Yi X., Wu R., Gan H., Shi Y., et al. Exome sequencing identifies NMNAT1 mutations as a cause of Leber congenital amaurosis. Nat. Genet. 2012;44:972–974. doi: 10.1038/ng.2370. [DOI] [PubMed] [Google Scholar]
- 32.Koenekoop R.K., Wang H., Majewski J., Wang X., Lopez I., Ren H., Chen Y., Li Y., Fishman G.A., Genead M., et al. Mutations in NMNAT1 cause Leber congenital amaurosis and identify a new disease pathway for retinal degeneration. Nat. Genet. 2012;44:1035–1039. doi: 10.1038/ng.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perrault I., Hanein S., Zanlonghi X., Serre V., Nicouleau M., Defoort-Delhemmes S., Delphin N., Fares-Taie L., Gerber S., Xerri O., et al. Mutations in NMNAT1 cause Leber congenital amaurosis with early-onset severe macular and optic atrophy. Nat. Genet. 2012;44:975–977. doi: 10.1038/ng.2357. [DOI] [PubMed] [Google Scholar]
- 34.Falk M.J., Zhang Q., Nakamaru-Ogiso E., Kannabiran C., Fonseca-Kelly Z., Chakarova C., Audo I., Mackay D.S., Zeitz C., Borman A.D., et al. NMNAT1 mutations cause Leber congenital amaurosis. Nat. Genet. 2012;44:1040–1045. doi: 10.1038/ng.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siemiatkowska A.M., Schuurs-Hoeijmakers J.H., Bosch D.G., Boonstra F.N., Riemslag F.C., Ruiter M., de Vries B.B., den Hollander A.I., Collin R.W., Cremers F.P. Nonpenetrance of the most frequent autosomal recessive Leber congenital amaurosis mutation in NMNAT1. JAMA Ophthalmol. 2014;132:1002–1004. doi: 10.1001/jamaophthalmol.2014.983. [DOI] [PubMed] [Google Scholar]
- 36.Valle D., Kaiser-Kupfer M.I., Del Valle L.A. Gyrate atrophy of the choroid and retina: Deficiency of ornithine aminotransferase in transformed lymphocytes. Proc. Natl. Acad. Sci. USA. 1977;74:5159–5161. doi: 10.1073/pnas.74.11.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitchell G.A., Brody L.C., Looney J., Steel G., Suchanek M., Dowling C., Der Kaloustian V., Kaiser-Kupfer M., Valle D. An initiator codon mutation in ornithine-delta-aminotransferase causing gyrate atrophy of the choroid and retina. J. Clin. Investig. 1988;81:630–633. doi: 10.1172/JCI113365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McWilliam P., Farrar G.J., Kenna P., Bradley D.G., Humphries M.M., Sharp E.M., McConnell D.J., Lawler M., Sheils D., Ryan C., et al. Autosomal dominant retinitis pigmentosa (ADRP): Localization of an ADRP gene to the long arm of chromosome 3. Genomics. 1989;5:619–622. doi: 10.1016/0888-7543(89)90031-1. [DOI] [PubMed] [Google Scholar]
- 39.Bandah-Rozenfeld D., Mizrahi-Meissonnier L., Farhy C., Obolensky A., Chowers I., Pe’er J., Merin S., Ben-Yosef T., Ashery-Padan R., Banin E., et al. Homozygosity mapping reveals null mutations in FAM161A as a cause of autosomal-recessive retinitis pigmentosa. Am. J. Hum. Genet. 2010;87:382–391. doi: 10.1016/j.ajhg.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collin R.W., van den Born L.I., Klevering B.J., de Castro-Miro M., Littink K.W., Arimadyo K., Azam M., Yazar V., Zonneveld M.N., Paun C.C., et al. High-resolution homozygosity mapping is a powerful tool to detect novel mutations causative of autosomal recessive RP in the dutch population. Investig. Ophthalmol. Vis. Sci. 2011;52:2227–2239. doi: 10.1167/iovs.10-6185. [DOI] [PubMed] [Google Scholar]
- 41.Collin R.W., Littink K.W., Klevering B.J., van den Born L.I., Koenekoop R.K., Zonneveld M.N., Blokland E.A., Strom T.M., Hoyng C.B., den Hollander A.I., et al. Identification of a 2 Mb human ortholog of Drosophila eyes shut/spacemaker that is mutated in patients with retinitis pigmentosa. Am. J. Hum. Genet. 2008;83:594–603. doi: 10.1016/j.ajhg.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Audo I., Bujakowska K.M., Leveillard T., Mohand-Said S., Lancelot M.E., Germain A., Antonio A., Michiels C., Saraiva J.P., Letexier M., et al. Development and application of a next-generation-sequencing (NGS) approach to detect known and novel gene defects underlying retinal diseases. Orphanet J. Rare Dis. 2012;7:8. doi: 10.1186/1750-1172-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nikopoulos K., Gilissen C., Hoischen A., van Nouhuys C.E., Boonstra F.N., Blokland E.A., Arts P., Wieskamp N., Strom T.M., Ayuso C., et al. Next-generation sequencing of a 40 Mb linkage interval reveals TSPAN12 mutations in patients with familial exudative vitreoretinopathy. Am. J. Hum. Genet. 2010;86:240–247. doi: 10.1016/j.ajhg.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abu-Safieh L., Alrashed M., Anazi S., Alkuraya H., Khan A.O., Al-Owain M., Al-Zahrani J., Al-Abdi L., Hashem M., Al-Tarimi S., et al. Autozygome-guided exome sequencing in retinal dystrophy patients reveals pathogenetic mutations and novel candidate disease genes. Genome Res. 2013;23:236–247. doi: 10.1101/gr.144105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Audo I., Bujakowska K., Orhan E., Poloschek C.M., Defoort-Dhellemmes S., Drumare I., Kohl S., Luu T.D., Lecompte O., Zrenner E., et al. Whole-exome sequencing identifies mutations in gpr179 leading to autosomal-recessive complete congenital stationary night blindness. Am. J. Hum. Genet. 2012;90:321–330. doi: 10.1016/j.ajhg.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeitz C., Jacobson S.G., Hamel C.P., Bujakowska K., Neuille M., Orhan E., Zanlonghi X., Lancelot M.E., Michiels C., Schwartz S.B., et al. Whole-exome sequencing identifies LRIT3 mutations as a cause of autosomal-recessive complete congenital stationary night blindness. Am. J. Hum. Genet. 2013;92:67–75. doi: 10.1016/j.ajhg.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carss K.J., Arno G., Erwood M., Stephens J., Sanchis-Juan A., Hull S., Megy K., Grozeva D., Dewhurst E., Malka S., et al. Comprehensive rare variant analysis via whole-genome sequencing to determine the molecular pathology of inherited retinal disease. Am. J. Hum. Genet. 2017;100:75–90. doi: 10.1016/j.ajhg.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haer-Wigman L., van Zelst-Stams W.A.G., Pfund R., van den Born L.I., Klaver C.C.W., Verheij J.B.G.M., Hoyng C.B., Breuning M.H., Boon C.J.F., Kievit A.J., et al. Diagnostic exome sequencing identifies a genetic cause in 50% of 266 Dutch patients with visual impairment. Investig. Ophthalmol. Vis. Sci. 2017;25:591–599. doi: 10.1038/ejhg.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Combs R., McAllister M., Payne K., Lowndes J., Devery S., Webster A.R., Downes S.M., Moore A.T., Ramsden S., Black G., et al. Understanding the impact of genetic testing for inherited retinal dystrophy. Eur. J. Hum. Genet. EJHG. 2013;21:1209–1213. doi: 10.1038/ejhg.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.El Shamieh S., Neuille M., Terray A., Orhan E., Condroyer C., Demontant V., Michiels C., Antonio A., Boyard F., Lancelot M.E., et al. Whole-exome sequencing identifies KIZ as a ciliary gene associated with autosomal-recessive rod-cone dystrophy. Am. J. Hum. Genet. 2014;94:625–633. doi: 10.1016/j.ajhg.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bujakowska K.M., Fernandez-Godino R., Place E., Consugar M., Navarro-Gomez D., White J., Bedoukian E.C., Zhu X., Xie H.M., Gai X., et al. Copy-number variation is an important contributor to the genetic causality of inherited retinal degenerations. Genet. Med. 2017;19:643–651. doi: 10.1038/gim.2016.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eisenberger T., Neuhaus C., Khan A.O., Decker C., Preising M.N., Friedburg C., Bieg A., Gliem M., Charbel Issa P., Holz F.G., et al. Increasing the yield in targeted next-generation sequencing by implicating cnv analysis, non-coding exons and the overall variant load: The example of retinal dystrophies. PLoS ONE. 2013;8:e78496. doi: 10.1371/journal.pone.0078496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bainbridge J.W., Smith A.J., Barker S.S., Robbie S., Henderson R., Balaggan K., Viswanathan A., Holder G.E., Stockman A., Tyler N., et al. Effect of gene therapy on visual function in Leber’s congenital amaurosis. N. Engl. J. Med. 2008;358:2231–2239. doi: 10.1056/NEJMoa0802268. [DOI] [PubMed] [Google Scholar]
- 54.Hauswirth W.W., Aleman T.S., Kaushal S., Cideciyan A.V., Schwartz S.B., Wang L., Conlon T.J., Boye S.L., Flotte T.R., Byrne B.J., et al. Treatment of Leber congenital amaurosis due to RPE65 mutations by ocular subretinal injection of adeno-associated virus gene vector: Short-term results of a phase I trial. Hum. Gene Ther. 2008;19:979–990. doi: 10.1089/hum.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maguire A.M., Simonelli F., Pierce E.A., Pugh E.N., Jr., Mingozzi F., Bennicelli J., Banfi S., Marshall K.A., Testa F., Surace E.M., et al. Safety and efficacy of gene transfer for Leber’s congenital amaurosis. N. Engl. J. Med. 2008;358:2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Russell S., Bennett J., Wellman J.A., Chung D.C., Yu Z.F., Tillman A., Wittes J., Pappas J., Elci O., McCague S., et al. Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: A randomised, controlled, open-label, phase 3 trial. Lancet (Lond. Engl.) 2017;390:849–860. doi: 10.1016/S0140-6736(17)31868-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.MacLaren R.E., Groppe M., Barnard A.R., Cottriall C.L., Tolmachova T., Seymour L., Clark K.R., During M.J., Cremers F.P., Black G.C., et al. Retinal gene therapy in patients with choroideremia: Initial findings from a phase 1/2 clinical trial. Lancet (Lond. Engl.) 2014;383:1129–1137. doi: 10.1016/S0140-6736(13)62117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edwards T.L., Jolly J.K., Groppe M., Barnard A.R., Cottriall C.L., Tolmachova T., Black G.C., Webster A.R., Lotery A.J., Holder G.E., et al. Visual acuity after retinal gene therapy for choroideremia. N. Engl. J. Med. 2016;374:1996–1998. doi: 10.1056/NEJMc1509501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koenekoop R.K., Sui R., Sallum J., van den Born L.I., Ajlan R., Khan A., den Hollander A.I., Cremers F.P., Mendola J.D., Bittner A.K., et al. Oral 9-cis retinoid for childhood blindness due to Leber congenital amaurosis caused by RPE65 or LRAT mutations: An open-label phase 1b trial. Lancet (Lond. Engl.) 2014;384:1513–1520. doi: 10.1016/S0140-6736(14)60153-7. [DOI] [PubMed] [Google Scholar]
- 60.Scholl H.P., Moore A.T., Koenekoop R.K., Wen Y., Fishman G.A., van den Born L.I., Bittner A., Bowles K., Fletcher E.C., Collison F.T., et al. Safety and proof-of-concept study of oral QLT091001 in retinitis pigmentosa due to inherited deficiencies of retinal pigment epithelial 65 protein (RPE65) or lecithin:Retinol acyltransferase (LRAT) PLoS ONE. 2015;10:e0143846. doi: 10.1371/journal.pone.0143846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clinical Trials. [(accessed on 17 March 2018)]; Available online: http://clinicaltrials.gov.
- 62.Slijkerman R.W., Song F., Astuti G.D., Huynen M.A., van Wijk E., Stieger K., Collin R.W. The pros and cons of vertebrate animal models for functional and therapeutic research on inherited retinal dystrophies. Prog. Retin. Eye Res. 2015;48:137–159. doi: 10.1016/j.preteyeres.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 63.Sung C.H., Davenport C.M., Nathans J. Rhodopsin mutations responsible for autosomal dominant retinitis pigmentosa. Clustering of functional classes along the polypeptide chain. J. Biol. Chem. 1993;268:26645–26649. [PubMed] [Google Scholar]
- 64.Bujakowska K.M., Zhang Q., Siemiatkowska A.M., Liu Q., Place E., Falk M.J., Consugar M., Lancelot M.E., Antonio A., Lonjou C., et al. Mutations in IFT172 cause isolated retinal degeneration and bardet-biedl syndrome. Hum. Mol. Genet. 2015;24:230–242. doi: 10.1093/hmg/ddu441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sangermano R., Khan M., Cornelis S.S., Richelle V., Albert S., Garanto A., Elmelik D., Qamar R., Lugtenberg D., van den Born L.I., et al. ABCA4 midigenes reveal the full splice spectrum of all reported noncanonical splice site variants in stargardt disease. Genome Res. 2018;28:100–110. doi: 10.1101/gr.226621.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sangermano R., Bax N.M., Bauwens M., van den Born L.I., De Baere E., Garanto A., Collin R.W., Goercharn-Ramlal A.S., den Engelsman-van Dijk A.H., Rohrschneider K., et al. Photoreceptor progenitor mRNA analysis reveals exon skipping resulting from the ABCA4 c.5461-10T→C mutation in Stargardt disease. Ophthalmology. 2016;123:1375–1385. doi: 10.1016/j.ophtha.2016.01.053. [DOI] [PubMed] [Google Scholar]
- 67.Tucker B.A., Mullins R.F., Streb L.M., Anfinson K., Eyestone M.E., Kaalberg E., Riker M.J., Drack A.V., Braun T.A., Stone E.M. Patient-specific iPSC-derived photoreceptor precursor cells as a means to investigate retinitis pigmentosa. Elife. 2013;2:e00824. doi: 10.7554/eLife.00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parfitt D.A., Lane A., Ramsden C.M., Carr A.J., Munro P.M., Jovanovic K., Schwarz N., Kanuga N., Muthiah M.N., Hull S., et al. Identification and correction of mechanisms underlying inherited blindness in human iPSC-derived optic cups. Cell Stem Cell. 2016;18:769–781. doi: 10.1016/j.stem.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lukovic D., Artero Castro A., Delgado A.B., Bernal Mde L., Luna Pelaez N., Diez Lloret A., Perez Espejo R., Kamenarova K., Fernandez Sanchez L., Cuenca N., et al. Human iPSC derived disease model of MERTK-associated retinitis pigmentosa. Sci. Rep. 2015;5:12910. doi: 10.1038/srep12910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yoshida T., Ozawa Y., Suzuki K., Yuki K., Ohyama M., Akamatsu W., Matsuzaki Y., Shimmura S., Mitani K., Tsubota K., et al. The use of induced pluripotent stem cells to reveal pathogenic gene mutations and explore treatments for retinitis pigmentosa. Mol. Brain. 2014;7:45. doi: 10.1186/1756-6606-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Albert S., Garanto A., Sangermano R., Khan M., Bax N.M., Hoyng C.B., Zernant J., Lee W., Allikmets R., Collin R.W.J., et al. Identification and rescue of splice defects caused by two neighboring deep-intronic ABCA4 mutations underlying stargardt disease. Am. J. Hum. Genet. 2018;102:517–527. doi: 10.1016/j.ajhg.2018.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peng Y.Q., Tang L.S., Yoshida S., Zhou Y.D. Applications of CRISPR/CAS9 in retinal degenerative diseases. Int. J. Ophthalmol. 2017;10:646–651. doi: 10.18240/ijo.2017.04.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li P., Kleinstiver B.P., Leon M.Y., Prew M.S., Navarro-Gomez D., Greenwald S.H., Pierce E.A., Joung J.K., Liu Q. Allele-specific CRISPR-Cas9 genome editing of the single-base P23H mutation for rhodopsin-associated dominant retinitis pigmentosa. CRISPR J. 2018;1 doi: 10.1089/crispr.2017.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kajiwara K., Berson E., Dryja T. Digenic retinitis pigmentosa due to mutations at the unlinked peripherin/RDS and ROM1 loci. Science (N. Y.) 1994;264:1604–1608. doi: 10.1126/science.8202715. [DOI] [PubMed] [Google Scholar]
- 75.Beales P.L., Badano J.L., Ross A.J., Ansley S.J., Hoskins B.E., Kirsten B., Mein C.A., Froguel P., Scambler P.J., Lewis R.A., et al. Genetic interaction of BBS1 mutations with alleles at other BBS loci can result in non-Mendelian Bardet-Biedl syndrome. Am. J. Hum. Genet. 2003;72:1187–1199. doi: 10.1086/375178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Badano J.L., Leitch C.C., Ansley S.J., May-Simera H., Lawson S., Lewis R.A., Beales P.L., Dietz H.C., Fisher S., Katsanis N. Dissection of epistasis in oligogenic Bardet-Biedl syndrome. Nature. 2006;439:326–330. doi: 10.1038/nature04370. [DOI] [PubMed] [Google Scholar]
- 77.Vithana E.N., Abu-Safieh L., Pelosini L., Winchester E., Hornan D., Bird A.C., Hunt D.M., Bustin S.A., Bhattacharya S.S. Expression of PRPF31 mRNA in patients with autosomal dominant retinitis pigmentosa: A molecular clue for incomplete penetrance? Investig. Ophthalmol. Vis. Sci. 2003;44:4204–4209. doi: 10.1167/iovs.03-0253. [DOI] [PubMed] [Google Scholar]
- 78.Venturini G., Rose A.M., Shah A.Z., Bhattacharya S.S., Rivolta C. CNOT3 is a modifier of PRPF31 mutations in retinitis pigmentosa with incomplete penetrance. PLoS Genet. 2012;8:e1003040. doi: 10.1371/journal.pgen.1003040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sobreira N., Schiettecatte F., Valle D., Hamosh A. Genematcher: A matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Genematcher. [(accessed on 17 March 2018)]; Available online: https://genematcher.org/
- 81.ERDC Webpage. [(accessed on 17 March 2018)]; Available online: http://www.erdc.info/
- 82.Astuti G.D.N., van den Born L.I., Khan M.I., Hamel C.P., Bocquet B., Manes G., Quinodoz M., Ali M., Toomes C., McKibbin M., et al. Identification of inherited retinal disease-associated genetic variants in 11 candidate genes. Genes. 2018;9:21. doi: 10.3390/genes9010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nishiguchi K.M., Rivolta C. Genes associated with retinitis pigmentosa and allied diseases are frequently mutated in the general population. PLoS ONE. 2012;7:e41902. doi: 10.1371/journal.pone.0041902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gustafson K., Duncan J.L., Biswas P., Soto-Hermida A., Matsui H., Jakubosky D., Suk J., Telenti A., Frazer K.A., Ayyagari R. Whole Genome sequencing revealed mutations in two independent genes as the underlying cause of retinal degeneration in an Ashkenazi Jewish pedigree. Genes. 2017;8:210. doi: 10.3390/genes8090210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Llavona P., Pinelli M., Mutarelli M., Marwah V.S., Schimpf-Linzenbold S., Thaler S., Yoeruek E., Vetter J., Kohl S., Wissinger B. Allelic Expression imbalance in the human retinal transcriptome and potential impact on inherited retinal diseases. Genes. 2017;8:283. doi: 10.3390/genes8100283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.El Shamieh S., Méjécase C., Bertelli M., Terray A., Michiels C., Condroyer C., Fouquet S., Sadoun M., Clérin E., Liu B., et al. Further Insights into the ciliary gene and protein KIZ and its murine ortholog PLK1S1 Mutated in rod-cone dystrophy. Genes. 2017;8:277. doi: 10.3390/genes8100277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dockery A., Stephenson K., Keegan D., Wynne N., Silvestri G., Humphries P., Kenna P.F., Carrigan M., Farrar G.J. Target 5000: Target Capture sequencing for inherited retinal degenerations. Genes. 2017;8:304. doi: 10.3390/genes8110304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Comander J., Weigel-DiFranco C., Maher M., Place E., Wan A., Harper S., Sandberg M.A., Navarro-Gomez D., Pierce E.A. The Genetic basis of pericentral retinitis pigmentosa—A Form of mild retinitis pigmentosa. Genes. 2017;8:256. doi: 10.3390/genes8100256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Di Iorio V., Karali M., Brunetti-Pierri R., Filippelli M., Di Fruscio G., Pizzo M., Mutarelli M., Nigro V., Testa F., Banfi S., Simonelli F. Clinical and Genetic evaluation of a cohort of pediatric patients with severe inherited retinal dystrophies. Genes. 2017;8:280. doi: 10.3390/genes8100280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Porto F.B.O., Jones E.M., Branch J., Soens Z.T., Maia I.M., Sena I.F.G., Sampaio S.A.M., Simões R.T., Chen R. Molecular Screening of 43 brazilian families diagnosed with leber congenital amaurosis or early-onset severe retinal dystrophy. Genes. 2017;8:355. doi: 10.3390/genes8120355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Brandl C., Schulz H.L., Charbel Issa P., Birtel J., Bergholz R., Lange C., Dahlke C., Zobor D., Weber B.H.F., Stöhr H. Mutations in the Genes for interphotoreceptor matrix proteoglycans, IMPG1 and IMPG2, in Patients with vitelliform macular lesions. Genes. 2017;8:170. doi: 10.3390/genes8070170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McGuigan D.B., Heon E., Cideciyan A.V., Ratnapriya R., Lu M., Sumaroka A., Roman A.J., Batmanabane V., Garafalo A.V., Stone E.M., et al. EYS Mutations causing autosomal recessive retinitis pigmentosa: Changes of Retinal structure and function with disease progression. Genes. 2017;8:178. doi: 10.3390/genes8070178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Roosing S., Cremers F.P.M., Riemslag F.C.C., Zonneveld-Vrieling M.N., Talsma H.E., Klessens-Godfroy F.J.M., den Hollander A.I., van den Born L.I. A Rare form of retinal dystrophy caused by hypomorphic nonsense mutations in CEP290. Genes. 2017;8:208. doi: 10.3390/genes8080208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tracewska-Siemiątkowska A., Haer-Wigman L., Bosch D.G.M., Nickerson D., Bamshad M.J., University of Washington Center for Mendelian Genomics. van de Vorst M., Rendtorff N.D., Möller C., Kjellström U., et al. An Expanded multi-organ disease phenotype associated with mutations in YARS. Genes. 2017;8:381. doi: 10.3390/genes8120381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Littink K.W., Stappers P.T.Y., Riemslag F.C.C., Talsma H.E., van Genderen M.M., Cremers F.P.M., Collin R.W.J., van den Born L.I. Autosomal recessive NRL Mutations in patients with enhanced S-Cone Syndrome. Genes. 2018;9:68. doi: 10.3390/genes9020068. [DOI] [PMC free article] [PubMed] [Google Scholar]