Abstract
Background
Malignant glioma (MG), the most common primary brain tumor in adults, is extremely aggressive and uniformly fatal. Several treatment strategies have shown significant preclinical promise in murine models of glioma; however, none have produced meaningful clinical responses in human patients. We hypothesize that introduction of an additional preclinical animal model better approximating the complexity of human MG, particularly in interactions with host immune responses; will bridge the existing gap between these two stages of testing. Here, we characterize the immunologic landscape and gene expression profiles of spontaneous canine glioma and evaluate its potential for serving as such a translational model.
Methods
RNA in situ hybridization, flowcytometry, and RNA sequencing were used to evaluate immune cell presence and gene expression in healthy and glioma-bearing canines.
Results
Similar to human MGs, canine gliomas demonstrated increased intratumoral immune cell infiltration (CD4+, CD8+ and CD4+Foxp3+ T-cells). The peripheral blood of glioma-bearing dogs also contained a relatively greater proportion of CD4+Foxp3+ regulatory T-cells and plasmacytoid dendritic cells. Tumors were strongly positive for PD-L1 expression and glioma-bearing animals also possessed a greater proportion of immune cells expressing the immune checkpoint receptors CTLA-4 and PD-1. Analysis of differentially expressed genes in our canine populations revealed several genetic changes paralleling those known to occur in human disease.
Conclusion
Naturally occurring canine glioma has many characteristics closely resembling human disease, particularly with respect to genetic dysregulation and host immune responses to tumors, supporting its use as a translational model in the preclinical testing of prospective anti-glioma therapies proven successful in murine studies.
Keywords: Malignant Glioma, Glioblastoma, spontaneous canine glioma, immunotherapy, gene expression
INTRODUCTION
Malignant gliomas (MGs) are the most frequently diagnosed, and most lethal, primary brain tumors in adults. With an extremely poor prognosis and near 100% recurrence rate, these tumors remain one of the biggest therapeutic challenges of the modern era.1,2 A variety of immunotherapy-based regimens have shown significant antitumor efficacy in preclinical evaluations, with some resulting in complete and sustained tumor eradication in murine models of glioma.3–5 However none have come close to challenging the current standard of care for human patients suffering from this devastating illness. Additionally, toxicities initially encountered in human phase I/II trials are often not initially identified in preclinical mouse studies. Although the murine model remains an effective, economical, and vital preclinical model for performing initial molecular studies to understand this disease process, it has poorly predicted the human response to treatment with prospective anti-glioma therapies. The discrepancy between the success of preclinical studies and subsequent uniform failure of these therapies in large-scale phase III human clinical trials likely stems from the limited capacity of the murine model of glioma to fully approximate the intricate complexity of human disease.6
Human MGs are highly heterogeneous tumors, arising and continuously evolving in response to selective pressures within each unique tumor microenvironment (TME), including endogenous antitumor immune responses. A hallmark adaptation of human MG is the induction of a profoundly immunosuppressive environment that cripples naturally generated antitumor immune responses and limits the effectiveness of immunotherapies.7,8 A critical component of tumor development and progression, is the complex interactions between tumor cells and the host immune system. This critical component is poorly modeled by the xenograft murine tumors that are established in immunodeficient host.
Dogs are known to spontaneously develop brain tumors with similar clinical and histopathological features to human disease.9 In particular, brachycephalic breeds like Boxers, English Bulldogs, and Boston Terriers are disproportionately prone to developing malignant gliomas.10,11 Spontaneously arising in genetically diverse and immunocompetent populations, canine gliomas may more accurately model the complexity of human disease, introducing the possibility of using canine studies to bridge the translational gap that currently exists between preclinical murine studies and human clinical trials. The larger dog brain size is also conducive to imaging studies monitoring disease progression and permits surgical tumor resection, allowing evaluation of treatment regimens more representative of those employed in human patients. Some groups have used a canine model of glioma for testing various treatment and drug delivery strategies12–14 highlighting the translational importance of this model.
In order to integrate this model into the current anti-glioma therapy development pipeline, canine glioma must be thoroughly studied and defined in comparison to human disease. There have been various studies categorizing genetic changes specific to canine gliomas;15,16 for a comprehensive review on this topic, please refer to the paper by Bentley et al.6 However, the interactions between the canine immune system and glioma tumors, systemically as well as within the tumor microenvironment, have not been described. Here, we further evaluate and characterize the immunologic and genetic expression profiles of spontaneous canine glioma, providing a foundation for future immunotherapy studies.
MATERIALS AND METHODS
Canine Patients
Canine tumor and normal brain tissue samples were collected at Purdue University IACUC protocol # 1404001056. The dogs were aged 3.0 to 9.6 years (median 6.7 years) and all were brachycephalic, except for two normal controls (Table 1). Of the 10 canine tumor samples, 7 were collected during surgery and 3 were collected during necropsy. Normal brain tissue was collected from dogs euthanized for unrelated reasons. Samples were immediately flash-frozen in liquid nitrogen and transferred to a −80° C freezer, with additional tissue placed in formalin. Tumor biopsy specimens submitted in formalin were used for histologic classification and grading.17
Table 1.
Table of glioma histological type/normal control and dog breed
| Canine | Breed | Age | Diagnosis |
|---|---|---|---|
| 1 | Dogue de Bordeaux | 4.5 yr. | oligodendroglioma III |
|
|
|||
| 2 | Boxer | 7.0 yr. | oligodendroglioma III |
|
|
|||
| 3 | Boxer | 6.3 yr. | oligodendroglioma III |
|
|
|||
| 4 | Boston terrier | 9.6 yr. | oligodendroglioma III |
|
|
|||
| 5 | Boston terrier | 8.8 yr. | oligodendroglioma II |
|
|
|||
| 6 | English Bulldog | 7.2 yr. | oligodendroglioma III |
|
|
|||
| 7 | Boxer | 7.8 yr. | oligodendroglioma III |
|
|
|||
| 8 | Boxer | 5.8 yr. | oligodendroglioma III |
|
|
|||
| 9 | Mixed Breed | 5.8 yr. | oligodendroglioma III |
|
|
|||
| 10 | Boxer | 3.0 yr. | oligodendroglioma III |
|
|
|||
| 11 | Boxer | 8.2 yr. | Normal Brain |
|
|
|||
| 12 | Chesapeake Bay Retriever | 3.7 yr. | Normal Brain |
|
|
|||
| 13 | Siberian Husky | 2.0 yr. | Normal Brain |
Immunohistochemistry
Gliomas were classified on the basis of hematoxylin and eosin-stained (H&E) sections and immunohistochemistry for glial fibrillary acidic protein (GFAP) and Olig2. A rabbit polyclonal antibody to recombinant mouse Olig2 (AB9610, EMD Millipore, Billerica, MA) was used for Olig2 immunohistochemistry as previously described.18 Normal astrocytes and oligodendrocytes in non-neoplastic canine cerebrum were used as positive controls for GFAP and Olig2, respectively.
RNA in Situ Hybridization
Detection of CD4, CD8, FoxP3, PD-1, PD-L1 and CTLA-4 mRNA expression in canine tumor samples was performed with RNA in situ hybridization using an RNAscope Multiplex Fluorescent Reagent Kit v2 (Cat. No. 323100, ACD). 4 µm sections of tissue were deparaffinized by twice incubating tissue slides in xylene for 5 minutes followed by twice incubation in 100% EtOH for 2 minutes. After slides were air-dried for 5 minutes, they were treated with 5–8 drops of RNAscope Hydrogen Peroxide, incubated for 10 minutes at room temperature, and washed twice in fresh distilled water. Afterwards, slides were submerged in a beaker containing RNAscope 1× Target Retrieval Reagents at temperatures between 98–102°C for 15 minutes and washed with distilled water. A hydrophobic barrier was drawn around the tumor sample on the slide using an Immedge hydrophobic barrier pen (Cat. No. H4000, Vector Laboratories). Each slide was treated with 5 drops of RNAscope Protease Plus and incubated in the ACD HybEZ Oven at 40° for 30 minutes.
Target probes were designed using custom software as described previously.19 GenBank accession numbers and probe regions are as follows: CD4 (GenBank NM_001003252.1; nucleotides 420–1373), CD8a (GenBank NM_001002935.2; nucleotides 41–900), FoxP3 (GenBank NM_001168461.1; nucleotides 427–1382), PDCD1 (PD-1) (GenBank NM_001314097.1; nucleotides 75–756), CD274 (PD-L1) (GenBank NM_001291972.1; nucleotides 283–1237), and CTLA-4 (GenBank NM_001003106.1; nucleotides 689–1761). Probes were mixed and slides were stained according to manufacturer’s (ACD) instructions. Fluorophores used were TSA Plus fluorescein (Cat. No. NEL741001KT, PerkinElmer), TSA Plus Cyanine 3 (Cat. No. NEL744001KT, PerkinElmer) and TSA Plus Cyanine 5 (Cat. No. NEL745001KT, PerkinElmer). Slides were counterstained with DAPI and treated with Prolong Gold antifade mounting medium (Cat. No. P36930, Life Technologies). Images were captured using a confocal microscope (Olympus Fluoview FV1200) at 20× and 60× magnification. Fluorescent dots present on images captured at 60× magnification were counted to estimate mRNA expression in each slide sample.
Flowcytometry
Leukocytes were isolated from samples of canine brain tissue and peripheral blood using a previously established and published protocol20–22 and single cell suspension was created. Cells were stained with anti-canine CD3 (Serotec, NC0522562) conjugated with anti-mouse IgG DyLight 549 (BioRad, STAR117D549GA), anti-canine CD4 PE/Cy7 (eBioscience, 25-5040-42), anti-canine CD8a eFluor 450 (eBioscience, 48-5080-42), anti-mouse/rat FoxP3 APC (eBioscience, 17-5773-82), anti-human CD152-FITC (MyBioSource.com, MBS666569), antihuman PD-1 biotinylated antibody (R&D Systems, BAF1086) conjugated with streptavidin FITC (eBioscience, 11-4317-87), anti-canine CD45 eFluor 450 (eBioscience, 48-5450-42), anticanine CD11c (BioRad, MCA17785) conjugated with anti-mouse IgG FITC (BioRad, STAR117F), anti-human CD123 PE (eBioscience, 12-1239-41), anti-human CD83 APC (MACS, 130-098-889), and anti-human PD-L1 (CD274) PE/Cy7 (BD Bioscience, BDB558017). Cells were analyzed by Flowcytometry, using a BD LSRFOrtessa X-20 cell analyzer, and data was analyzed with FlowJo V.10.2 software.
RNA extraction
Total RNA was purified from 25 mg brain tissue samples from 2 healthy and 4 glioma-bearing canines using a Qiagen RNeasy Mini Kit (74106), according to manufacturer’s instructions. A NanoDrop ND-1000 spectrophotometer was used to determine sample concentration and RNA quality. RNA integrity was assessed with standard denaturing agarose gel electrophoresis. Stranded total RNA library preparations were prepared using the Illumina TruSeq mRNA Library Prep Kit single index.
Screening for differentially expressed genes
A comparison of gene expression in canine glioma and healthy brain tissue was performed using package edge R of R.23 Volcano plot filtering, with screening thresholds of a false discovery rate (FDR) < 0.01 and fold change (FC) >2, was used to identify differentially expressed genes (DEG).
Functional enrichment analysis
Pathway analysis was performed to determine the significant biological pathways influenced by previously identified DEGs. DEGs were clustered into three groups according to the relative changes in expression between glioma and healthy tissue. Biological functional analysis was performed using gene ontology (GO) and the latest Kyoto Encyclopedia of Genes and Genomes (KEGG) databases through the Database for Annotation, Visualization, and Integrated Discovery (DAVID).24,25 A cutoff of FDR of <0.05 was set for selected GO and KEGG terms enriched in groups of DEGs.
Statistical methods
The expression of mRNAs between canine glioma and healthy brain tissues was compared using a paired t-test (p-value of <0.05 was considered statistically significant). Fisher’s exact test was used to select significant pathways (p <0.05) in pathway analysis. FDRs were calculated from Benjamini Hochberg FDR to correct p-values. All other data are presented as Mean ± SEM. Comparisons between two groups were conducted using Student’s t-test or Mann Whitney test as appropriate, and differences between more than two groups were assessed using ANOVA with Tukey’s post-hoc test. All analyses were conducted using GraphPad Prism version 4.0 (GraphPad Software, Inc.). All reported p-values were two sided and were considered to be statistically significant at * p<0.05, **p<0.01, ***p<0.001.
RESULTS
Canine glioma histology
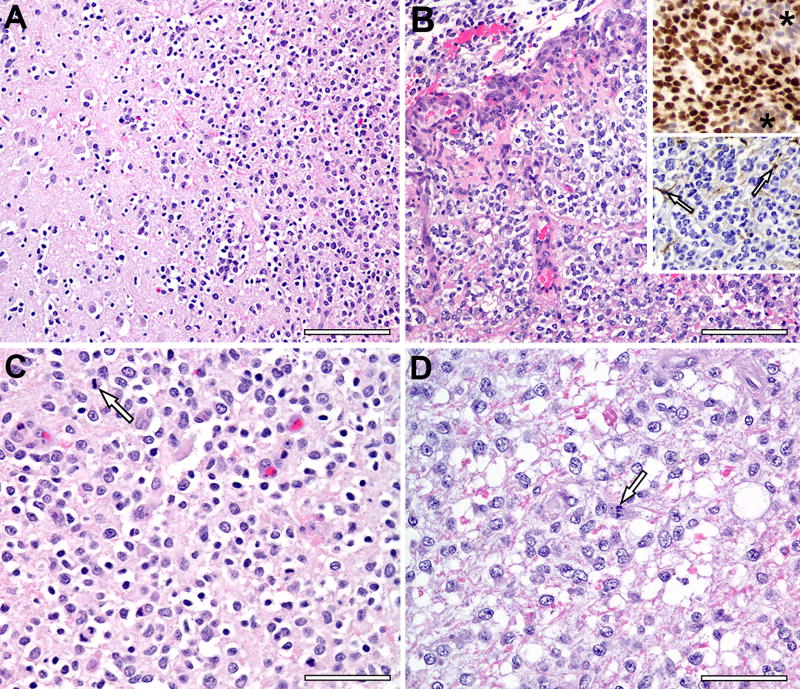
Tumors were classified using microscopic features on H&E and Olig2 and GFAP immunohistochemistry and graded according to current canine tumor classification guidelines.12 Histologic analysis of canine glioma tissue samples (n=10) demonstrated features consistent with oligodendroglioma, nine of which were grade III anaplastic oligodendrogliomas (Fig. 1). These findings are consistent with prior data showing that, in contrast to the grade II oligodendrogliomas more commonly seen in humans, canine oligodendrogliomas tend to be anaplastic grade III tumors.6 Histopathological features including frequent mitotic figures, microvascular proliferation, and the presence of necrosis were used to recognize these grade III anaplastic tumors. Neoplastic cells were identified based on histologic features and by excluding normal cells such as neurons, endothelial cells and glia, and the percentage of neoplastic cells positive for each of Olig2 and GFAP was counted manually. All oligodendrogliomas exhibited strong Olig2 nuclear immunoreactivity (85% – 100% of neoplastic cells) and stained negative for GFAP. The grade III oligodendrogliomas were infiltrative, poorly demarcated from adjacent neuroparenchyma, and formed microvascular proliferations around foci of necrosis. Neoplastic cells had variable nuclear diameters and chromatin content with occasional mitotic figures.
Figure 1. Histology of Canine Glioma Samples.
A) Grade III oligodendroglioma (upper right) invades adjacent cerebral parenchyma. Scale bar, 100 µm. (B) Microvascular proliferation is prominent around a necrotic focus (top) in Grade III oligodendroglioma. Scale bar, 100 µm. Upper inset depicts Olig2 immunohistochemistry with nuclear labeling in neoplastic cells but no Olig2 expression by cells of the microvascular proliferation (asterisks). Lower inset depicts processes of a few GFAP–positive astrocytes (arrows), but the neoplastic cells are negative. (C) Most neoplastic cells in this Grade III oligodendroglioma have a hyperchromatic nucleus with occasional mitotic figures (arrow) and a clear perinuclear ‘halo’, an artifact of delayed fixation. Scale bar, 50 µm. (D) Neoplastic cells in this Grade III oligodendroglioma have a larger hypochromatic nucleus with occasional mitotic figures (arrow) and pale eosinophilic cytoplasm with indistinct cell borders. Scale bar, 50 µm. (n = 10)
Immune cell infiltrations in canine glioma
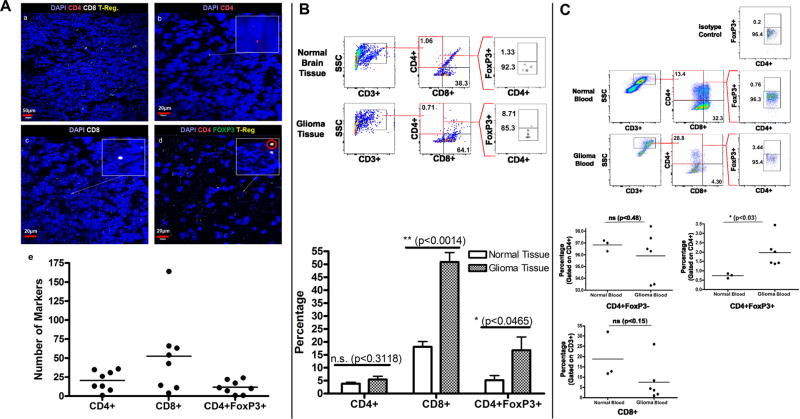
RNA in situ hybridization demonstrated the presence of CD4+ (20.375/high power field +/− 4.705), CD8+ (52.375/high power field +/−18.114), and CD4+FoxP3+ regulatory (Treg) T-cells (11.625/high power field +/− 3.106) within canine glioma tissue (Fig. 2A). These findings were confirmed in fresh canine glioma and normal brain tissue, with flow cytometry showing intratumoral infiltration with CD4+ (5.43% +/− 1.27), CD8+ (50.83% +/− 3.67), and regulatory (16.76% +/− 5.10) T-cells. We also found a significantly higher frequency of CD8+ (p < 0.0014) and Treg (p < 0.0465) cell infiltration in canine glioma compared to healthy canine brain (Fig. 2B).
Figure 2. T-cell Presence in Canine Glioma and Peripheral Blood.
A) RNA in situ hybridization of tumor samples stained with DAPI and three RNA probe conjugated markers, cyanine 3 [CD4], cyanine 5 [CD8] and fluorescein [FoxP3]. Yellow markers, a combination of green [FoxP3] and red [CD4], are indicative of Treg cells. (a) Staining for CD4, CD8, and FoxP3 shown at 20× magnification. Scale bar, 50 µm. (b) Staining for CD4 at 60×. Scale bar, 20 µm. (c) Staining for CD8 at 60×. Scale bar, 20 µm. (d) Staining for Treg (CD4 and FoxP3) shown at 60× magnification. Scale bar, 20 µm. (e) A scatter plot showing the number of CD4, CD8 and Treg markers counted on slide samples at 60× magnification. (n = 8) B) Gating strategy for CD4+, CD8+ and CD4+FoxP3+ Treg cells in normal brain tissue and high-grade canine glioma. The bar diagram represents the percentage of CD4+, CD8+, and CD4+FoxP3+ T cells in normal brain tissue (n = 3) vs. high grade glioma (n = 6). C) CD8+, CD4+ and CD4+Foxp3+ T cells in the peripheral blood of normal dogs compared to dogs with glioma.
Glioma-bearing canines also possessed significantly greater proportions of circulating Treg cells (0.737% +/− 0.073 normal vs. 1.972% +/− 0.324 glioma; p<0.03) (Fig. 2C).
Immune checkpoint expression in healthy and glioma-bearing canines
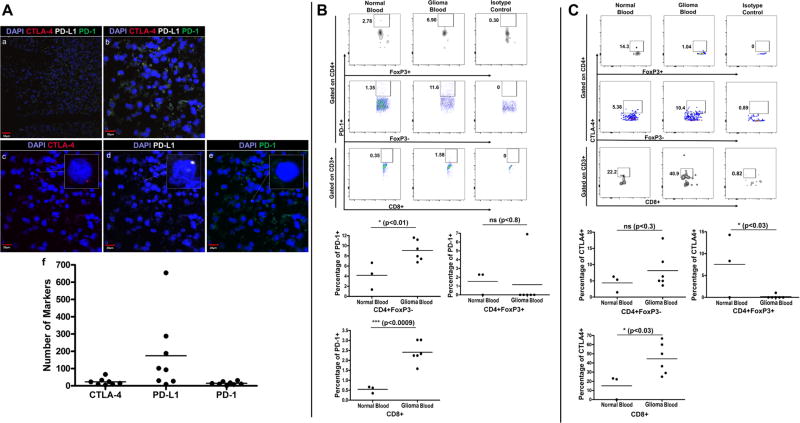
We evaluated the presence of cytotoxic T-lymphocyte antigen-4 (CTLA-4) and programmed cell death-1 (PD-1) and its ligand, PD-L1 in the glioma microenvironment using RNA in situ hybridization. We observed prominent PD-L1 staining in canine glioma tissue (173.875/high power field +/− 76.170) and the presence of CTLA-4 (23.00/high power field +/− 6.982) and PD-1 (14.875/high power field +/− 3.340) expressing cells (Fig. 3A).
Figure 3. Immune Checkpoint Markers on Tumor-Infiltrating and Circulating Immune Cells.
A) RNA in situ hybridization of tumor samples stained with DAPI and three RNA probe conjugated markers, cyanine 3 [CTLA-4], cyanine 5 [PD-L1] and fluorescein [PD-1]. (a) Staining for CTLA-4, PD-L1, and PD-1 shown at 20× magnification. Scale bar, 50 µm. (b) Staining for CTLA-4, PD-L1, and PD-1 shown at 60× magnification. Scale bar, 20 µm. (c) Staining for CTLA-4 at 60×. Scale bar, 20 µm. (d) Staining for PD-L1 at 60×. Scale bar, 20 µm. (e) Staining for PD-1 at 60×. Scale bar, 20 µm. (f) Scatter plot showing number of CTLA-4, PD-L1 and PD-1 markers counted on slide samples at 60× magnification. (n = 8) B) Gating strategy for analyzing PD-1 expression on CD4+, CD8+ and CD4+FoxP3+ T cells in peripheral blood of normal dogs (n = 3) vs. dogs with glioma (n = 6) and scatter plots representing the percentage of T-cells expressing PD-1. C) Gating strategy for analyzing CTLA-4 expression on CD4+, CD8+ and CD4+FoxP3+ T cells in peripheral blood of normal (n = 3) and glioma-bearing dogs (n = 6) and scatter plots representing the percentage of T-cells expressing CTLA-4.
Flow cytometric analysis was performed on peripheral blood lymphocytes to further define immune checkpoint expression. Compared to healthy controls, there was significantly greater PD-1 expression on CD4+ (4.157% +/− 1.536 normal vs. 9.052% +/− 0.826 glioma; p<0.01) and CD8+ (0.543% +/− 0.097 normal vs. 2.408% +/− 0.225 glioma; p<0.0009) T-cells from glioma-bearing canines (Fig. 3B). Tumor-bearing animals possessed a significantly greater proportion of CTLA-4-expressing CD8+ T-cells (15.10% +/− 7.554 normal vs. 44.52% +/− 6.951 glioma; p<0.03) and relatively more CTLA-4-expressing CD4+ T-cells, although this difference did not reach statistical significance (Fig. 3C). We also observed fewer CTLA-4-expressing peripheral blood Treg cells in glioma-bearing dogs (7.543% +/− 4.147 normal vs. 0.1733% +/− 0.1733 glioma; p<0.03). The average peripheral blood Treg PD-1 expression did not differ between our two canine populations.
Dendritic cell populations in canine glioma
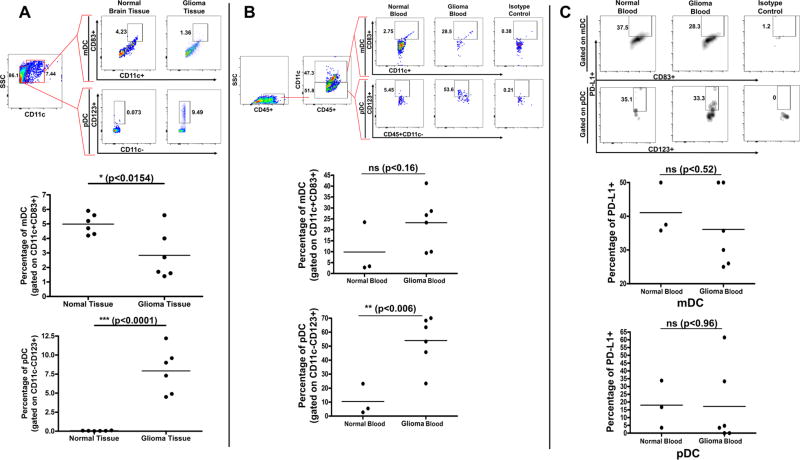
Using flow cytometry, we compared the presence of plasmacytoid (pDC) and myeloid (mDC) dendritic cells in the brain and peripheral blood of glioma-bearing and healthy dogs. In comparison to healthy canine brain, canine glioma tissue contained a significantly greater presence of pDCs (0.0650% +/− 0.011 normal vs. 7.917% +/− 1.204 glioma; p<0.0001) and fewer mDCs (4.983% +/− 0.2845 normal vs. 2.833% +/− 0.6805 glioma; p<0.0154) (Fig 4A). There was also a significant increase in circulating pDCs in glioma-bearing canines (10.43% +/− 6.385 normal vs. 54.02% +/− 7.217 glioma; p<0.006) and a trend towards increased mDC presence, though this was not statistically significant (p<0.16) (Fig 4B).
Figure 4. pDC and mDC Populations in Brain Tissue and Peripheral Blood of Normal and Glioma-Bearing Canine Patients.
A) Gating strategy for analyzing pDCs and mDCs from normal brain tissue (n = 6) and canine glioma (n = 6). Scatter plots representing the percentage of pDCs and mDCs B) Gating strategy for analyzing pDCs and mDCs in the peripheral blood of normal (n = 3) and glioma-bearing dogs (n = 6). Scatter plots representing the percentage of pDCs and mDCs C) Gating strategy for analyzing PD-L1 expression on pDCs and mDCs in peripheral blood of normal dogs (n = 3) vs. dogs with glioma (n = 6) and scatter plots representing the percentage of DCs expressing PD-L1.
Similar to human patients,26 PD-L1 expression was detected on peripheral blood mDCs and pDCs in tumor-bearing and healthy canines, though there were no significant differences in expression frequency or density on either DC subtype (Fig 4C).
Canine expression of genes associated with human glioma
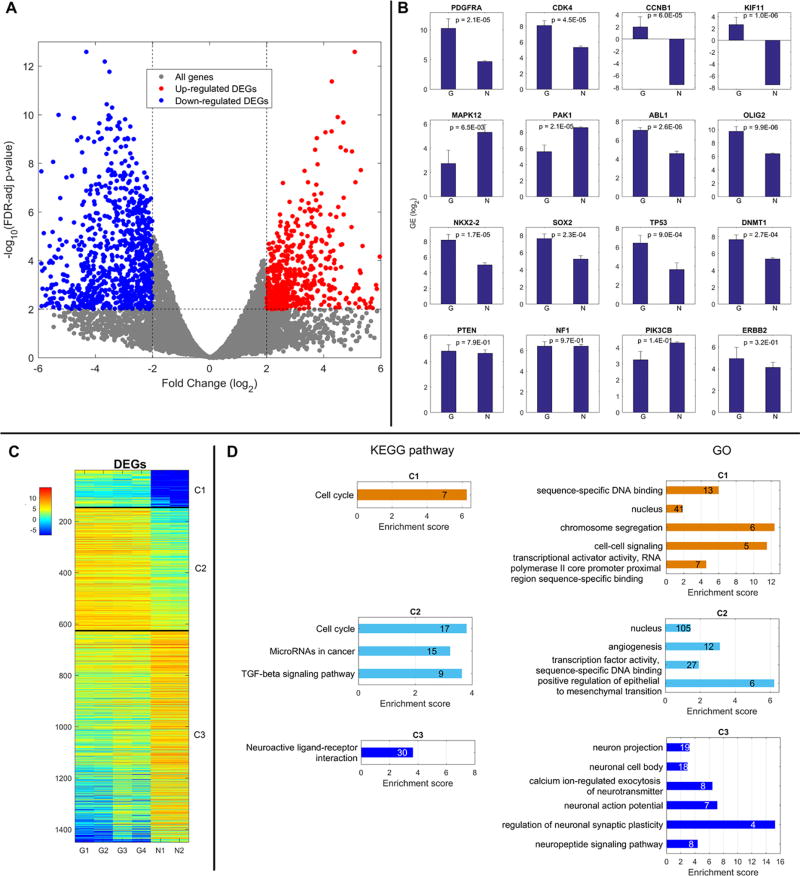
Gene expression in canine glioma and normal canine brain was evaluated with mRNA sequencing. Volcano plot analysis identified 1450 differentially expressed genes (DEGs) between these populations (Fig. 5A). Of these DEGs, 626 were upregulated (red dots) and 824 were downregulated (blue dots) in glioma tissue relative to healthy controls. We selected a subset of genes associated with human glioma and evaluated their expression in our canine populations; their relative expression, on a logarithmic scale, is depicted in (Fig 5B).
Figure 5. DEG Expression Patterns and Corresponding Biological Functions/Pathways.
A) Volcano Plot showing cutoffs of FDR-adjusted P-value and FC for all genes and DEGs. B) Gene expression in normal and glioma canine brain tissue of a set of genes associated with human glioma. C) Heat Map of Gene Expression for 3 Groups of DEGs D) Significantly over-represented KEGG pathways and GO functions in 3 groups of DEGs identified (n = 2 normal; 4 glioma).
DEG analyses revealed that similar to human MG, canine glioma possess genetic mutations impacting the retinoblastoma (RB), TP53, and receptor tyrosine kinase (RTK)/PI3K pathways. Canine gliomas also exhibited amplification of neurodevelopmental genes, OLIG2, NKX2-2, and SOX2, commonly overexpressed in human astrocytomas and oligodendrogliomas. Other genes upregulated in tumors of both species were CCNB1, a mitosis-related cyclin, and KIF11, a mitotic kinesin promoting tumor invasion and proliferation.27 Additionally, increased expression of DNA methyltransferase DNMT1 in both species suggests the importance of epigenetic changes in oncogenesis.28,29 In human GBM, overexpression of DNMT1 has been correlated with reduced MGMT expression30 and repression of the p53 pathway.31
We also identified several genes with canine patterns of expression opposite those seen in human glioma. PAK1, which promotes Akt signaling, angiogenesis, and GBM invasiveness32 and MAPK12, which represses cell proliferation33 are two kinases commonly upregulated in human GBM, that were downregulated in our canine oligodendroglioma.
Functional pathway analysis of canine DEGs
DEGs were categorized into one of three groups based on relative changes in expression between healthy tissue and glioma. Groups C1 and C2 contained genes overexpressed in canine glioma. Genes expressed only in glioma tissue were assigned to C1, and genes expressed in both populations were assigned to C2. C3 contained the 824 genes that were downregulated in glioma as compared to healthy tissue. A depiction of this hierarchal clustering is shown in (Fig 5C). A list of all the genes and involved pathways is provided as part of the supplementary file.
Gene ontology and KEGG pathway enrichment analyses were performed on clustered DEGs to categorize them within biologically relevant functions or pathways (Fig. 5D). A comprehensive list of all genes affected is provided in the supplementary data. DEGs in clusters C1 and C2 were evaluated with both methods. Common pathways identified as impacted by genes overexpressed in canine glioma include those enhancing transcriptional activity, cell cycle progression, and angiogenesis. Others promoted TGF-beta signaling and epithelial to mesenchymal transition, both of which have strong implications in tumor development. There was also a prevalent upregulation of genes related to microRNAs previously associated with cancer, further highlighting the importance of epigenetic changes in oncogenesis. Analysis of the C3 cluster showed that genes downregulated in glioma includes those supporting neuronal maintenance and function. Healthy brain tissue, primarily composed of neurons, would be expected to have greater expression of genes related to normal neuronal function as compared to gliomas, which are principally formed of malignant glial cells. The relatively reduced expression of neuron-related genes in canine glioma tissue clearly reflects the different primary cell populations within these samples and supports the overall validity of this analysis.
DISCUSSION
Human MGs are highly aggressive, uniformly fatal brain tumors critically in need of more effective treatment strategies. One of the factors underlying the persistent difficulties to develop survival-prolonging treatments is the lack of a preclinical model capable of reliably predicting treatment efficacy in human patients. Although essential for studying basic mechanisms driving glioma progression, murine tumor models fail to fully encompass the complexity of human disease. Utilization of a higher-level spontaneous animal model better approximating human disease is expected to more accurately predict the efficacy of prospective anti-glioma treatments following murine studies, thereby improving the possibility of successful transition to human clinical trials.6,34 In contrast to mice, dogs spontaneously develop MGs with similar histopathologic features, radiologic presentations, and clinical responses to conventional antitumor therapies as human disease.35,36 In this study, we for the first time characterize the immunologic landscape of canine glioma and further evaluate its potential to approximate human disease.
Given the importance of host immune responses to human MG progression as well as response to treatment, especially immunotherapeutic treatment strategies, it is imperative that the influence of these complex interactions be accounted for at some stage of preclinical testing. A hallmark of human GBM is the induction of an immunosuppressive state that reduces the effectiveness of endogenous antitumor immune responses, and consequently the therapeutic potential of immunotherapies. In this study, we demonstrate that canine tumors possess many of the same immunologic characteristics as their human counterparts. In addition to establishing the presence of tumor infiltrating lymphocytes (TILs) in canine glioma, we show that pDCs are the major antigen-presenting cell in the canine glioma microenvironment. Furthermore, we characterize the expression of immune checkpoints within the brain tissue and peripheral blood of healthy and glioma-bearing canines. Given the similarities we have identified between canine and human glioma immune profiles, we predict that glioma-bearing dogs will respond similarly to treatment, particularly with immunotherapies, as human patients.
Human gliomas contain prominent CD4+ and CD8+ T-cell infiltrates.37,38 Similar to the trend we observed in glioma-bearing dogs, the majority of T-cells within human glioma are CD8+ T-cells, the primary effectors of host antitumor immune responses.37 Also characteristic of human MG is an intratumoral accumulation of immunosuppressive regulatory T-cells. Treg cells have a strong inhibitory impact on dendritic cell maturation and the activation, proliferation, and function of CD4+ and CD8+ lymphocytes,26 and their intratumoral accumulation is associated with a poor prognosis.39,40 In addition to intratumoral Treg cell infiltrates, human MG patients have been shown to possess elevated levels of circulating Treg cells (8.56%) as compared to healthy volunteers (0.48%).41 We observed an accumulation of Treg cells within the tumors and peripheral blood of glioma-bearing canines as compared to healthy controls, supporting the assertion that Treg cells contribute similarly to glioma-induced immunosuppression in canines.
In addition to accumulating inhibitory leukocytes, human MGs suppress antitumor responses through enhanced activation of CTLA-4 and PD-1 immune checkpoints, endogenous negative regulators of T-cell activity. Binding of PD-1 on activated T-cells to PD-L1, often overexpressed by tumor cells, inhibits T-cell proliferation and cytokine release, leading to T-cell anergy, apoptosis, or Treg cell development. Activation of CTLA-4, induced on naïve T-cells, prevents initial T-cell activation. Consistent with a recent observation that 88% of human GBM express PD-L1,37 we found elevated PD-L1 staining in all canine glioma samples. Not expressed by healthy human tissues, elevated PD-L1 expression in canine tumors indicates that canine gliomas employ similar methods of immunosuppression as human tumors. Increased expression of CTLA-4 and PD-1 receptors, characteristically seen on intratumoral37,39 and peripheral blood38,42 lymphocytes of human MG patients, was observed in the lymphocytes of our glioma-bearing canines. In comparing immune checkpoint expression on different lymphocyte populations we did find a relatively reduced expression of CTLA-4 on canine glioma-bearing Tregs, which is in contrast to reports of elevated Treg CTLA-4 expression in human MG patients (76.8%) compared to healthy controls (50%).41 This discrepancy could indicate a different role for, or timing of, immunosuppression via this pathway in human and canine glioma.
Although MGs are highly genetically diverse, there are several pathways dysregulated in the majority of human tumors, such as PI3K, RAS, etc. In a recent study of core genetic mutations in human GBM, 87%, 78%, and 88% of analyzed samples harbored somatic mutations of the RB, TP53, and RTK/PI3K signaling pathways, respectively.43 One of the most common mutations affecting the RB pathway in human GBM is amplification of CDK4 (14%), a cyclin-dependent kinase promoting cell cycle progression.43 Upregulated in our canine tumor samples, amplification of CDK4 is also associated with progression of human anaplastic oligodendrogliomas.44 Altered p53 pathway signaling in human GBM occurs through direct mutation of p53 (35%), though also occurs via ARF deletions (49%) and/or amplification of MDM2 (14%) or MDM4 (7%).43 Direct P53 inactivation is less commonly seen in human oligodendrogliomas,44 and TP53 mRNA expression was relatively upregulated in our canine oligodendroglioma, consistent with historical evaluations.16 This overexpression may be related to additional functions of p53, although these tumors may possess other mutations allowing escape of p53-dependent growth control. One such mutation seen in human tumors and our canine glioma is overexpression of OLIG2, a neurodevelopmental transcription factor. OLIG2 directly represses the activity of cell cycle inhibitor p21 (CDKN1A)45 and antagonizes p53 function by impacting post-translational protein modification.46 Other neurodevelopmental transcription factors overexpressed in human oligodendrogliomas and GBM subtypes as well as our canine glioma are NKX2-247 and SOX2.48 Also commonly observed in human GBM are activating mutations of the growth-promoting receptors EGFR (57%) and PDGFRA (10%).48,49 PDGFRA amplification also occurs in highly anaplastic human oligodendrogliomas displaying features of WHO grade IV malignancies44 and was observed in our canine tumors as well. Other genes associated with the PI3K pathway in humans that were not differentially regulated in our canine populations are ERBB2, PTEN, and NF1.43 PTEN loss is less common in human anaplastic oligodendrogliomas44 and rarely occurs in canine gliomas.50 These results imply that human and canine gliomas arise from dysregulation of common pathways and suggest that they may respond similarly to treatment. Future analyses must assess downstream signaling and the influence of epigenetic mutations like MGMT methylation on disease progression and treatment response. Studies with larger sample sizes may be useful in identifying less commonly mutated pathways and may lead to the identification of novel molecularly defined subclasses with relevant clinical applications, as has been done in human disease.
This study has identified clear commonalities between the immunologic response profiles and genetic mutations observed in human and canine gliomas. Arising from commonly dysregulated cellular pathways and displaying similar interactions with host immune systems, spontaneous canine glioma possess many of the same complex characteristics found in human disease that may render them a more appropriate model to predict therapeutic efficacy of anti-glioma treatments, particularly immunotherapies. Incorporation of spontaneous canine glioma as a transitional model for testing emerging anti-glioma treatments proven successful in preclinical murine studies has potential to significantly improve the success of subsequent human clinical trials, leading to the development of more effective treatments for MG in both human and dogs.
Supplementary Material
IMPORTANCE OF THE STUDY.
MGs are highly infiltrative, heterogeneous, and treatment-resistant tumors. Underlying the failure of promising pre-clinical treatment strategies to successfully treat human disease is the lack of ability of the artificial mouse model to fully capture the complexity of human MG and therefore poor capacity to predict adverse effect and therapeutic success in human patients. Canine brain tumors that spontaneously develop in immunocompetent hosts and share similar environmental stressors as human tumors provide an attractive intermediate platform for evaluating the effectiveness of anti-glioma treatment strategies, especially immunotherapies, proven successful in preclinical murine studies. To substantiate the validity of this model, we for the first time describe the immunologic landscape and gene expression profiles of spontaneous canine glioma. Understanding the immune responses generated against canine glioma will help better integrate this model into the anti-glioma therapy development process.
Acknowledgments
This work was supported by the NIH K08NS092895 grant (MD). The bioinformatics analysis was done by the Collaborative Core for Cancer Bioinformatics (C3B) shared by IU Simon Cancer Center (Grant P30CA082709) and Purdue University Center for Cancer Research (Grant P30CA023168) and supported by Walther Cancer Foundation.
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
References
- 1.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. The New England journal of medicine. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. The Lancet Oncology. 2009;10(5):459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 3.Zeng J, See AP, Phallen J, et al. Anti-PD-1 blockade and stereotactic radiation produce long-term survival in mice with intracranial gliomas. International journal of radiation oncology, biology, physics. 2013;86(2):343–349. doi: 10.1016/j.ijrobp.2012.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reardon DA, Gokhale PC, Klein SR, et al. Glioblastoma Eradication Following Immune Checkpoint Blockade in an Orthotopic, Immunocompetent Model. Cancer immunology research. 2016;4(2):124–135. doi: 10.1158/2326-6066.CIR-15-0151. [DOI] [PubMed] [Google Scholar]
- 5.Vom Berg J, Vrohlings M, Haller S, et al. Intratumoral IL-12 combined with CTLA-4 blockade elicits T cell-mediated glioma rejection. The Journal of experimental medicine. 2013;210(13):2803–2811. doi: 10.1084/jem.20130678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bentley RT, Ahmed AU, Yanke AB, Cohen-Gadol AA, Dey M. Dogs are man's best friend: in sickness and in health. Neuro-oncology. 2017;19(3):312–322. doi: 10.1093/neuonc/now109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dix AR, Brooks WH, Roszman TL, Morford LA. Immune defects observed in patients with primary malignant brain tumors. Journal of neuroimmunology. 1999;100(1–2):216–232. doi: 10.1016/s0165-5728(99)00203-9. [DOI] [PubMed] [Google Scholar]
- 8.Walker DG, Chuah T, Rist MJ, Pender MP. T-cell apoptosis in human glioblastoma multiforme: implications for immunotherapy. Journal of neuroimmunology. 2006;175(1–2):59–68. doi: 10.1016/j.jneuroim.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 9.Lipsitz D, Higgins RJ, Kortz GD, et al. Glioblastoma multiforme: clinical findings, magnetic resonance imaging, and pathology in five dogs. Veterinary pathology. 2003;40(6):659–669. doi: 10.1354/vp.40-6-659. [DOI] [PubMed] [Google Scholar]
- 10.Snyder JM, Shofer FS, Van Winkle TJ, Massicotte C. Canine intracranial primary neoplasia: 173 cases (1986–2003) Journal of veterinary internal medicine. 2006;20(3):669–675. doi: 10.1892/0891-6640(2006)20[669:cipnc]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 11.Song RB, Vite CH, Bradley CW, Cross JR. Postmortem evaluation of 435 cases of intracranial neoplasia in dogs and relationship of neoplasm with breed, age, body weight. Journal of veterinary internal medicine. 2013;27(5):1143–1152. doi: 10.1111/jvim.12136. [DOI] [PubMed] [Google Scholar]
- 12.Joshi AD, Botham RC, Schlein LJ, et al. Synergistic and targeted therapy with a procaspase-3 activator and temozolomide extends survival in glioma rodent models and is feasible for the treatment of canine malignant glioma patients. Oncotarget. 2017;8(46):80124–80138. doi: 10.18632/oncotarget.19085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dickinson PJ, LeCouteur RA, Higgins RJ, et al. Canine spontaneous glioma: a translational model system for convection-enhanced delivery. Neuro-oncology. 2010;12(9):928–940. doi: 10.1093/neuonc/noq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Debinski W, Dickinson P, Rossmeisl JH, Robertson J, Gibo DM. New agents for targeting of IL-13RA2 expressed in primary human and canine brain tumors. PloS one. 2013;8(10):e77719. doi: 10.1371/journal.pone.0077719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dickinson PJ, York D, Higgins RJ, LeCouteur RA, Joshi N, Bannasch D. Chromosomal Aberrations in Canine Gliomas Define Candidate Genes and Common Pathways in Dogs and Humans. Journal of neuropathology and experimental neurology. 2016;75(7):700–710. doi: 10.1093/jnen/nlw042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stoica G, Kim HT, Hall DG, Coates JR. Morphology, immunohistochemistry, and genetic alterations in dog astrocytomas. Veterinary pathology. 2004;41(1):10–19. doi: 10.1354/vp.41-1-10. [DOI] [PubMed] [Google Scholar]
- 17.Higgins RBA, Dickinson P, Sisó-Llonch S. Tumors of the nervous system. Ames, Iowa: Wiley-Blackwell; 2016. [Google Scholar]
- 18.Bentley RT, Burcham GN, Heng HG, et al. A comparison of clinical, magnetic resonance imaging and pathological findings in dogs with gliomatosis cerebri, focusing on cases with minimal magnetic resonance imaging changes(double dagger) Veterinary and comparative oncology. 2016;14(3):318–330. doi: 10.1111/vco.12106. [DOI] [PubMed] [Google Scholar]
- 19.Wang F, Flanagan J, Su N, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. The Journal of molecular diagnostics: JMD. 2012;14(1):22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dey M, Chang AL, Miska J, et al. Dendritic Cell-Based Vaccines that Utilize Myeloid Rather than Plasmacytoid Cells Offer a Superior Survival Advantage in Malignant Glioma. Journal of immunology. 2015;195(1):367–376. doi: 10.4049/jimmunol.1401607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dey M, Chang AL, Wainwright DA, et al. Heme oxygenase-1 protects regulatory T cells from hypoxia-induced cellular stress in an experimental mouse brain tumor model. Journal of neuroimmunology. 2014;266(1–2):33–42. doi: 10.1016/j.jneuroim.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wainwright DA, Chang AL, Dey M, et al. Durable therapeutic efficacy utilizing combinatorial blockade against IDO, CTLA-4, and PD-L1 in mice with brain tumors. Clinical cancer research: an official journal of the American Association for Cancer Research. 2014;20(20):5290–5301. doi: 10.1158/1078-0432.CCR-14-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 25.Huang da W, Sherman BT, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID gene ID conversion tool. Bioinformation. 2008;2(10):428–430. doi: 10.6026/97320630002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rabinovich GA, Gabrilovich D, Sotomayor EM. Immunosuppressive strategies that are mediated by tumor cells. Annual review of immunology. 2007;25:267–296. doi: 10.1146/annurev.immunol.25.022106.141609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venere M, Horbinski C, Crish JF, et al. The mitotic kinesin KIF11 is a driver of invasion, proliferation, and self-renewal in glioblastoma. Science translational medicine. 2015;7(304):304ra143. doi: 10.1126/scitranslmed.aac6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ning X, Shi Z, Liu X, et al. DNMT1 and EZH2 mediated methylation silences the microRNA-200b/a/429 gene and promotes tumor progression. Cancer letters. 2015;359(2):198–205. doi: 10.1016/j.canlet.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 29.Gomori E, Pal J, Kovacs B, Doczi T. Concurrent hypermethylation of DNMT1, MGMT and EGFR genes in progression of gliomas. Diagnostic pathology. 2012;7:8. doi: 10.1186/1746-1596-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rahman WF, Rahman KS, Nafi SN, Fauzi MH, Jaafar H. Overexpression of DNA methyltransferase 1 (DNMT1) protein in astrocytic tumour and its correlation with O6-methylguanine-DNA methyltransferase (MGMT) expression. International journal of clinical and experimental pathology. 2015;8(6):6095–6106. [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Bian EB, He XJ, et al. Epigenetic repression of long non-coding RNA MEG3 mediated by DNMT1 represses the p53 pathway in gliomas. International journal of oncology. 2016;48(2):723–733. doi: 10.3892/ijo.2015.3285. [DOI] [PubMed] [Google Scholar]
- 32.Aoki H, Yokoyama T, Fujiwara K, et al. Phosphorylated Pak1 level in the cytoplasm correlates with shorter survival time in patients with glioblastoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2007;13(22 Pt 1):6603–6609. doi: 10.1158/1078-0432.CCR-07-0145. [DOI] [PubMed] [Google Scholar]
- 33.Zhang D, Li Y, Wang R, et al. Inhibition of REST Suppresses Proliferation and Migration in Glioblastoma Cells. International journal of molecular sciences. 2016;17(5) doi: 10.3390/ijms17050664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LeBlanc AK, Mazcko C, Brown DE, et al. Creation of an NCI comparative brain tumor consortium: informing the translation of new knowledge from canine to human brain tumor patients. Neuro-oncology. 2016;18(9):1209–1218. doi: 10.1093/neuonc/now051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paoloni M, Khanna C. Translation of new cancer treatments from pet dogs to humans. Nature reviews Cancer. 2008;8(2):147–156. doi: 10.1038/nrc2273. [DOI] [PubMed] [Google Scholar]
- 36.Schiffman JD, Breen M. Comparative oncology: what dogs and other species can teach us about humans with cancer. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2015;370(1673) doi: 10.1098/rstb.2014.0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berghoff AS, Kiesel B, Widhalm G, et al. Programmed death ligand 1 expression and tumor-infiltrating lymphocytes in glioblastoma. Neuro-oncology. 2015;17(8):1064–1075. doi: 10.1093/neuonc/nou307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kmiecik J, Poli A, Brons NH, et al. Elevated CD3+ and CD8+ tumor-infiltrating immune cells correlate with prolonged survival in glioblastoma patients despite integrated immunosuppressive mechanisms in the tumor microenvironment and at the systemic level. Journal of neuroimmunology. 2013;264(1–2):71–83. doi: 10.1016/j.jneuroim.2013.08.013. [DOI] [PubMed] [Google Scholar]
- 39.Jacobs JF, Idema AJ, Bol KF, et al. Regulatory T cells and the PD-L1/PD-1 pathway mediate immune suppression in malignant human brain tumors. Neuro-oncology. 2009;11(4):394–402. doi: 10.1215/15228517-2008-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jacobs JF, Idema AJ, Bol KF, et al. Prognostic significance and mechanism of Treg infiltration in human brain tumors. Journal of neuroimmunology. 2010;225(1–2):195–199. doi: 10.1016/j.jneuroim.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 41.El Andaloussi A, Lesniak MS. An increase in CD4+CD25+FOXP3+ regulatory T cells in tumor-infiltrating lymphocytes of human glioblastoma multiforme. Neuro-oncology. 2006;8(3):234–243. doi: 10.1215/15228517-2006-006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei B, Wang L, Zhao X, Du C, Guo Y, Sun Z. The upregulation of programmed death 1 on peripheral blood T cells of glioma is correlated with disease progression. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35(4):2923–2929. doi: 10.1007/s13277-013-1376-9. [DOI] [PubMed] [Google Scholar]
- 43.Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reifenberger G, Louis DN. Oligodendroglioma: toward molecular definitions in diagnostic neuro-oncology. Journal of neuropathology and experimental neurology. 2003;62(2):111–126. doi: 10.1093/jnen/62.2.111. [DOI] [PubMed] [Google Scholar]
- 45.Ligon KL, Huillard E, Mehta S, et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53(4):503–517. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mehta S, Huillard E, Kesari S, et al. The central nervous system-restricted transcription factor Olig2 opposes p53 responses to genotoxic damage in neural progenitors and malignant glioma. Cancer cell. 2011;19(3):359–371. doi: 10.1016/j.ccr.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rousseau A, Nutt CL, Betensky RA, et al. Expression of oligodendroglial and astrocytic lineage markers in diffuse gliomas: use of YKL-40, ApoE, ASCL1, and NKX2-2. Journal of neuropathology and experimental neurology. 2006;65(12):1149–1156. doi: 10.1097/01.jnen.0000248543.90304.2b. [DOI] [PubMed] [Google Scholar]
- 48.Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reifenberger G, Collins VP. Pathology and molecular genetics of astrocytic gliomas. Journal of molecular medicine. 2004;82(10):656–670. doi: 10.1007/s00109-004-0564-x. [DOI] [PubMed] [Google Scholar]
- 50.Thomas R, Duke SE, Wang HJ, et al. 'Putting our heads together': insights into genomic conservation between human and canine intracranial tumors. Journal of neuro-oncology. 2009;94(3):333–349. doi: 10.1007/s11060-009-9877-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.