A 5-year-old male presented to the Undiagnosed Diseases Network (UDN) with a history of global developmental delay, postnatal microcephaly, hypotonia, jerking movements concerning for seizures, minimal speech, severe gastroesophageal reflux disease (GERD), dysmorphic features, and partial agenesis of the corpus callosum on brain magnetic resonance imaging (MRI).
The patient was born at term following an uncomplicated pregnancy. Birth weight and length were in the 30th and 20th percentile respectively; head circumference at birth was unknown but not reported to be abnormal. He had poor feeding in the newborn period and GERD was diagnosed in the first month of life. Weight gain and height progressed normally. Head circumference measured at the 6th percentile at five weeks of age and fell below the 3rd percentile at three months.
Reduced muscle tone and truncal hypotonia were first appreciated at six months of age, as were mild motor delays. His first words were spoken at 11 months of age, but he was slow to gain vocabulary. Of note, he had no regression of developmental milestones.
Unexplained spasms concerning for seizures began at 12 months of age. Thought to correlate with gastrointestinal discomfort and gas, the episodes were characterized by extension and body stiffening followed by a scream, hyperextension of arms, head turn, and opisthotonic posturing. In the absence of epileptiform activity on electroencephalogram (EEG), his neurologists favored myoclonic spasms over seizures. Over time, the spells evolved to be more consistent with dyskinetic and choreiform movement. In addition to his myoclonic spasms, the patient also experienced myoclonic jerks characterized by downward/inward hyperextension of the arms. Trials of various anticonvulsants showed only marginal effect. He had intermittent hand stereotypies.
The patient’s medical history was significant for intermittent esotropia, mild obstructive sleep apnea, and dysmorphic features including: mild pectus excavatum, epicanthal folds, widely-spaced teeth, high-arched palate, telecanthus, and broad mouth. Throughout his workup, the patient underwent extensive diagnostic imaging and laboratory studies (Table 1; available at www.jpeds.com). There was no reported family history of similarly affected individuals. Both medical and biochemical geneticists who evaluated him indicated that his constellation of features was suggestive of an underlying genetic etiology, but not consistent with any known syndrome. With all evaluations failing to reveal an underlying etiology, the medical geneticist recommended whole exome sequencing (WES). Unfortunately, access to WES on a clinical basis was unavailable due to lack of insurance coverage by a commercial insurance provider. As a result, the child’s rare disease remained undiagnosed and the family was left without answers.
Table 1, online only.
Summary of Diagnostic Evaluations
| Exam Type | Exam | Result |
|---|---|---|
|
| ||
| Genetics labs | Karyotype (550 bands) | Normal male - 46,XY |
| Array CGH | Normal male–arr(1–22)x2,(XY)x1 | |
| Fragile X | Normal–32 CGG repeats | |
| Prader Willi-Angelman methylation | Normal | |
| 7q11.23 FISH for Williams syndrome | Normal | |
|
| ||
| Biochemical labs | Amino acids, plasma (2012) | Normal |
| Carnitine, plasma (2012) | Total carnitine: 80nmol/mL (nl 36–68), Free carnitine: 71nmol/mL (nl 27–49) | |
| Acylcarnitine (2012) | Normal acylcarnitine profile | |
| Pyruvate (2012) | Slightly elevated: 1.6 (nl 0.7–1.4) | |
| Lactate 2012 | Slightly elevated: 3.3 (nl 0.6–3.2) | |
| Organic acids, urine (2012) | Pattern not consistent with specific IEM | |
| Prolactin (2012) | Normal | |
| Carbohydrate def. transferrin (2013) | Normal | |
| CSF Cell count (2013) | Normal | |
| CSF Glucose (2013) | Normal | |
| CSF Protein (2013) | Low: <10 (nl 15–60 mg/dL) | |
| CSF Neurotransmitters (2013) | Slightly low homovanillic acid: 258 (nl 294-1115nmol/L) | |
| Amino acids, urine (2013) | Normal | |
| Amino acids, plasma (repeat - 2013) | Normal | |
| Creatine, plasma (2013) | High: 119.2 (nl 28–102umol/L) | |
| Guanidinoacetate, plasma (2013) | Elevated: 4.4 (nl 0.3–1.6umol/L); not in the range associated with classic guanidinoacetate methyltransferase deficiency | |
| Oligosaccharides, urine (2013) | Broad light band of material within an intermediate migration. Could be the result of medication. Pattern was observed once in a patient with glycogen storage disease but was not a consistent finding. | |
| Homocysteine (2013) | Normal | |
| Guanidinoacetate, plasma and urine (repeat 2013) | Elevated in plasma: 2.1 (nl 0.3– 1.6umol/L) | |
| Creatine, plasma and urine (2013) | Slight elevation in urine: 913 (nl 20– 900mmole/mole creatine) | |
| Acylcarnitine (2013) | Normal | |
| Carnitine (2013) | Normal | |
| Creatine kinase (2013) | Normal | |
| CSF amino acids | Normal | |
| Free T3 (2016) | Normal | |
| Reverse T3 (2016) | Slightly elevated: 24.5 (nl 8.3– 22.9ng/dL) | |
|
| ||
| Diagnostic Radiology | CT Head (9mo) | Normal |
| MRI Brain (11mo) | Partial agenesis of the anterior corpus callosum; Delayed myelination of frontal lobes for patient's age | |
| MRI Brain (22mo) | Hypoplastic rostrum of the corpus callosum; Incomplete myelination of the subcortical white matter of the bilateral frontal and temporal lobes | |
| MRI Brain (27mo) | Abnormal appearance of the white matter of the cerebral hemispheres most prominently involving the frontal lobes; Prominent perivascular spaces throughout the bilateral cerebral hemispheres; Hypoplastic genu of the corpus callosum. | |
| MR Spectroscopy (27mo) | NAA appears to be below normal for age; creatine is readily observed in all spectra; no evidence for elevated lactate | |
| FL Barium Swallow | Question of one episode of aspiration with solid Consistency (cookie) | |
| FL Upper GI exam | Normal | |
| XR Chest | Normal | |
|
| ||
| Other studies | EEGs, multiple | No epileptiform activity or focal abnormalities |
| Sleep study (36mo) | Mild obstructive sleep apnea | |
| pH probe | Confirmed GERD | |
| Echocardiogram | Normal | |
The Undiagnosed Diseases Network
Approximately 25–30 million individuals in the United States are living with a rare disease.(1) Many children with rare diseases remain undiagnosed throughout life, leading to excess medical care, expensive diagnostic odysseys, and frustration for patients and their families.(2,3) Advances in genomic technology have allowed for more comprehensive genetic analyses of patients with rare diseases.
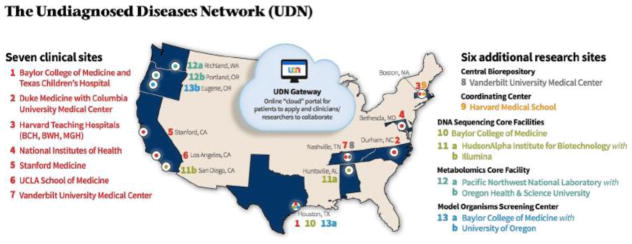
In an effort to better characterize patients with rare and undiagnosed diseases, the National Institutes of Health launched a single-site project, the Undiagnosed Diseases Program, to improve our understanding of the etiology of these disorders. Following initial success, the program expanded to encompass additional clinical and research institutions, thus establishing the Undiagnosed Diseases Network.(4) The UDN is a network of investigators across 13 institutions designated to serve public need by bringing expertise in clinical diagnostics, translational research, and multi-omics technologies to solve medical mysteries (Figure 1).
Figure 1.
Geographical representation and structure of Undiagnosed Diseases Network clinical and research sites as of September 28th, 2017.
Delineated by the NIH Common Fund, the UDN’s main objectives are 3-fold: (1) to improve the level of diagnosis and care for patients with undiagnosed diseases; (2) to facilitate research into the etiology of undiagnosed diseases; and (3) to create an integrated and collaborative research community to identify improved options for optimal patient management. With these goals in mind, the UDN began accepting applications in September 2015.
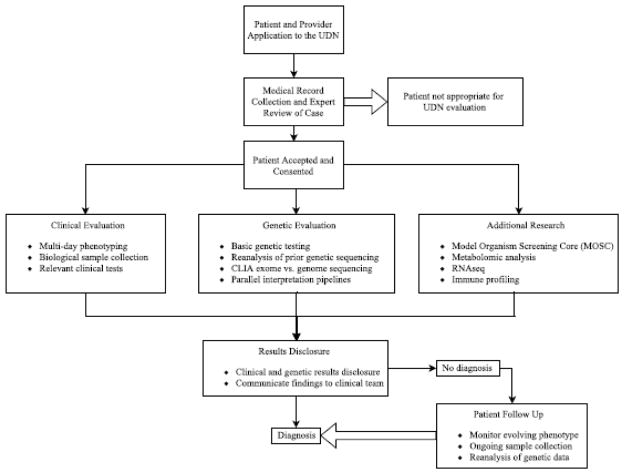
The UDN accepts applications from both pediatric and adult patients. Applicants are eligible if they have a condition with at least one objective clinical finding that remains undiagnosed despite thorough evaluation by a healthcare provider. The most common disease domains of applicants include neurology, musculoskeletal, and allergy/immunology. Participation in the UDN requires consent to store and share information and biomaterials among both UDN centers and collaborating research institutions. A UDN-wide committee of clinicians reviews each patient prior to acceptance into the study. Patients are seen at one of the seven clinical sites by expert clinicians for an evaluation that often spans several days. Specific evaluations and additional research studies are determined on a case-by-case basis by UDN clinician-scientists (Figure 2).
Figure 2.
Sample workflow of patient application and participation in the UDN.
Teams of clinicians and medical researchers join together to conduct precise clinical evaluations, analyze genomic data, and pursue state-of-the-art follow-up studies to understand complex disease mechanisms. Ultimately, the UDN aims to reduce the burden of undiagnosed diseases on patients, families, and providers. In this report, we use the case example above to illustrate the function and mission of the UDN.
Evaluation at a clinical site of the UDN
Initial evaluation
Upon exhausting all clinically available diagnostic evaluations, the treating medical geneticist referred the patient to the UDN with the hope of revealing a unifying diagnosis. The patient’s parents submitted an application to the UDN via the online portal (http://gateway.undiagnosed.hms.harvard.edu) with the required short medical practitioner referral letter. The patient’s application, one of 1918 applications received by the UDN, was then subject to detailed medical record review and discussion with a multi-disciplinary team of experts at one of the seven UDN clinical sites. The patient’s application was one of 824 accepted to date. The site expert review panel and the network-wide panel accepted this patient’s case for enrollment in the UDN.
Once accepted and regardless of socioeconomic status, patients receive the benefit of extensive clinical evaluations with appropriate specialists in addition to access to clinical and translational research studies including genomic testing when appropriate. The initial encounter for this patient consisted of an in-person research study consent with a genetic counselor and clinical research coordinator, and blood and urine sample collection for genomic and metabolomic analyses. DNA was extracted from blood from the patient and his unaffected parents, who served as controls, to perform trio WES analysis via the CLIA-certified exome sequencing core (Table II; available at www.jpeds.com). Clinical variant interpretation by the sequencing lab was guided by American College of Medical Genetics and Genomics recommendations.(5) In parallel, research personnel at the clinical site applied a variety of computational algorithms for genomic analysis with the goal of increasing the likelihood of finding a molecular diagnosis for this patient. Relevant findings on genetic testing would guide subsequent clinical evaluations with physicians.
Table 2; online only.
Whole exome sequencing metrics for proband, mother, and father.
| Proband | Mother | Father | |
|---|---|---|---|
| Bases covered at >20x | 35,361,835 | 35,227,910 | 35,335,258 |
| Average coverage | 140x | 130x | 134x |
| Number of reads | 113,645,796 | 104,581,354 | 109,752,884 |
| Percent of reads aligned | 98.85% | 98.86% | 98.74% |
Genetic and clinical evaluations
Trio WES of this patient and his unaffected parents revealed two variants: a de novo heterozygous truncating variant in the FOXG1 gene (c.624C>A; p.Tyr208X; NM_005249.4; GRCh37) and a maternally inherited heterozygous missense variant in the SCN5A gene (c.3911C>T; p.Thr1304Met; NM_198056.2; GRCh37). The FOXG1 variant had been previously reported in a patient with FOXG1 syndrome.(6) The SCN5A variant had been previously reported in patients with a variety of inherited cardiac arrhythmia syndromes, including long QT syndrome,(7–10) Brugada syndrome,(11) sudden infant death syndrome (SIDS),(12) and lone atrial fibrillation,(13) some of which carry a risk of sudden cardiac death.
Interpretation of FOXG1 variant
Clinical evaluation of the FOXG1 variant included deep phenotyping by a medical geneticist and a neurologist specialized in movement disorders. From there, close clinical comparisons were made to investigate if FOXG1 variant was sufficient to explain the patient’s constellation of features (Table 3). Team discussion with referring providers led to a consensus that the evidence supported a diagnosis of FOXG1 syndrome.
Table 3.
Clinical correlation of patient characteristics with FOXG1 syndrome
| Clinical Findings | Patient | Published FOXG1 cases19,25 |
|---|---|---|
| Pregnancy/Birth Parameters | Normal pregnancy/birth | Normal pregnancy/birth |
|
| ||
| Growth Parameters | Microcephaly noted at 3 months (−5.2 SD at present) | Deceleration of OFC noted before 1yo Microcephaly |
|
| ||
| Development | Absent speech | Absent speech |
| Sits/stands with support | Late or absent sitting/walking | |
| Absent walking | ||
|
| ||
| Brain abnormalities | Partial agenesis of the corpus callosum Hypomyelination of the subcortical white matter involving frontal and temporal loves Prominent perivascular spaces |
Hypogenesis/agenesis of corpus callosum Reduced white matter volume Simplified gyral pattern, pachygyria |
|
| ||
| Behavior/sleep | Insomnia | Poor sleep |
| Excess irritability and crying | ||
| Inappropriate laughing | ||
|
| ||
| Motor/dyskinesias | Episodic myoclonic jerks Complex spells of brief dystonic posturing +/− repetitive myoclonus Tongue protrusions Hand stereotypies Drooling |
Dyskinesias (chorea, dystonia) Spasticity Stereotypic hand movements Drooling Bruxism |
|
| ||
| Seizures/Epilepsy | Infantile spasms | Seizures |
| EEGs: no epileptic activity | ||
|
| ||
| Gastrointestinal/Respiratory | Severe GERD | GERD |
| Constipation | Constipation | |
| Feeding difficulty | Feeding difficulties | |
| Obstructive sleep apnea | Obstructive sleep apnea | |
| Aspiration | ||
|
| ||
| Musculoskeletal | Truncal hypotonia | Hypotonia |
| Pectus excavatum | Kyphosis | |
| Pes planus | ||
|
| ||
| Ophthalmologic | Strabismus | Strabismus |
| Nystagmus | ||
FOXG1 syndrome, or congenital Rett syndrome, is a rare autosomal dominant neurodevelopmental disorder characterized by global developmental delay without regression, hypotonia, movement disorders, GERD, and microcephaly. Brain imaging often reveals hypogenesis or agenesis of the corpus callosum, delayed myelination, and cortical malformations. Symptom onset is usually within the first months of life.
The FOXG1 gene was first associated with disease in a series of patients diagnosed with the congenital variant of Rett syndrome, for whom prior analysis of MECP2 or CDKL5 was uninformative.(14) Since then, approximately 80 cases of have been reported in the literature.(14–28) Although initially designated as “congenital variant Rett syndrome” due to overlap of many clinical findings and molecular mechanisms,(29) current understanding of the phenotype leaves most clinicians favoring “FOXG1 syndrome.” Frameshift, truncating, nonsense and missense variants have been identified throughout the FOXG1 gene in affected individuals.(25) Chromosomal deletions encompassing FOXG1 and its regulatory regions at 14q12 have also been described.(15–17,19,30–34) Thus far, all reported pathogenic variants have occurred de novo. However, there are families reported with multiple affected children, suggesting germline mosaicism.(35)
Interpretation of SCN5A variant
Pathogenic variants in the SCN5A gene (OMIM: 600163) cause primary arrhythmia syndromes and dilated cardiomyopathy in an autosomal dominant inheritance pattern with incomplete penetrance and variable expressivity, at times presenting as sudden cardiac death.(36) Clinical cardiovascular genetic counselor review of the SCN5A variant revealed conflicting case data, insufficient segregation data in a published family,(7) and higher-than-expected frequency in general population control samples (ExAC allele frequency: 35/126,116 European Non-Finnish alleles),(37,38) all of which bring into question the pathogenicity of the variant. Although the SCN5A variant was reported as medically actionable by the sequencing laboratory, the clinical cardiovascular genetics expert team considered it a variant of uncertain significance, citing lack of sufficient evidence of pathogenicity. As such, use of the SCN5A variant to guide predictive risk-assessment for family members was not recommended unless additional evidence supporting pathogenicity was obtained.
The uncertainty of the reported SCN5A variant warranted a baseline evaluation by a clinical electrophysiologist to assess for any clinical manifestations of these conditions. This was especially important in light of prior seizure-like episodes which at initial UDN evaluation were unclear to be all myoclonic spasms or distinct seizure episodes with possible cardiac etiology.(39) The patient was asymptomatic from a cardiovascular standpoint and an electrocardiogram, echocardiogram, and 48-hour rhythm monitoring were all within normal limits for age. Although there was no evidence for SCN5A-related disease at that time, due to the age-dependent penetrance of the syndromes associated with pathogenic variants in this gene, we recommended consideration of ongoing periodic cardiac surveillance including electrocardiograms, echocardiograms, and rhythm monitoring to mitigate risk of sudden cardiac death. We also recommended the patient’s mother seek a cardiology evaluation because the SCN5A variant was inherited maternally.
Throughout the cardiology work-up of the patient, the clinical providers communicated openly with the UDN team to understand the interpretation of the SCN5A variant and relay back their clinical assessments.
The UDN clinicians and team of referring providers disclosed the molecular diagnosis of FOXG1 syndrome and the SCN5A variant of uncertain significance to the patient’s family at a joint visit. The patient’s clinical care transitioned back to the referring providers.
Role of the Undiagnosed Diseases Network
This case epitomizes the diagnostic odyssey and potential value of comprehensive clinical and research evaluations for patients with undiagnosed diseases. Prior to enrollment in the UDN, this patient and his family went four years without an explanation for his clinical symptoms despite clinical evaluations by a wide range of providers. The differential diagnosis was broad and included serine biosynthesis disorders, Coffin-Lowry syndrome, Allan-Herndon-Dudley syndrome, Sandifer syndrome, and Angelman syndrome. Unfortunately, insurance barriers limited access to additional diagnostic testing (specifically, WES) that could have aided providers in pinpointing a diagnosis earlier.
Impact of molecular diagnostic testing
An important outcome of achieving a genetic diagnosis for this patient was newly informed management of his neurologic symptoms. Prior to evaluation through the UDN and for much of his early life, the etiology of his involuntary movements was not well understood. In fact, only recently have researchers been able to better delineate the movement disorders associated with FOXG1 syndrome.(40) Following molecular diagnosis, our patient established care with a movement disorder specialist and began a regimen of medication (clonazepam), which has led to a marked reduction of symptoms.
Although the evolution of his abnormal movement spells is still ongoing, he is now cared for by a neurologist who is familiar with FOXG1 syndrome and its associated neurologic features. Although it occurs for a minority of patients, identification of secondary findings reported on WES is a real possibility. The “medically actionable” SCN5A variant, upon closer interpretation by genetics professionals and in light of cardiology evaluations, was not found to correlate with overt clinical disease. This case demonstrates the UDN’s approach to genetic findings of uncertain significance by modeling the importance of appropriate referrals, clinical correlation, and review of all reported genetic findings on WES.
Advancing understanding of rare disease
The patient’s thorough clinical phenotyping and molecular testing was supplemented by additional computational tools and resources utilized in the UDN which could have pointed clinical providers to a more specific differential diagnosis for our patient. Such tools include publicly available automated genotype-phenotype tools such as PhenoTips. Integrated into the UDN workflow, the PhenoTips software uses Human Phenotype Ontology terms describing the clinical presentation of patients.(41) The algorithm creates an output suggesting additional diagnostic testing and clinical features to consider. It also matches the inputted terms against Online Mendelian Inheritance in Man to prioritize syndromes which have the most phenotypic similarity. The output list of relevant syndromes can guide clinical and molecular analysis, including if there should be consideration of two distinct genetic conditions. In our patient’s case, FOXG1 syndrome was second on the PhenoTips automated differential and was used in both clinical and research curation of the patient’s whole exome sequencing data.
Clinical correlation of genotype and phenotype for our patient remained challenging even when more than 80 cases of FOXG1 syndrome had been reported in the literature. In general, understanding the clinical characteristics of a rare disease is even more arduous with a single patient. To address this challenge, the UDN harnesses emerging social media tools such as Matchmaker Exchange (http://www.matchmakerexchange.org/)(42) and participant webpages to promote data sharing and to connect individuals with similar phenotypes or genotypes. The efforts to match individuals with overlapping clinical features and candidate genes have resulted in characterization of new gene-disease relationships through small, cohort-based studies.(43)
Similarly, the UDN encourages patient and family participation in rare disease organizations which seek to provide support to their members, and to promote awareness and interest in the research community. The UDN referred the patient’s family to the International FOXG1 Foundation (foxg1.org), an organization for ~300 families and their ‘foxes,’ which has a strong focus on member connection, education, and engagement of the research community. In many cases, these patient and parent-run organizations are able to encourage research, by recruiting prominent clinical and research scientists to act as advisory members, promoting awareness of current research efforts, and fostering relationships between institutions and scientists with similar interests. The UDN recognizes that increased connections between rare disease advocates, clinicians and patients alike, will improve our understanding of undiagnosed diseases.
Integration of clinical and research evaluations
The UDN’s ability to both provide clinical care consultations and support ongoing research sets it apart from other multi-institution initiatives into rare disease. Beyond the clinical and scientific resources available, a patient’s successful participation in the UDN relies on open communication and collaboration with the referring clinical providers. The relationship between clinical providers and members of the UDN is one that promotes the two-way sharing of information. From the initial referral letter, to the team discussions on clinical and research findings, to involvement in the ultimate results disclosure and follow up, the referring providers are invited to give input throughout the process, as illustrated in our patient’s case.
Because the UDN is not structured to follow patients long term for clinical care, there is a natural transition of care back to the referring providers. As patients continue to be cared for by their home clinicians, any new clinical information is communicated back to the research teams. The UDN prioritizes team-based care, multidisciplinary research collaborations, and partnerships with treating physicians.
Opportunities for translational research
Although this patient’s case highlights 3 main strengths of the UDN, in depth phenotypic evaluation, thorough genomic evaluation, and coordination of patient care, other patients benefit from the additional research opportunities the UDN provides. For cases in which WES does not provide a molecular diagnosis, resources exist within the UDN to investigate the relationship between candidate genes and disease. The UDN’s wide-reaching clinical and research network allows for the characterization and validation of candidate variants and genes. For example, the Model Organisms Screening Core can develop mouse or zebrafish knockout models of candidate genes.(44) Studies of functional effects of uncertain genetic variants including in-vitro assays utilizing patient-derived cells(45) and mRNA transcriptional analysis(46) can provide further clues into disease mechanisms to support emerging gene-disease relationships. Other multi-omics profiling includes metabolomics, which can characterize baseline trends of thousands of metabolites in an affected individual compared with unaffected controls, in addition to healthy versus disease states, to suggest aberrant molecular pathways that may be contributing to disease. Patients without strong suspicion for a genetic etiology, such as those with unexplained immunologic conditions, may benefit from detailed immune profiling as part of their participation in the UDN. Through these cutting-edge techniques, the investigators that make up the UDN can more precisely diagnosis and understand the mechanisms of rare diseases.
Implications for Pediatric Practice
This case report highlights ongoing state-of-the-art approaches the UDN employs to solve medical mysteries. Collaborations within and outside of the UDN, among expert clinicians, biomedical researchers, informaticians, geneticists, and engaged patients and their families, are expanding the spectrum of diagnosed diseases and benefiting various patient populations spanning multiple disease domains. The UDN is one of the first and largest initiatives to pioneer the integration of translational research and emerging diagnostic techniques into clinical care for patients with undiagnosed diseases. The lessons learned from the UDN will conceivably inform the wider medical community as such tools and technologies are increasingly available to practicing clinical providers. Efforts are underway within the UDN to increase collaboration with clinical providers and patients nationwide.
Acknowledgments
Supported by the National Institutes of Health (NIH) Common Fund through the Office of Strategic Coordination/Office of the NIH Director under Award Number(s) U01HG007708 U01HG007942 and U01HG007530. E.A. is a Founder of Personalis Inc, Deep Cell Inc, and advisor to Genome Medical and SequenceBio. M.W. has ownership interest in Personalis Inc. P.F. serves as an Associate Editor for The Journal of Pediatrics. The other authors declare no conflicts of interest.
Patient and his parents; Members of the Undiagnosed Diseases Network: David R. Adams, Mercedes E. Alejandro, Patrick Allard, Mahshid S. Azamian, Carlos A. Bacino, Ashok Balasubramanyam, Hayk Barseghyan, Gabriel F. Batzli, Alan H. Beggs,, Hugo J. Bellen, Anna Bican, David P. Bick, Camille L. Birch, Devon Bonner, Braden E. Boone, Bret L. Bostwick, Lauren C. Briere, Donna M. Brown, Matthew Brush, Elizabeth A. Burke, Lindsay C. Burrage, Shan Chen, Gary D. Clark, Terra R. Coakley, Joy D. Cogan, Cynthia M. Cooper, Heidi Cope, William J. Craigen, Precilla D'Souza, Mariska Davids, Jean M. Davidson, Jyoti G. Dayal, Esteban C. Dell'Angelica, Shweta U. Dhar, Ani Dillon, Katrina M. Dipple, Laurel A. Donnell-Fink, Naghmeh Dorrani, Daniel C. Dorset, Emilie D. Douine, David D. Draper, David J. Eckstein, Lisa T. Emrick, Christine M. Eng, Ascia Eskin, Cecilia Esteves, Tyra Estwick, Liliana Fernandez, Brent L. Fogel, Noah D. Friedman, William A. Gahl, Emily Glanton, Rena A. Godfrey, David B. Goldstein, Sarah E. Gould, Jean-Philippe F. Gourdine, Catherine A. Groden, Andrea L. Gropman, Melissa Haendel, Rizwan Hamid, Neil A. Hanchard, Lori H. Handley, Matthew R. Herzog, Ingrid A. Holm, Jason Hom, Ellen M. Howerton, Howard J. Jacob, Mahim Jain, Yong-hui Jiang, Jean M. Johnston, Angela L. Jones, David M. Koeller, Isaac S. Kohane, Jennefer N. Kohler, Donna M. Krasnewich, Elizabeth L. Krieg, Joel B. Krier, Jennifer E. Kyle, Seema R. Lalani, C. Christopher Lau, Jozef Lazar, Brendan H. Lee, Hane Lee, Shawn E. Levy, Richard A. Lewis, Sharyn A. Lincoln, Allen Lipson, Sandra K. Loo, Joseph Loscalzo, Richard L. Maas, Ellen F. Macnamara, Calum A. MacRae, Valerie V. Maduro, Marta M. Majcherska, May Christine V. Malicdan, Laura A. Mamounas, Teri A. Manolio, Thomas C. Markello, Ronit Marom, Julian A. Martínez-Agosto, Shruti Marwaha, Thomas May, Allyn McConkie-Rosell, Colleen E. McCormack, Alexa T. McCray, Jason D. Merker, Thomas O. Metz, Matthew Might, Paolo M. Moretti, John J. Mulvihill, Jennifer L. Murphy, Donna M. Muzny, Michele E. Nehrebecky, Stan F. Nelson, J. Scott Newberry, John H. Newman, Sarah K. Nicholas, Donna Novacic, Jordan S. Orange, J. Carl Pallais, Christina GS. Palmer, Jeanette C. Papp, Neil H. Parker, Loren DM. Pena, John A. Phillips III, Jennifer E. Posey, John H. Postlethwait, Lorraine Potocki, Barbara N. Pusey, Amy K. Robertson, Lance H. Rodan, Jill A. Rosenfeld, Jacinda B. Sampson, Susan L. Samson, Kelly Schoch, Molly C. Schroeder, Daryl A. Scott, Prashant Sharma, Vandana Shashi, Edwin K. Silverman, Janet S. Sinsheimer, Kevin S. Smith, Ariane G. Soldatos, Rebecca C. Spillmann, Kimberly Splinter, Joan M. Stoler, Nicholas Stong, Jennifer A. Sullivan, David A. Sweetser, Cynthia J. Tifft, Camilo Toro, Alyssa A. Tran, Tiina K. Urv, Zaheer M. Valivullah, Eric Vilain, Tiphanie P. Vogel, Daryl M. Waggott, Colleen E. Wahl, Nicole M. Walley, Chris A. Walsh, Michael F. Wangler, Patricia A. Ward, Katrina M. Waters, Bobbie-Jo M. Webb-Robertson, Monte Westerfield, Anastasia L. Wise, Lynne A. Wolfe, Elizabeth A. Worthey, Shinya Yamamoto, Yaping Yang, Guoyun Yu, Diane B. Zastrow, Chunli Zhao, Allison Zheng
Acknowledgements are available at www.jpeds.com:
Abbreviations
- UDN
Undiagnosed Diseases Network
- WES
whole exome sequencing
- GERD
gastroesophageal reflux disorder
- MRI
magnetic resonance imaging
- EEG
electroencephalograms
Footnotes
Learn more at: https://undiagnosed.hms.harvard.edu/
Ethics statement: The clinical trial was approved by the Institutional Review Board of Stanford University and the NIH (NIH Study Reference Number 15-HG-0130), and written informed consent and assent was obtained from the patient and family members.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Haffner ME, Whitley J, Moses M. Two decades of orphan product development. Nature. 2002;1(October 2002):821–5. doi: 10.1038/nrd919. [DOI] [PubMed] [Google Scholar]
- 2.Spillmann RC, McConkie-Rosell A, Pena L, Jiang Y-H, Schoch K, Walley N, et al. A window into living with an undiagnosed disease: illness narratives from the Undiagnosed Diseases Network. Orphanet J Rare Dis [Internet] 2017;12(1):71. doi: 10.1186/s13023-017-0623-3. Available from: http://ojrd.biomedcentral.com/articles/10.1186/s13023-017-0623-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carmichael N, Tsipis J, Windmueller G, Mandel L, Estrella E. “Is it Going to Hurt”: The Impact of the Diagnostic Odyssey on Children and Their Families. J Genet Couns. 2015;24(2):325–35. doi: 10.1007/s10897-014-9773-9. [DOI] [PubMed] [Google Scholar]
- 4.Gahl WA, Wise AL, Ashley EA. The Undiagnosed Diseases Network of the National Institutes of Health: A National Extension. JAMA : the journal of the American Medical Association. 2015;314(17):1797–8. doi: 10.1001/jama.2015.12249. [DOI] [PubMed] [Google Scholar]
- 5.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med [Internet] 2015;17(5):405–23. doi: 10.1038/gim.2015.30. Available from: http://dx.doi.org/10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mencarelli MA, Spanhol-Rosseto A, Artuso R, Rondinella D, De Filippis R, Bahi-Buisson N, et al. Novel FOXG1 mutations associated with the congenital variant of Rett syndrome. J Med Genet. 2009;47(1):49–53. doi: 10.1136/jmg.2009.067884. [DOI] [PubMed] [Google Scholar]
- 7.Wattanasirichaigoon D, Vesely MR, Duggal P, Levine JC, Blume ED, Wolff GS, et al. Sodium channel abnormalities are infrequent in patients with long QT syndrome: Identification of two novel SCN5A mutations. Am J Med Genet. 1999;86(5):470–6. doi: 10.1002/(sici)1096-8628(19991029)86:5<470::aid-ajmg13>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 8.Kapa S, Tester DJ, Salisbury Ba, Harris-Kerr C, Pungliya MS, Alders M, et al. Genetic testing for long-qt syndrome: Distinguishing pathogenic mutations from benign variants. Circulation. 2009;120(18):1752–60. doi: 10.1161/CIRCULATIONAHA.109.863076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Priori S, Napolitano C, Schwartz P, Bloise R, Crotti L, Ronchetti E. The Elusive Link Between LQT3 and Brugada syndrome: The role of Flecainide Challenge. Circulation. 2000:11–3. doi: 10.1161/01.cir.102.9.945. [DOI] [PubMed] [Google Scholar]
- 10.Kapplinger JD, Tester DJ, Salisbury Ba, Carr JL, Harris-Kerr C, Pollevick GD, et al. Spectrum and prevalence of mutations from the first 2,500 consecutive unrelated patients referred for the FAMILION long QT syndrome genetic test. Hear Rhythm [Internet] 2009;6(9):1297–303. doi: 10.1016/j.hrthm.2009.05.021. Available from: http://dx.doi.org/10.1016/j.hrthm.2009.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geena K, Ye Chan K, Kang IS, Jinyoung S, June H, Young Keun O. A pediatric case of Brugada syndrome diagnosed by fever-provoked ventricular tachycardia. Korean J Pediatr [Internet] 2014;57(8):374–8. doi: 10.3345/kjp.2014.57.8.374. Available from: http://dx.doi.org/10.3345/kjp.2014.57.8.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnestad M, Crotti L, Rognum TO, Insolia R, Pedrazzini M, Ferrandi C, et al. Prevalence of long-QT syndrome gene variants in sudden infant death syndrome. Circulation. 2007;115(3):361–7. doi: 10.1161/CIRCULATIONAHA.106.658021. [DOI] [PubMed] [Google Scholar]
- 13.Olesen MS, Yuan L, Liang B, Hols AG, Nielsen N, Nielsen JB, et al. High prevalence of long QT syndrome-associated SCN5A variants in patients with early-onset lone atrial fibrillation. Circ Cardiovasc Genet. 2012;5(4):450–9. doi: 10.1161/CIRCGENETICS.111.962597. [DOI] [PubMed] [Google Scholar]
- 14.Ariani F, Hayek G, Rondinella D, Artuso R, Mencarelli MA, Spanhol-Rosseto A, et al. FOXG1 Is Responsible for the Congenital Variant of Rett Syndrome. Am J Hum Genet. 2008;83(1):89–93. doi: 10.1016/j.ajhg.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papa FT, Mencarelli MA, Caselli R, Katzaki E, Sampieri K, Meloni I, et al. A 3 Mb deletion in 14q12 causes severe mental retardation, mild facial dysmorphisms and Rett-like features. Am J Med Genet Part A. 2008;146(15):1994–8. doi: 10.1002/ajmg.a.32413. [DOI] [PubMed] [Google Scholar]
- 16.Jacob FD, Ramaswamy V, Andersen J, Bolduc FV. Atypical Rett syndrome with selective FOXG1 deletion detected by comparative genomic hybridization: case report and review of literature. Eur J Hum Genet [Internet] 2009;17(12):1577–81. doi: 10.1038/ejhg.2009.95. Available from: http://dx.doi.org/10.1038/ejhg.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mencarelli MA, Kleefstra T, Katzaki E, Papa FT, Cohen M, Pfundt R, et al. 14q12 Microdeletion syndrome and congenital variant of Rett syndrome. Eur J Med Genet [Internet] 2009;52(2–3):148–52. doi: 10.1016/j.ejmg.2009.03.004. Available from: http://dx.doi.org/10.1016/j.ejmg.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 18.Bahi-Buisson N, Nectoux J, Girard B, Van Esch H, De Ravel T, Boddaert N, et al. Revisiting the phenotype associated with FOXG1 mutations: Two novel cases of congenital Rett variant. Neurogenetics. 2010;11(2):241–9. doi: 10.1007/s10048-009-0220-2. [DOI] [PubMed] [Google Scholar]
- 19.Kortüm F, Das S, Flindt M, Morris-Rosendahl DJ, Stefanova I, Goldstein A, et al. The core FOXG1 syndrome phenotype consists of postnatal microcephaly, severe mental retardation, absent language, dyskinesia, and corpus callosum hypogenesis. J Med Genet. 2011;48(6):396–406. doi: 10.1136/jmg.2010.087528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olson HE, Tambunan D, Lacoursiere C, Goldenberg M, Pinsky R, Martin E, et al. Mutations in epilepsy and intellectual disability genes in patients with features of Rett syndrome. Am J Med Genet Part A. 2015;167(9):2017–25. doi: 10.1002/ajmg.a.37132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das DK, Jadhav V, Ghattargi VC, Udani V. Novel mutation in Forkhead box G1 (FOXG1) gene in an Indian patient with Rett syndrome. Gene. 2014;538(1):109–12. doi: 10.1016/j.gene.2013.12.063. [DOI] [PubMed] [Google Scholar]
- 22.Hadzsiev K, Polgar N, Bene J, Komlosi K, Karteszi J, Hollody K, et al. Analysis of Hungarian patients with Rett syndrome phenotype for MECP2, CDKL5 and FOXG1 gene mutations. J Hum Genet [Internet] 2011;56(3):183–7. doi: 10.1038/jhg.2010.156. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21160487. [DOI] [PubMed] [Google Scholar]
- 23.Seltzer LE, Ma M, Ahmed S, Bertrand M, Dobyns WB, Wheless J, et al. Epilepsy and outcome in FOXG1-related disorders. Epilepsia. 2014;55(8):1292–300. doi: 10.1111/epi.12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Bruyn C, Vanderhasselt T, Tanyalçin I, Keymolen K, Van Rompaey KL, De Meirleir L, et al. Thin genu of the corpus callosum points to mutation in FOXG1 in a child with acquired microcephaly, trigonocephaly, and intellectual developmental disorder: A case report and review of literature. Eur J Paediatr Neurol. 2014;18(3):420–6. doi: 10.1016/j.ejpn.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 25.Mitter D, Pringsheim M, Kaulisch M, Plümacher KS, Schröder S, Warthemann R, et al. FOXG1 syndrome: genotype–phenotype association in 83 patients with FOXG1 variants. Genet Med [Internet] 2017;0(March) doi: 10.1038/gim.2017.75. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28661489%0Ahttp://www.nature.com/doifinder/10.1038/gim.2017.75. [DOI] [PubMed] [Google Scholar]
- 26.Le Guen T, Fichou Y, Nectoux J, Bahi-Buisson N, Rivier F, Boddaert N, et al. A missense mutation within the fork-head domain of the forkhead box G1 Gene (FOXG1) affects its nuclear localization. Hum Mutat. 2011;32(2):2026–35. doi: 10.1002/humu.21422. [DOI] [PubMed] [Google Scholar]
- 27.Philippe C, Amsallem D, Francannet C, Lambert L, Saunier A, Verneau F, et al. Phenotypic variability in Rett syndrome associated with FOXG1 mutations in females. J Med Genet. 2010;47(1):59–65. doi: 10.1136/jmg.2009.067355. [DOI] [PubMed] [Google Scholar]
- 28.Takahashi S, Matsumoto N, Okayama a, Suzuki N, Araki a, Okajima K, et al. FOXG1 mutations in Japanese patients with the congenital variant of Rett syndrome. Clin Genet. 2012;82(6):569–73. doi: 10.1111/j.1399-0004.2011.01819.x. [DOI] [PubMed] [Google Scholar]
- 29.Dastidar SG, Bardai F, Ma C, Pirce V, Rawat V, Verma P, et al. Isoform-sepcific toxicity of Mecp2 in postmitotic neurons: Supression of neurotoxicity by FoxG1. J Neurosci. 2012;32(8):2846–55. doi: 10.1523/JNEUROSCI.5841-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ellaway CJ, Ho G, Bettella E, Knapman A, Collins F, Hackett A, et al. 14q12 microdeletions excluding FOXG1 give rise to a congenital variant Rett syndrome-like phenotype. Eur J Hum Genet [Internet] 2012;21(September 2011):1–6. doi: 10.1038/ejhg.2012.208. Available from: http://dx.doi.org/10.1038/ejhg.2012.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fryssira H, Tsoutsou E, Psoni S, Amenta S, Liehr T, Anastasakis E, et al. Partial monosomy14q involving FOXG1 and NOVA1 in an infant with microcephaly, seizures and severe developmental delay. Mol Cytogenet [Internet] 2016;9(1):55. doi: 10.1186/s13039-016-0269-1. Available from: http://molecularcytogenetics.biomedcentral.com/articles/10.1186/s13039-016-0269-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumakura A, Takahashi S, Okajima K, Hata D. A haploinsufficiency of FOXG1 identified in a boy with congenital variant of Rett syndrome. Brain Dev [Internet] 2014;36(8):725–9. doi: 10.1016/j.braindev.2013.09.006. Available from: http://dx.doi.org/10.1016/j.braindev.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 33.Perche O, Haddad G, Menuet A, Callier P, Marcos M, Briault S, et al. Dysregulation of FOXG1 pathway in a 14q12 microdeletion case. Am J Med Genet Part A. 2013;161(12):3072–7. doi: 10.1002/ajmg.a.36170. [DOI] [PubMed] [Google Scholar]
- 34.Takagi M, Sasaki G, Mitsui T, Honda M, Tanaka Y, Hasegawa T. A 2.0Mb microdeletion in proximal chromosome 14q12, involving regulatory elements of FOXG1, with the coding region of FOXG1 being unaffected, results in severe developmental delay, microcephaly, and hypoplasia of the corpus callosum. Eur J Med Genet [Internet] 2013;56(9):526–8. doi: 10.1016/j.ejmg.2013.05.012. Available from: http://dx.doi.org/10.1016/j.ejmg.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 35.McMahon KQ, Papandreou A, Ma M, Barry BJ, Mirzaa GM, Dobyns WB, et al. Familial recurrences of FOXG1-related disorder: Evidence for mosaicism. Am J Med Genet Part A [Internet] 2015;(167A):3096–102. doi: 10.1002/ajmg.a.37353. Available from: http://doi.wiley.com/10.1002/ajmg.a.37353. [DOI] [PMC free article] [PubMed]
- 36.Giudicessi JR, Ackerman MJ. Determinants of incomplete penetrance and variable expressivity in heritable cardiac arrhythmia syndromes. Transl Res [Internet] 2013;161(1):1–14. doi: 10.1016/j.trsl.2012.08.005. Available from: http://dx.doi.org/10.1016/j.trsl.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature [Internet] 2016;536(7616):285–91. doi: 10.1038/nature19057. Available from: http://www.nature.com/doifinder/10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whiffin N, Minikel E, Walsh R, Donnell-luria AO, Ing AY, Barton PJR, et al. Using & high resolution variant frequencies to empower clinical genome interpretation. 2016 doi: 10.1038/gim.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zaidi A, Clough P, Cooper P, Scheepers B, Fitzpatrick AP. Misdiagnosis of epilepsy: Many seizure-like attacks have a cardiovascular cause. J Am Coll Cardiol [Internet] 2000;36(1):181–4. doi: 10.1016/s0735-1097(00)00700-2. Available from: http://dx.doi.org/10.1016/S0735-1097(00)00700-2. [DOI] [PubMed] [Google Scholar]
- 40.Papandreou A, Schneider RB, Augustine EF, Ng J, Mankad K, Meyer E, et al. Delineation of the movement disorders associated with FOXG1 mutations. Neurology. 2016;86:1794–800. doi: 10.1212/WNL.0000000000002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Girdea M, Dumitriu S, Fiume M, Bowdin S, Boycott KM, Chénier S, et al. PhenoTips: Patient phenotyping software for clinical and research use. Hum Mutat. 2013;34(8):1057–65. doi: 10.1002/humu.22347. [DOI] [PubMed] [Google Scholar]
- 42.Philippakis AA, Azzariti DR, Beltran S, Brookes AJ, Brownstein CA, Brudno M, et al. The Matchmaker Exchange: A Platform for Rare Disease Gene Discovery. Hum Mutat. 2015;36(10):915–21. doi: 10.1002/humu.22858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schoch K, Meng L, Szelinger S, Bearden DR, Stray-Pedersen A, Busk OL, et al. A Recurrent De Novo Variant in NACC1 Causes a Syndrome Characterized by Infantile Epilepsy, Cataracts, and Profound Developmental Delay. Am J Hum Genet. 2017;100(2):343–51. doi: 10.1016/j.ajhg.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chao HT, Davids M, Burke E, Pappas JG, Rosenfeld JA, McCarty AJ, et al. A Syndromic Neurodevelopmental Disorder Caused by De Novo Variants in EBF3. Am J Hum Genet. 2017;100(1):128–37. doi: 10.1016/j.ajhg.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bashamboo A, Donohoue PA, Vilain E, Rojo S, Calvel P, Seneviratne SN, et al. A recurrent p.Arg92Trp variant in steroidogenic factor- 1 (NR5A1) can act as a molecular switch in human sex development. Hum Mol Genet. 2015;25(16):3446–53. doi: 10.1093/hmg/ddw186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shashi V, Pena LDM, Kim K, Burton B, Hempel M, Schoch K, et al. De Novo Truncating Variants in ASXL2 Are Associated with a Unique and Recognizable Clinical Phenotype. Am J Hum Genet. 2016;99(4):991–9. doi: 10.1016/j.ajhg.2016.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]