Abstract
Eukaryotic mRNA metabolism regulates its stability, localization, and translation using complementarity with counter-part RNAs. To modulate their stability, small and long noncoding RNAs can establish complementarity with their target mRNAs. Although complementarity of small interfering RNAs and microRNAs with target mRNAs has been studied thoroughly, partial complementarity of long noncoding RNAs (lncRNAs) with their target mRNAs has not been investigated clearly. To address that research gap, our lab investigated whether the sequence complementarity of two lncRNAs, lincRNA-p21 and OIP5-AS1, influenced the quantity of target RNA expression. We predicted a positive correlation between lncRNA complementarity and target mRNA quantity. We confirmed this prediction using RNA affinity pull down, microarray, and RNA-sequencing analysis. In addition, we utilized the information from this analysis to compare the quantity of target mRNAs when two lncRNAs, lincRNA-p21 and OIP5-AS1, are depleted by siRNAs. We observed that human and mouse lincRNA-p21 regulated target mRNA abundance in complementarity-dependent and independent manners. In contrast, affinity pull down of OIP5-AS1 revealed that changes in OIP5-AS1 expression influenced the amount of some OIP5-AS1 target mRNAs and miRNAs, as we predicted from our sequence complementarity assay. Altogether, the current study demonstrates that partial complementarity of lncRNAs and mRNAs (even miRNAs) assist in determining target RNA expression and quantity.
Introduction
Post transcription gene regulation mainly occurs by complementary interaction of RNA with RNA-binding proteins (RBPs) and target RNA species [1–3]. RBPs mainly utilize RNA-binding domains such as RNA recognition motifs, the K Homology (KH) domain, and Zinc Finger motifs to interact with target mRNAs or with microRNAs (miRNAs) [3, 4]. Differential binding of RBPs to target RNAs determines the ability of these RBPs to modulate RNA processing, decay, localization, and translation [3]. Complementarity between regulatory RNAs and target RNAs also contributes to gene expression regulation. For instance, perfect complementarity of small interfering RNAs (siRNA) with target mRNAs promotes mRNA decay, while partial complementarity of miRNAs accelerates mRNA decapping, deadenylation, and translation repression [5]. Recent findings suggest that complementary binding of long noncoding RNAs (lncRNAs) to miRNAs and mRNAs have functional consequences as well [6, 7]. Those changes include miRNA-mediated lncRNA decay, lncRNA-dependent mRNA decay/translation, and lncRNA-associated miRNA decay/sequestration [6–8] Altogether, complementary interactions of regulatory factors with target RNAs reveals their critical functions in post-transcriptional gene expression regulation.
Although complementarity of lncRNAs to miRNAs and mRNAs has been reported recently, little is known about the consequences of RNA complementarity on target RNA abundance and stability. Complementarity of small regulatory RNAs to mRNAs can be predicted by numerous in silico programs and high through-put analysis with mechanistic and statistical details in complementarity-based free-energy calculation [9, 10]. This kind of success is achievable due to the intrinsic simplicity of siRNA and miRNAs when interacting with target mRNAs. Due to the complex nature of lncRNAs and mRNAs sequences and interactions, predicting and experimentally testing complementarity is more difficult than assessing mRNA-miRNA interactions. Previously, it was feasible only to investigate lncRNA-mRNA or lncRNA-lncRNA interactions one-by-one [11]. Recent studies of Alu-containing lncRNAs with target mRNAs, however, have established complementarity-based gene expression regulation [12] and permitted further profiling of genome-wide RNA complementarity by in vivo cross-linking analysis [13–15]. Taken together, studies of lncRNA complementarity are mutually informing with classic small RNA complementarity.
Herein, we provide our analysis of lncRNA complementarity to target mRNAs and miRNAs with functional outcomes. We first analyzed complementarity of human and mouse lincRNA-p21 for the quantity of target mRNAs containing repeats of certain RNA sequences. We also utilized additional lncRNA, OIP5-AS1 for regulation of target mRNA and miRNA abundance. We believe our studies provide a fundamental basis of lncRNA complementarity to target RNAs for regulation of their steady-state expression levels.
Material and Methods
Cell culture, transfection, small interfering RNAs, and plasmids
Human HeLa cells and Mouse Embryonic Fibroblasts (MEF) were cultured in DMEM (Invitrogen) supplemented with 10% (v/v) FBS and antibiotics. Cells were transfected (Lipofectamine 2000, Invitrogen) with siRNAs targeting sequence of TTCTCCGAACGTGTCACGT for GFP mRNA as control, CTGCAAGGCCGCATGATGA for human lincRNA-p21 [7] or OIP5-AS1 GGCTGAGTTTCATTTGAAACAGGTG for human OIP5-AS1 [16]. Plasmids that expressed mouse lincRNA-p21-MS2, OIP5-AS1-MS2, and MS2-GST were transfected at 1–2 µg/ml. Transfected cells were generally analyzed 48 h later.
RNA affinity pull down analysis
RNA affinity pull down was performed as described previously [7, 11, 16]. Acidic phenol was used for purification of RNAs from the MST-GST pellets for microarray analysis or small RNA-sequencing [17]. Trizol (from Invitrogen) was used to extract total RNA for subsequent high through-put analysis. For human lincRNA-p21 and OIP-5 AS1 sequences, we utilized the information from a previous publication (7) and from an NCBI Reference sequence lacking BC067230 annotation in UCSC.
cDNA microarray analysis
The RNA in pull down pellets was isolated with phenol-chloroform and analyzed using mRNA microarrays. Microarray experiments were performed following the protocols of the Gene Expression and Genomics Unit in NIA/NIH. Raw microarray data were filtered by P values of _0.02, normalized by Z-score transformation, and tested for significant differences in signal intensity by Z-test. To exclude possible outliers, the sample quality was analyzed by scatter plot, principal component analysis, and gene sample Z-score-based hierarchy clustering. Transcripts were considered to be significantly changed when they had Z-test P values of < 0.05, Z-ratio absolute values of 1.5 in both directions, a multiple comparison correction false-discovery rate of _0.30, a positive Z-score signal of average intensity, and an independent one-way analysis of variance (ANOVA) on sample group analysis P value of 0.05.
miRNA sequencing analysis
Briefly, in a total reaction volume of 20 µl, 2 µg of total RNA were ligated to 100 pmol adenylated 3′ adapter containing a unique pentamer barcode (App (Barcode)TCGTATGCCGTCTTCTGCTTGT), 1 µg Rnl2 (1–249) K227Q (the plasmid for expression of recombinant ligase is available at www.addgene.org) in 50 mM Tris–HCl, pH 7.6, 10 mM MgCl2, 10 mM 2-mercaptoethanol, 0.1 mg/ml acetylated bovine serum albumin (BSA) (Sigma, St Louis, MO, USA) and 15% dimethyl sulfoxide (DMSO) for 16 h on ice. Following 3′ adapter ligation, 21 barcoded samples were pooled, and products were purified on a 15% denaturing polyacylamide gel. Small RNAs measuring 45–50 nt in length were excised from the gel, then eluted and ligated to 100 pmol 5′ oligoribonucleotide adapter (GUUCAGAGUUCUACAGUCCGACGAUC) in a 20 µl reaction volume using 1 µg T4 RNA ligase 1 (Rnl1) (Thermo Fisher, Glen Burnie, MD,USA) in 50 mM Tris–HCl, pH 7.6, 10 mM MgCl2, 10 mM 2-mercaptoethanol, 0.1 mg/ml acetylated BSA, 0.2 mM adenosine triphosphate and 15% DMSO for 1 h at 37 °C. Ligated small RNAs were purified on a 12% polyacrylamide gel, reverse transcribed using SuperScript III Reverse Transcriptase (Life Technologies, Carlsbad, CA, USA), and amplified by PCR using appropriate primers (forward primer: AATGATACGGCGACCACCGACAGGTTCAGAGTTCTACAGTCCGA; reverse primer: CAAGCAGAAGACGGCATACGA). cDNA libraries were sequenced on an Illumina HiSeq 2000 instrument at the Rockefeller University Genomics Resource Center.
RNA Complementarity analysis
Complementarity of lncRNAs and target transcripts was analyzed using methods we reported in a previous study [18]. For mouse lincRNA-p21 complementarity, we utilized the BLAST for "Optimize for Somewhat similar sequences" with "alignment parameters": Expect threshold = 1000000, word size = 7, and filter masker = none. We parsed the results to obtain a predetermined sufficient number of hits: E value <=200, Word Size >= 15, and match reverse/complimentary strand.
Copy number analysis
To calculate human and mouse lincRNA-p21 copy number per cell, we utilized a real-time PCR (RT-PCR) assay in which a standard curve was generated from their expression plasmids. Serial dilutions were performed using known quantities of human or mouse lincRNA-p21 to generate standards with copy numbers from 30–300,000. The standard curve was generated by plotting 2-Ct against copy number. HeLa and MEF were cultured as described above and harvested in 1X PBS at 70% confluency. Harvested cells were counted using a hemocytometer (Hausser Scientific). Total RNA was extracted using Trizol (Invitrogen), and 0.5 µg of RNA was reverse transcribed using Maxima H Minus Reverse Transcriptase (Thermo Scientific). cDNA was diluted 1:10 in DEPC-treated water, and 2.5 µL was used for qPCR with 2.5 µL SYBR (Kapa) and 5 µL forward and reverse primer mix. The copy number per 2.5 µL of cDNA was calculated by substituting 2-Ct into the trend line equations for human or mouse lincRNA-p21 standard curves, followed by calculations to account for dilutions and cell number.
Results
Human lincRNA-p21 maintains expression of mRNAs with sequence repeats
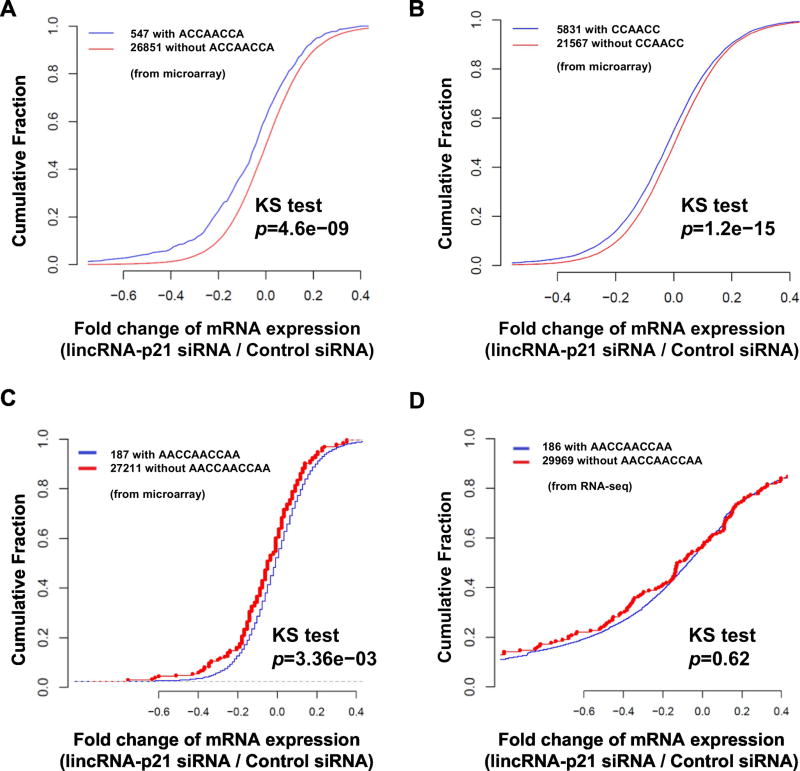
To study the complementarity of lncRNAs with mRNAs, we first focused on human lincRNA-p21 having Alu-repeats [12], one of the principal RNA sequences contributing to RNA complementarity [19]. We hypothesized that Alu-repeats in human lincRNA-p21 affect the steady-state levels of mRNAs containing certain repeat sequences, possibly by influencing their stability or transcription. To test this hypothesis, we depleted human lincRNA-p21 in HeLa cells, profiled transcriptome by cDNA microarray, and compared the abundance of mRNAs containing ACCAACCA and CCAACC sequences (on the informed assumption that these may serve as motifs for interacting with a part of lincRNA-p21 sequences). We compared the abundance of 547 mRNAs with ACCAACCA sequences and 5831 mRNAs with CCAACC sequences to the quantity of remaining mRNAs in cumulative fractions. Our results showed that ACCAACCA- and CCAACC-containing mRNAs are less abundant than the remaining mRNAs when human lincRNA-p21 is depleted (p=4.6e−09 and 1.2e−15 by Kolmogorov–Smirnov, KS, test) (Figures 1A and 1B; Tables S1 and S2). We then extended the repeat sequences to AACCAACCAA and observed, after human lincRAN-p21 depletion, that the quantity of AACCAACCAA-containing mRNAs decreased slightly when compared with the expression of remaining mRNAs (Figure 1C). We confirmed this microarray result by analyzing the cumulative fraction plot from RNA-seq (Figure 1D). Considering that human lincRNA-p21 may have a low copy number per cell, we calculated the copy number of human lincRNA-p21 in HeLa cells and found that 53 molecules of lincRNA-p21 could exist per cell (Figure S1A). In contrast, mouse lincRNA-p21 merely exists in a basal condition in MEF (Figure S1B). These results demonstrated that human lincRNA-p21 is required to achieve steady-state levels of mRNAs containing CCAACC and ACCAACCA sequences.
Figure 1. Human lincRNA-p21 affects expression of ACCAACCA and CCAACC -containing mRNAs.
(a, b, and c) Cumulative Fraction Plots of mRNAs with or without CCAACCA, ACCAACCA, and AACCAACCAA when human lincRNA-p21 is silenced in HeLa cells from the microarray result. (d) Cumulative Fraction Plot of mRNAs with or without AACCAACCAA when human lincRNA-p21 is silenced in HeLa cells from total RNA-seq data.
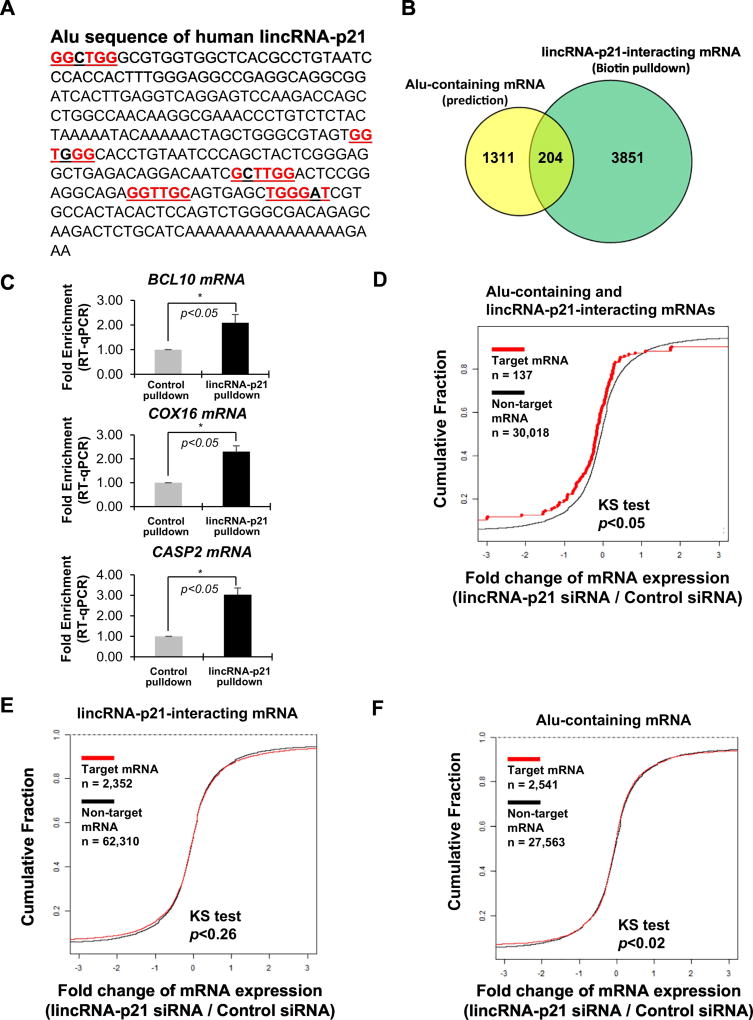
Next, we searched the human lincRNA-p21 sequence and identified GGTTGG sequences as a part of the Alu-repeat, which could establish hybridization with mRNAs containing CCAACC and ACCAACCA sequences (Figure 2A). To determine if Alu-repeat-containing mRNAs establish a biochemical complex with human lincRNA-p21, we performed an affinity pull down of human lincRNA-p21 from HeLa cells using an anti-sense DNA oligo against human lincRNA-p21, followed by a cDNA microarray analysis. Our comparison demonstrated that 204 mRNAs (out of a total of 4055 mRNAs) overlapped with mRNAs containing Alu-repeats (1515 mRNAs in total) (Figure 2B). We also validated that several mRNAs including BCL10, COX16, and CASP2 mRNA directly interacted with human lincRNA-p21 by PAR-CLIP (Photoactivatable Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation), followed by qPCR analysis (Figure 2C). The cumulative fraction analysis showed that Alu-repeat-containing mRNAs identified from human lincRNA-p21 pull down (n=137) were less abundant than the remaining mRNAs (n=30,018) (Figure 2D) when lincRNA-p21 was depleted. However, we did not observe a clear difference among all human lincRNA-p21-bound mRNAs and Alu-repeat mRNAs (Figures 2E and 2F). We also observed similar changes using RNA-sequencing data (Figures S2A, S2B, and S2C). Our results demonstrated that human lincRNA-p21 regulates the abundance of a subset of mRNAs using its sequence complementarity.
Figure 2. Human lincRNA-p21 affects abundance of a subset of Alu-repeats containing target mRNAs.
(a) Alu sequence of human lincRNA-p21 with complementary sequences to CCAACC highlighted in red. (b) Comparison of mRNAs from Alu-containing mRNAs and affinity pull down of human lincRNA-p21 in HeLa cell lysates. (c) PAR-CLIP qPCR analysis of mRNAs identified from human lincRNA-p21 pull down and microarray analysis. *p<0.05 (student’s t-test), compared with control condition. (d, e, and f) Cumulative Fraction Plots of both Alu-repeats-containing mRNAs and lincRNA-p21 pull down targets (d), pull down target mRNAs only (e), or Alu-repeats containing mRNAs only (f) along with the rest of mRNAs after human lincRNA-p21 depletion.
Since we previously reported that human lincRNA-p21 is targeted by let-7b miRNA for its decay, we hypothesized that human lincRNA-p21 attenuates target mRNA decay by influencing accessibility of let-7b to target mRNAs. To test this possibility, after silencing human lincRNA-p21, we compared the quantity of AGO-target versus non-target mRNAs (Figure S2D), and the quantity of let-7b-target versus non-target mRNAs in HeLa cells (Figure S2E). Our CDF analysis showed that, when human lincRNA-p21 was depleted, AGO-target and let-7b-target mRNAs were more abundant than the remaining mRNAs. Unexpectedly, these findings suggest that human lincRNA-p21 may facilitate miRNA-mediated mRNA decay in global scale. Taken together, our results indicate that human lincRNA-p21 maintains expression levels of a subset of mRNAs possessing CCAACC and Alu-repeat sequences by direct interaction through sequence complementarity.
Mouse lincRNA-p21 does not contain target-specific sequence complementarity
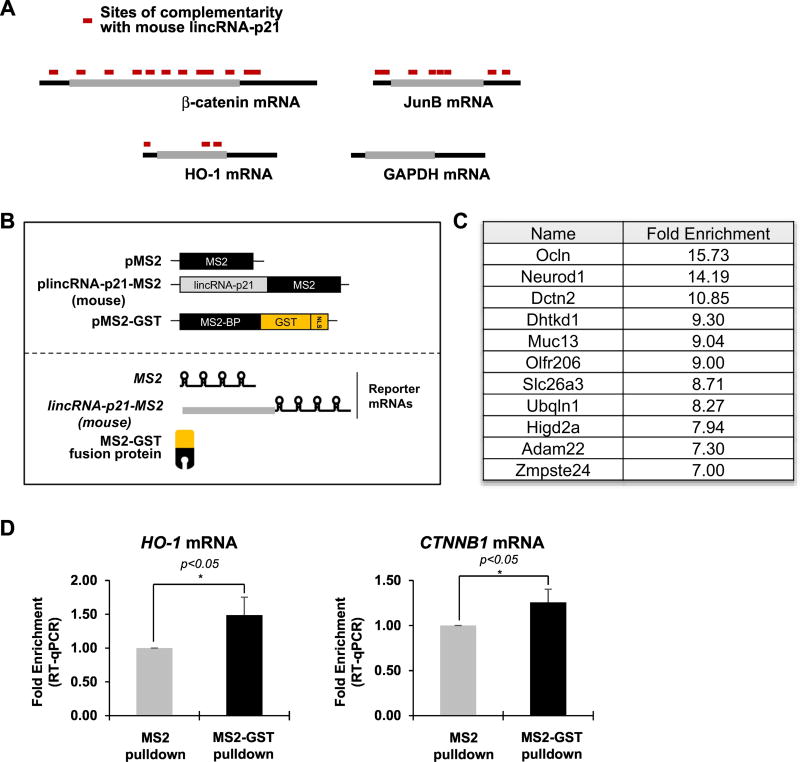
Next, we analyzed mouse lincRNA-p21 and its impact on target mRNA abundance. Lack of conservation and the absence of Alu-repeats in mouse lincRNA-p21 required that we rely on a complementarity prediction, as occurred in our previous study [7]. As in our prior analysis of partial complementarity of human lincRNA-p21 to its target mRNAs for translation regulation, we tested whether mouse lincRNA-p21 in the current study interacted with the homologous mouse mRNAs [7]. We observed 10 complementary sites on beta catenin mRNA (Ctnnb1), 6 complementary sites on JunB (Junb), and 3 complementary sites on HO-1 (Hmox1) mRNA, but found no GAPDH (Gapdh) mRNA (Figure 3A and Table S3). Because mouse lincRNA-p21 has a very low copy number in Mouse Embryonic Fibroblasts (0.04 copies per cell, Figure S1B), we performed affinity pull down after exogenous expression of mouse lincRNA-p21 with MS2 aptamer, followed by cDNA microarray analysis (Figure 3B). In that assay, we identified 266 mRNAs that co-purified with mouse lincRNA-p21 in mouse embryonic fibroblasts (Figure 3C). We validated some of those mRNAs by PAR-CLIP and qPCR analysis (Figure 3D). These results indicate that mouse lincRNA-p21 may interact with target mRNAs to establish an RNA-RNA complex.
Figure 3. Mouse lincRNA-p21 is associated a subset of mRNA.
(a) Schematic of target mRNAs containing partial complementarity with mouse lincRNA-p21. (b) Schematic of affinity pull down using MS2 aptamer for mouse lincRNA-p21 enrichment. (c) List of representative mRNAs enriched in mouse lincRNA-p21 pull down and cDNA microarray from mouse embryonic fibroblasts. (d) Existence of RNA-RNA interaction was validated by MS2 pulldown and UV crosslinking followed by qPCR analysis. *p<0.05 (student’s t-test), compared with control condition.
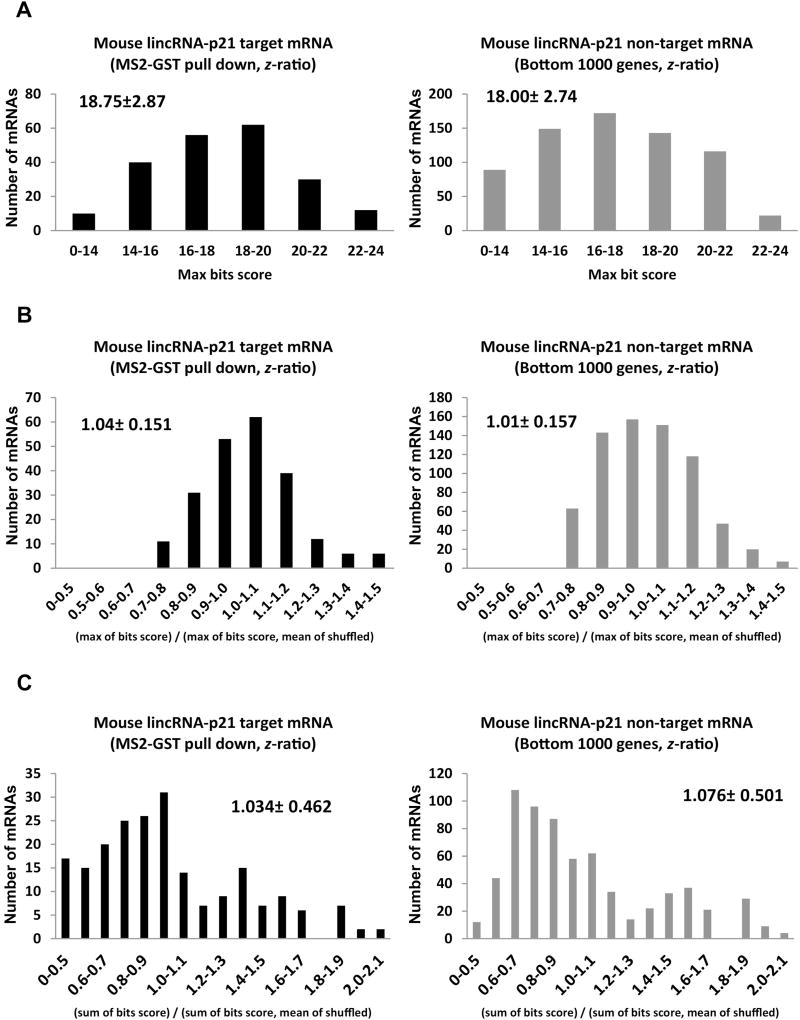
For analysis of sequence complementarity in mouse target mRNA abundance, we utilized 266 target mRNAs and 1000 non-target mRNAs from the bottom for in silico prediction and statistics. We considered target mRNAs having at least 14 nt matches and took the maximum bit score with 18.75 ± 2.87 matches for target mRNAs and 18.00 ± 2.74 for non-target mRNAs (Figure 4A). We also shuffled each mRNA sequence 100 times and blasted them against mouse lincRNA-p21, took the mean of the max bit scores for the 100 shuffled mRNA sequences, and then took the ratio (max bit score for original mRNA seq)/(mean of shuffled max bit scores). Again, our results show 1.04 ± 0.151 for target mRNAs similar to 1.01 ± 0.157 for nonZealy et al., page 8 target mRNAs (Figure 4B). Alternatively, we tried to take the mean of the sum of bit scores for the 100 shuffled mRNA sequences and determined the ratio (sum of bit scores for original mRNA seq)/(mean of sum of shuffled max bit scores) 1.034 ± 0.462 or 1.076 ± 0.501 for target or non-target mRNAs, respectively (Figure 4C). These results demonstrated that there were no preferences for mouse lincRNA-p21 in target mRNAs complementarity from transcripts profiled by affinity pull down in mouse cells.
Figure 4. Mouse lincRNA-p21 does not have complementarity with mRNAs identified from affinity pull down.
Analysis of partial complementarity with 266 target mRNAs and 1000 non-target with max bits score (a), max bits score/mean of shuffled (b), and sum of bit score/mean of shuffled (c) with mRNAs having match length greater than 14.
Next, we narrowed down the complementary sequences with E-value <15 and repeated the analysis. After utilizing 266 target mRNAs and 266 non-target mRNAs from the bottom, we identified 21.6 ± 2.67 Max bits score for target mRNAs and 21.5 ± 1.86 for non-target mRNAs (Figure S3A). We had similar observations for max of bits score / mean of shuffled (1.01 ± 0.150 and 1.00 ± 0.126) and sum of bits score / mean of shuffled (1.04 ± 0.50 and 0.99 ± 0.34) (Figures S3B and S3C). In addition, when we analyzed the top (the first) 1000 target and non-target mRNAs, there was no significant difference in mRNA complementarity (Figures S4A, S4B, and S4C). These results revealed that there is no target-specific sequence complementarity of mouse lincRNA-p21.
Human OIP5-AS1 promotes target mRNA decay by stabilizing target miRNAs
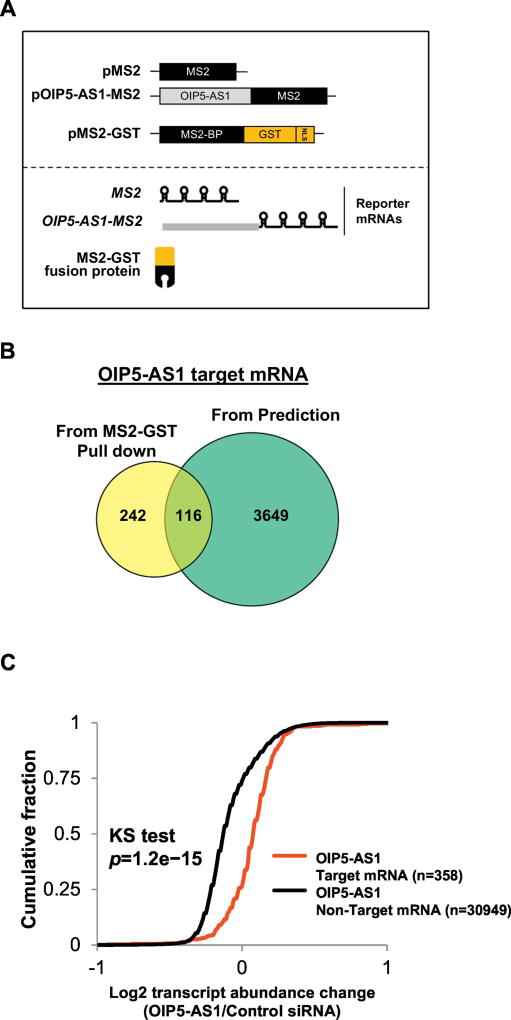
Because the effect of sequence complementarity may vary in each lncRNA, we examined an additional lncRNA, OIP5-AS1, a zebrafish homolog in humans [11, 16, 20]. After identifying human OIP5- AS1 target mRNAs by MS2-GST pull down analysis [11], we compared this profile to mRNAs containing sequence complementarity (Figures 5A, 5B, Tables S5, S6) as performed previously [18]. From this comparison, we identified 3765 mRNAs (18166 complementary sequences in total) having sequence complementarity with human OIP5-AS1 and 358 mRNAs which co-purified with human OIP5-AS1 in HeLa cells. Of those, 116 mRNAs overlapped in both assays. We also compared the influence of human OIP5-AS1 depletion in HeLa cells on the abundance of target and non-target mRNAs (Figure 5C Table S7). The result has shown that 358 target mRNAs are more abundant after human OIP5-AS1 depletion compared to 30949 non-target mRNAs. This result demonstrates that human OIP5-AS1 can decrease the abundance of target mRNAs via sequence complementarity.
Figure 5. Human OIP5-AS1 decreases steady-state level of target mRNAs.
(a) Schematic of human OIP5-AS1 pull down in HeLa cells (b) Comparison of mRNAs identified from complementarity prediction and affinity pull down analysis (c) Cumulative Fraction plot of mRNAs targeted by OIP5-AS1 identified from affinity pull down analysis when OIP5-AS1 is depleted in HeLa. The microarray of OIP5-AS1 pull down and total RNA is average of three replicates.
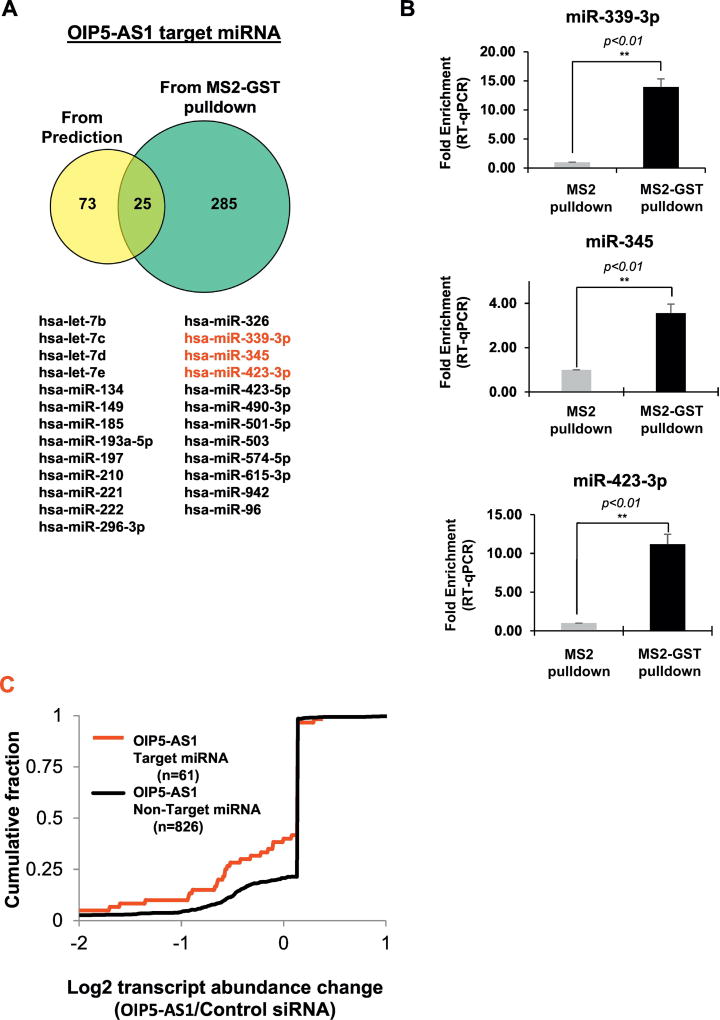
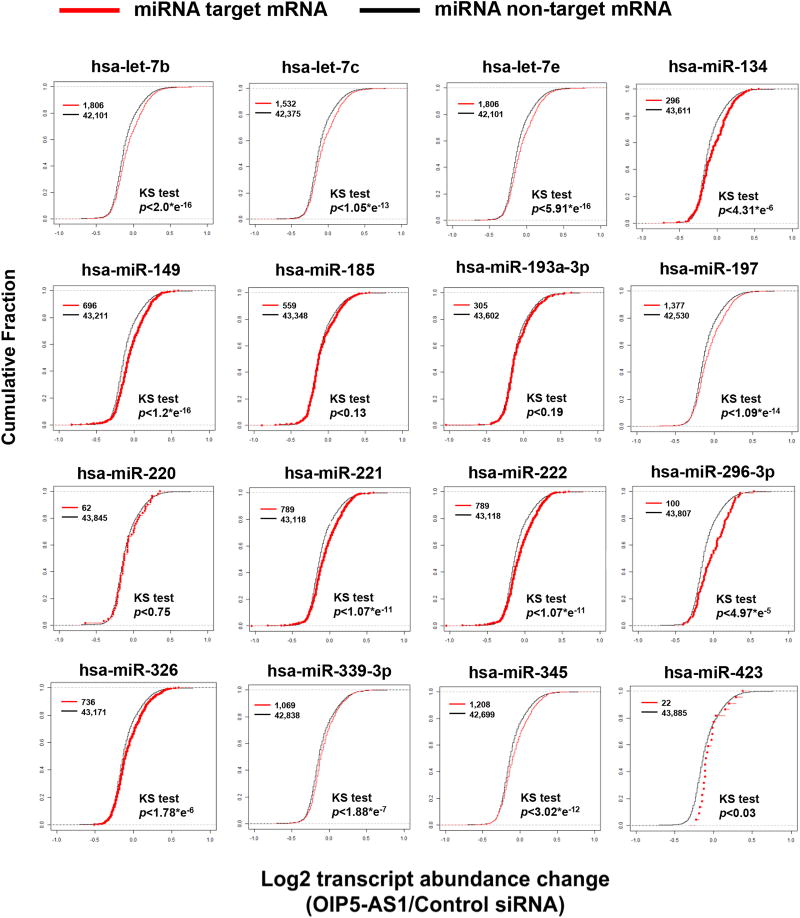
Next, we hypothesized that since human OIP5-AS1 interacts with several miRNAs (10), OIP5-AS1 may affect expression of miRNAs. We first compared miRNAs predicted to target OIP5-AS1 with those identified from affinity pull down of OIP5-AS1 followed by small RNA sequencing (Figure 6A). Our comparison has shown that 310 miRNAs are co-purified with human OIP5-AS1 from HeLa cell lysates, whereas 98 miRNAs are predicted to target OIP5-AS1 with 25 miRNAs in common (Table S8). We confirmed some of the target miRNAs by PAR-CLIP and qPCR after MS2 pull down (Figure 6B). Then, we plotted the cumulative fraction of mature miRNAs after OIP5-AS1 depletion in HeLa cells with two groups: 61 miRNAs interacting with OIP5-AS1 and 826 other miRNAs. Interestingly, we observed OIP5-AS1 target miRNAs are less abundant after OIP5-AS1 depletion in HeLa cells (Figure 6C, Table S9). We further analyzed the abundance of mRNAs targeted by OIP5-AS1-interacting miRNAs upon OIP5-AS1 depletion, and observed that OIP5-AS1 depletion increases abundance of mRNAs targeted by those miRNAs (Figure 7). Our results indicate that OIP5-AS1 interacts with target mRNAs to modulate their abundance.
Figure 6. Human OIP5-AS1 increases abundance of miRNAs.
(a) Comparison of miRNAs identified from OIP5-AS1 pull down and computational prediction (b) Validation of OIP5-AS1 target miRNAs by MS2-pulldown and UV crosslinking followed by qPCR analysis. **p<0.01 (student’s t-test), compared with control condition. (c) Cumulative fraction plot of miRNAs identified from OIP5-AS1 pull down when it is depleted in HeLa. The miRNA sequencing of OIP5-AS1 pull down and small RNA is average of two replicates.
Figure 7. miRNAs interacting with OIP5-AS1 affect abundance of target mRNAs.
Cumulative Fraction plot of mRNAs targeted by 16 miRNAs identified from OIP5-AS1 pull down when OIP5-AS1 is depleted in HeLa cells.
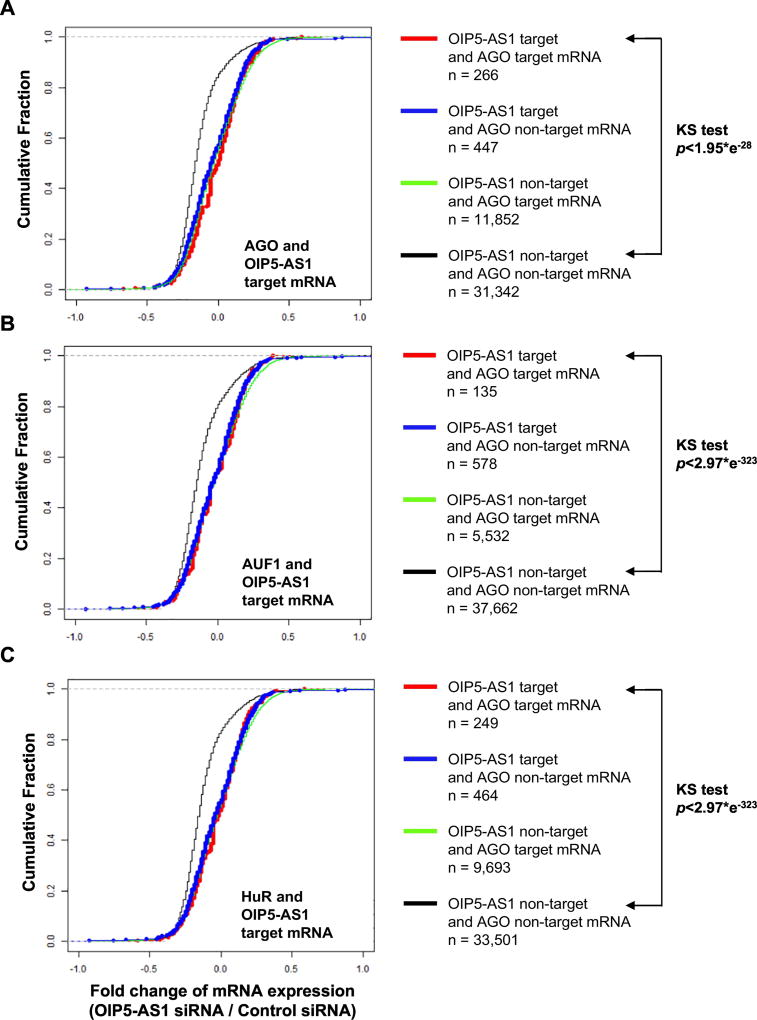
Next, we tested the possible mechanisms of how OIP5-AS1 regulates abundance of target mRNAs. To further analyze the involvement of miRNA-mediated mRNA decay, we plotted the cumulative fraction of both AGO and OIP5-AS1 target mRNAs. Our result showed that both AGO and OIP5-AS1 target mRNAs are more abundant than non-target mRNAs when OIP5-AS1 is silenced (Figure 8A). We also tested the abundance of mRNAs targeted by positive regulators of miRNA-mediated mRNA decay such as AUF1 and HuR [8, 17]. Interestingly, mRNAs targeted by AUF1 and HuR along with OIP5-AS1 exhibited higher steady state levels than non-target mRNAs (Figures 8B and 8C), when OIP-5-AS1 is depleted, suggesting that AUF1 and HuR are involved in OIP5-AS1-mediated target mRNA decay. This result demonstrates that OIP5-AS1 enhances miRNA steady-state level and promotes miRNA-mediated mRNA decay.
Figure 8. Protein factors of miRNA-mediated gene silencing are involved in abundance of mRNAs targeted by OIP5-AS1.
(a,b,c) Cumulative Fraction analysis of mRNAs targeted by AGO (a), AUF1 (b), or HuR (c) in combination of OIP5-AS1 when OIP5-AS1 is depleted in HeLa cells.
Discussion
The results described above have shown that lncRNA complementarity has functional consequences on steady-state levels of target RNA species. Our findings are exemplified by human lincRNA-p21 with Alurepeats, which has a preferential effect on Alu-repeat-containing target mRNAs (Figures 1 and 2), and by human OIP5-AS1, whose presence decreases target mRNA abundance but increases target miRNA levels (Figures 5, 6, 7, and 8). We believe our data will provide a fundamental basis for exploring the role of lncRNA complementarity in target transcript regulation.
Target mRNA complementarity with lncRNAs
Our analysis of Alu-repeats in human lincRNA-p21 and mRNAs revealed that Alu-repeat and CCAACC sequence-containing mRNAs were sensitive to lincRNA-p21 depletion as indicated by decrease of steady-state levels (Figures 1A and 1B). The presence of Alu-repeats in human lincRNA-p21 [12] suggests that this change was due in part to hybridization of Alu-repeats in human lincRNA-p21 that target mRNAs through partial complementarity (Figure 2D). Although our microarray and RNA-seq analysis after siRNA transfection showed that the abundance of complementary target mRNAs changes, we cannot rule out an offtarget effect of siRNAs contributing to miRNA abundance, non-specifically [21]. It is also possible that the interaction between CCAACC DNA sequence and nuclear lincRNA-p21 could affect transcription of target mRNAs. We also discovered 204 mRNAs out of 1505 Alu-containing RNAs identified from human lincRNA-p21 pull down, demonstrating the presence of target mRNAs in biochemical complex in human cells (Figures 2B and 2C). Although analysis of all 4055 mRNAs identified from pull down profiling revealed that there was no difference in their steady state levels after human lincRNA-p21 depletion, only a subset of those target RNAs were Alu-containing mRNAs (204 out of 4055). Similar changes were observed in mouse lincRNA-p21 with very little correlation between pull down analysis and complementarity prediction (Figures 3 and 4; Figures S3 and S4). Thus, it is very plausible that not all biochemical interactions in human and mouse cells result in the functional consequence of changing target mRNA abundance. LncRNAs could also regulate target mRNA translation, and our previous study [7] showed that human lincRNA-p21 suppresses translation of its target mRNA. It remains to be investigated whether CCTTCC sequence-containing mRNAs are also subjected to translational suppression by human lincRNAp21.
Target miRNA complementarity and stability regulation
Our profiling also identified OIP5-AS1 as a major regulator of target mRNA and miRNA abundance. First, we observed that OIP5-AS1-bound mRNAs were likely more abundant than non-target mRNAs once OIP5-AS1 was depleted (Figure 5C). Prediction of RNA complementarity between OIP5-AS1 and mRNAs has shown that less than 10% of mRNAs were indeed identified as real interactors in pull down assay (Figure 5B). Although we need to determine whether changes of steady-state levels resulted from a differential decay rate, a possible explanation could be that OIP5-AS1 depletion by siRNA mainly decreased the abundance of target miRNAs (Figure 6C). This finding implies that OIP5-AS1-target mRNAs were susceptible to miRNA-mediated decay. A follow up study should address how OIP5-AS1 promotes decay of target miRNAs, and how target mRNAs are degraded by miRNAs via deadenylation-mediated 3’-to-5’ decay. Altogether, we have shown that complementarity of lncRNA with target mRNAs could affect their steadystate level globally; however, this effect might come from an indirect interaction.
RNA complementarity with evolutionary conservation
lncRNA conservation across species remains a central issue for understanding lncRNA functionality [22]. A principal concern is why there is little evolutionary pressure to preserve lncRNA sequences, even in closely related species. When considering the possibility of RNA complementarity in cross-species conservation, we speculate that lncRNAs may have evolved to have less complementarity with mRNAs if lncRNAs are localized exclusively in the nucleus. Considering that they have a significant portion of transcripts located in the cytoplasm [7, 11, 16], lincRNA-p21 and OIP5-AS1 could have evolved to decrease the degree of complementarity to target transcripts and to minimize the detrimental effects on mRNAs and miRNAs. In contrast to numerous mRNAs interacting with non-conserved lincRNA-p21 and OIP5-AS1 in cytoplasm, nuclear-retained lncRNAs such as NEAT1 and MALAT1 are highly conserved across species. Although a portion of their transcripts is located in chromatin, NEAT1 and MALAT1 may be less likely to associate with mature mRNAs in the nucleus. Our complementarity assay of mouse and human NEAT1 and MALAT1 against mouse and human transcriptomes identified a small number of mRNAs (Tables S10, S11, S12, and S13), which supports our speculation regarding a minimum interaction between nuclear lncRNAs and mRNAs with higher conservation. Altogether, our results suggest that the evolutionary conservation of lncRNA sequences may explain their possible complementarity with target mRNAs.
In summary, we have shown that RNA complementarity in lncRNA may contribute to an abundance of target mRNAs and miRNAs. Our results from studies involving human lincRNA-p21 indicate that it regulates target mRNA abundance through sequence complementarity. In addition, the abundance of Alurepeat-containing mRNAs changed after human lincRNA-p21 depletion. Our analysis of mouse lincRNA-p21 indicates that there is no significant difference in complementarity between target and non-target mRNAs. However, we observed that a subset of OIP5-AS1-interacting mRNAs have a partial sequence complementarity. Further studies on other lncRNAs will offer an opportunity to investigate whether lncRNA complementarity in general influences mRNA metabolism.
Based on the findings reported in this study, we can utilize the concept of complementarity-based gene expression regulation in post-transcriptional gene expression. Although we did not investigate the effect of lncRNA complementarity on mRNA translation [7, 23] and protein output, future studies could determine the functional consequence of RNA complementarity on efficiency of mRNA translation and maintenance of steady-state protein levels affecting human physiology and pathobiology.
Supplementary Material
Research highlights.
Global changes of lncRNAs target mRNA abundance are measured.
Human lincRNA-p21 may regulate a subset of target mRNAs abundance in complementary dependent manner.
Mouse lincRNA-p21 does not regulate target mRNAs abundance based on sequence complementarity
Human OIP5-AS1 may regulate target mRNAs abundance by stabilizing target miRNAs.
LncRNAs could modulate many of target mRNAs abundance in complementary-dependent and independent manner.
Acknowledgments
We appreciate Andrea J Kriz, Supriyo De, Yongqing Zhang, and Xuebing Wu for computational analysis. We also thank Markus Hafner for miRNA sequencing and Jiyoung Kim for pulldown analysis. We appreciate the MUSC Center for Academic Excellence/Writing Center and the faculty reviewer for consultant editing for manuscript preparation.
Funding
RWZ, MF, SD, DM, HT, HM, JCC, ESL, KWM, and JHY were supported by start-up funds from the Medical University of South Carolina. JHY was supported by NIH/NIA intramural research program. S.H.K was supported by American Society of Nephrology Career Development award and Center for Genomic Medicine Discovery grant, MUSC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015;16:421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 2.Yoon JH, Abdelmohsen K, Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol. 2013;425:3723–3730. doi: 10.1016/j.jmb.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gehring NH, Wahle E, Fischer U. Deciphering the mRNP Code: RNA-Bound Determinants of Post-Transcriptional Gene Regulation. Trends in Biochemical Sciences. 2017;42:369–382. doi: 10.1016/j.tibs.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 4.Yoon JH, Jo MH, White EJ, De S, Hafner M, Zucconi BE, Abdelmohsen K, Martindale JL, Yang X, Wood WH, 3rd, Shin YM, Song JJ, Tuschl T, Becker KG, Wilson GM, Hohng S, Gorospe M. AUF1 promotes let-7b loading on Argonaute 2. Genes Dev. 2015;29:1599–1604. doi: 10.1101/gad.263749.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoon J-H, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Seminars in Cell & Developmental Biology. 2014;34:9–14. doi: 10.1016/j.semcdb.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoon J-H, Abdelmohsen K, Srikantan S, Yang X, Martindale Jennifer L, De S, Huarte M, Zhan M, Becker Kevin G, Gorospe M. LincRNA-p21 Suppresses Target mRNA Translation. Molecular Cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoon J-H, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga-Yamanaka K, White EJ, Orjalo AV, Rinn JL, Kreft SG, Wilson GM, Gorospe M. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun. 2013;4:2939. doi: 10.1038/ncomms3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peterson SM, Thompson JA, Ufkin ML, Sathyanarayana P, Liaw L, Congdon CB. Common features of microRNA target prediction tools. Frontiers in Genetics. 2014;5:23. doi: 10.3389/fgene.2014.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dweep H, Sticht C, Gretz N. In-Silico Algorithms for the Screening of Possible microRNA Binding Sites and Their Interactions. Current Genomics. 2013;14:127–136. doi: 10.2174/1389202911314020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J, Noh JH, Lee SK, Munk R, Sharov A, Lehrmann E, Zhang Y, Wang W, Abdelmohsen K, Gorospe M. LncRNA OIP5-AS1/cyrano suppresses GAK expression to control mitosis. Oncotarget. 2017:49409–49420. doi: 10.18632/oncotarget.17219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chillón I, Pyle AM. Inverted repeat Alu elements in the human lincRNA-p21 adopt a conserved secondary structure that regulates RNA function. Nucleic Acids Research. 2016;44:9462–9471. doi: 10.1093/nar/gkw599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu Z, Zhang Qiangfeng C, Lee B, Flynn Ryan A, Smith Martin A, Robinson James T, Davidovich C, Gooding Anne R, Goodrich Karen J, Mattick John S, Mesirov Jill P, Cech Thomas R, Chang Howard Y. RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell. 2016:1267–1279. doi: 10.1016/j.cell.2016.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aw Jong Ghut A, Shen Y, Wilm A, Sun M, Lim Xin N, Boon K-L, Tapsin S, Chan Y-S, Tan C-P, Sim Adelene YL, Zhang T, Susanto Teodorus T, Fu Z, Nagarajan N, Wan Y. In Vivo Mapping of Eukaryotic RNA Interactomes Reveals Principles of Higher-Order Organization and Regulation. Molecular Cell. 2016;62:603–617. doi: 10.1016/j.molcel.2016.04.028. [DOI] [PubMed] [Google Scholar]
- 15.Sharma E, Sterne-Weiler T, O’Hanlon D, Blencowe Benjamin J. Global Mapping of Human RNA-RNA Interactions. Molecular Cell. 2016;62:618–626. doi: 10.1016/j.molcel.2016.04.030. [DOI] [PubMed] [Google Scholar]
- 16.Kim J, Abdelmohsen K, Yang X, De S, Grammatikakis I, Noh JH, Gorospe M. LncRNA OIP5-AS1/cyrano sponges RNA-binding protein HuR. Nucleic Acids Research. 2016;44:2378–2392. doi: 10.1093/nar/gkw017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Min KW, Jo MH, Shin S, Davila S, Zealy RW, Kang SI, Lloyd LT, Hohng S, Yoon JH. AUF1 facilitates microRNA-mediated gene silencing. Nucleic Acids Res. 2017:6064–6073. doi: 10.1093/nar/gkx149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abdelmohsen K, Panda AC, Kang M-J, Guo R, Kim J, Grammatikakis I, Yoon J-H, Dudekula DB, Noh JH, Yang X, Martindale JL, Gorospe M. 7SL RNA represses p53 translation by competing with HuR. Nucleic Acids Research. 2014;42:10099–10111. doi: 10.1093/nar/gku686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3[prime] UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ulitsky I, Shkumatava A, Jan Calvin H, Sive H, Bartel David P. Conserved Function of lincRNAs in Vertebrate Embryonic Development despite Rapid Sequence Evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Putzbach W, Gao QQ, Patel M, van Dongen S, Haluck-Kangas A, Sarshad AA, Bartom ET, Kim K-YA, Scholtens DM, Hafner M, Zhao JC, Murmann AE, Peter ME. Many si/shRNAs can kill cancer cells by targeting multiple survival genes through an off-target mechanism. eLife. 2017;6 doi: 10.7554/eLife.29702. advanced online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnsson P, Lipovich L, Grandér D, Morris KV. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochimica et Biophysica Acta (BBA) - General Subjects. 2014;1840:1063–1071. doi: 10.1016/j.bbagen.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, Forrest ARR, Carninci P, Biffo S, Stupka E, Gustincich S. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.