Abstract
Background and purpose
Hypoxia is a known prognostic factor in head and neck cancer. Hypoxia imaging PET radiotracers such as 18F-FMISO are promising but not widely available. The aim of this study was therefore to design a surrogate for 18F-FMISO TBRmax based on 18F-FDG PET and contrast-enhanced CT radiomics features, and to study its performance in the context of hypoxia-based patient stratification.
Methods
121 lesions from 75 head and neck cancer patients were used in the analysis. Patients received pre-treatment 18F-FDG and 18F-FMISO PET/CT scans. 79 lesions were used to train a cross-validated LASSO regression model based on radiomics features, while the remaining 42 were held out as an internal test subset.
Results
In the training subset, the highest AUC (0.873 ± 0.008) was obtained from a signature combining CT and 18F-FDG PET features. The best performance on the unseen test subset was also obtained from the combined signature, with an AUC of 0.833, while the model based on the 90th percentile of 18F-FDG uptake had a test AUC of 0.756.
Conclusion
A radiomics signature built from 18F-FDG PET and contrast-enhanced CT features correlates with 18F-FMISO TBRmax in head and neck cancer patients, providing significantly better performance with respect to models based on 18F-FDG PET only. Such a biomarker could potentially be useful to personalize head and neck cancer treatment at centers for which dedicated hypoxia imaging PET radiotracers are unavailable.
Keywords: FMISO, FDG, hypoxia, radiomics, PET, CT, head and neck cancer, PET/CT
1. Introduction
Head and neck squamous cell carcinomas (HNSCC) treated with chemoradiotherapy can reach overall survival rates of nearly 90% when human papillomavirus (HPV)-positive (HPV+) tumors dominate the cohort [1, 2, 3]. However, head and neck tumors with regions of insufficient oxygenation –a condition known as hypoxia– are still associated with poor prognosis [4]. Studies have shown that hypoxia triggers angiogenesis, increases radioresistance and reduces the effectiveness of surgery, an alleged consequence of hypoxia selected pressure for tumor aggressiveness and metastasis [5, 6, 7, 8]. This has motivated the development of HNSCC chemoradiotherapy protocols that stratify patients based on the assessment of tumor hypoxia [9, 10, 11]. Major alterations of the treatment are being tested in clinical trials, such as the reduction of the radiation dose to non-hypoxic lesions from the standard 70 Gy to 30 Gy, thereby significantly reducing the risk of normal tissue toxicity [12, 13].
The preferred technique to detect hypoxia in this context is currently positron emission tomography (PET) combined with a hypoxia radiotracer such as 18F–Fluoromisonidazole (18F-FMISO) [14, 15, 16]. Previous studies have shown that 18F-FMISO PET is a prognostic biomarker both before and during treatment [17, 18, 19]. However, 18F-FMISO is still an investigational agent that requires an Institutional Review Board (IRB) with an Investigational New Drug approval from the FDA. The task of extending existing hypoxia-based HNSCC trials to the routine clinical setting would be simpler if an equivalent biomarker could be obtained from standard imaging techniques.
Hypoxia has been shown to increase 18F-Fluorodeoxyglucose (18F-FDG) uptake in cancer cell lines [20]. This is not however the only contribution to the 18F-FDG signal, and studies assessing the overall correlation between 18F-FDG and the level of hypoxia obtain conflicting results [21, 22]. In this study we sought radiomics features to explore the hypothesis that an image-based biomarker obtained from the combination of 18F-FDG PET scans and their accompanying contrast-enhanced CT scans correlates with 18F-FMISO-assessed hypoxia, with contrast-enhanced CT scans acting as surrogates for blood flow [23]. In particular, we analyze its utility for the task of discriminating with enough specificity a subset of candidate patients for dose de-escalation, using only imaging techniques employed routinely in the clinical setting. To maximize the information extracted, we quantify the images using radiomics features [24] and develop the biomarker using machine learning.
2. Materials and methods
2.1. Dataset and hypoxia definition
Patients were treated for head and neck cancer with chemoradiation at Memorial Sloan Kettering Cancer Center (MSKCC) under a de-escalation trial (IRB #04-070; NTC00606294 on clinicaltrials.gov). To be included in the present retrospective analysis, patients had to have received pre-treatment 18F-FDG static PET/CT for treatment planning, as well as 18F-FMISO dynamic PET/CT. Scans were performed in a GE Medical Systems PET/CT scanner with the patient immobilized with a customized mask. Prior to the 18F-FDG PET/CT scan patients were given 100 ml of omnipaque contrast. The 18F-FMISO PET/CT scan consisted of a 30-minute dynamic scan followed by two 10-minute static scans obtained approximately 90 and 150 minutes post-injection, respectively. The average time between the two PET scans was 6 days, with a standard deviation of 5 days. All patients with the available data treated between January 2011 and February 2016 were included in the analysis.
The lesions were delineated by experienced physicians on the contrast-enhanced CT for radiotherapy treatment planning. 18F-FDG uptake was quantified in terms of the body-weight standardized uptake value (SUV). 18F-FMISO uptake was normalized by the uptake measured in the ipsilateral (with respect to the lesion) jugular vein. The level of hypoxia of a lesion was defined in terms of its maximum tumor-to-blood uptake ratio (TBRmax) on the last static scan. In particular, in this study a lesion was considered to be hypoxic if TBRmax > 1.41.
2.2. Radiomics features
Radiomics features were extracted from both 18F-FDG PET and CT scans, including itensity features (from PET and CT), shape features (CT), and texture features (CT). Intensity features also included variables based on two-dimensional volume-intensity histograms, for example the volume spanned by voxels with intensities above 70 Hounsfield units (HU), denoted V>70. Features were extracted in Matlab using the CERR Radiomics Imaging Quantification toolbox (RIQ, [25, 26], April 2017 version). All code used to derive features is open source and available for download. Table S1 in the Supplementary Material includes a full list of the radiomics features computed.
In CT scans, radiomics features were extracted for each lesion from four different volumes of interest, denoted vGTV (gross tumor volume, GTV), vCT∼ (GTV after filtering out voxels outside of HU ∈ [−100, 150]), v↑ (SUV > 42% SUVmax) and v↓ (SUV < 42% SUVmax), respectively, as shown in Figure 1.
Figure 1.
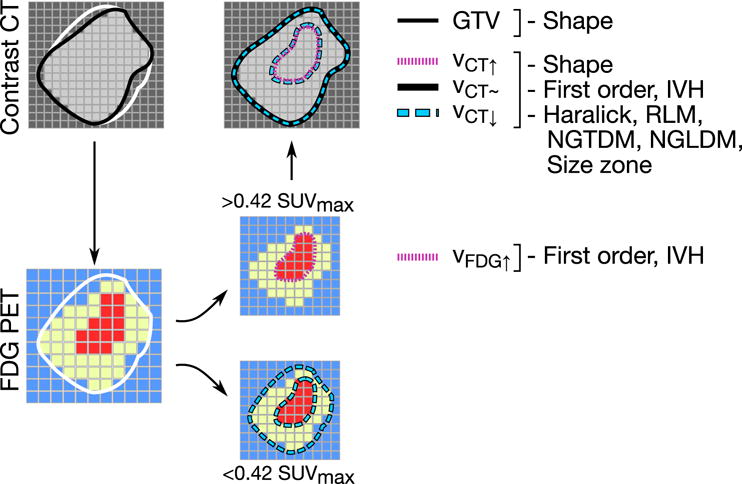
Feature extraction pipeline, indicating the volumes used to compute each family of features. IVH = Intensity Volume Histogram, RLM=Run Length Matrix, NGTDM=Neighborhood Gray-Tone Difference Matrix, Neighboring Gray-Level Dependence Matrix.
For 18F-FDG PET features, the volume of interest was defined as the region within the GTV with SUV > 42% SUVmax [27]. We use the intersection between the fixed-threshold contour and the GTV to avoid the overestimation of small lesion boundaries [28].
2.3. Data analysis
All lesions with a volume larger than 10 cm3 were considered for the analysis. The dataset was divided into a training subset comprising approximately 65% of the lesions, and an internal test subset which has held out and only used to test the final models. TBRmax was used as the continuous response variable to predict with a supervised learning model. However, as the ultimate goal was stratification, the response was dichotomized by classifying lesions with TBRmax > 1.4 as hypoxic, and the performance was evaluated in terms of the area under the receiver operating characteristic curve (AUC).
First, the training dataset was tested for any univariable associations between clinical predictors and the lesions TBRmax. Correlations between numerical predictors and TBRmax were measured in terms of the Spearman correlation coefficient, while associations with categorical predictors were assessed using balanced one-way ANOVA. p-values were corrected for multiple-testing using the Benjamini-Hochberg procedure.
Figure 2 illustrates the radiomics modelling process, which was performed in steps: (1) feature selection based on cross-validated LASSO regression; (2) multivariable linear model building, based on the selected features; and (3) evaluation of the best linear model on the hold-out test set. The process was performed independently for CT and 18F-FDG PET features. Steps (2) and (3) were also performed for the combined set of selected PET and CT features.
Figure 2.
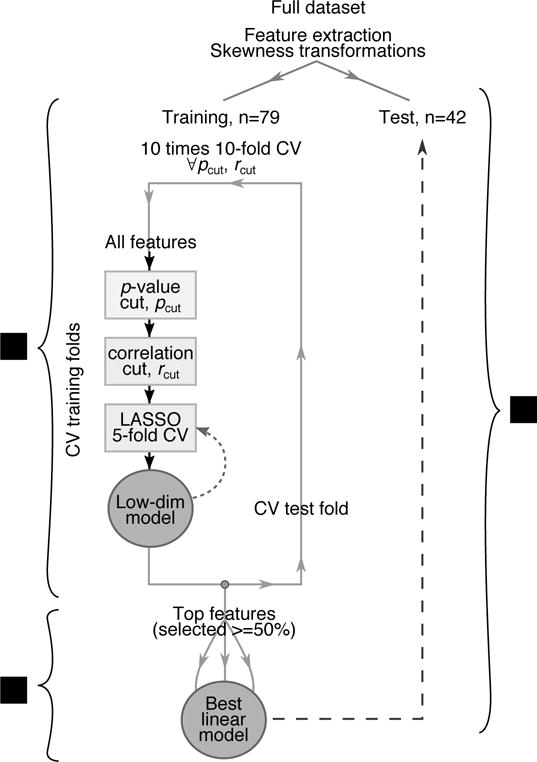
Flow chart of the steps followed to derive the predictive model, including (1) feature selection, (2) model building, and (3) testing. The curved, dashed line indicates the addition of interaction terms between the selected features to assess the importance of non-linearity.
The feature selection step is based on a LASSO linear model embedded in a 10-fold cross-validation loop reshuffled 10 times. In each fold features were pre-selected by applying a univariable p-value cut and removing those that were highly correlated. The remaining features were used in a LASSO linear model with nested 5-fold cross-validation to determine the regularization parameter yielding the smallest mean square error. The selected features were used again as input to a second LASSO model, this time also including bilinear interaction terms, to assess the non-linearity of the problem. The whole process, outlined in Figure 2, was repeated multiple times to scan over possible values of the pre-selection cuts (p = 0.01, 0.02,… 0.1, Spearman r = 0.5, 0.6… 0.8) and find the ones that maximized the cross-validation AUC.
For the model building step, only features selected in 50% of the 100 cross-validation runs or more were used. Multiple 1- and 2-variable linear regression models were created by taking all possible combinations of the selected features. An optimistic bound on the expected performance of the models was determined in terms of the mean AUC obtained from 10-fold cross-validation reshuffled 10 times.
For each category (PET, CT, or PET+CT) only the linear model with the best AUC was evaluated on the test dataset. The final model coefficients were determined by fitting to the entire training subset.
To assess whether the test AUCs of the three models were significantly better than a model based only on (the 90th percentile of the 18F-FDG SUV, used here as a robust variant of the maximum SUV), we computed 1000 bootstrap replicas of the test dataset, calculated the corresponding AUCs, and derived a p-value based on the two-sided Wilcoxon rank sum test.
Further details about the definition of the radiomics features and the data analysis procedure can be found in the supplementary material.
3. Results
75 patients satisfying all the requisites were identified, adding up to 121 lesions in total. A randomly chosen subset of 79 lesions were used for training, and the remaining 42 were held out for testing purposes. Patient characteristics are listed in Table 1. None of them were found to be significantly different between the training and testing datasets.
Table 1.
Characteristics of the lesions used in the analysis. For tumor subsite, HPV and p16 status, p-values are based on the Fisher exact test. For the mean and standard deviations of the 18F-FDG maximum SUV and 18F-FMISO TBRmax, p-values are based on the 2-sample t-test.
| Type |
Ntrain (Fraction) |
Ntest (Fraction) |
p-value |
|---|---|---|---|
| Subsite | |||
|
| |||
| Base of Tongue | 40 (0.51) | 21 (0.5) | 1 |
| Hypopharynx | 1 (0.01) | 1 (0.02) | 1 |
| Supraglottic larynx | 2 (0.03) | 1 (0.02) | 1 |
| Tonsil | 32 (0.41) | 19 (0.45) | 0.7 |
| Unknown | 4 (0.05) | 0 (0) | 0.3 |
|
| |||
| HPV status | |||
|
| |||
| HPV+ | 45 (0.57) | 27 (0.64) | 0.6 |
| HPV− | 11 (0.14) | 7 (0.17) | 0.8 |
| Unknown | 23 (0.29) | 8 (0.19) | 0.3 |
|
| |||
| p16 status | |||
|
| |||
| p16+ | 57 (0.72) | 34 (0.81) | 0.4 |
| p16− | 7 (0.09) | 4 (0.1) | 1 |
| Unknown | 15 (0.19) | 4 (0.1) | 0.2 |
|
| |||
| Type | Training Mean (SD) |
Test Mean (SD) |
p-value |
|
| |||
| 18F-FMISO TBRmax | 1.91 (0.73) | 1.76 (0.56) | 0.3 |
| 18F-FDG SUVmax | 13.64 (5.76) | 13.74 (5.23) | 0.9 |
3.1. Association with clinical variables
TBRmax was tested for association with HPV status, p162 status and tumor subsite, and no significant correlation was found. Clinical variables were therefore not included in the radiomics models.
3.2. Model training
CT model
The optimal pre-selection cuts were pcut = 0.02 and rcut = 0.6. Two variables were selected by the LASSO model 50% of the time or more: the long-run high-grey-level emphasis of the v↑ region, denoted ; and V>70 of the v↓ region, denoted . The specific definition of that was selected was sensitive to texture directionality; in particular, it used the run-length matrix calculated along the direction that yielded the maximum result in that tumor. The resulting cross-validated AUC was 0.78, as shown in Table 2. Adding interaction terms did not improve the result. Using these two features only in a multivariable regression model yielded a mean cross-validation AUC of 0.841, higher than using any of the two features alone.
Table 2.
Performance of the models derived from the training subset assessed in terms of the mean and standard deviation (SD) of the cross-validation AUC. AUCs are also given for the test subset, indicating the bootstrap p-value of the difference with respect to the model with only .
| Modality | Feature pre-selection | Interaction terms | AUC | |
|---|---|---|---|---|
| Training | mean ± SD | |||
|
| ||||
| CT | None | No | 0.78 ± 0.03 | |
| None | Yes | 0.78 ± 0.03 | ||
|
| ||||
|
ε↑,
|
No | 0.841 ± 0.007 | ||
|
| ||||
| PET | None | No | 0.852 ± 0.008 | |
| None | Yes | 0.853 ± 0.007 | ||
|
| ||||
|
|
No | 0.855 ± 0.005 | ||
|
| ||||
| , | No | 0.861 ± 0.007 | ||
|
| ||||
| PET + CT | , | No | 0.873 ± 0.008 | |
|
| ||||
| Test | value (p) | |||
|
| ||||
| PET |
|
No | 0.756 | |
| PET | , | No | 0.767 (p < 0.001) | |
| CT |
ε↑,
|
No | 0.828 (p < 0.0001) | |
| PET + CT | , | No | 0.833 (p < 0.0001) | |
PET model
The optimal pre-selection cuts were pcut = 0.01 and rcut = 0.5. The maximum SUV, here defined in a robust way via the 90th percentile, , was selected 100% of the time. In addition, the skewness of the SUV distribution, , was selected exactly 50% of the time. The cross-validated AUC was 0.852, with no improvement gained from adding interactions. Using these two features only in a multivariable regression model resulted in a mean cross-validation AUC of 0.861, higher than using any of the two features alone.
Combined CT and PET model
All the possible combinations between the top two CT and PET features were considered as predictors in a multivariable linear regression model. The highest performance was achieved when combining and , with an AUC of 0.873. Adding interaction terms did not improve the performance.
All the intermediate performance results can be found in Table I of the supplementary material.
3.3. Model testing
Four multivariable regression models were tested on the unseen test subset: (i) and , (ii) , (iii) and , (iv) and . They were trained on the entire training dataset. The performance obtained in terms of AUC was 0.828, 0.736, 0.767 and 0.833, respectively, as shown in Table 2. The highest performance was achieved by the combined and model, in agreement with what was observed in the training dataset. The discriminative power of the two features can be seen in Figure 3a, while the correlation between the linear combination of the two features and 18F-FMISO TBRmax is shown in Figure 3b. The AUC of the combined model was significantly higher than the AUC of the model based only on (p < 0.0001).
Figure 3.
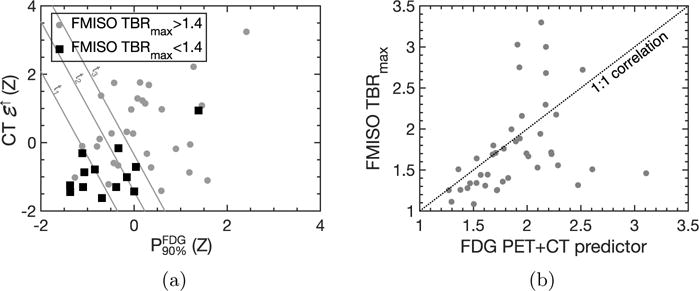
(a) Scatter plot of against for the test dataset. The markers are color coded according to their TBRmax value. The grey lines result from applying fixed thresholds to the model derived from the training dataset. The resulting classifications have the following false positive and true positive rates for TBRmax > 1.4: t1 FPR=0.07, TPR=0.72; t2 FPR=0.31, TPR=0.76; t3 FPR=0.46, TPR=0.97. Threshold t1 would result in 6 normoxic lesions out of 13 being classified as such, with only one hypoxic lesion being misclassified as normoxic. Threshold t2 would result in 8 normoxic lesions being correctly classified, with 7 hypoxic lesions being incorrectly classified as normoxic. Finally, threshold t3 would classify 12 out of 13 normoxic lesions correctly, but it would incorrectly classify 9 hypoxic lesions as normoxic. (b) Correlation plot of the combined and signature versus F-FMISO TBRmax in the test dataset (Spearman ρ = 0.57, p < 0.01). For values of the PET+CT predictor below 2.0, the Spearman ρ is 0.72, p < 0.01.
4. Discussion
This study explores the possibility of obtaining a surrogate hypoxia biomarker derived from conventional imaging techniques. The proposed signature is based on 18F-FDG PET and its companion contrast-enhanced CT scan, and reaches an AUC of 0.83 in the test dataset.
We used TBRmax to quantify hypoxia as it minimizes uptake normalization errors and is highly correlated with the tumor-to-muscle ratio that is commonly used in clinical trials [9, 13, 30]. It has been argued that for a more reliable determination of tumor hypoxia a kinetic analysis of dynamic PET data may be needed [31], as the raw 18F-FMISO uptake is expected to be affected by factors such as the distribution volume [32]. An interesting follow-up study would be to find predictors of the irreversible binding rate of 18F-FMISO, as determined from kinetic analysis.
We found that the most predictive 18F-FDG PET features were the 90th percentile and the skewness of the SUV distribution, denoted and respectively. captures the bias of the distribution towards either the higher or lower ends of the SUV spectrum, and its role could be to capture the fraction of hot (and potentially hypoxic) voxels. While remains important in models including CT features, the contribution of becomes negligible.
We studied features extracted from CT scans independently, but the analysis was in fact not completely agnostic to 18F-FDG PET, as the volumes of interest included high-SUV and low-SUV subregions of the GTV. The two top CT features were the volume taken by voxels with HU> 70 within the low-SUV subvolume (denoted ) and the long-run high-grey-level emphasis calculated along the direction with the maximum value within the high-SUV subvolume (denoted ). By measuring the volume of high contrast-CT density voxels, is adding a blood flow dimension to 18F-FDG uptake. Visual examination of the lesions shows that those with low tend to have low SUV, CT-hypodense cores such as the one in Figure 4c; it is therefore identifying lesions as necrotic (negligible 18F-FMISO TBRmax due to limited blood flow) or viable (non negligible TBRmax). On the other hand, is sensitive to successions of voxels (‘runs’) with high CT density within the high-SUV subregion. This suggests that it could be capturing the presence of blood vessels, as in the examples shown in 4a (dense vasculature) and 4b (single blood vessel). The focus on high-18F-FDG uptake regions could be helping reveal whether the uptake is due to perfusion or hypoxia.
Figure 4.
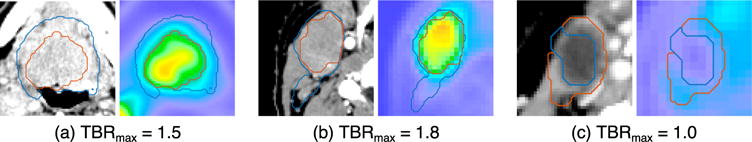
Representative slices from the CT and 18F-FDG PET scans of three lesions with a diverse range of radiomics features and TBRmax. Each subfigure corresponds to a different lesion, with the CT scan on the left panel and the 18F-FDG PET scan on the right panel. The main features for each lesion are: (a) TBRmax=1.5, =35 cc, =9735, =18.2, =0.48; (b) TBRmax=1.8, =6 cc, =10980, =11.5, = 0.17; (c) TBRmax=1.0, =0 cc, =5702, =1.9, =0.65. The white contour indicates v↓, and the dark grey contour indicates v↑.
This is not the first time that CT features have been found to be associated with hypoxia. Studies have shown that hypodense neck lymph nodes in routine contrast-enhanced CT scans have hypoxic conditions [23, 33]. Nodal CT density has also been shown to be a prognostic factor in head and neck cancer [34, 35]. More recently, Panth et al found significant differences in the time evolution of CT radiomics features between mice whose hypoxic fraction was reduced due to induced GADD34 over-expression and a control group [36]. In a study by Ganeshan et al, texture features obtained from contrast-enhanced CT images showed significant correlation with the average intensity of tumor staining with pimonidazole in NSCLC patients [37].
Our results suggest that features extracted from routine contrast-enhanced CT features can significantly improve the ability of 18F-FDG PET to indicate hypoxia, at least to the extent that it can be measured by 18F-FMISO TBRmax. A large number of radiomics features were considered in the analysis, to maximize the amount of information extracted from the images. The final model only contains two radiomics features, and , both of which have plausible interpretations.
As seen in Figure 3b, there are outliers in the prediction. Such deviations are expected, as there are multiple situations that can lead to enhanced 18F-FDG uptake in the absence of hypoxia, including high levels of metabolism, active inflammation, or proliferation rate. Similarly, it has been shown that variations in the distribution volume of 18F-FMISO can result in particular values of TBR being associated to different levels of hypoxia [32].
From a clinical standpoint, the operating point of the radiomics biomarker would need to be optimized in order to achieve the desired specificity and senbiomarker, assuming sitivity. Figure 3a shows three possible thresholds for the proposed radiomics biomarker, assuming that lesions with TBRmax > 1.4 are defined as hypoxic. In particular, threshold t1 has a specificity of 93%, and would therefore satisfy the criterion of having a minimal number of false positives included in the de-escalation candidate subset. It may be possible to improve the performance by deriving a higher-order classifier, as opposed to the linear function proposed here; this would however require a larger dataset to avoid overfitting. Our results are only the first step, and suggest that such larger scale effort would be worthwhile.
Key limitations of this study are that it is single institution –although internal validation was successful–, and that the CT imaging protocol was not designed for close monitoring of contrast delivery. Although we believe the results are likely to have broader validity, a multi-institutional study is needed in order to definitively establish this signature (or one like it) to the point that it could be reliably used for patient management. Lastly, the 18F-FMISO endpoint itself, TBRmax, could be somewhat noisy, which limits the ability to predict its value. Viewed from this perspective, the correlation achieved is promising.
5. Conclusion
We have identified image features derived from conventional imaging that correlate with the magnitude of hypoxia in head and neck cancer patients. In particular, our results show that a combined radiomics biomarker based on 18F-FDG PET and contrast-enhanced CT can emulate 18F-FMISO TBRmax-based stratification with significantly higher accuracy than 18F-FDG PET alone. After validation on large multi-institutional cohorts, such a biomarker could potentially be useful for head and neck cancer patient stratification in situations where 18F-FMISO is not available.
Supplementary Material
Acknowledgments
MCO is supported by a Junior Research Fellowship from Trinity College, University of Cambridge. This research was funded in part through NIH grant #1 R01 CA157770-01A1 as well as NIH/NCI Cancer Center Support Grant P30 CA008748. The sponsors were not involved at any stage of the study design, data collection and analysis, manuscript preparation or submission.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
The authors declare that they have no conflicts of interest.
Typical thresholds include tumor-to-blood or tumor-to-muscle ratios of 1.2 or 1.4. This study uses 1.4 to obtain a roughly balanced dataset, that is, with positive and negative classes of comparable size.
p16 is a tumor suppressor gene which has been suggested to be correlated with treatment response and survival [29].
Bibliography
- 1.Setton J, Caria N, Romanyshyn J, Koutcher L, Wolden SL, Zelefsky MJ, et al. Intensity-modulated radiotherapy in the treatment of oropharyngeal cancer: An update of the memorial sloan-kettering cancer center experience. International Journal of Radiation Oncology Biology Physics. 2012;82:291. doi: 10.1016/j.ijrobp.2010.10.041. https://www.ncbi.nlm.nih.gov/pubmed/21167652. [DOI] [PubMed] [Google Scholar]
- 2.Fakhry C, Zhang Q, Nguyen-Tan PF, Rosenthal D, El-Naggar A, Garden AS, et al. Human papillomavirus and overall survival after progression of oropharyngeal squamous cell carcinoma. Journal of clinical oncology. 2014;32:3365. doi: 10.1200/JCO.2014.55.1937. http://ascopubs.org/doi/abs/10.1200/jco.2014.55.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tân PF, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Eng J Med. 2010;363:24. doi: 10.1056/NEJMoa0912217. URL http://www.ncbi.nlm.nih.gov/pubmed/22115925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clavo B, Robaina F, Fiuza D, Ruiz A, Lloret M, Rey-Baltar D, et al. Predictive value of hypoxia in advanced head and neck cancer after treatment with hyperfractionated radio-chemotherapy and hypoxia modification. Clinical and Translational Oncology. 2017;19:419. doi: 10.1007/s12094-016-1541-x. URL http://www.ncbi.nlm.nih.gov/pubmed/27527617http://link.springer.com/10.1007/s12094-016-1541-x. [DOI] [PubMed] [Google Scholar]
- 5.Isa AY, Ward TH, West CML, Slevin NJ, Homer JJ. Hypoxia in head and neck cancer. The British journal of radiology. 2006;79:791. doi: 10.1259/bjr/17904358. URL http://www.ncbi.nlm.nih.gov/pubmed/16854964. [DOI] [PubMed] [Google Scholar]
- 6.Facciabene A, Peng X, Hagemann IS, Balint K, Barchetti A, Wang LP, et al. Tumour hypoxia promotes tolerance and angiogenesis via CCL28 and Treg cells. Nature. 2011;475:226. doi: 10.1038/nature10169. URL https://www.ncbi.nlm.nih.gov/pubmed/21753853. [DOI] [PubMed] [Google Scholar]
- 7.Dachs GU, Tozer GM. Hypoxia modulated gene expression. Angiogenesis, metastasis and therapeutic exploitation. 2000;36:1649. doi: 10.1016/s0959-8049(00)00159-3. URL https://www.ncbi.nlm.nih.gov/pubmed/10959051. [DOI] [PubMed] [Google Scholar]
- 8.Nordsmark M, Bentzen SM, Rudat V, Brizel D, Lartigau E, Stadler P, et al. Prognostic value of tumor oxygenation in 397 head and neck tumors after primary radiation therapy. An international multi-center study. Radiotherapy and Oncology. 2005;77:18. doi: 10.1016/j.radonc.2005.06.038. URL https://www.ncbi.nlm.nih.gov/pubmed/16098619. [DOI] [PubMed] [Google Scholar]
- 9.Lee N, Schoder H, Beattie B, Lanning R, Riaz N, McBride S, et al. Strategy of Using Intratreatment Hypoxia Imaging to Selectively and Safely Guide Radiation Dose De-escalation Concurrent With Chemotherapy for Locoregionally Advanced Human Papillomavirus–Related Oropharyngeal Carcinoma, International. Journal of Radiation Oncology*Biology*Physics. 2016;96:9. doi: 10.1016/j.ijrobp.2016.04.027. URL http://www.sciencedirect.com/science/article/pii/S0360301616301778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Welz S, Mönnich D, Pfannenberg C, Nikolaou K, Reimold M, La Fougère C, et al. Prognostic value of dynamic hypoxia PET in head and neck cancer: Results from a planned interim analysis of a randomized phase II hypoxia-image guided dose escalation trial. Radiotherapy and Oncology. 2017 doi: 10.1016/j.radonc.2017.04.004. URL http://www.ncbi.nlm.nih.gov/pubmed/28434798. [DOI] [PubMed]
- 11.Pigorsch SU, Wilkens JJ, Kampfer S, Kehl V, Hapfelmeier A, Schläger C, et al. Do selective radiation dose escalation and tumour hypoxia status impact the loco-regional tumour control after radio-chemotherapy of head & neck tumours? The ESCALOX protocol. Radiation Oncology. 2017;12:45. doi: 10.1186/s13014-017-0776-1. URL http://www.ncbi.nlm.nih.gov/pubmed/28249612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vordermark D, Horsman MR. Molecular Radio-Oncology. Vol. 198. Springer; 2016. pp. 123–142. URL http://www.ncbi.nlm.nih.gov/pubmed/27318684http://link.springer.com/10.1007/978-3-662-49651-0{_}6. [DOI] [PubMed] [Google Scholar]
- 13.Riaz N, Sherman EJ, Katabi N, Leeman JE, Higginson DS, Boyle J, et al. A personalized approach using hypoxia resolution to guide curative-intent radiation dose-reduction to 30 Gy: A novel de-escalation paradigm for HPV-associated oropharynx cancers (OPC) Journal of Clinical Oncology. 2017;35:6076. URL http://ascopubs.org/doi/abs/10.1200/JCO.2017.35.15_suppl.6076. [Google Scholar]
- 14.Bartlett RM, Beattie BJ, Naryanan M, Georgi JC, Chen Q, Carlin SD, et al. Image-Guided PO2 Probe Measurements Correlated with Parametric Images Derived from 18F-Fluoromisonidazole Small-Animal PET Data in Rats. Journal of Nuclear Medicine. 2012;53:1608. doi: 10.2967/jnumed.112.103523. URL http://www.ncbi.nlm.nih.gov/pubmed/22933821http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC3784982http://jnm.snmjournals.org/cgi/doi/10.2967/jnumed.112.103523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carlin S, Pugachev A, Sun X, Burke S, Claus F, O’Donoghue J, et al. In vivo characterization of a reporter gene system for imaging hypoxia-induced gene expression. Nuclear medicine and biology. 2009;36:821. doi: 10.1016/j.nucmedbio.2009.06.006. URL http://www.ncbi.nlm.nih.gov/pubmed/9720294http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC2754273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muzi M, Peterson LM, O’Sullivan JN, Fink JR, Rajendran JG, McLaughlin LJ, et al. 18F-Fluoromisonidazole Quantification of Hypoxia in Human Cancer Patients Using Image-Derived Blood Surrogate Tissue Reference Regions. Journal of Nuclear Medicine. 2015;56:1223. doi: 10.2967/jnumed.115.158717. URL http://www.ncbi.nlm.nih.gov/pubmed/26112020http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC4526410http://jnm.snmjournals.org/cgi/doi/10.2967/jnumed.115.158717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rasey JS, Koh WJ, Evans ML, Peterson LM, Lewellen TK, Graham MM, et al. Quantifying regional hypoxia in human tumors with positron emission tomography of [18F]fluoromisonidazole: a pretherapy study of 37 patients. International journal of radiation oncology, biology, physics. 1996;36:417. doi: 10.1016/s0360-3016(96)00325-2. URL http://www.ncbi.nlm.nih.gov/pubmed/8892467. [DOI] [PubMed] [Google Scholar]
- 18.Lee N, Nehmeh S, Schöder H, Fury M, Chan K, Ling CC, et al. Prospective Trial Incorporating Pre-/Mid-Treatment [18F]-Misonidazole Positron Emission Tomography for Head-and-Neck Cancer Patients Undergoing Concurrent Chemoradiotherapy, International. Journal of Radiation Oncology*Biology*Physics. 2009;75:101. doi: 10.1016/j.ijrobp.2008.10.049. URL http://www.sciencedirect.com/science/article/pii/S0360301608036948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zschaeck S, Haase R, Abolmaali N, Perrin R, Stützer K, Appold S, et al. Spatial distribution of FMISO in head and neck squamous cell carcinomas during radio-chemotherapy and its correlation to pattern of failure. Acta Oncologica. 2015;54:1355. doi: 10.3109/0284186X.2015.1074720. URL http://www.ncbi.nlm.nih.gov/pubmed/26398663http://www.tandfonline.com/doi/full/10.3109/0284186X.2015.1074720. [DOI] [PubMed] [Google Scholar]
- 20.Clavo AC, Brown RS, Wahl RL. Fluorodeoxyglucose uptake in human cancer cell lines is increased by hypoxia. Journal of nuclear medicine. 1995;36:1625. URL http://www.ncbi.nlm.nih.gov/pubmed/7658223. [PubMed] [Google Scholar]
- 21.Lopci E, Grassi I, Chiti A, Nanni C, Cicoria G, Toschi L, et al. PET radiopharmaceuticals for imaging of tumor hypoxia: a review of the evidence. American journal of nuclear medicine and molecular imaging. 2014;4:365. URL http://www.ncbi.nlm.nih.gov/pubmed/24982822. [PMC free article] [PubMed] [Google Scholar]
- 22.Thorwarth D, Eschmann SM, Holzner F, Paulsen F, Alber M. Combined uptake of [18F]FDG and [18F]FMISO correlates with radiation therapy outcome in head-and-neck cancer patients. Radiotherapy and Oncology. 2006;80:151. doi: 10.1016/j.radonc.2006.07.033. URL http://linkinghub.elsevier.com/retrieve/pii/S0167814006003124. [DOI] [PubMed] [Google Scholar]
- 23.Hermans R. Tumour oxygenation measurements using computed tomography and magnetic resonance imaging. Cancer Imaging. 2002;2:138. URL https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4448600/ [Google Scholar]
- 24.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2016;278:563. doi: 10.1148/radiol.2015151169. URL http://pubs.rsna.org/doi/10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Computational environment for radiological research. http://github.com/cerr/CERR.
- 26.Apte A, Veeraraghavan H, Oh J, Kijewski P, Deasy J. SU-E-J-253: The Radiomics Toolbox in the Computational Environment for Radiological Research (CERR) Medical Physics. 2015;42:3324. URL http://doi.wiley.com/10.1118/1.4924339. [Google Scholar]
- 27.Lim R, Eaton A, Lee NY, Setton J, Ohri N, Rao S, et al. 18F-FDG PET/CT metabolic tumor volume and total lesion glycolysis predict outcome in oropharyngeal squamous cell carcinoma., Journal of nuclear medicine: official publication. Society of Nuclear Medicine. 2012;53:1506. doi: 10.2967/jnumed.111.101402. URL http://www.ncbi.nlm.nih.gov/pubmed/22895812. [DOI] [PubMed] [Google Scholar]
- 28.Foster B, Bagci U, Mansoor A, Xu Z, Mollura DJ. A review on segmentation of positron emission tomography images. Computers in biology and medicine. 2014;50:76. doi: 10.1016/j.compbiomed.2014.04.014. URL http://www.ncbi.nlm.nih.gov/pubmed/24845019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Romagosa C, Simonetti S, López-Vicente L, Mazo A, Lleonart ME, Castellvi J, et al. p16Ink4a overexpression in cancer: a tumor suppressor gene associated with senescence and high-grade tumors. Oncogene. 2011;30:2087. doi: 10.1038/onc.2010.614. URL http://www.nature.com/doifinder/10.1038/onc.2010.614. [DOI] [PubMed] [Google Scholar]
- 30.Grkovski M, Lee NY, Schöder H, Carlin SD, Beattie BJ, Riaz N, et al. Monitoring early response to chemoradiotherapy with 18F-FMISO dynamic PET in head and neck cancer. European Journal of Nuclear Medicine and Molecular Imaging. 2017 doi: 10.1007/s00259-017-3720-6. URL http://www.ncbi.nlm.nih.gov/pubmed/28540417. [DOI] [PMC free article] [PubMed]
- 31.Thorwarth D, Eschmann SM, Scheiderbauer J, Paulsen F, Alber M. Kinetic analysis of dynamic 18F-fluoromisonidazole PET correlates with radiation treatment outcome in head-and-neck cancer. BMC cancer. 2005;5:152. doi: 10.1186/1471-2407-5-152. URL http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1325034{&}tool=pmcentrez{&}rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grkovski M, Schöder H, Lee N, Carlin SD, Beattie BJ, Riaz N, et al. Multiparametric imaging of tumor hypoxia and perfusion with 18F-FMISO dynamic PET in head and neck cancer. Journal of Nuclear Medicine p jnumed.116.188649. 2017 doi: 10.2967/jnumed.116.188649. URL http://www.ncbi.nlm.nih.gov/pubmed/28183993. [DOI] [PMC free article] [PubMed]
- 33.Lartigau E, Le Ridant AM, Lambin P, Weeger P, Martin L, Sigal R, et al. Oxygenation of head and neck tumors. Cancer. 1993;71:2319. doi: 10.1002/1097-0142(19930401)71:7<2319::aid-cncr2820710724>3.0.co;2-c. URL http://www.ncbi.nlm.nih.gov/pubmed/8453553. [DOI] [PubMed] [Google Scholar]
- 34.Grabenbauer GG, Steininger H, Meyer M, Fietkau R, Brunner T, Heinkelmann P, et al. Nodal CT density and total tumor volume as prognostic factors after radiation therapy of stage III/IV head and neck cancer., Radiotherapy and oncology. Journal of the European Society for Therapeutic Radiology and Oncology. 1998;47:175. doi: 10.1016/s0167-8140(98)00016-4. URL http://www.ncbi.nlm.nih.gov/pubmed/9683366. [DOI] [PubMed] [Google Scholar]
- 35.Munck JN, Cvitkovic E, Piekarski JD, Benhamou E, Recondo G, Bachouchi M, et al. Computed tomographic density of metastatic lymph nodes as a treatment-related prognostic factor in advanced head and neck cancer. Journal of the National Cancer Institute. 1991;83:569. doi: 10.1093/jnci/83.8.569. URL http://www.ncbi.nlm.nih.gov/pubmed/1848639. [DOI] [PubMed] [Google Scholar]
- 36.Panth KM, Leijenaar RT, Carvalho S, Lieuwes NG, Yaromina A, Dubois L, et al. Is there a causal relationship between genetic changes and radiomics-based image features? An in vivo preclinical experiment with doxycycline inducible GADD34 tumor cells. Radiotherapy and Oncology. 2015;116:462. doi: 10.1016/j.radonc.2015.06.013. URL http://linkinghub.elsevier.com/retrieve/pii/S0167814015002959. [DOI] [PubMed] [Google Scholar]
- 37.Ganeshan B, Goh V, Mandeville HC, Ng QS, Hoskin PJ, Miles KA. NonSmall Cell Lung Cancer: Histopathologic Correlates for Texture Parameters at CT. Radiology. 2013;266:326. doi: 10.1148/radiol.12112428. URL http://www.ncbi.nlm.nih.gov/pubmed/23169792. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


