Abstract
Microfluidic organ-on-a-chip models of human intestine have been developed and used to study intestinal physiology and pathophysiology. In this article, we review this field and describe how microfluidic Intestine Chips offer new capabilities not possible with conventional culture systems or organoid cultures, including the ability to analyze contributions of individual cellular, chemical, and physical control parameters one-at-a-time; to coculture human intestinal cells with commensal microbiome for extended times; and to create human-relevant disease models. We also discuss potential future applications of human Intestine Chips, including how they might be used for drug development and personalized medicine.
Keywords: Organs-on-Chips, Gut-on-a-Chip, Intestine-on-a-Chip, Microfluidic
Abbreviations used in this paper: ECM, extracellular matrix; IBD, inflammatory bowel disease; PD, pharmacodynamics; PDMS, polydimethylsiloxane; PK, pharmacokinetics; 3D, 3-dimensional
Summary.
Organs-on-chips are microfluidic cell culture systems that recapitulate the structure, function, physiology, and pathology of living human organs in vitro. In this article, we review recent development of various human intestine-on-a-chip models and their potential value for disease modeling, drug discovery, and personalized medicine.
The major organ function of the human intestine is to carry out digestion, absorption, secretion, and motility,1 in addition to establishing a protective epithelial barrier between this digestive environment and the body. In addition, intestines regulate systemic physiology by metabolizing drugs2; communicate with other organs, such as the liver3, 4 and pancreas,5 via portal flow; and they contain an enteric nervous system that forms a part of the gut-brain axis.6, 7 The intestine is also the major site at which commensal microbes of the gut microbiome live and interact with gut lymphoid tissues and the host immune system, which contributes significantly to intestinal homeostasis.8, 9 For example, the gut microbiome and its metabolites (eg, short-chain fatty acids) have been recently shown to play a central role in the maintenance of intestinal health, immune modulation, and the development of both enteral and nonenteral diseases.10, 11 However, analysis of gut microbiome interactions with human intestinal cells has been limited to genetic or metagenomics analysis because it has not been possible to coculture these microbes with living epithelium for more than about 1 day using conventional culture models or even more sophisticated intestinal organoid cultures. Thus, there have been great efforts to develop experimental in vitro or ex vivo models of human intestine that permit analysis of intestinal pathophysiology both in the presence and absence of living microbiome.
The most common in vitro intestine models used to study barrier function or model drug absorption involve culturing an established human intestinal epithelial cell line (eg, Caco-212, 13 or HT-2914, 15 cells) on extracellular matrix (ECM)-coated, porous membranes within Transwell insert culture devices. Although these models are most commonly used by the pharmaceutical industry, this 2-dimensional (2D) culture format fails to recapitulate physiological 3-dimensional (3D) intestinal cell and tissue morphology or re-establish other key intestinal differentiated functions (eg, mucus production, villi formation, cytochrome P-450-based drug metabolism).16, 17 These conventional static models also cannot support the coculture of commensal microbiome with human intestinal cells, which are critical for gut physiology,16 because the bacteria rapidly overgrow and contaminate the human cell cultures within a day. Several ex vivo models, such as the everted sac18 or the Ussing chamber,19, 20 have been developed for drug transport assays; however, their expected lifespan (<8 hours) is not sufficient to enable many studies on normal intestinal physiology, develop intestinal disease models, or study clinically relevant host-microbiome crosstalk.
Although it had been technically challenging to culture primary human intestinal epithelial cells, intestinal 3D organoid cultures derived from either intestinal crypts containing endogenous intestine cells or from induced pluripotent stem cells have revolutionized the field by maintaining stem cell niches and supporting differentiation of various differentiated intestinal epithelial cell subtypes in vitro.21, 22 When cultured within a 3D ECM gel in medium containing Wnt, R-spondin, noggin, and other growth factors, small intestinal organoids (enteroids) also spontaneously undergo villus-crypt morphologic organization and intestinal histogenesis.22 Each organoid line derived from an intestinal tissue biopsy of an individual patient can be grown, frozen, and revived for multiple reuses, which can potentially be used to establish biobanks23, 24 and develop multiplexed screening platforms for validating new drug candidates and to advance personalized medicine.25 However, organoids are also limited in that they lack other supporting cell and tissue types found within the living intestine, such as endothelium-lined blood vessels and immune cells, which are important for drug transport, pharmacokinetic (PK) analysis, and disease modeling. They also do not experience fluid flows and cyclic mechanical deformations similar to those experienced in a peristalsing intestine that contribute significantly to intestinal health and function. Furthermore, because each enteroid forms a closed lumen when cultured within surrounding ECM gel, it is experimentally difficult to sample or manipulate luminal components (eg, microbial cells, nutrients, drugs, or toxins). This structure also significantly limits the ability of researchers to study many critical intestinal functions (eg, absorption, drug PK, or drug metabolism), in addition to critical host-microbiome interactions.26
These challenges have recently been overcome by the development of microfluidic Organ Chip models of human intestine. Organ Chips are microfluidic cell culture devices, originally fabricated using methods adapted from computer microchip manufacturing (eg, soft lithography), which contain continuously perfused chambers inhabited by living cells arranged to simulate tissue- and organ-level physiology.27 Over the past 5 years, Organ Chip models of intestine have been engineered with increasing complexity that also include neighboring channels lined by human microvascular endothelium, and commensal microbes, immune cells, and pathogenic bacteria, and some permit application of cyclic mechanical forces that mimic peristalsis-like deformations experienced by living intestine in vivo (Figure 1). Next are review various types of engineered in vitro models that emulate the structure, function, physiology, and pathology of the living human intestine (Table 1). Also considered are the implications of this work for development of more complex disease models, drug development, and personalized medicine in the future.
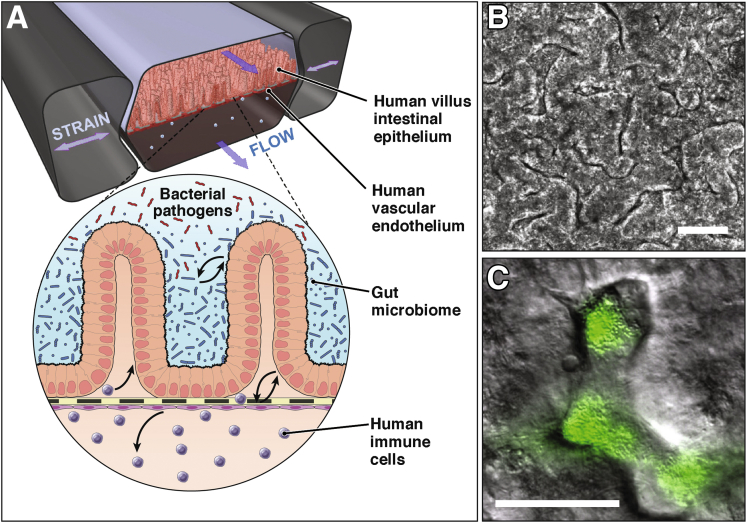
Figure 1.
The mechanically active human Gut Chip. (A) Human villus intestinal epithelium and vascular endothelium are lined on opposite sides of a flexible porous membrane under fluid flows and peristalsis-like strains. A zoom-in schematic shows the intestinal microenvironment undergoing complex crosstalk between commensal gut microbiome, bacterial pathogens, and immune cells in parenchymal and vascular channels, respectively. Figure modified with permission from Reference 75. (B) Villus morphogenesis of human Caco-2 intestinal epithelium in the Gut Chip under physiologically controlled motions and flow. Figure modified with permission from References 17 and 42. (C) An overlaid image of the coculture of green fluorescent protein–labeled Escherichia coli and microengineered villi in the Gut Chip. Bars = 50 μm.
Table 1.
Design Characteristics of Microfluidic Intestine Models
| Model | TEER | Absorption | Coculture | Microbiome | Differentiation | Peristalsis | Drug metabolism | Crypt-villus axis | Oxygen modulation | Disease modeling |
|---|---|---|---|---|---|---|---|---|---|---|
| Static | ||||||||||
| Transwell | Yes12 | Yes12, 13 | No | Yes14, 15 (<24 h) | No | No | No | No | No | No |
| Organoid | No | Yes95 | No | Yes96 (<1 h) | Yes21 | No | Yes97 | Yes21 | No | Yes22 |
| Ex vivo | Yes19, 98 | Yes19, 20 | No | Yes97 (<3 h) | Yes19 | No | Yes100 | Yes19 | Yes99 | Yes98 |
| Scaffold | No | No | No | No | Yes40 | No | No | Yes40 | No | No |
| Microfluidic | ||||||||||
| 2-channel | Yes32 | Yes30 | Yes37 | No | No | No | Yes39 | Yes39 | No | No |
| Ex vivo | No | Yes99 | No | No | Yes101 | No | No | Yes101 | No | Yes101 |
| Multichannel (HuMiX) | Yes41 | No | No | Yes41 (<24 h) | No | No | No | No | Yes41 | No |
| Gut Chip | Yes16, 42 | Yes16 | No | Yes16, 42 (>7 d) | Yes17, 42 | Yes16, 17, 42 | Yes16, 17 | Yes16, 17, 42 | No | Yes42 |
TEER, transepithelial electrical resistance.
Microfluidic Intestine Chip Models
Microfluidic devices containing hollow microchannels less than 1 mm in width support laminar fluid flow and control of nanoliter to microliter scale fluid volumes, and thus, they are amenable to use for culture of living cells. By using a syringe or a peristaltic pump, culture medium may be perfused at desired flow rates through each microchannel, which can mimic the dynamic ranges of fluid flows and associated shear stresses on the cell surface that are observed in the human intestinal lumen,28, 29 and in the blood capillaries. This fluidic control also enables delivery of nutrients, growth factors, drug compounds, or even toxins to the intestinal epithelium grown on the microfluidic channels in a highly regulated spatiotemporal manner.
Most of the Intestine Chips contain 2 hollow channels separated by a common porous, ECM-coated polyester or polycarbonate membrane, which had immortalized human intestinal epithelial cells cultured on 1 of its surfaces.30 The epithelial monolayer formed in this device could be probed from both the apical and basal sides of the epithelium, and this enabled quantification of tight junction barrier function31, 32 and absorption of nutrients33 and drugs.30, 34 Some devices integrated multiple chambers with different types of cells to measure PK and pharmacodynamics (PD) properties of drug compounds in vitro.35, 36, 37 However, the cells were seeded at a low density in these studies and they were perfused only through the apical channel; as a result, the monolayer of the immortalized intestinal cells could only be cultured for a limited time (<5 days). A more recent study demonstrated real-time barrier integrity assessment capabilities in a membrane-free culture of perfused intestinal epithelium tubes.38
To better mimic the 3D form of human intestine using cell monolayers cultured under microfluidic flow, micromolding methods have been used to form polymeric scaffolds (eg, Ca-alginate or collagen gel) into villus-shaped structures.39, 40 Culturing human Caco-2 intestinal epithelial cells on these crenulated surfaces was shown to promote formation of a similarly crenulated epithelium, and this was accompanied by increased absorption of drug compounds and improved cellular metabolic enzyme cytochrome P-450 activity in response to apical fluid shear stress.39 However, this design did not enable analysis of intestinal barrier function because the solid polymer material blocked the abluminal surface of the epithelium.
A multichannel Intestine Chip called HuMiX also has been described that separates a luminal microbial compartment from layers of Caco-2 intestinal epithelium by nanoporous membranes.41 This model was shown to facilitate survival of a host-microbiome ecosystem containing Caco-2 cells and anaerobic human gut bacteria (eg, Bacteroides caccae) with constant perfusion of culture medium.41 However, these studies were carried out in the absence of mechanical deformations similar to those associated with peristalsis that can critically influence the microbial growth in the intestine.42 This experimental model is also limited by the physical separation of microbial and host epithelial cells by a nanoporous membrane, and the fact that the cocultures can only survive for short times (<24 hours), which may restrict its use for long-term profiling of host-microbial interactions and intestinal pathophysiology.
Mechanically Active Gut Chip Model
A more sophisticated, microfluidic 2-channel Gut Chip model has been developed that enables human intestinal epithelium, capillary endothelium, immune cells, and even commensal microbial cells to grow, coexist, and interact while experiencing physiologically relevant fluid flow and peristalsis-like mechanical deformations in vitro.42 We use herein “Gut Chip” to refer to previously published human intestine model using Caco-2 cells that used this term.16, 17, 42 The Gut Chip is made of a flexible, gas-permeable, silicone polymer (polydimethylsiloxane [PDMS]) that is crystal clear so that it allows high-resolution imaging by phase contrast, differential interference contrast, or immunofluorescence confocal microscopy. It contains 2 parallel microchannels (<1 mm wide) separated by a thin (∼20 μm), ECM-coated, flexible, porous PDMS membrane, and it is surrounded on both sides by hollow, full height, side chambers (Figure 1A).16, 17, 42, 43 Intestinal epithelial cells are cultured on the top surface of the membrane within the upper parenchymal channel, and microvascular endothelial cells are grown on the lower surface of the same membrane within the lower vascular channel to recreate the intestinal tissue-tissue interface. Importantly, pneumatic application of cyclic suction to the hollow side chambers results in outward deformation of the vertical side walls and attached horizontal ECM-coated membrane with the adherent cell layers, thereby mimicking cyclic mechanical deformations similar to those experienced by the intestinal tissues during peristalsis (Figure 1A).
Under these physiological conditions, human Caco-2 intestinal epithelial cells, which characteristically grow as flattened cells within a planar monolayer in conventional 2-dimensional cultures, spontaneously undergo villus morphogenesis inside this mechanically dynamic Gut Chip (Figure 1B).17 The microengineered villi are lined by all 4 lineages of differentiated small intestinal cells (absorptive, goblet, enteroendocrine, and Paneth), which exhibit columnar cell morphology similar to that observed in the living intestine.17 Intestinal villus morphogenesis on-chip is also accompanied by establishment of a crypt-villus axis, including restriction of proliferative cells to the basal crypts and their upward migration, drug metabolizing activity, mucus production, and glucose reuptake.17
Importantly, because the Gut Chip has continuous fluid flow, villi formation, and mucus production, it is also possible to coculture living commensal microbes (eg, Lactobacillus rhamnosus GG)16 or the VSL#3 clinical probiotic formulation containing 8 different microbial strains42) in direct contact with the intestinal epithelial cells within the lumen of the parenchymal channel for weeks in vitro without compromising barrier integrity or intestinal cell function (Figure 1C).16, 42 In fact, barrier function was shown to increase when the intestinal epithelium was cocultured with L rhamnosus GG.16 In addition, transcriptomic analyses targeting approximately 23,000 human genes revealed that the in vivo relevant fluid flow and physical deformations dramatically changed the gene expression profiles compared with the static Transwell cultures, and that mechanically active Gut Chips that also contained a mixture of commensal microbes (VSL#3 probiotic formulation) showed the highest level of genetic similarity to normal human ileum.
Taken together, these data suggest that the Gut Chip that emulates the dynamic human intestinal microenvironment more faithfully mimics intestinal function than previously described in vitro intestine models. Pathogenic bacteria, such as enteroinvasive E coli, also can be integrated into this model system, with or without commensal microbes of the normal microbiome, to study how interplay between these microbes and human intestinal epithelial cells contributes to intestinal homeostasis and intestinal disease development.42
Indeed, the Gut Chip was used to demonstrate complex immune-microbiome inflammatory interactions germane to chronic inflammatory diseases, such as inflammatory bowel disease (IBD).42 Addition of lipopolysaccharide endotoxin to the luminal microchannel also was shown to induce secretion of the proinflammatory cytokines, interleukin-1β, -6, -8, and tumor necrosis factor-α into the lower vascular channel. Moreover, this was accompanied by increased expression of intercellular adhesion molecule-1 on the endothelium, villus blunting, and compromise of intestinal barrier function, but only in the presence of white blood cells (eg, peripheral blood mononuclear cells) that were infused through the capillary microchannel. Moreover, additional experiments confirmed that all 4 cytokines were required to be present at the concentrations measured within the capillary channel to induce this intestinal injury response. Interestingly, additional studies revealed that cessation of cyclic peristalsis-like mechanical motions caused an increase in bacterial overgrowth in the Gut Chip, which closely resembled the small intestinal bacterial overgrowth that can result from cessation of peristalsis in patients with ileus. Thus, by being able to control flow independently of mechanical deformations, this study revealed that the past clinical assumption that small intestinal bacterial overgrowth results from cessation of fluid flow when peristalsis is inhibited44 is incorrect; instead, it is the lack of mechanical deformations that drives this bacterial overgrowth response. Thus, the innovative modularity of the microfluidic Gut Chip (Figure 1A) can be leveraged to gain new insights into intestinal pathophysiology, and to understand disease mechanisms in ways that are not possible using conventional in vitro gut models or simpler microfluidic Intestine Chips.
Human Primary Intestine Models
Although human Intestine Chips lined by established intestinal epithelial cell lines (eg, Caco-2 cells) have been shown to mimic many physiological and pathophysiological functions of the human intestine, they are still limited by the fact that they use immortalized cells that were originally isolated from a human tumor. For example, these cultures might be particularly difficult to use in studies where genome fidelity is important (eg, analysis of intestinal cancer formation). In vitro intestine models have been created using human fetal intestinal tissue explants, but they deteriorate within 24 hours of culture.45, 46 Human intestinal organoids offer another way to study normal human intestinal stem cell differentiation and villus morphogenesis in vitro; however, they do not recreate a physiologically relevant physical microenvironment (fluid flow, peristalsis-like deformations) and their enclosed lumens limit their value for transport studies and coculture with pathogens.
To overcome these challenges, human enteroids47, 48 or colonoids49 created from patient biopsies have been enzymatically fragmented and the released intestinal stem cells plated on Transwell inserts to create primary human intestinal monolayers, which permit sampling from both the apical and basolateral sides in vitro. These models showed establishment of intact polarized monolayers with apical villin and basolateral Na+/K+-ATPase expression47 and successful cultivation of multiple human norovirus strains in the presence of bile.48
Although static Transwell cultures offer better access to the apical lumen of these organoids, they still lack other organ-level features of the intestine, including an endothelium-lined vascular compartment, flow, cyclic mechanical deformation, or immune cells, which have been shown to be important for mimicking intestinal physiology and pathophysiology in the human Gut Chip.42 Organoid cultures also have been modified to address some of these challenges. For example, application of mechanical forces to ECMs containing organoids has shown to modulate their phenotype and influence primary intestine cell differentiation.50 In addition, miniaturized bioreactors have been developed to generate fluid flows that facilitate nutrient and oxygen uptake, and fluid shear stresses that convey physiologically relevant mechanical signals to cells in organoid cultures.51
Most recently, organoid and organ chip approaches have been combined to develop a microfluidic primary human Intestine Chip model. Human enteroids cultured from patient duodenal biopsies were enzymatically fragmented and the released intestinal stem cells were plated on the ECM-coated porous membrane of a 2-channel PDMS microfluidic device with primary human intestinal microvascular endothelial cells on the opposite side of the same membrane within a parallel channel.52 Much like the Caco-2 cell-lined Gut Chip, with application of fluid flow and peristalsis-like deformations, the intestinal epithelial cells undergo villus histogenesis with multilineage differentiation. Interestingly, although this differentiation process is similar to that observed within the intestinal organoids, transcriptomic analysis revealed that the mechanically active Intestine Chip more closely mimics the proliferation and host defense response to infection functions of the human duodenum from which the organoids were originally isolated.
Thus, this primary Intestine Chip may be useful as a research tool for many applications where normal intestinal function is crucial, including studies of metabolism, nutrition, and cancer progression, as well as drug absorption and PK/PD. It also may offer a novel method for emerging disciplines, such as personalized precision medicine,53, 54 which are in constant search for faithful and robust tools for disease modeling and drug discovery. Genetic polymorphisms in drug transporters, drug-metabolizing enzymes, and drug targets have been linked to the efficacy, dosage, and toxicity profile in humans.55 Hence, possible applications for this model range from studying specific pathologic mechanisms (eg, inflammation, infection, and nutrient malabsorption) to drug discovery in disease-specific and patient-specific contexts.
Limitations of Human Intestine Chip Models
Although microfluidic Intestine Chip models faithfully mimic many different phenotypes and responses of human intestine, they only incorporate limited components of the 4-layered intestinal wall, and these missing features might play a significant role in some disorders. For example, smooth muscle cells express Toll-like receptors 1–9 and can regulate neuronal integrity by modulating glial-derived neurotrophic factor production.56 This is important because the enteric nervous system regulates intestinal secretion, blood flow, and gut motility.57 Subtle alterations in transmitter production and release have been linked to functional and inflammatory diseases of the gastrointestinal tract.58, 59, 60 Thus, adding more components, such as muscle and nervous system cells, should be considered in the future for relevant applications.
Although PDMS holds many favorable properties for manufacturing microfluidic organ devices, it also has the potential drawback of adsorbing small and hydrophobic molecules.61, 62 To overcome this, surface modification of PDMS or alternative polymeric (eg, polyurethane, styrenic block copolymers)63 or ECM-based materials64 could potentially be used in studies designed to predict drug bioavailability or absorption. Alternatively, when using Organ Chips to model drug PK and PD, PDMS adsorption can be taken into account using computational modeling for most compounds.65
In addition, most of the past work on Intestine Chips has been carried out in academic laboratories, and thus, the results and robustness of the models can vary from laboratory to laboratory. However, the recent emergence of multiple companies that are beginning to commercialize Organ Chip technology offers great hope that issues relating to scale up of manufacturing, robustness, cost, and ease of use will be resolved over time. These challenges must be overcome before human Intestine Chips become a common tool used in academic and industrial research laboratories around the world.
Disease Models Using Human Intestine Chips
Only a small body of work has been published on disease models using microfluidic intestine devices. The 2-channel human Gut Chip was used to coculture multiple commensal microbes in contact with living human intestinal epithelial cells and to analyze how gut microbiome, inflammatory cells, and peristalsis-associated mechanical deformations independently contribute to intestinal bacterial overgrowth and inflammation, which are associated with IBD.42, 66 Recent studies suggest that the same Gut Chip model can be used to model the radiation injury response of human intestine (eg, cell death, villus blunting, compromised barrier function) when exposed to γ-radiation, and prevention of these responses using a potential prophylactic radiation countermeasure drug.67 In addition, the recently developed primary human Intestine Chip also has been used to model differential sensitivity of intestinal responses to bacteria with different levels of virulence (Kasendra et al, unpublished data).
A potentially important application of the microfluidic Intestine Chip models is to study various types of enteropathies for which current in vitro models have only a limited ability to recapitulate, such as those associated with environmental enteric dysfunction,68 colorectal cancer,69 cystic fibrosis,70 bacterial infectious diseases,71 and celiac disease.72 We believe the likelihood that Intestine Chips will be able to meet this challenge is high given the successful modeling of other complex diseases (eg, pulmonary edema,73 asthma,74 chronic obstructive pulmonary disease,74, 75 or Barth syndrome76) with other microfluidic Organ Chips. Moreover, we have previously demonstrated that the Gut Chip can be used to model key features of human intestinal diseases, including IBD and small intestinal bacterial overgrowth caused by ileus.42
In addition to recapitulating disease phenotypes, human Organ Chips offer a unique way to get insight into the molecular, genetic, and biophysical basis of disease based on their ability to incorporate different levels of cellular complexity, and to independently control various critical parameters (eg, chemical gradients, mechanical forces, cell types, and ECM).42, 77
In past Gut Chip disease modeling studies, induction of the pathologic phenotype was accomplished by exposure to specific pathogenic organisms, toxins, or inflammatory mediators (eg, cytokines), with combination of 2 or more of these factors being required to produce disease responses.42 For example, both lipopolysaccharide endotoxin and immune cells had to be added simultaneously in the Gut Chip to produce villus blunting and compromise intestinal barrier function, and neither could induce this response on their own.42 Inclusion of additional components, such as microbiome,16, 42 pathogenic bacteria,42 mechanical cues,17, 42 ECM, connective tissue cells, neuronal cells, organ-specific immune cells, and/or hormonal signals might be needed to model other specific phenotypes that result from complex interplay among several signaling pathways. This goal also might be achieved by creating Intestine Chips with cells isolated from intestinal biopsies taken from patients with specific diseases. This approach also could facilitate development of robust personalized disease models for improved drug screening and matching.78, 79
Intestine Chip Models for Developmental Therapeutics
Intestine Chip–based disease models can be exploited to identify new therapeutic targets, repurpose existing drugs, test new therapies, and carry out PK/PD testing in vitro. Although extensive testing with conventional culture models and animal experiments are commonly carried out before performing clinical trials, these models often fail to predict efficacy and safety of novel drugs, which results in loss of enormous amounts of money, time, and effort.80 In addition, drug toxicities and efficacies greatly vary depending on the individual patient, given both genetic81 and environmental influences (eg, nutrients,82, 83 drug intake,83, 84 gut microbiome85, 86). To meet this challenge, there has been increased interest in pursuing a personalized medicine approach to develop therapeutics that are tailored for an individual patient’s genetic background. Primary intestine models can facilitate this approach by engineering artificial intestine microenvironments using the patient’s own cells (epithelial, endothelial, immune, connective tissue, or neural) and gut microbiome. Because the intestine is a major site for drug metabolism (both by cytochrome P-450 enzymes in intestinal epithelial cells and by the gut microbiome87), creation of Intestine Chips can facilitate study of drug breakdown both alone and when coupled to other Organ Chips that also contribute to drug metabolism (eg, Liver Chip) and clearance (eg, Kidney Chip) in a human Body-on-a-Chip configuration.36, 37
Importantly, the gut microbiome in a mouse or other laboratory animal is remarkably different from that in a human (eg, ∼85% of gut microbiome that colonizes in human does not exist in mice88), and thus, creation of human Intestine Chips with human microbiome could have a major positive impact on this field. Intestine Chips that contain human intestinal microbiome should better recapitulate metabolism of oral drugs, and hence, testing of novel drug compounds on these platforms could lead to results that inform clinical trials design in a helpful way. We envision that integrating patient-specific intestinal organoids, local immune cells, and the commensal microbiome into Intestine Chips will create a highly defined and controllable translation platform that should accelerate discovery of new drug and personalized precision therapeutics.
It is important to note that commensal microbes also can be developed as probiotic therapeutics for treatment of human intestinal diseases. For example, fecal microbiota transplantation containing commensal microbes has already been approved by the US Food and Drug Administration for treatment of Clostridium difficile infections by restoring the normal gut microbiome in these patients.89, 90 Probiotic formulations also have been used to treat inflammation associated with IBD, although the effectiveness and mechanism of action have not been fully validated.91 Importantly, the probiotic L rhamnosus GG was shown to increase intestinal barrier function when cocultured in the lumen of the intestinal epithelial channel of the human Gut Chip,16 and the VSL#3 probiotic formulation suppressed villus blunting and loss of barrier function induced by infection with pathogenic E. coli in this model.42 Thus, the Intestine Chip approach also may be useful for discovery of new microbiome-based therapeutics, such as genetically engineered commensal bacteria.92 By integrating circulating and organ-specific immune cells into Intestine Chips, they might be useful for in vitro development of new mucosal vaccines.93, 94
Conclusions
In this article, we reviewed recently developed microfluidic human Intestine Chip models and described the advantages they offer relative to commonly used 2D and 3D in vitro culture systems, including their ability to more closely emulate the structure, function, physiology, and pathology of the living human intestine. Microfluidic Organ Chip systems provide unparalleled independent control over multiple key biologic, chemical, molecular, cellular and mechanical parameters within the intestinal microenvironment, thereby enabling researchers to apply a synthetic biology approach at the cell, tissue, and organ levels that can lead to new insights into intestinal physiology and disease mechanisms. By harnessing these unique capabilities, Intestine Chips can be applied to analyze the molecular processes underlying various enteropathies, and to advance development of new therapies. Creation of personalized Intestine Chips containing stem cells, microbiome, and immune cells from the same patient will offer a powerful new approach to define patient-specific drug responses and toxicities, and hence to advance precision medicine. This field is still in its infancy, and there remains much to be done in terms of increasing the robustness of these models and validating their value for drug development and personalized medicine. However, the advantages they provide over conventional culture systems, and even enteroid cultures, are becoming increasingly clear.
Author contributions
Amir Bein, Woojung Shin, Sasan Jalili-Firoozinezhad, Min Hee Park, Alexandra Sontheimer-Phelps, Alessio Tovaglieri, Angeliki Chalkiadaki, Hyun Jung Kim, and Donald E. Ingber designed and drafted the manuscript. Amir Bein, Woojung Shin, Hyun Jung Kim, and Donald E. Ingber designed the figure and revised the manuscript.
Footnotes
Conflicts of interest This author discloses the following: Donald E. Ingber is a founder, holds equity, and chairs the scientific advisory board of Emulate Inc. The remaining authors disclose no conflicts.
Funding This work was supported by grants from DARPA, Food and Drug Administration, Gates Foundation, and the Wyss Institute for Biologically Inspired Engineering at Harvard University. Woojung Shin is a recipient of the Graduate Student Fellowship by Alternatives in Scientific Research of The International Foundation for Ethical Research. Hyun Jung Kim is a recipient of 2016 Innovator Awards (No. 2016-1141) by Kenneth Rainin Foundation.
Contributor Information
Hyun Jung Kim, Email: hyunjung.kim@utexas.edu.
Donald E. Ingber, Email: don.ingber@wyss.harvard.edu.
References
- 1.Silverthorn D.U., Ober W.C., Garrison C.W., Silverthorn A.C., Johnson B.R. Pearson/Benjamin Cummings; San Francisco: 2009. Human physiology: an integrated approach. [Google Scholar]
- 2.Benet L.Z., Wu C.-Y., Hebert M.F., Wacher V.J. Intestinal drug metabolism and antitransport processes: a potential paradigm shift in oral drug delivery. J Control Release. 1996;39:139–143. [Google Scholar]
- 3.Moore F.A., Moore E.E., Poggetti R., McAnena O.J., Peterson V.M., Abernathy C.M., Parsons P.E. Gut bacterial translocation via the portal vein: a clinical perspective with major torso trauma. J Trauma Acute Care Surg. 1991;31:629–638. doi: 10.1097/00005373-199105000-00006. [DOI] [PubMed] [Google Scholar]
- 4.Bloemen J.G., Venema K., van de Poll M.C., Damink S.W.O., Buurman W.A., Dejong C.H. Short chain fatty acids exchange across the gut and liver in humans measured at surgery. Clin Nutr. 2009;28:657–661. doi: 10.1016/j.clnu.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 5.Ahuja M., Schwartz D.M., Tandon M., Son A., Zeng M., Swaim W., Eckhaus M., Hoffman V., Cui Y., Xiao B. Orai1-mediated antimicrobial secretion from pancreatic acini shapes the gut microbiome and regulates gut innate immunity. Cell Metab. 2017;25:635–646. doi: 10.1016/j.cmet.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cryan J.F., Dinan T.G. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13:701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 7.Mayer E.A. Gut feelings: the emerging biology of gut–brain communication. Nat Rev Neurosci. 2011;12:453–466. doi: 10.1038/nrn3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garrett W.S., Gordon J.I., Glimcher L.H. Homeostasis and inflammation in the intestine. Cell. 2010;140:859–870. doi: 10.1016/j.cell.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Round J.L., Mazmanian S.K. The gut microbiome shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong J.M., De Souza R., Kendall C.W., Emam A., Jenkins D.J. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 2006;40:235–243. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- 11.Smith P.M., Howitt M.R., Panikov N., Michaud M., Gallini C.A., Bohlooly-y M., Glickman J.N., Garrett W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hidalgo I.J., Raub T.J., Borchardt R.T. Characterization of the human colon carcinoma cell line (Caco-2) as a model system for intestinal epithelial permeability. Gastroenterology. 1989;96:736–749. [PubMed] [Google Scholar]
- 13.Artursson P., Karlsson J. Correlation between oral drug absorption in humans and apparent drug permeability coefficients in human intestinal epithelial (Caco-2) cells. Biochem Biophys Res Commun. 1991;175:880–885. doi: 10.1016/0006-291x(91)91647-u. [DOI] [PubMed] [Google Scholar]
- 14.Pinto M.G.V., Gómez M.R., Seifert S., Watzl B., Holzapfel W.H., Franz C.M. Lactobacilli stimulate the innate immune response and modulate the TLR expression of HT29 intestinal epithelial cells in vitro. Int J Food Microbiol. 2009;133:86–93. doi: 10.1016/j.ijfoodmicro.2009.05.013. [DOI] [PubMed] [Google Scholar]
- 15.Eveillard M., Fourel V., Bare M.C., Kernéis S., Coconnier M.H., Karjalainen T., Bourlioux P., Servin A.L. Identification and characterization of adhesive factors of Clostridium difficile involved in adhesion to human colonic enterocyte-like Caco-2 and mucus-secreting HT29 cells in culture. Mol Microbiol. 1993;7:371–381. doi: 10.1111/j.1365-2958.1993.tb01129.x. [DOI] [PubMed] [Google Scholar]
- 16.Kim H.J., Huh D., Hamilton G., Ingber D.E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip. 2012;12:2165–2174. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- 17.Kim H.J., Ingber D.E. Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr Biol. 2013;5:1130–1140. doi: 10.1039/c3ib40126j. [DOI] [PubMed] [Google Scholar]
- 18.Alam M.A., Al-Jenoobi F.I., Al-mohizea A.M. Everted gut sac model as a tool in pharmaceutical research: limitations and applications. J Pharm Pharmacol. 2012;64:326–336. doi: 10.1111/j.2042-7158.2011.01391.x. [DOI] [PubMed] [Google Scholar]
- 19.Rozehnal V., Nakai D., Hoepner U., Fischer T., Kamiyama E., Takahashi M., Yasuda S., Mueller J. Human small intestinal and colonic tissue mounted in the Ussing chamber as a tool for characterizing the intestinal absorption of drugs. European Journal of Pharmaceutical Sciences. 2012;46:367–373. doi: 10.1016/j.ejps.2012.02.025. [DOI] [PubMed] [Google Scholar]
- 20.Smith P., Mirabelli C., Fondacaro J., Ryan F., Dent J. Intestinal 5-fluorouracil absorption: Use of Ussing chambers to assess transport and metabolism. Pharm Res. 1988;5:598–603. doi: 10.1023/a:1015950215230. [DOI] [PubMed] [Google Scholar]
- 21.Sato T., Van Es J.H., Snippert H.J., Stange D.E., Vries R.G., Van Den Born M., Barker N., Shroyer N.F., Van De Wetering M., Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung P., Sato T., Merlos-Suárez A., Barriga F.M., Iglesias M., Rossell D., Auer H., Gallardo M., Blasco M.A., Sancho E. Isolation and in vitro expansion of human colonic stem cells. Nat Med. 2011;17:1225–1227. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- 23.van de Wetering M., Francies H.E., Francis J.M., Bounova G., Iorio F., Pronk A., van Houdt W., van Gorp J., Taylor-Weiner A., Kester L. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sato T., Clevers H. SnapShot: growing organoids from stem cells. Cell. 2015;161:1700–1700.e1. doi: 10.1016/j.cell.2015.06.028. [DOI] [PubMed] [Google Scholar]
- 25.Fatehullah A., Tan S.H., Barker N. Organoids as an in vitro model of human development and disease. Nat Cell Biol. 2016;18:246–254. doi: 10.1038/ncb3312. [DOI] [PubMed] [Google Scholar]
- 26.Park G.-S., Park M.H., Shin W., Zhao C., Sheikh S., Oh S.J., Kim H.J. Emulating host-microbiome ecosystem of human gastrointestinal tract in vitro. Stem Cell Rev Rep. 2017;13:321–334. doi: 10.1007/s12015-017-9739-z. [DOI] [PubMed] [Google Scholar]
- 27.Bhatia S.N., Ingber D.E. Microfluidic organs-on-chips. Nat Biotechnol. 2014;32:760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- 28.Vickerman V., Blundo J., Chung S., Kamm R. Design, fabrication and implementation of a novel multi-parameter control microfluidic platform for three-dimensional cell culture and real-time imaging. Lab Chip. 2008;8:1468–1477. doi: 10.1039/b802395f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanaka Y., Yamato M., Okano T., Kitamori T., Sato K. Evaluation of effects of shear stress on hepatocytes by a microchip-based system. Meas Sci Technol. 2006;17:3167–3170. [Google Scholar]
- 30.Gao D., Liu H., Lin J.-M., Wang Y., Jiang Y. Characterization of drug permeability in Caco-2 monolayers by mass spectrometry on a membrane-based microfluidic device. Lab Chip. 2013;13:978–985. doi: 10.1039/c2lc41215b. [DOI] [PubMed] [Google Scholar]
- 31.ávan der Meer A.D., JungáKim H., ávan der Helm M.W., den Berg A. Measuring direct current trans-epithelial electrical resistance in organ-on-a-chip microsystems. Lab Chip. 2015;15:745–752. doi: 10.1039/c4lc01219d. [DOI] [PubMed] [Google Scholar]
- 32.Maoz B.M., Herland A., Henry O.Y.F., Leineweber W., Yadid M., Doyle J., Mannix R., Kujala V., Fitzgerald E.A., Parker K.K. Organs-on-chips with combined multi-electrode array and transepithelial electrical resistance measurement capabilities. Lab Chip. 2017;17:2294–2302. doi: 10.1039/c7lc00412e. [DOI] [PubMed] [Google Scholar]
- 33.Imura Y., Asano Y., Sato K., Yoshimura E. A microfluidic system to evaluate intestinal absorption. Anal Sci. 2009;25:1403–1407. doi: 10.2116/analsci.25.1403. [DOI] [PubMed] [Google Scholar]
- 34.Pocock K., Delon L., Bala V., Rao S., Priest C., Prestidge C.A., Thierry B. Intestine-on-a-chip microfluidic model for efficient in vitro screening of oral chemotherapeutic uptake. ACS Biomater Sci Eng. 2017;3:951–959. doi: 10.1021/acsbiomaterials.7b00023. [DOI] [PubMed] [Google Scholar]
- 35.Kimura H., Ikeda T., Nakayama H., Sakai Y., Fujii T. An on-chip small intestine–liver model for pharmacokinetic studies. J Lab Autom. 2015;20:265–273. doi: 10.1177/2211068214557812. [DOI] [PubMed] [Google Scholar]
- 36.Maschmeyer I., Lorenz A.K., Schimek K., Hasenberg T., Ramme A.P., Hübner J., Lindner M., Drewell C., Bauer S., Thomas A. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip. 2015;15:2688–2699. doi: 10.1039/c5lc00392j. [DOI] [PubMed] [Google Scholar]
- 37.Esch M.B., Mahler G.J., Stokol T., Shuler M.L. Body-on-a-chip simulation with gastrointestinal tract and liver tissues suggests that ingested nanoparticles have the potential to cause liver injury. Lab Chip. 2014;14:3081–3092. doi: 10.1039/c4lc00371c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trietsch S.J., Naumovska E., Kurek D., Setyawati M.C., Vormann M.K., Wilschut K.J., Lanz H.L., Nicolas A., Ng C.P., Joore J., Kustermann S., Roth A., Hankemeier T., Moisan A., Vulto P. Membrane-free culture and real-time barrier integrity assessment of perfused intestinal epithelium tubes. Nat Commun. 2017;8:262. doi: 10.1038/s41467-017-00259-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shim K.-Y., Lee D., Han J., Nguyen N.-T., Park S., Sung J.H. Microfluidic gut-on-a-chip with three-dimensional villi structure. Biomed Microdevices. 2017;19:37. doi: 10.1007/s10544-017-0179-y. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y., Gunasekara D.B., Reed M.I., DiSalvo M., Bultman S.J., Sims C.E., Magness S.T., Allbritton N.L. A microengineered collagen scaffold for generating a polarized crypt-villus architecture of human small intestinal epithelium. Biomaterials. 2017;128:44–55. doi: 10.1016/j.biomaterials.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shah P., Fritz J.V., Glaab E., Desai M.S., Greenhalgh K., Frachet A., Niegowska M., Estes M., Jäger C., Seguin-Devaux C. A microfluidics-based in vitro model of the gastrointestinal human–microbe interface. Nat Commun. 2016;7:11535. doi: 10.1038/ncomms11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim H.J., Li H., Collins J.J., Ingber D.E. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc Natl Acad Sci. 2016;113:E7–E15. doi: 10.1073/pnas.1522193112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huh D., Kim H.J., Fraser J.P., Shea D.E., Khan M., Bahinski A., Hamilton G.A., Ingber D.E. Microfabrication of human organs-on-chips. Nat Protoc. 2013;8:2135–2157. doi: 10.1038/nprot.2013.137. [DOI] [PubMed] [Google Scholar]
- 44.Bures J., Cyrany J., Kohoutova D., Forstl M., Rejchrt S., Kvetina J., Vorisek V., Kopacova M. Small intestinal bacterial overgrowth syndrome. World J Gastroenterol. 2010;16:2978–2990. doi: 10.3748/wjg.v16.i24.2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsilingiri K., Sonzogni A., Caprioli F., Rescigno M. A novel method for the culture and polarized stimulation of human intestinal mucosa explants. J Vis Exp. 2013;75:4368. doi: 10.3791/4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yissachar N., Zhou Y., Ung L., Lai N.Y., Mohan J.F., Ehrlicher A., Weitz D.A., Kasper D.L., Chiu I.M., Mathis D., Benoist C. An intestinal organ culture system uncovers a role for the nervous system in microbe-immune crosstalk. Cell. 2017;168:1135–1148. doi: 10.1016/j.cell.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Foulke-Abel J., In J., Kovbasnjuk O., Zachos N.C., Ettayebi K., Blutt S.E., Hyser J.M., Zeng X.L., Crawford S.E., Broughman J.R., Estes M.K., Donowitz M. Human enteroids as an ex-vivo model of host-pathogen interactions in the gastrointestinal tract. Exp Biol Med. 2014;239:1124–1134. doi: 10.1177/1535370214529398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ettayebi K., Crawford S.E., Murakami K., Broughman J.R., Karandikar U., Tenge V.R., Neill F.H., Blutt S.E., Zeng X.L., Qu L., Kou B., Opekun A.R., Burrin D., Graham D.Y., Ramani S., Atmar R.L., Estes M.K. Replication of human noroviruses in stem cell-derived human enteroids. Science. 2016;353:1387–1393. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y., DiSalvo M., Gunasekara D.B., Dutton J., Proctor A., Lebhar M.S., Williamson I.A., Speer J., Howard R.L., Smiddy N.M., Bultman S.J., Sims C.E., Magness S.T., Allbritton N.L. Self-renewing monolayer of primary colonic or rectal epithelial cells. Cell Mol Gastroenterol Hepatol. 2017;4:165–182. doi: 10.1016/j.jcmgh.2017.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gjorevski N., Sachs N., Manfrin A., Giger S., Bragina M.E., Ordonez-Moran P., Clevers H., Lutolf M.P. Designer matrices for intestinal stem cell and organoid culture. Nature. 2016;539:560–564. doi: 10.1038/nature20168. [DOI] [PubMed] [Google Scholar]
- 51.Qian X., Nguyen H.N., Song M.M., Hadiono C., Ogden S.C., Hammack C., Yao B., Hamersky G.R., Jacob F., Zhong C., Yoon K.J., Jeang W., Lin L., Li Y., Thakor J., Berg D.A., Zhang C., Kang E., Chickering M., Nauen D., Ho C.Y., Wen Z., Christian K.M., Shi P.Y., Maher B.J., Wu H., Jin P., Tang H., Song H., Ming G.L. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kasendra M, Tovaglieri A, Sontheimer-Phelps A, Jalili-Firoozinezhad S, Bein A, Chalkiadaki A, Scholl W, Zhang C, Rickner H, Richmond CA, Li H, Breault DT, Ingber DE. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci Rep (in press).https://doi.org/10.1038/s41598-018-21201-7. [DOI] [PMC free article] [PubMed]
- 53.Ma L., Barker J., Zhou C., Li W., Zhang J., Lin B., Foltz G., Kublbeck J., Honkakoski P. Towards personalized medicine with a three-dimensional micro-scale perfusion-based two-chamber tissue model system. Biomaterials. 2012;33:4353–4361. doi: 10.1016/j.biomaterials.2012.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gaignebet L., Kararigas G. En route to precision medicine through the integration of biological sex into pharmacogenomics. Clin Sci (Lond) 2017;131:329–342. doi: 10.1042/CS20160379. [DOI] [PubMed] [Google Scholar]
- 55.Evans W.E., Relling M.V. Moving towards individualized medicine with pharmacogenomics. Nature. 2004;429:464–468. doi: 10.1038/nature02626. [DOI] [PubMed] [Google Scholar]
- 56.Brun P., Gobbo S., Caputi V., Spagnol L., Schirato G., Pasqualin M., Levorato E., Palu G., Giron M.C., Castagliuolo I. Toll like receptor-2 regulates production of glial-derived neurotrophic factors in murine intestinal smooth muscle cells. Mol Cell Neurosci. 2015;68:24–35. doi: 10.1016/j.mcn.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 57.Grundy D., Schemann M. Enteric nervous system. Curr Opin Gastroenterol. 2007;23:121–126. doi: 10.1097/MOG.0b013e3280287a23. [DOI] [PubMed] [Google Scholar]
- 58.Geboes K., Collins S. Structural abnormalities of the nervous system in Crohn's disease and ulcerative colitis. Neurogastroenterol Motil. 1998;10:189–202. doi: 10.1046/j.1365-2982.1998.00102.x. [DOI] [PubMed] [Google Scholar]
- 59.Vasina V., Barbara G., Talamonti L., Stanghellini V., Corinaldesi R., Tonini M., De Ponti F., De Giorgio R. Enteric neuroplasticity evoked by inflammation. Auton Neurosci. 2006;126-127:264–272. doi: 10.1016/j.autneu.2006.02.025. [DOI] [PubMed] [Google Scholar]
- 60.Villanacci V., Bassotti G., Nascimbeni R., Antonelli E., Cadei M., Fisogni S., Salerni B., Geboes K. Enteric nervous system abnormalities in inflammatory bowel diseases. Neurogastroenterol Motil. 2008;20:1009–1016. doi: 10.1111/j.1365-2982.2008.01146.x. [DOI] [PubMed] [Google Scholar]
- 61.Halldorsson S., Lucumi E., Gómez-Sjöberg R., Fleming R.M. Advantages and challenges of microfluidic cell culture in polydimethylsiloxane devices. Biosens Bioelectron. 2015;63:218–231. doi: 10.1016/j.bios.2014.07.029. [DOI] [PubMed] [Google Scholar]
- 62.Gomez-Sjoberg R., Leyrat A.A., Houseman B.T., Shokat K., Quake S.R. Biocompatibility and reduced drug absorption of sol-gel-treated poly(dimethyl siloxane) for microfluidic cell culture applications. Anal Chem. 2010;82:8954–8960. doi: 10.1021/ac101870s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Domansky K., Sliz J.D., Wen N., Hinojosa C., Thompson G., Fraser J.P., Hamkins-Indik T., Hamilton G.A., Levner D., Ingber D.E. SEBS elastomers for fabrication of microfluidic devices with reduced drug absorption by injection molding and extrusion. Microfluid Nanofluid. 2017;21:107. [Google Scholar]
- 64.Mondrinos M.J., Yi Y.S., Wu N.K., Ding X., Huh D. Native extracellular matrix-derived semipermeable, optically transparent, and inexpensive membrane inserts for microfluidic cell culture. Lab Chip. 2017;17:3146–3158. doi: 10.1039/c7lc00317j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Prantil-Baun R., Novak R., Debarun D., Somayaji M.R., Andrzej P., Ingber D.E. Physiologically based pharmacokinetic and pharmacodynamic analysis enabled by microfluidically linked organs-on-chips. Annu Rev Pharmacol Toxicol. 2018;58:37–64. doi: 10.1146/annurev-pharmtox-010716-104748. [DOI] [PubMed] [Google Scholar]
- 66.Lee J., Choi J.-H., Kim H.J. Human gut-on-a-chip technology: will this revolutionize our understanding of IBD and future treatments? Expert Rev Gastroenterol Hepatol. 2016;10:883–885. doi: 10.1080/17474124.2016.1200466. [DOI] [PubMed] [Google Scholar]
- 67.Jalili-Firoozinezhad S, Prantil-Baun R, Jiang A, Potla R, Mammoto T, Weaver JC, Ferrante TC, Kim HJ, Cabral JMS, Levy O, and Ingber DE. Modeling radiation injury-induced cell death and countermeasure drug responses in a human Gut-on-a-Chip. Cell Death Dis (in press). https://doi.org/10.1038/s41419-018-0304-8. [DOI] [PMC free article] [PubMed]
- 68.Brown E.M., Wlodarska M., Willing B.P., Vonaesch P., Han J., Reynolds L.A., Arrieta M.-C., Uhrig M., Scholz R., Partida O. Diet and specific microbial exposure trigger features of environmental enteropathy in a novel murine model. Nat Commun. 2015;6:7806. doi: 10.1038/ncomms8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pereira J.F., Awatade N.T., Loureiro C.A., Matos P., Amaral M.D., Jordan P. The third dimension: new developments in cell culture models for colorectal research. Cell Mol Life Sci. 2016;73:3971–3989. doi: 10.1007/s00018-016-2258-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fisher J.T., Zhang Y., Engelhardt J.F. Comparative biology of cystic fibrosis animal models. Cystic Fibrosis. Methods in Molecular Biology (Methods and Protocols). 2011;742:311–334. doi: 10.1007/978-1-61779-120-8_19. Humana Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tarr P.I., Gordon C.A., Chandler W.L. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet. 2005;365:1073–1086. doi: 10.1016/S0140-6736(05)71144-2. [DOI] [PubMed] [Google Scholar]
- 72.Stoven S., Murray J.A., Marietta E.V. Latest in vitro and in vivo models of celiac disease. Expert Opin Drug Discov. 2013;8:445–457. doi: 10.1517/17460441.2013.761203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huh D., Leslie D.C., Matthews B.D., Fraser J.P., Jurek S., Hamilton G.A., Thorneloe K.S., McAlexander M.A., Ingber D.E. A human disease model of drug toxicity–induced pulmonary edema in a lung-on-a-chip microdevice. Sci Transl Med. 2012;4:159ra47. doi: 10.1126/scitranslmed.3004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Benam K.H., Villenave R., Lucchesi C., Varone A., Hubeau C., Lee H.-H., Alves S.E., Salmon M., Ferrante T.C., Weaver J.C., Bahinski A., Hamilton G.A., Ingber D.E. Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat Meth. 2016;13:151–157. doi: 10.1038/nmeth.3697. [DOI] [PubMed] [Google Scholar]
- 75.Benam K.H., Villenave R., Lucchesi C., Mazur M., Hamilton G., Ingber D. Development of a human COPD model-on-a-chip to mimic disease exacerbation (a small airway-on-a-chip model) Eur Respir J. 2014;44(Suppl 58):P3340. [Google Scholar]
- 76.Wang G., McCain M.L., Yang L., He A., Pasqualini F.S., Agarwal A., Yuan H., Jiang D., Zhang D., Zangi L. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med. 2014;20:616–623. doi: 10.1038/nm.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ingber D.E. Reverse engineering human pathophysiology with organs-on-chips. Cell. 2016;164:1105–1109. doi: 10.1016/j.cell.2016.02.049. [DOI] [PubMed] [Google Scholar]
- 78.Mack D.L., Guan X., Wagoner A., Walker S.J., Childers M.K. Disease-in-a-dish: the contribution of patient-specific induced pluripotent stem cell technology to regenerative rehabilitation. Am J Phys Med Rehabil. 2014;93(Suppl 3):S155–S168. doi: 10.1097/PHM.0000000000000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Astolfi M., Peant B., Lateef M.A., Rousset N., Kendall-Dupont J., Carmona E., Monet F., Saad F., Provencher D., Mes-Masson A.M., Gervais T. Micro-dissected tumor tissues on chip: an ex vivo method for drug testing and personalized therapy. Lab Chip. 2016;16:312–325. doi: 10.1039/c5lc01108f. [DOI] [PubMed] [Google Scholar]
- 80.Perel P., Roberts I., Sena E., Wheble P., Briscoe C., Sandercock P., Macleod M., Mignini L.E., Jayaram P., Khan K.S. Comparison of treatment effects between animal experiments and clinical trials: systematic review. BMJ. 2007;334:197. doi: 10.1136/bmj.39048.407928.BE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pirmohamed M., Park B.K. Genetic susceptibility to adverse drug reactions. Trends Pharmacol Sci. 2001;22:298–305. doi: 10.1016/s0165-6147(00)01717-x. [DOI] [PubMed] [Google Scholar]
- 82.Drasar B.S., Montgomery F., Tomkins A.M. Diet and faecal flora in three dietary groups in rural northern Nigeria. Epidemiology & Infection. 1986;96:59–65. doi: 10.1017/s0022172400062537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Everett J.R., Loo R.L., Pullen F.S. Pharmacometabonomics and personalized medicine. Ann Clin Biochem. 2013;50:523–545. doi: 10.1177/0004563213497929. [DOI] [PubMed] [Google Scholar]
- 84.Pirmohamed M. Pharmacogenetics: past, present and future. Drug Discov Today. 2011;16:852–861. doi: 10.1016/j.drudis.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 85.Clayton T.A., Baker D., Lindon J.C., Everett J.R., Nicholson J.K. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci. 2009;106:14728–14733. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nicholson J.K., Holmes E., Wilson I.D. Opinion: gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol. 2005;3:431–438. doi: 10.1038/nrmicro1152. [DOI] [PubMed] [Google Scholar]
- 87.Haiser H.J., Gootenberg D.B., Chatman K., Sirasani G., Balskus E.P., Turnbaugh P.J. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science. 2013;341:295–298. doi: 10.1126/science.1235872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nguyen T.L.A., Vieira-Silva S., Liston A., Raes J. How informative is the mouse for human gut microbiota research? Dis Model Mech. 2015;8:1–16. doi: 10.1242/dmm.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bakken J.S., Borody T., Brandt L.J., Brill J.V., Demarco D.C., Franzos M.A., Kelly C., Khoruts A., Louie T., Martinelli L.P. Treating Clostridium difficile infection with fecal microbiota transplantation. Clin Gastroenterol Hepatol. 2011;9:1044–1049. doi: 10.1016/j.cgh.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kassam Z., Lee C.H., Yuan Y., Hunt R.H. Fecal microbiota transplantation for Clostridium difficile infection: systematic review and meta-analysis. Am J Gastroenterol. 2013;108:500–508. doi: 10.1038/ajg.2013.59. [DOI] [PubMed] [Google Scholar]
- 91.Sheil B., Shanahan F., O'mahony L. Probiotic effects on inflammatory bowel disease. J Nutr. 2007;137:819S–824S. doi: 10.1093/jn/137.3.819S. [DOI] [PubMed] [Google Scholar]
- 92.Steidler L., Hans W., Schotte L., Neirynck S., Obermeier F., Falk W., Fiers W., Remaut E. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science. 2000;289:1352–1355. doi: 10.1126/science.289.5483.1352. [DOI] [PubMed] [Google Scholar]
- 93.Lycke N. Recent progress in mucosal vaccine development: potential and limitations. Nat Rev Immunol. 2012;12:592–605. doi: 10.1038/nri3251. [DOI] [PubMed] [Google Scholar]
- 94.Neutra M.R., Kozlowski P.A. Mucosal vaccines: the promise and the challenge. Nat Rev Immunol. 2006;6:148–158. doi: 10.1038/nri1777. [DOI] [PubMed] [Google Scholar]
- 95.Zietek T., Rath E., Haller D., Daniel H. Intestinal organoids for assessing nutrient transport, sensing and incretin secretion. Sci Rep. 2015;5:16831. doi: 10.1038/srep16831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang Y.G., Wu S., Xia Y., Sun J. Salmonella-infected crypt-derived intestinal organoid culture system for host–bacterial interactions. Physiol Rep. 2014;2:e12147. doi: 10.14814/phy2.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lu W., Rettenmeier E., Paszek M., Yueh M.-F., Tukey R.H., Trottier J., Barbier O., Chen S. Crypt organoid culture as an in vitro model in drug metabolism and cytotoxicity studies. Drug Metab Dispos. 2017;45:748–754. doi: 10.1124/dmd.117.075945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Madsen K., Cornish A., Soper P., McKaigney C., Jijon H., Yachimec C., Doyle J., Jewell L., De Simone C. Probiotic bacteria enhance murine and human intestinal epithelial barrier function. Gastroenterology. 2001;121:580–591. doi: 10.1053/gast.2001.27224. [DOI] [PubMed] [Google Scholar]
- 99.Worton K., Candy D., Wallis T., Clarke G., Osborne M., Haddon S., Stephen J. Studies on early association of Salmonella typhimurium with intestinal mucosa in vivo and in vitro: relationship to virulence. J Med Microbiol. 1989;29:283–294. doi: 10.1099/00222615-29-4-283. [DOI] [PubMed] [Google Scholar]
- 100.Sjöberg Å., Lutz M., Tannergren C., Wingolf C., Borde A., Ungell A.-L. Comprehensive study on regional human intestinal permeability and prediction of fraction absorbed of drugs using the Ussing chamber technique. Eur J Pharm Sci. 2013;48:166–180. doi: 10.1016/j.ejps.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 101.Dawson A., Dyer C., Macfie J., Davies J., Karsai L., Greenman J., Jacobsen M. A microfluidic chip based model for the study of full thickness human intestinal tissue using dual flow. Biomicrofluidics. 2016;10:064101. doi: 10.1063/1.4964813. [DOI] [PMC free article] [PubMed] [Google Scholar]