Abstract
Rnd1, a member of Rho GTPases, was found to be downregulated in human malignancies and downregulation of Rnd1 promotes tumor invasion via various mechanisms. However, the role of Rnd1 in hepatocellular carcinoma (HCC) progression remains unclear. In this study, our results demonstrated that Rnd1 was downregulated in HCC cells and in human HCC tissues. Low expression of Rnd1 was associated with aggressive clinic-pathologic characteristics, such as vascular invasion, and poor prognosis in patients who underwent curative surgery for HCC. Overexpression of Rnd1-suppressed cell growth, migration, invasion, and EMT processes in vitro and in vivo. Furthermore, Rnd1 blocked HCC progression by restricting EMT process through inhibition of the Raf/MEK/ERK cascade, and this was correlated with a reduction in RhoA activity. Combination of Rnd1 overexpression with sorafenib, a Raf signaling pathway inhibitor, showed a more potent inhibition on HCC metastasis. Moreover, epigenetic inhibitors (5-Aza and SAHA) increased the expression of Rnd1, and potentiated sorafenib-induced toxicity in HCC cells. In a conclusion, Rnd1-suppressed EMT-mediated metastasis of HCC by reducing the activity of the RhoA/Raf/MEK/ERK signaling pathway, functioning as a favorable anti-metastasis target for HCC patients. Rnd1 overexpression in combination with sorafenib may result in enhanced anti-metastasis efficacy in HCC.
Introduction
Liver cancer, mostly hepatocellular carcinoma (HCC), is the fifth most prevalent malignance and the second-leading cause of cancer-related mortality among men worldwide1. Surgical resection is the primary treatment for HCC. However, the 5-year recurrence rate is as high as 70%, primarily due to intrahepatic and extrahepatic metastasis2. Decades of research have brought insight into the progression of tumors with the identification of a set of genes correlated with metastasis and exploration of therapeutic targets to prolong patients’ survival3,4. However, there are still many unexplored factors in the progression of HCC metastasis. To further reveal the driving events of HCC metastasis is of great significance.
Epithelial–mesenchymal transition (EMT) is a critical step in invasion-metastasis cascades5. During this process, tumor cells reversibly transform from an epithelial phenotype to a mesenchymal phenotype, followed by enhanced cell invasive capabilities. And EMT is characterized by downregulation of the epithelial marker, E-cadherin, and upregulation of mesenchymal markers, such as N-cadherin and vimentin6. Our previous studies had revealed that EMT played a key role in chemo-resistance of HCC and could be modulated by microRNAs, genes, and drugs7,8.
Rho GTPases belongs to the family of small G proteins, which acts as molecular switches by cycling between active state (GTP-bound) and inactive state (GDP-bound). Only in the active state, Rho GTPases could combine to a wide variety of targeted proteins to generate a response in the modulation of cytoskeleton dynamics and gene transcription9. Through this process, Rho GTPases play a pivotal role in the regulation of cellular physiological functions, such as cell polarity, adhesion, locomotion, and invasion, as well as cell proliferation and survival10. Moreover, the crosstalk among different Rho GTPases has been shown to regulate the process of EMT11. Deregulation of Rho GTPases has also been validated to be intricately involved in tumor progression10. As a member of the Rho family of GTPases, Rnd1 is located at chromosome 12q12-q13, the deletion of which is regularly observed in pancreatic cancer and adenoid cystic carcinoma12,13. Besides, it had been shown that inactivation of Rnd1 could drive breast cancer initiation and progression14. Nevertheless, whether Rnd1 plays a role in HCC progression remains undetermined. In the present study, we aimed to investigate the epigenetic regulation and the underlying mechanisms of Rnd1 in HCC.
Results
Downregulation of Rnd1 is associated with aggressive clinic pathological features and poor prognosis in HCC
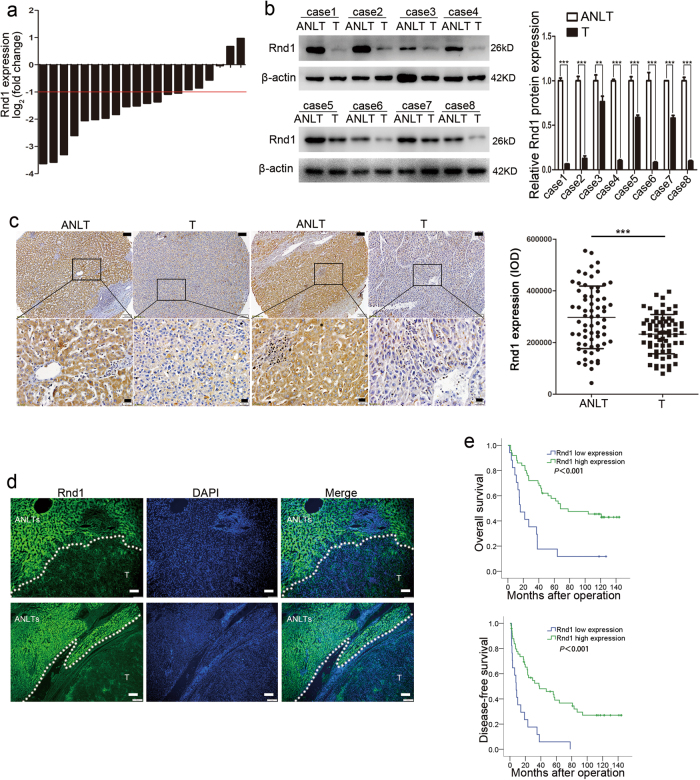
The mRNA expression level of Rnd1 was compared in 20 paired HCC tissue and adjacent non-tumor liver tissue (ANLT). The results showed that the mean level of Rnd1 mRNA in 14 HCC cases was significantly lower (more than twofold; i.e., log2 [fold change] > 1) than that in the ANLTs (70.0%; Fig. 1a). Consistent with these findings, Rnd1 protein level was decreased in HCC tissues compared with ANLT by western blot, immunohistochemistry, and immunofluorescence assay (Fig. 1b–d).
Fig. 1. Rnd1 expression is downregulated in HCC and predicts poor prognosis.
a Rnd1 mRNA expression in 20 paired HCC tissues (T) and adjacent non-tumor liver tissues (ANLTs). Expression level of Rnd1 was normalized to GAPDH. Fold changes were analyzed using the formula: 2−(∆∆ CT [HCC/ANLT]). Red line indicates fold change of Rnd1 equal to 2. b Protein expression of Rnd1 in select HCC tissues (T; n = 8) and adjacent non-tumor liver tissues (ANLTs; n = 8). Densitometry of western blot in panel. Data represent means ± S.D. of three independent experiments, significant difference was tested by Student’s t-test. **P < 0.01, ***P < 0.001. c Representative IHC images of Rnd1 expression in HCC tissues (T) and adjacent non-tumor liver tissues (ANLTs). Scale bar: 100 μm (upper), 20 μm (lower). Rnd1 expression was quantitated based on integrated optical density (IOD). Results were expressed as mean ± S.D., n = 67 patients. Significant difference was tested by Student’s t-test. ***P < 0.001. d Representative IF images of Rnd1 expression in HCC tissues (T) and adjacent non-tumor liver tissues (ANLTs); boundaries between T and ANLT tissue are indicated by a dashed line. Scale bar: 100 μm. e Kaplan–Meier analysis indicates that low expression of Rnd1 is correlated with poor overall survival and disease-free survival in 67 cases of HCC patients. **P < 0.01
To illustrate the clinical relevance of Rnd1 expression in HCC, the cutoff finder software was used to determine the optimal cutoff points to separate patients into high and low expression group, based on integral optical density (IOD). According to this criteria, associations between Rnd1 expression and clinicopathologic characteristics of HCC were evaluated, the results showed that patients with low Rnd1 expression had a large tumor size, high incidence of microvascular invasion, and poor cell differentiation in tumor tissue (Supplementary table 1). Similarly, HCC cases with vascular invasion had lower expression of Rnd1 than those without vascular invasion in the TCGA data sets (P = 0.019, Supplementary Figure 1). Survival analysis suggested that patients with low Rnd1 expression in HCC tissue had shorter overall survival (OS) and disease-free survival (DFS) time than those with high expression (Fig. 1e). Multivariate analysis identified Rnd1 expression as an independent risk factor for OS and DFS (Table 1). These results suggested that low Rnd1 expression was associated with invasive characteristics and poor prognosis in HCC patients.
Table 1.
Univariate and multivariate analyses of risk factors associated with overall survival and disease-free survival of HCC patients in training cohort
| Variables | Overall Survival | Disease-free survival | |||||||
|---|---|---|---|---|---|---|---|---|---|
| n | Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | |||||
| HR(95% CI) | P | HR(95% CI) | P | HR(95% CI) | P | HR(95% CI) | P | ||
| Sex | |||||||||
| Male | 58 | 1 | 0.433 | NA | 1 | 0.385 | NA | ||
| Female | 9 | 0.689(0.271–1.751) | 0.685(0.292–1.609) | ||||||
| Age, years | |||||||||
| ≤60 | 53 | 1 | 0.574 | NA | 1 | 0.321 | NA | ||
| >60 | 14 | 1.226(0.603–2.494) | 1.389(0.725–2.660) | ||||||
| HBV | |||||||||
| Negative | 8 | 1 | 0.040 | 1 | NS | 1 | 0.216 | NA | |
| Positive | 59 | 4.443(1.072–18.414) | 2.588(0.589–11.017) | 1.716(0.729–4.041) | |||||
| Liver cirrhosis | |||||||||
| Absence | 8 | 1 | 0.051 | NA | 1 | 0.454 | NA | ||
| Presence | 59 | 4.128(0.996–17.105) | 1.386(0.590–3.259) | ||||||
| Tumor size, cm | |||||||||
| ≤5 cm | 47 | 1 | <0.001 | 1 | 0.003 | 1 | 0.004 | 1 | 0.042 |
| >5 cm | 20 | 3.432(1.833–6.426) | 2.919(1.443–5.903) | 2.383(1.320–4.302) | 1.934(1.024–3.655) | ||||
| AFP, ng/ml | |||||||||
| <20 | 27 | 1 | 0.646 | NA | 1 | 0.634 | NA | ||
| ≥20 | 40 | 1.153(0.625–2.125) | 1.146(0.653–2.013) | ||||||
| Capsulation formation formation formation | |||||||||
| Absence | 31 | 1 | 0.162 | NA | 1 | 0.055 | NA | ||
| Presence | 36 | 0.652(0.357–1.188) | 0.582(0.335–1.012) | ||||||
| Microvascular invasion | |||||||||
| Presence | 12 | 1 | <0.001 | 1 | NS | 1 | <0.001 | 1 | NS |
| Absence | 55 | 6.204(2.976–12.933) | 2.050(0.773–5.434) | 4.893(2.378–10.069) | 1.641(0.648–4.156) | ||||
| Edmondson-Steiner grade | |||||||||
| I & II | 43 | 1 | 0.001 | 1 | NS | 1 | <0.001 | 1 | 0.008 |
| III & IV | 24 | 2.731(1.487–5.015) | 2.117(0.967–4.634) | 3.235(1.809–5.785) | 2.423(1.262–4.652) | ||||
| BCLC stage | |||||||||
| 0 & A | 9 | 1 | 0.020 | 1 | 0.042 | 1 | 0.195 | NA | |
| B & C | 58 | 10.589(1.453–77.186) | 8.104(1.081–60.767) | 1.760(0.748–4.144) | |||||
| TNM stage | |||||||||
| Early (I & II) | 59 | 1 | 0.083 | NA | 1 | 0.549 | NA | ||
| Late (III & IV) | 8 | 1.876(0.921–3.822) | 1.247(0.606–2.566) | ||||||
| Rnd1 expression | |||||||||
| Low | 17 | 1 | 0.001 | 1 | 0.038 | 1 | <0.001 | 1 | 0.044 |
| High | 50 | 0.318(0.167–0.607) | 0.447(0.209–0.955) | 0.297(0.160–0.550) | 0.450(0.207–0.978) | ||||
HR hazard risk ratio, CI confidence interval, NA not applicable, NS not significant
*Significant results (P<0.05) are given in bold
Downregulation of Rnd1 facilitates growth, migration, and metastasis of HCC cells in vitro and in vivo
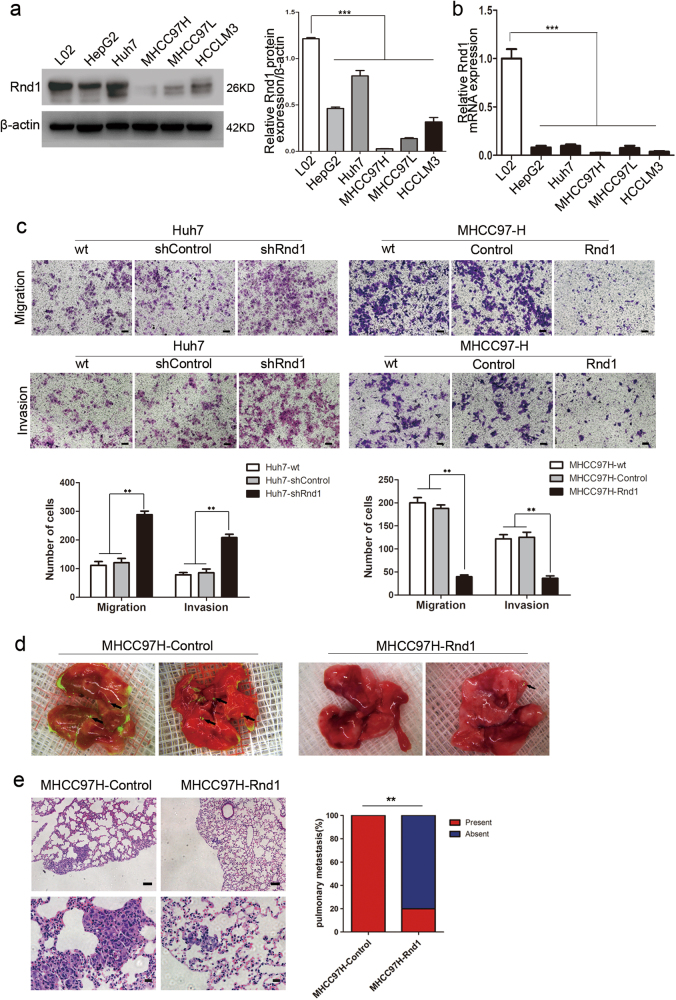
As Rnd1 expression is associated with microvascular invasion in human HCC, we focused on the effects of Rnd1 on migration and invasion of HCC cells. First, we measured the mRNA and protein expression of Rnd1 by real-time PCR and western blot in a normal liver cell line (L02) and in a series of HCC cell lines with varying metastatic potential (MHCC97H, HCCLM3, MHCC97L, HepG2, and Huh7)15. Compared with L02, Rnd1 expression was lower in the HCC cells (Fig. 2a, b). We used short hairpin RNA (shRNA) to interfere the expression of Rnd1 in the Huh7 cell line (termed Huh7-shControl/Huh7-shRnd1; Supplementary Figure 2a) and used complementary DNA (cDNA) to stably overexpress Rnd1 in MHCC97H cell line (termed MHCC97H-Control/MHCC97H-Rnd1; Supplementary Figure 2b). Rnd1 showed mainly cytoplasmic expression in MHCC97H cells by immunofluorescence (Supplementary Figure 2c).
Fig. 2. Attenuated Rnd1 expression promotes the migration and invasion of HCC cells in vitro and in vivo.
a, b Western blot and Real-time PCR analysis of the Rnd1 protein and mRNA expression, respectively, in one normal liver cell line (L02) and five HCC cell lines (Huh7, HepG2, MHCC97L, MHCC97H, and HCCLM3). Panels represent mean ± S.D. (n = 3) from three independent experiments plated in triplicate. Significance was determined using one-way ANOVA. ***P < 0.001. c The impact of Rnd1 knockdown and overexpression on cell migration and invasion, as evaluated by transwell assays with or without Matrigel. Representative images of migrated or invasive cells are shown. Scale bar: 100 μm. The number of cells passed through the membrane was counted and compared in the graphs below, the results are expressed as the mean ± S.D. (n = 3) of three independent experiments, significance was determined using one-way ANOVA. **P < 0.01. d Representative fluorescence images of lungs derived from orthotopic xenograft tumor models. e Representative HE staining images for lung metastasis. Scale bar: 100 μm (upper), 20 μm (lower). The percentage of mice with or without metastatic nodules in the lungs was calculated and compared (5 mice in each group). Significance was determined using χ2 test. **P < 0.01
A transwell assay with or without Matrigel showed that migration and invasion were increased in Huh7-shRnd1 cells, and were decreased in MHCC97H-Rnd1 cells, compared to corresponding control groups (Fig. 2c). Furthermore, wound-healing assays demonstrated that Rnd1 expression is reversed related with the speed of wound closure (Supplementary Figure 3). These results suggested that Rnd1 negatively regulated tumor migration and invasion of HCC cells in vitro. Additionally, Rnd1 also suppressed cell proliferation and arrested the cell cycle in the G2/M phase (Supplementary Figure 4a-d), but had no effect on the apoptosis of HCC cells in vitro (Supplementary Figure 4e). Moreover, in order to reduce the possibility of off-target effects, we designed another three shRNA plasmids to interfere Rnd1 expression in Huh7 cells. Two working shRNA plasmids inhibited Rnd1 expression and their effects of knockdown could be overcome by expressing Rnd1 cDNA (Supplementary Figure 5a, b). Besides, the transfection of them also promoted the proliferation, migration and invasion of Huh7 cells in vitro (Supplementary Figure 5c-e).
We established an orthotopic xenograft tumor models to verify the effects of Rnd1 on HCC progression in vivo. Forty days after tumor incubation, the tumor volume in the Huh7-shRnd1 group was larger than that of the Huh7-control group (P = 0.01; Supplementary Figure 6a), and the tumor volume in MHCC97H-Rnd1 group was smaller than that of the MHCC97H group (P < 0.01; Supplementary Figure 6b). All mice in the MHCC97H-control group developed lung metastasis, whereas only one mouse in the MHCC97H-Rnd1 group (Fig. 2d, e). These results indicated that Rnd1-suppressed HCC growth and metastasis in vivo.
Downregulation of Rnd1 induces EMT of HCC cells
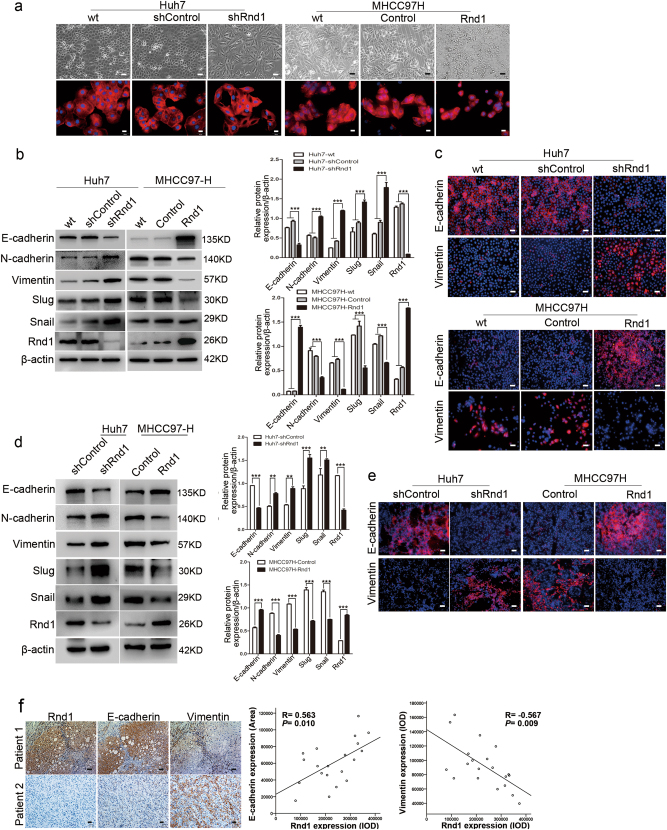
Because the Rho GTPase family is involved in modulating cytoskeletal reconstruction9, we therefore evaluated the impact of Rnd1 on cytoskeleton remodeling of HCC cells. Rnd1 silencing induced spindle-like mesenchymal morphology in Huh7 cells (Fig. 3a), and overexpression of Rnd1 induced a change from a mesenchymal-like shape to a cobblestone-like epithelial morphology in MHCC97H cells (Fig. 3a). As the EMT process is critical in tumor metastasis and there is a close relationship between EMT and cytoskeletal reconstruction16,17, the expression of epithelial and mesenchymal markers and related molecules which could lead to EMT in HCC cells were evaluated. Western blot analysis confirmed reduced epithelial marker (E-cadherin) and increased mesenchymal markers (N-cadherin, Vimentin, slug, and snail) accompanied with Rnd1 knockdown in Huh7 cells (Fig. 3b). Whereas increased Rnd1 manifested the opposite effects in MHCC97H cells (Fig. 3b). Moreover, immunofluorescence assay demonstrated that Rnd1 expression had a positive and negative effect on the fluorescence intensity of E-cadherin and vimentin, respectively (Fig. 3c).
Fig. 3. Low expression of Rnd1 induces EMT in HCC cells.
a (upper) Representative phase-contrast images of Huh7-shRnd1 cells, MHCC97H-Rnd1 cells, and their respective wild type and control cells. a (lower) Representative IF images of cytoskeleton in Huh7-shRnd1 cells, MHCC97H-Rnd1 cells, and their respective wild type and control cells. Scale bar: 50 μm (upper), 20 μm (lower). b Western blot analysis of E-cadherin, N-cadherin, vimentin, Slug, and Snail protein expression in HCC cells with knockdown or overexpression of Rnd1 and their corresponding wild type and control cells. Densitometry of western blot in panel. Data are mean ± S.D. (n = 3) and are representative of three independent experiments. Significance was determined using two-way ANOVA. ***P < 0.001. c IF staining for E-cadherin and vimentin in Huh7, Huh7-shControl, Huh7-shRnd1, MHCC97H, MHCC97H-Control, and MHCC97H-Rnd1 cells. 4′,6-diamidino-2-phenylindole (DAPI) was used to stain nuclei. Scale bar: 50 μm. d Western blot analysis of E-cadherin, N-cadherin, vimentin, slug, snail, and Rnd1 expression in HCC tissues derived from orthotopic xenograft tumor models. Densitometry of western blot in panel. Data represent means ± S.D. (n = 3) of three independent experiments. Significance was determined using two-way ANOVA. *P < 0.05, **P < 0.01, ***P < 0.001. e Representative IF images of E-cadherin and vimentin from Huh7-shControl, Huh7-shRnd1, MHCC97H-Control, and MHCC97H-Rnd1 tumors in nude mice. Scale bar: 50 μm. f A positive association between Rnd1 and E-cadherin, however, a negative association between Rnd1 and vimentin is observed in serial slices of hepatoma tissue (n = 20; R = 0.563, P = 0.010 and R = −0.567, P = 0.009, respectively). Scale bar: 100 μm (upper), 20 μm (lower)
In the orthotopic xenograft models, tissues derived from Huh7-shRnd1 tumors expressed lower E-cadherin protein levels but higher levels of mesenchymal markers compared with the control group (Fig. 3d, e). In contrast, opposite effects of E-cadherin and mesenchymal markers were observed in tissues derived from MHCC97H-Rnd1 tumors compared to controls (Fig. 3d, e). Additionally, IHC for serial slices of human tumor HCC tissue demonstrated that Rnd1 expression is positively correlated with E-cadherin (R = 0.563, P = 0.010) and negatively correlated with vimentin expression (R = −0.567, P = 0.009; Fig. 3f). These results indicated that Rnd1 might act as an inhibitor of EMT in HCC.
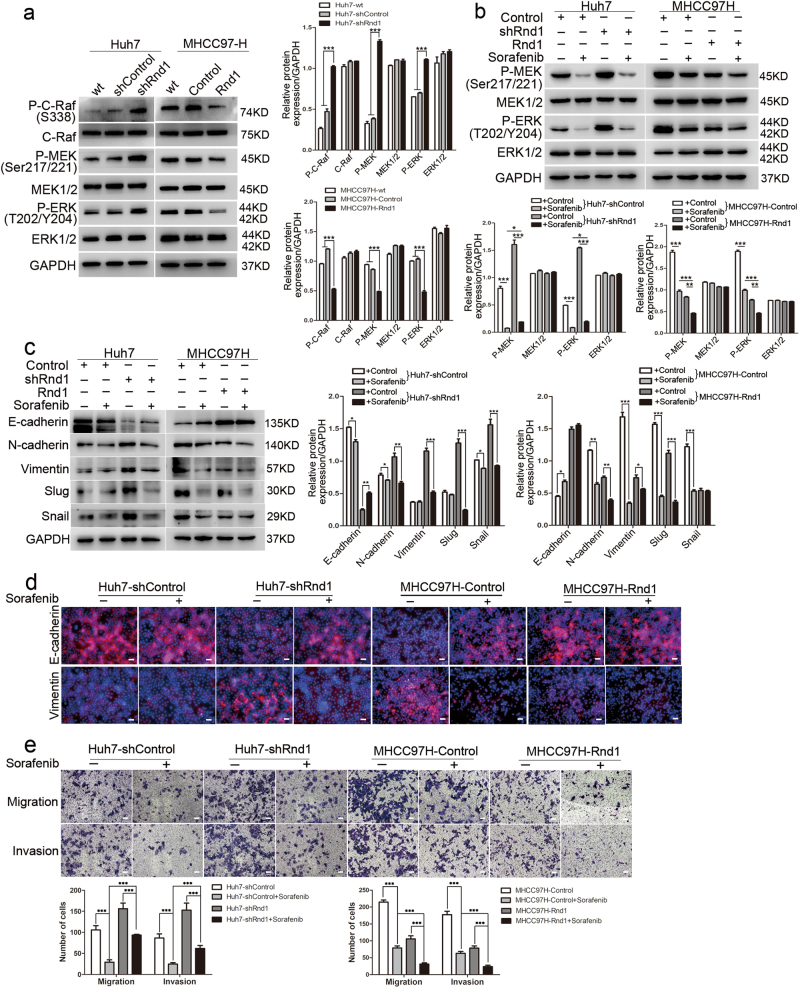
Rnd1 inhibits the Raf/MEK/ERK signaling pathway in HCC
It was reported that Rnd1 restricted the activity of MEK and ERK in breast cancers14, and hyper-activation of Raf/MEK/ERK signaling induces EMT in cancer cells18,19. Consistently, we found that knockdown of Rnd1 in Huh7 cells significantly increased the phosphorylation levels of Raf, MEK, and ERK (Fig. 4a), whereas ectopic expression of Rnd1 in MHCC97H cells decreased Raf, MEK, and ERK phosphorylation (Fig. 4a). To determine the necessity of Raf/MEK/ERK pathway in Rnd1-medicated functions, we blocked the Raf/MEK/ERK pathway with sorafenib, a serine/threonine kinase inhibitor that targets c-Raf and both mutant and wild-type B-Raf20. The phosphorylation of MEK and ERK in Huh7-shControl and MHCC97H-Rnd1 cells with higher-expression of Rnd1 was inhibited more pronounced by sorafenib than in Huh7-shRnd1 and MHCC97H-Control cells with lower expression of Rnd1 (Fig. 4b). Moreover, Sorafenib restrained the EMT-promoting effect of Rnd1 silencing in Huh7-shRnd1 and MHCC97H-Control cells (Fig. 4c, d). Transwell assays showed that sorafenib had a more pronounced effect on repressing the invasiveness and migratory ability of HCC cells with higher expression of Rnd1 than the cells with lower expression of Rnd1 (Fig. 4e). These results suggested that there was a synergism between Rnd1 and sorafenib in suppressing the activity of Raf/MEK/ERK signaling pathway, as well as the migration and invasion ability of HCC cells.
Fig. 4. Silencing of Rnd1 induces EMT in HCC cells by the activating Raf/MEK/ERK signaling pathway.
a Western blot analysis of the phosphorylation of Raf, MEK, and ERK in Huh7-shRnd1 cells, MHCC97H-Rnd1 cells, and their corresponding wild type and control cells. All data for the densitometric analysis of the western blots are presented as mean ± S.D. (n = 3) of three independent experiments. Significance was determined using two-way ANOVA. ***P < 0.001. b Western blot analysis of the phosphorylation levels of Raf, MEK, and ERK in Huh7-shControl/Huh7-shRnd1 and MHCC97H-Control/ MHCC97H-Rnd1 cells after treated with sorafenib for 24 h. Densitometry of western blot in panel. Data are mean ± S.D. (n = 3) and are representative of three independent experiments. Significance was determined using two-way ANOVA. ***P < 0.001. c Western blot for E-cadherin, N-cadherin, vimentin, Slug, and Snail revealed that sorafenib reverses EMT in Huh7-shRnd1 and MHCC97H-Control cells after 72 h treatment. Densitometry of western blot in panel. Data are mean ± S.D. (n = 3) and are representative of three independent experiments. Significance was determined using two-way ANOVA. ***P < 0.001. d Representative IF images of E-cadherin and vimentin in Huh7-control/+sorafenib, Huh7-shRnd1/+sorafenib, MHCC97H-control/+sorafenib, and MHCC97H-Rnd1/+sorafenib. Scale bar: 50 μm. e Representative images of transwell assays with or without Matrigel® for Huh7-shRnd1 cells, MHCC97H-Rnd1 cells, and their corresponding control cells with or without sorafenib treatment. Scale bar: 100 μm. Panel: quantification of migration and invasion of cells, data represent means ± S.D. (n = 3) of three independent experiments, significance was determined using one-way ANOVA. ***P < 0.001
The MEK inhibitor U0126 was also used to further assess the relationship between Rnd1 and Raf/MEK/ERK pathway. Western blotting demonstrated that the high expression of p-ERK induced by Rnd1 downregulation was suppressed by U0126 (Supplementary Figure 7a). And the mesenchymal-like properties were also attenuated, as indicated by the changes of protein level and fluorescence intensity of EMT markers (Supplementary Figure 7b, c). Furthermore, Transwell assay showed that U0126 inhibited cell migration and invasion, and this effect was more pronounced in Rnd1 overexpressing cells (Supplementary Figure 7d). Collectively, these data suggested that a close interaction existed between Rnd1 and the Raf/MEK/ERK pathway.
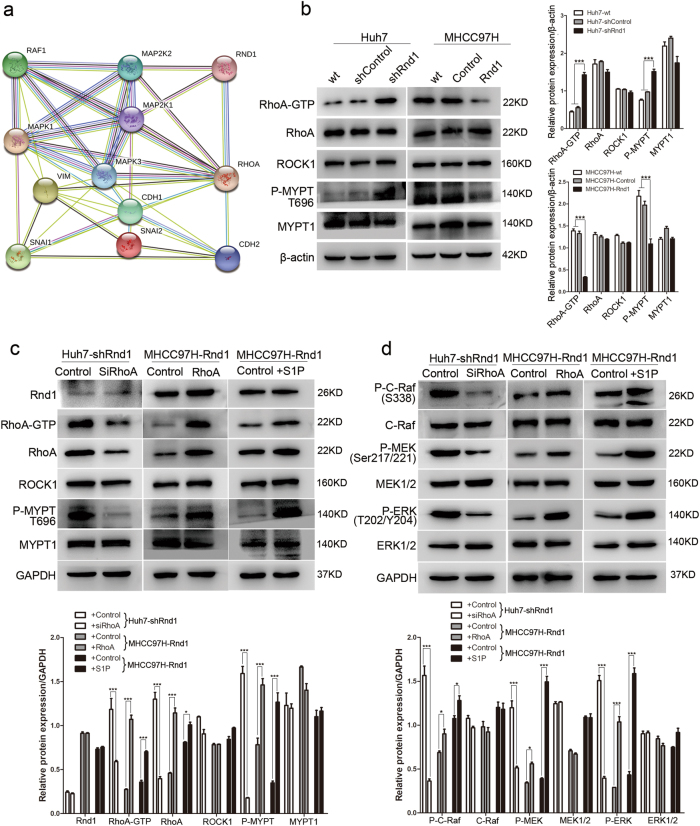
Rnd1 regulates the Raf/MEK/ERK pathway by modulating the activity of RhoA
STRING 10.0 software was used to constructed the protein–protein interaction networks, which revealed that Rnd1 had a strong connection with RhoA (Supplementary Figure 8), a protein that plays a key role in EMT by regulating cytoskeleton organization, stress fiber bundling, and focal adhesion21–23. The STRING database also exhibited that RhoA could interact with the Raf/MEK/ERK signaling pathway (Fig. 5a). So we hypothesized that Rnd1 might modulate Raf/MEK/ERK signaling by interacting with RhoA. Since RhoA cycles between an active GTP-bound form and an inactive GDP-bound form24, therefore we examined the level of RhoA activation by using a RhoA-GTP pull-down assay. Knockdown of Rnd1 in Huh7 cells upregulated RhoA-GTP expression; in contrast, overexpression of Rnd1 in MHCC97H cells downregulated RhoA-GTP expression (Fig. 5b). ROCK1 (rho-associated, coiled-coil-containing protein kinase 1), a main downstream mediator of Rho GTPase signaling, regulates the cytoskeleton by stimulating phosphorylation of MYPT1 (myosin-associated phosphatase type 1)25–27, and phosphorylated MYPT1 is regarded as an indicator of ROCK1 activation28. The phosphorylation level of MYPT1 in Huh7-shRnd1 and MHCC97H-Control cells was higher than in the Huh7-shControl and MHCC97H-Rnd1 cells (Fig. 5b), suggesting that high expression of Rnd1 suppressed the activation of the RhoA/ROCK1/MYPT1 signaling axis.
Fig. 5. Attenuated Rnd1 expression activates Raf/MEK/ERK signaling by enhancing the activity of RhoA.
a The interaction diagram of Rnd1 with RhoA, Raf/MEK/ERK, and EMT related molecules, as analyzed by STRING 10.0. b Western blot analysis of RhoA-GTP pull-down assay, indicating the degree of RhoA activation, and protein expression of ROCK1 and P-MYPT(T696), in Huh7-shRnd1 cells, MHCC97H-Rnd1 cells, and their corresponding wild type and control cells. c Changes in protein expression of Rnd1, RhoA-GTP, ROCK1, and P-MYPT (T696) in HCC cells with modulation of Rnd1 treated with RhoA siRNA, ectopic expression of RhoA, or stimulated with S1P. d Changes in protein expression of P-C-Raf (S338), P-MEK (Ser217/221), and P-ERK (T202/Y204) in HCC cells with modulation of Rnd1 treated with RhoA siRNA, ectopic expression of RhoA, or stimulated with S1P. All data for the densitometric analysis of the western blots are presented as mean ± S.D. (n = 3) of three independent experiments. Significance was determined using two-way ANOVA. *P < 0.01, ***P < 0.001
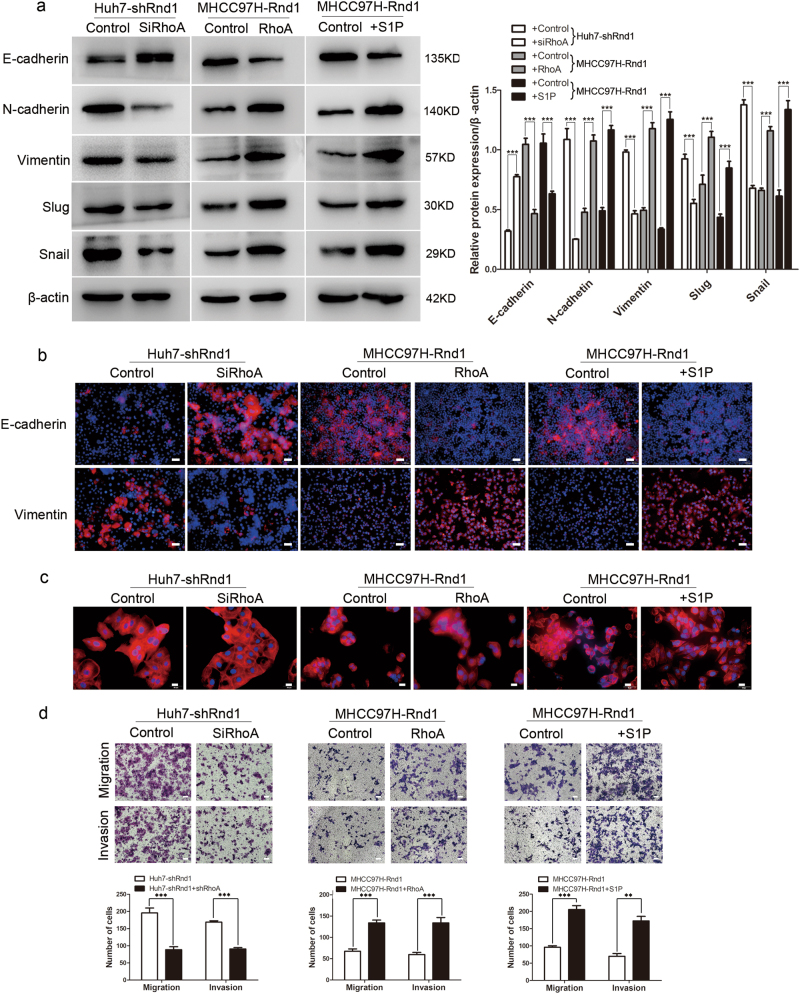
To assess the importance of RhoA in Rnd1-mediated signal pathway and functions, we modulated the expression of RhoA in HCC cells. Inhibition of RhoA by siRNA in Huh7-shRnd1 cells had no effects on the expression of Rnd1, but reduced the expression of RhoA, RhoA-GTP, and P-MYPT1 (Fig. 5c), and downregulated the phosphorylation of Raf, MEK, and ERK (Fig. 5d). Furthermore, RhoA knockdown increased E-cadherin expression and reduced the expression of mesenchymal markers in Huh7-shRnd1 cells (Fig. 6a, b). Additionally, the shape of Huh7-shRnd1-siRhoA cells changed from a spindle-like appearance to a cobblestone-like appearance (Fig. 6c). Huh7-shRnd1-siRhoA cells also exhibited reduced migration and invasive ability (Fig. 6d). These results indicated that the inhibition RhoA reversed the activation of the Raf/MEK/ERK pathway and EMT that was induced by Rnd1 silencing.
Fig. 6. Rnd1 mediates the migration, invasion, and EMT of HCC cells by regulating RhoA activity.
a Western blot analysis of E-cadherin, N-cadherin, vimentin, Slug, and Snail in HCC cells with modulation of Rnd1 treated with RhoA siRNA, ectopic expression of RhoA, or stimulated with S1P. Densitometry of western blot in panel. Data are mean ± S.D. (n = 3) and are representative of three independent experiments. Significance was determined using two-way ANOVA. ***P < 0.001. b Representative IF images of E-cadherin and vimentin in HCC cells with modulation of Rnd1 treated with RhoA siRNA, ectopic expression of RhoA, or stimulated with S1P. Scale bar: 50 μm. c The cytoskeleton of HCC cells with modulation of Rnd1 treated with RhoA siRNA, ectopic expression of RhoA, or stimulated with S1P. Scale bar: 20 μm. d Migration and invasion were assessed using transwell assays with or without Matrigel® in HCC cells with modulation of Rnd1 treated with RhoA siRNA, ectopic expression of RhoA, or stimulated with S1P. Scale bar: 100 μm. Panel: quantification of migration and invasion of cells. Data are mean ± S.D. (n = 3) and are representative of three independent experiments. Significance was determined using one-way ANOVA. ***P < 0.001
The results of rescue assays showed that ectopic expression of RhoA in MHCC97H-Rnd1 cells increased the activity of RhoA and the phosphorylation of MYPT1 (Fig. 5c), as well as the activity of Raf/MEK/ERK pathway (Fig. 5d). MHCC97H-Rnd1-RhoA cells exhibited downregulation of E-cadherin and upregulation of mesenchymal markers (Fig. 6a, b). Additionally, MHCC97H-Rnd1-RhoA cells presented a spindle-like morphology and exhibited increased migration and invasive ability (Fig. 6c, d). Treatment of MHCC97H-Rnd1 cells with the Rho-activator sphingosine-1-phosphate (S1P) dramatically induced activation of the RhoA/ROCK1/MYPT1 pathway (Fig. 5c), and resulted in the reactivation of the Raf/MEK/ERK pathway (Fig. 5d). After treatment with S1P, MHCC97H-Rnd1 cells presented a mesenchymal-like phenotype, based on cell shape and expression of key EMT molecules (Fig. 6a–c), and also exhibited augmented migratory and invasive ability (Fig. 6d). These observations suggested that the reactivation of RhoA abolished the inhibitory effect of Rnd1 on the Raf/MEK/ERK pathway and related EMT phenomena.
Epigenetic mechanisms for the inactivation of Rnd1 in HCC
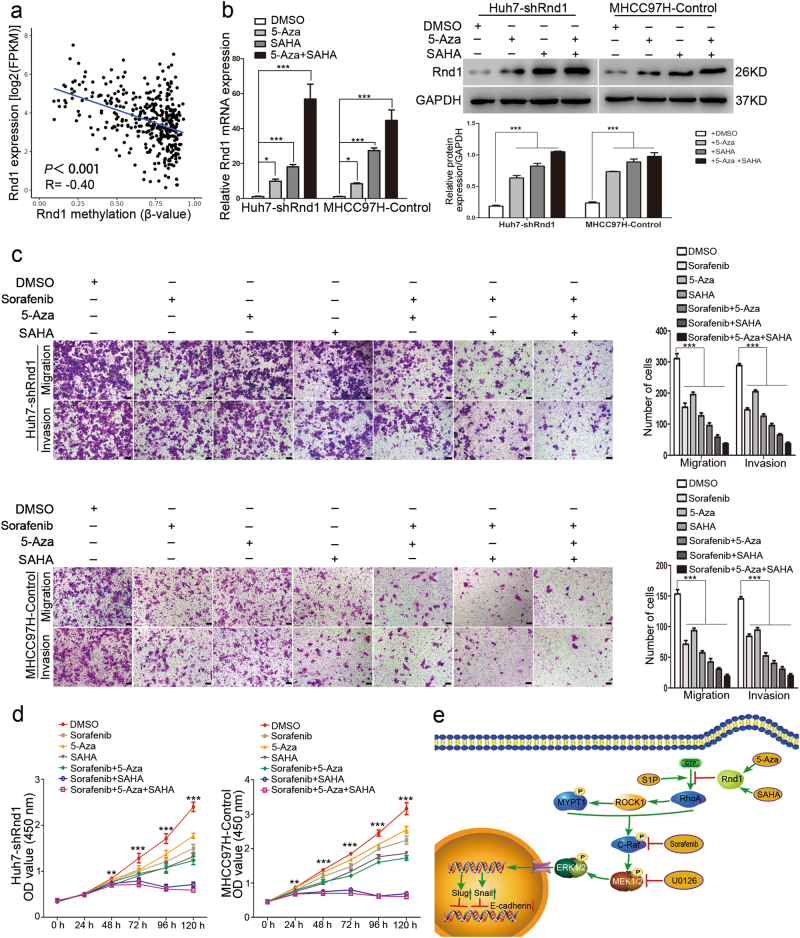
TCGA data sets were also employed to explore the underlying mechanisms of Rnd1 downregulation in HCC. The results revealed that Rnd1 expression was negatively correlated with promoter methylation in HCC (R = −0.40, P < 0.001; Fig. 7a), indicating that promoter methylation contributed to Rnd1 silencing in HCC. Furthermore, the results also exhibited that genetic alteration, including copy number variation and somatic mutation, had no influence on the expression of Rnd1 (Supplementary Figure 9a,b). Besides, these results were consistent with earlier research29. A recent study suggested that histone deacetylation was involved in the transcriptional repression of Rnd1;14 so the DNA methylation inhibitor 5-aza-2-deoxycytidine (5-Aza) and the histone deacetylase inhibitor SAHA were used to treat Huh7-shRnd1 and MHCC97H-Control cells with low expression of Rnd1. Western blot and Real-time PCR confirmed that 5-Aza, SAHA and both compounds could rescue Rnd1 expression in both cell lines (Fig. 7b). Next, transwell assays demonstrated that 5-Aza, SAHA, or both compounds could reinforce the inhibitory effect of sorafenib on the migration and invasion of HCC cells (Fig. 7c). CCK-8 assays also showed that the proliferating rate of Huh7-shRnd1 and MHCC97H-Control cells treated by sorafenib cooperated with 5-Aza, SAHA or both compounds was markedly slower as compared with sorafenib-treated and control cells (Fig. 7d). These results suggested that epigenetic inhibitors (5-aza and SAHA) potentiated sorafenib-induced toxicity in Rnd1-downregulated HCC cells.
Fig. 7. The contribution of epigenetic mechanisms to Rnd1 silencing in HCC.
a The plot showed the relationship between Rnd1 expression and promoter methylation in TCGA data set. n = 396. b Detection of Rnd1 by Real-time PCR and Western blot in Huh7-shRnd1 and MHCC97H-Control cells after incubated with either 5-Aza (10 µM), SAHA (5 µM), or both compounds for 24 h. All data represent mean ± S.D. (n = 3) for three independent experiments. Significance was determined using one-way ANOVA. *P < 0.01, ***P < 0.001. c The cell migration and invasion evaluated by transwell assays with or without Matrigel in Huh7-shRnd1 and MHCC97H-Control cells treated with DMSO, sorafenib, 5-Aza, SAHA, sorafenib+5-Aza, sorafenib+SAHA or their mixture. Scale bar: 100 μm. Panel: quantification of migration and invasion of cells. Data are mean ± S.D. (n = 3) and are representative of three independent experiments. Significance was determined using one-way ANOVA. ***P < 0.001. d Changes in the growth rate of Huh7-shRnd1 and MHCC97H-Control cells treated with DMSO, sorafenib, 5-Aza, SAHA, sorafenib+5-Aza, sorafenib+SAHA or their mixture. Data are mean ± S.D. (n = 4) and are representative of three independent experiments. Significance was determined using two-way ANOVA. **P < 0.01, ***P < 0.001. e Schematic of Rnd1 suppressing the Raf/MEK/ERK cascade by inhibiting the activation of RhoA and restraining EMT in HCC, and Rnd1 silencing could be overcome by 5-Aza and SAHA
Thus, we conclude that Rnd1 suppresses the activity of RhoA, leading to the inhibition of the Raf/MEK/ERK signaling pathway, and impairing the migration, invasion and EMT of HCC cells; in addition, Rnd1 silencing can be rescued by 5-Aza and SAHA in HCC cells (Fig. 7e).
Discussion
HCC is aggressive and displays a strong tendency to both recurrence and metastasis30. A better understanding of the underlying mechanisms for these aspects will lead to better treatments for HCC. In this study, Rnd1 was found to be downregulated in HCC tissues and cells. Through in vitro and in vivo experiments, it was found that Rnd1 regulated EMT progression by modulating the Raf/MEK/ERK pathway through altered RhoA activity, which in turn affected cell migration and invasion. Besides, both DNA methylation and histone deacetylation were key factors for Rnd1 downregulation in HCC cells. Collectively, Rnd1 acted as a promising anti-metastasis target for HCC patients.
Owning to the involvement of Rho GTPases in many essential physiological processes of normal cells, deregulation of Rho GTPases endow cells with the hallmarks of malignancy, including malignant transformation, cancer cell survival, and metastasis31. In this study, we revealed that Rnd1, a member of Rho GTPases, was downregulated in HCC tissues, and negatively correlated with malignant clinic pathological features and poor prognosis of HCC patients. We also verified that Rnd1 had potent inhibitory effects on cell proliferation, migration, invasion, and metastasis in HCC. Recent studies had demonstrated that Rnd1 expression was downregulated at the initiation and progression stages of breast cancer14. However, the exact role of Rnd1 in other types of tumors are still unclear. Our studies further confirmed the pleiotropic roles of Rnd1 in the progression of HCC.
EMT is an essential process for cancerous cells to acquire metastatic capability, following by the changes of cell shape32. Rnd1 had the function of modulating cytoskeleton and cell shape33. In this research, Rnd1-silenced cells acquired a mesenchymal-like phenotype with the downregulation of epithelial marker and the upregulation of mesenchymal markers, as well as the acquisition of elongated spindle shape. Conversely, HCC cells with Rnd1 overexpression acquired epithelial-like phenotype with upregulation of E-cadherin. These results were also verified by expressions of Rnd1 and EMT markers in xenografted tumors and HCC tissues. Therefore, Rnd1 inactivation-induced EMT in HCC.
The signaling mechanisms underlying the inhibitory effects on EMT and HCC progression mediated by Rnd1 were also investigated. Our results showed that Rnd1-restrained HCC invasion, metastasis, and EMT through inhibiting the activity of Raf/MEK/ERK signaling. Aberrant activation of Raf/MEK/ERK pathway is implicated in malignant transformation and progression of HCC and related with poor prognosis34. Furthermore, mounting evidences have showed that Raf/MEK/ERK signaling facilitates tumor invasion and metastasis by modulating EMT35,36. Therefore, molecular therapies had been developed to interfere this pathway for the treatment of HCC. The multi-kinase inhibitor sorafenib, an orally active inhibitor of Raf kinases and receptor tyrosine kinases, is the only approved systemic agent for patients with advanced HCC. Nevertheless, a considerable number of HCC patients still suffer from disease progression during sorafenib treatment, namely sorafenib resistance. Our results demonstrated that Rnd1 overexpression and sorafenib had synergistic inhibition effect on Raf/MEK/ERK signaling. Meanwhile, Rnd1 overexpression reinforced the inhibitory effect of sorafenib on cell migration, invasion and EMT. In our previous studies, sorafenib showed pro-metastasis effect in HCC37, but aspirin minimized this effect by increasing HTATIP2 expression through suppression of COX238. Previous experiments had shown that upregulation of COX2 was depended on the activation of MAPKs39. Rnd1 had the function of restraining the activity of Raf, MEK, and ERK. In this scenario, Rnd1 may act like aspirin and reduce the expression of COX2 leading to alleviate the pro-metastasis effects of sorafenib in HCC. These data suggested that exogenous-induced Rnd1 might help attenuate the sorafenib resistance in HCC and provide a novel approach for improving sorafenib efficiency.
Additionally, we also found that Rnd1 modulated the Raf/MEK/ERK signaling by connecting with RhoA. The results exhibited that Rnd1 was inversely correlated with the activity of RhoA. In addition, enhancing RhoA activity could not interfere Rnd1 expression, but could offset Rnd1-induced inhibition of cell migration, invasion, and EMT, as well as the activity of Raf/MEK/ERK pathway. Recently, Bist et al. showed that a dominant-negative form of RhoA could counteract ERK-induced NF-kB activation, but they also found that the activation of RhoA is dependent on ERK40. These findings suggested that the two pathways could interact with each other. According to our results, Rnd1 inhibited the metastasis and EMT of HCC by decreasing the activity of RhoA/ROCK1/MYPT1/Raf/MEK/ERK signaling axis. Thus, these findings identified a novel pathway restrained by Rnd1 for its potent malignancy suppressing activity.
Finally, TCGA analysis indicated that promoter methylation was inversely associated with Rnd1 expression in HCC. DNA methylation contributed to the tumor initiation and progression41. Therefore, our results indicated that promoter methylation-mediated silencing of Rnd1 might be involved in the process of tumorigenesis. Furthermore, we also found that epigenetic inhibitors elevated the expression of Rnd1 in HCC cells. Aberrant epigenetic modifications were associated with the activation of oncogenes, inactivation of tumor suppressors and acquisition of drug resistance42. Preclinical and clinical trials had been launched to test the anti-tumor effects of epigenetic inhibitors. However, the clinical efficacy of the inhibitors was limited due to their therapeutic targets were global, so they could not selectively efficient against solid tumors43. Considering that Rnd1 is frequently silenced in HCC, generating specific inhibitors for Rnd1 methylation may achieve stronger tumor-selective toxicity than traditional drugs.
In a summary, our study shows that Rnd1 is negatively associated with the aggressive characteristics and poor prognosis of HCC patients. Besides, Rnd1 upregulation suppresses the activity of the Raf/MEK/ERK pathway by inhibiting RhoA activation, restraining migration, invasion, and EMT of HCC cells. The combination of pharmaceutical upregulation of Rnd1 with conventional measures may serve as a new weapon for HCC treatment in the clinical application.
Materials and methods
Patients and tissue specimens
We collected 67 pairs of liver cancer (T) and adjacent non-tumor tissue (NT) from treatment-naive patients who underwent hepatectomy for HCC between May 1999 and November 2002 at the Liver Cancer Institute and Zhongshan Hospital (Fudan University, Shanghai, China). The research was approved by the research ethics committee of Zhongshan Hospital. Fresh HCC tissues were snap-frozen in liquid nitrogen and stored at −80 °C for construction of tissue microarray and immunohistochemistry (IHC). The clinical and pathological features of these patients are described in Table 1. The uniform guidelines for post-operative follow-up procedures were described previously44.
We obtained 20 pairs of fresh HCC tissue and matched non-tumor liver tissue from patients who underwent surgical treatment for HCC between 2014 and 2016; these tissues were used for quantitative real-time polymerase chain reaction (qRT-PCR) analysis. Additionally, eight pairs of tumor/non-tumor liver tissue samples from HCC patients who underwent operative treatment in 2016 were randomly selected for western blot analyses. All human materials were obtained with informed consent.
Cell lines
The human HCC cell lines, including HepG2, Huh7, and a normal liver cell line (L02) were purchased from the cell bank of the Chinese Academy of Sciences (Shanghai, China). HCCLM3, MHCC97H, and MHCC97L cell lines were established at the Liver Cancer Institute, Fudan University (Shanghai, China)45. All cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (FBS; Gibco, Grand Island, NY) and 100 U ml−1 penicillin and 50 mg ml−1 streptomycin, and incubated at 37 °C and 5% CO2 in a humidified atmosphere.
Lentiviral vector construction
Lentiviral-mediated Rnd1 pLKO/shRNAs were purchased from Genesent (Shanghai, China). The sequences of the three Rnd1-targeting lentiviral shRNA constructs were: shRNA1: 5′-GCTCTGAACTCATCTCTTCTA -3′; shRNA2: 5′-GAGGACAGAAATCCTAGATTA-3′; shRNA3: 5′-CCCTACTACGATAATGTCCGT-3′; shRNA4: 5′-GGAGCTTAGTCTCTGGGATAC-3′; shRNA5: 5′- GGAAACCTCATCTTTGCATAG -3′; shRNA6: 5′- GCGACTCGGATGCAGTATTAC -3′. The plasmid encoding Rnd1 (NM_014470.3) was synthesized by using pLenO-DCE-Puro Vector (biolink, Shanghai, China). Lenti-Rnd1 viral particles were produced by cotransfection of 293T cells with four plasmids: the pRSV-Rev packaging helper plasmid, pMDLg/pRRE, pMD2.G, and the pLenO-DCE Transfer Vector. Stably transfected clones were validated using qRT-PCR and western blotting.
Plasmid, siRNA construction and transfection
Full-length human RhoA (NM_ 001664.2) was cloned into the control plasmid DNA vector (pcDNA 3.1) and transfected into HCC cells using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA), according to the manufacturer’s instructions. Cells transfected with empty vector were used as controls. RhoA siRNA was purchased from Biotrend Company (Shanghai, China); the RhoA siRNA sequence was 5′-CCACAGUGUUUGAGAACUA-3′46. siRNA transfection of HCC cells was accomplished using Lipofectamine 2000. Cells transfected with non-targeting siRNA were used as controls.
Active RhoA pull-down assay
GTP loading of RhoA was examined using a RhoA Activation Assay Kit (NewEast Biosciences, USA), according to the manufacturer’s protocol. Briefly, when the density of HCC cells reached about 80–90% confluence, 1 ml ice-cold 1× Assay/Lysis Buffer was added to the culture plate and collected into assay tubes. Then, 1 μl of anti-active RhoA monoclonal antibody was added into assay tubes. Afterwards, 20 µl of resuspended protein A/G Agarose beads was added to each tube, and the cell lysate mixtures were gently agitated for 1 h at 4 °C. The beads were collected by centrifugation for 1 min at 5000 × g, and were washed three times with 0.5 ml 1× Assay/Lysis buffer. The bead pellet was resuspended in 20 µl of 2× reducing SDS-PAGE sample buffer, and the samples were boiled for 5 min. Equivalent amounts of samples were separated by SDS-PAGE on a 12% gel and transferred to nitrocellulose membranes. RhoA expression was queried using the anti-RhoA antibody provided by the kit.
Xenograft model of human HCC in nude mice
The male BALB/c nude mice were purchased from Shanghai SLAC Laboratory Animal Co. Ltd. (Shanghai, China) and housed under specific pathogen-free conditions. For the xenograft model, Huh7-shControl, Huh7-shRnd1, MHCC97H-Control and MHCC97H-Rnd1 cells (5 × 106 cells) were injected subcutaneously into the left upper flank regions of three nude mice (4 weeks of age), respectively. After 1 month, subcutaneous tumor tissues were resected and cut into 1 mm3 pieces, the tumor pieces were implanted into the livers of nude mice (n = 6 for each group) according to standard procedures described previously47. After 40 days, mice were killed and tumor tissues and lungs were harvested, photographed, and weighed. The tumor volume was calculated by the following formula: tumor volume (mm3) = = (longest diameter × shortest diameter2)/2, and the incidence of lung metastasis of each group was quantified by fluorescence microscopy and HE straining. All procedures strictly followed the guidelines established by the Shanghai Medical Experimental Animal Care Commission.
Cell migration assays, Matrigel® invasion assays and Scratch-healing assays
The Transwell Permeable Supports System (Corning) was employed to detect the effects of Rnd1 on cell migration and invasion. For assays of migration, 5 × 104 cells in 1% serum medium were added to the upper chamber; for assays of invasion, 1 × 105 cells in 1% serum medium were added to the Matrigel-coated upper chamber and 10% serum medium was added to the lower chamber. Cells were permitted to translocate for 24 or 48 h at 37 °C. After removal of the non-migrated or non-invaded cells, the remaining cells were fixed with methanol, stained with crystal violet. Three random views of each chamber were photographed by inverted microscopy (magnification: 100×), and the cell numbers were counted by Image-Pro Plus software. For the scratch-healing assay, Rnd1-modified HCC cells were added to 6-well plates and starved for 24 h in non-serum medium. After that, a pipette tip was used to make a straight scratch on the monolayer of cells. The scratch was randomly photographed under an inverted microscope at 0 and 48 h. The following formulas were used to quantify the results: Total scratched area (0 h) = scratch width (mm) × length (mm); cell migrated area (48 h) = length of cell migration (mm) × 2 × Length (mm); percent wound closure (%) = cell migrated area/total scratched area. All samples were prepared in triplicate on three independent experiments.
Colony formation assays, cell proliferation assays, and cell cycle analysis
Colony formation assays were carried out in accordance with previous experiments47. For cell proliferation assays, 3000 cells/well were seeded in 96-well plates, and cell proliferation was measured using the Cell Counting Kit-8 (CCK-8; Dojindo Laboratories, Kumamoto, Japan), reading absorbance at 450 nm according to the manufacturer’s instructions. For the cell cycle analysis, collected HCC cells were fixed in 70% ice-cold ethanol for 12 h; after that, HCC cells were strained with PI by using PI/RNase Staining Buffer (BD Biosciences, San Jose, CA) according to the manufacturer’s protocol. Results collection and analysis were performed with a flow cytometer (FACS Calibur; BD Biosciences, San Jose, CA). Experiments were repeated three times.
Immunofluorescence assays and TUNEL assays
HCC cells were seeded in glass bottom dishes. After cells attached to the bottom of the dishes, cells were fixed in 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. Afterwards, the cells were incubated with blocking buffer (5% BSA in PBS, pH 7.4) for 30 min. Cells were incubated with the appropriate primary antibodies overnight at 4 °C. On the following day cells were incubated with Alexa Fluor 555-labeled secondary antibodies (Beyotime, Haimen, China) for 1 h and DAPI was used to mark nuclei. For the TUNEL assays, after cells were incubated with blocking buffer, the TUNEL reaction mixture (Roche, Pleasanton, CA, USA) was used to treat cells for 1 h at 37 °C in the dark place. Subsequently, cells were strained with DAPI for nuclei visualization. Apoptotic cells appeared as red-stained cells. The cells were observed and photographed under a fluorescence microscope (Olympus, Tokyo, Japan). Data are shown from a typical experiment performed in triplicate.
Real-time PCR assay
Total RNA was extracted with TRIzol Reagent (Sigma, St. Louis, MO) and reverse-transcribed into cDNA using the PrimeScript RT reagent Kit (TaKaRa, Dalian, China), according to manufacturer’s instructions. Quantitative PCR was performed to quantify the mRNA expression of Rnd1 and GAPDH with the RT-PCR kit (TaKaRa, Dalian, China). Each sample was tested in triplicate in three independent experiments.
Western blot assay
Total protein was extracted in RIPA lysis buffer (Beyotime, Haimen, China), as we have previously described7. A total of 30 µg protein from whole cell extracts was subjected to SDS-PAGE and transferred to PVDF membrane (Millipore, Bedford, MA). Membranes were blocked with 5% nonfat milk for 1 h and then incubated with the appropriate primary antibodies overnight at 4 °C. The next day, membranes were incubated with the appropriate HRP-conjugated secondary antibodies for 1 h at room temperature. Blots were visualized with an ECL detection kit (Millipore, Bedford, MA) and analyzed using Image-Pro Plus 6.0 software (Media Cybernetics, Silver Spring, USA).
Immunohistochemistry
Formalin-fixed paraffin sections or tissue arrays containing tumor tissues from HCC patients were stained for Rnd1 using the streptavidin–peroxidase system (ORIGENE, Beijing, China). The IOD of positive staining was analyzed using Image-Pro Plus 6.0 software.
Statistical analysis
Data were expressed as mean ± S.D.S derived from at least three independent experiments. Data were analyzed using SPSS 18.0 (Chicago, IL USA). The Student’s t-test was used to analyze differences between two groups, and two-way ANOVA was used when more than two groups were compared. For the in vivo assay, the differences of tumor volumes were compared by unpaired t-test, and χ2 test was used to analyze the differences in the incidence of lung metastasis between groups. The correlations between Rnd1 and the clinical and pathological features were analyzed using the Spearman correlation test. OS and DFS curves were protracted using the Kaplan–Meier method and estimated by the log-rank test. Variables that were independently associated with OS and DFS were identified using the Cox’s proportional hazards regression model. A two-tailed value of P < 0 .05 was considered significant.
Electronic supplementary material
Acknowledgements
This work was supported by the National Natural Science Foundation of China [grant number 81372655] and the China National Key Projects for Infectious Disease [grant number 2012ZX10002-012].
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
These authors contributed equally: Cheng-Dong Qin, De-Ning Ma, Shi-Zhe Zhang.
Edited by V. Stambolic
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41419-018-0517-x).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Torre LA, et al. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Shindoh J, et al. Risk factors of post-operative recurrence and adequate surgical approach to improve long-term outcomes of hepatocellular carcinoma. HPB. 2013;15:31–39. doi: 10.1111/j.1477-2574.2012.00552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nevins JR, Potti A. Mining gene expression profiles: expression signatures as cancer phenotypes. Nat. Rev. Genet. 2007;8:601–609. doi: 10.1038/nrg2137. [DOI] [PubMed] [Google Scholar]
- 4.Lin KT, Shann YJ, Chau GY, Hsu CN, Huang CY. Identification of latent biomarkers in hepatocellular carcinoma by ultra-deep whole-transcriptome sequencing. Oncogene. 2016;35:5078. doi: 10.1038/onc.2016.62. [DOI] [PubMed] [Google Scholar]
- 5.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prieto-García E, Díaz-García CV, García-Ruiz I, Agulló-Ortuño MT. Epithelial-to-mesenchymal transition in tumor progression. Med. Oncol. 2017;34:122. doi: 10.1007/s12032-017-0980-8. [DOI] [PubMed] [Google Scholar]
- 7.Ma DN, et al. MicroRNA-26a suppresses epithelial-mesenchymal transition in human hepatocellular carcinoma by repressing enhancer of zeste homolog 2. J. Hematol. Oncol. 2016;9:1. doi: 10.1186/s13045-015-0229-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiong W, et al. Residual hepatocellular carcinoma after oxaliplatin treatment has increased metastatic potential in a nude mouse model and is attenuated by Songyou Yin. BMC Cancer. 2010;10:219. doi: 10.1186/1471-2407-10-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu. Rev. Cell. Dev. Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 10.Porter AP, Papaioannou A, Malliri A. Deregulation of Rho GTPases in cancer. Small GTPases. 2016;7:123–138. doi: 10.1080/21541248.2016.1173767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellenbroek SI, Collard JG. Rho GTPases: functions and association with cancer. Clin. Exp. Metastas. 2007;24:657–672. doi: 10.1007/s10585-007-9119-1. [DOI] [PubMed] [Google Scholar]
- 12.Aguirre AJ, et al. High-resolution characterization of the pancreatic adenocarcinoma genome. Proc. Natl Acad. Sci. USA. 2004;101:9067–9072. doi: 10.1073/pnas.0402932101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rutherford S, Hampton GM, Frierson HF, Moskaluk CA. Mapping of candidate tumor suppressor genes on chromosome 12 in adenoid cystic carcinoma. Lab. Invest. 2005;85:1076–1085. doi: 10.1038/labinvest.3700314. [DOI] [PubMed] [Google Scholar]
- 14.Okada T, et al. The Rho GTPase Rnd1 suppresses mammary tumorigenesis and EMT by restraining Ras-MAPK signalling. Nat. Cell. Biol. 2015;17:81–94. doi: 10.1038/ncb3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang XY, et al. αB-crystallin complexes with 14-3-3ζ to induce epithelial-mesenchymal transition and resistance to sorafenib in hepatocellular carcinoma. Hepatology. 2013;57:2235–2247. doi: 10.1002/hep.26255. [DOI] [PubMed] [Google Scholar]
- 16.Morris HT, Machesky LM. Actin cytoskeletal control during epithelial to mesenchymal transition: focus on the pancreas and intestinal tract. Br. J. Cancer. 2015;112:613–620. doi: 10.1038/bjc.2014.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heerboth S, et al. EMT and tumor metastasis. Clin. Transl. Med. 2015;4:6. doi: 10.1186/s40169-015-0048-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaowinn S, et al. Cancer upregulated gene 2 induces epithelial-mesenchymal transition of human lung cancer cells via TGF-β signaling. Oncotarget. 2017;8:5092–5110. doi: 10.18632/oncotarget.13867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Y, et al. p38 MAPK mediates epithelial-mesenchymal transition by regulating p38IP and Snail in head and neck squamous cell carcinoma. Oral. Oncol. 2016;60:81–89. doi: 10.1016/j.oraloncology.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 20.Liu L, et al. Sorafenib blocks the RAF/MEK/ERK pathway, inhibits tumor angiogenesis, and induces tumor cell apoptosis in hepatocellular carcinoma model PLC/PRF/5. Cancer Res. 2006;66:11851–11858. doi: 10.1158/0008-5472.CAN-06-1377. [DOI] [PubMed] [Google Scholar]
- 21.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Z, et al. IBP regulates epithelial-to-mesenchymal transition and the motility of breast cancer cells via Rac1, RhoA and Cdc42 signaling pathways. Oncogene. 2014;33:3374–3382. doi: 10.1038/onc.2013.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang SC, et al. MicroRNA-122 triggers mesenchymal-epithelial transition and suppresses hepatocellular carcinoma cell motility and invasion by targeting RhoA. PLoS ONE. 2014;9:e101330. doi: 10.1371/journal.pone.0101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen, L. K., Kholodenko, B. N. & von, K. A. Rac1 and RhoA: networks, loops and bistability. Small GTPases 1–6 (2016). [DOI] [PMC free article] [PubMed]
- 25.Gilkes DM, et al. Hypoxia-inducible factors mediate coordinated RhoA-ROCK1 expression and signaling in breast cancer cells. Proc. Natl Acad. Sci. Usa. 2014;111:E384–E393. doi: 10.1073/pnas.1321510111. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Jia Y, et al. Rho kinase mediates Porphyromonas gingivalis outer membrane vesicle-induced suppression of endothelial nitric oxide synthase through ERK1/2 and p38 MAPK. Arch. Oral. Biol. 2015;60:488–495. doi: 10.1016/j.archoralbio.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 27.Riento K, Ridley AJ. Rocks: multifunctional kinases in cell behaviour. Nat. Rev. Mol. Cell. Biol. 2003;4:446–456. doi: 10.1038/nrm1128. [DOI] [PubMed] [Google Scholar]
- 28.van Nieuw Amerongen GP, et al. Involvement of Rho kinase in endothelial barrier maintenance. Arterioscler. Thromb. Vasc. Biol. 2007;27:2332–2339. doi: 10.1161/ATVBAHA.107.152322. [DOI] [PubMed] [Google Scholar]
- 29.Komatsu H, et al. Attenuated RND1 expression confers malignant phenotype and predicts poor prognosis in hepatocellular carcinoma. Ann. Surg. Oncol. 2017;24:850–859. doi: 10.1245/s10434-016-5573-9. [DOI] [PubMed] [Google Scholar]
- 30.El-Serag HB. Hepatocellular carcinoma. N. Engl. J. Med. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 31.Orgaz JL, Herraiz C, Sanz-Moreno V. Rho GTPases modulate malignant transformation of tumor cells. Small GTPases. 2014;5:e29019. doi: 10.4161/sgtp.29019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat. Cell. Biol. 2014;16:488–494. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- 33.Oinuma I, Katoh H, Harada A, Negishi M. Direct interaction of Rnd1 with Plexin-B1 regulates PDZ-RhoGEF-mediated Rho activation by Plexin-B1 and induces cell contraction in COS-7 cells. J. Biol. Chem. 2003;278:25671–25677. doi: 10.1074/jbc.M303047200. [DOI] [PubMed] [Google Scholar]
- 34.Delire B, Stärkel P. The Ras/MAPK pathway and hepatocarcinoma: pathogenesis and therapeutic implications. Eur. J. Clin. Invest. 2015;45:609–623. doi: 10.1111/eci.12441. [DOI] [PubMed] [Google Scholar]
- 35.Li X, et al. PHLPP is a negative regulator of RAF1, which reduces colorectal cancer cell motility and prevents tumor progression in mice. Gastroenterology. 2014;146:1301–1312. doi: 10.1053/j.gastro.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeramian A, et al. Endometrial carcinoma: molecular alterations involved in tumor development and progression. Oncogene. 2013;32:403–413. doi: 10.1038/onc.2012.76. [DOI] [PubMed] [Google Scholar]
- 37.Zhang W, et al. Sorafenib down-regulates expression of HTATIP2 to promote invasiveness and metastasis of orthotopic hepatocellular carcinoma tumors in mice. Gastroenterology. 2012;143:1641–1649. doi: 10.1053/j.gastro.2012.08.032. [DOI] [PubMed] [Google Scholar]
- 38.Lu L, et al. Aspirin minimized the pro-metastasis effect of sorafenib and improved survival by up-regulating HTATIP2 in hepatocellular carcinoma. PLoS. One. 2013;8:e65023. doi: 10.1371/journal.pone.0065023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin CC, et al. Up-regulation of COX-2/PGE2 by endothelin-1 via MAPK-dependent NF-κB pathway in mouse brain microvascular endothelial cells. Cell. Commun. Signal. 2013;11:8. doi: 10.1186/1478-811X-11-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bist P, et al. Annexin-A1 controls an ERK-RhoA-NFκB activation loop in breast cancer cells. Biochem. Biophys. Res. Commun. 2015;461:47–53. doi: 10.1016/j.bbrc.2015.03.166. [DOI] [PubMed] [Google Scholar]
- 41.Yen CY, et al. DNA methylation, histone acetylation and methylation of epigenetic modifications as a therapeutic approach for cancers. Cancer Lett. 2016;373:185–192. doi: 10.1016/j.canlet.2016.01.036. [DOI] [PubMed] [Google Scholar]
- 42.Flavahan, W. A., Gaskell, E. & Bernstein, B. E. Epigenetic plasticity and the hallmarks of cancer. Science357, eaal2380 (2017). [DOI] [PMC free article] [PubMed]
- 43.Vaiopoulos AG, KCh A, Papavassiliou AG. Epigenetic modifications in colorectal cancer: molecular insights and therapeutic challenges. Biochim. Biophys. Acta. 2014;1842:971–980. doi: 10.1016/j.bbadis.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 44.Gao Q, et al. CXCR6 upregulation contributes to a proinflammatory tumor microenvironment that drives metastasis and poor patient outcomes in hepatocellular carcinoma. Cancer Res. 2012;72:3546–3556. doi: 10.1158/0008-5472.CAN-11-4032. [DOI] [PubMed] [Google Scholar]
- 45.Li Y, et al. Establishment of cell clones with different metastatic potential from the metastatic hepatocellular carcinoma cell line MHCC97. World J. Gastroenterol. 2001;7:630–636. doi: 10.3748/wjg.v7.i5.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reffay M, et al. Interplay of RhoA and mechanical forces in collective cell migration driven by leader cells. Nat. Cell. Biol. 2014;16:217–223. doi: 10.1038/ncb2917. [DOI] [PubMed] [Google Scholar]
- 47.Chang RM, Yang H, Fang F, Xu JF, Yang LY. MicroRNA-331-3p promotes proliferation and metastasis of hepatocellular carcinoma by targeting PH domain and leucine-rich repeat protein phosphatase. Hepatology. 2014;60:1251–1263. doi: 10.1002/hep.27221. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.