Abstract
Background
Size and methylation mosaicism are a common phenomenon in Fragile X syndrome (FXS). Here, the authors report a study on twelve fragile X males with atypical mosaicism, seven of whom presented with autism spectrum disorder.
Methods
A combination of Southern Blot and PCR analysis was used for CGG allele sizing and methylation. FMR1 mRNA and FMRP expression were measured by qRT-PCR and by Homogeneous Time Resolved Fluorescence methodology, respectively.
Results
DNA analysis showed atypical size- or methylation-mosaicism with both, full mutation and smaller (normal to premutation) alleles, as well as a combination of methylated and unmethylated alleles. Four individuals carried a deletion of the CGG repeat and portions of the flanking regions. The extent of methylation among the participants was reflected in the lower FMR1 mRNA and FMRP expression levels detected in these subjects.
Conclusion
Decreased gene expression is likely the main contributor to the cognitive impairment observed in these subjects; although the presence of a normal allele did not appear to compensate for the presence of the full mutation, it correlated with better cognitive function in some but not all of the reported cases emphasizing the complexity of the molecular and clinical profile in FXS.
Keywords: FMR1mRNA, FMRP, transcription, fragile X syndrome, methylation, mosaicism, deletion
INTRODUCTION
Fragile X syndrome (FXS) is the most common form of inherited intellectual disability and the leading monogenic cause of autism spectrum disorder (ASD). Patients with FXS exhibit mild to severe impairment of higher cognitive function and are also affected by a broad spectrum of clinical, physical and behavioral problems. These include hyper-responsiveness to sensory stimuli, hyperactivity, impulsive behavior, gaze aversion, anxiety and shyness (1).
The most common cause of FXS is the expansion of a CGG trinucleotide repeat in the 5′ untranslated region (UTR) of the fragile X mental retardation 1 (FMR1) gene (2), which leads to DNA methylation, aberrant heterochromatinization and subsequent silencing of the FMR1 gene. As a consequence, the encoded gene product, FMR1 protein (FMRP), is not produced and its absence gives rise to fragile X syndrome (3, 4)(3, 4)(3, 4)(3, 4). The length of the CGG repeat defines the allele classes, which, according to the American College of Medical Genetics (5) guidelines, are defined as follows: normal, for alleles with between 6 and 44 CGG repeats; intermediate/gray zone, for alleles with 45 and 54 CGG repeats; premutation, for alleles ranging from 55 to 200 repeat and full mutation, for alleles with more than 200 CGG repeats.
Methylation of full mutation alleles occurs early in embryonic development and may play a role in the stability of the expanded repeats (6, 7). The hypermethylated state of the fragile X full mutation is found associated locally with histone deacetylation and chromatin remodeling (8), and with transcriptional silencing of the gene (3). FMR1 gene silencing due to DNA methylation in the promoter is associated with hypoacetylation of histone H3 and H4, methylation of repressive histone post-translational modifications such as lysine 9 of histone 3 (H3K9) and lysine 27 of histone 3 (H3K27), and demethylation of lysine 4 of histone 3 (H3K4) (8-10). A DNA methylation boundary was reported 650-800 nucleotides upstream the CGG repeat, separating a hypermethylated region upstream of the boundary from a downstream unmethylated FMR1 promoter region. This boundary is generally present in normal and unmethylated full mutation cells but is lost in individuals with FXS as the promoter region becomes hypermethylated (11-13); however, the significance of this boundary is not known at present. Importantly, the methylation profile of FMR1 gene in patients with FXS varies across cell types and the extend of methylation appears to correlate with their clinical phenotypes (14-17).
The expanded CGG repeat within the FMR1 gene predisposes the repeat to instability, which can result in “size mosaicism”, operationally defined as the presence of both premutation and full mutation repeat expansions within or across cell types (18-20). Size mosaicism comprising both full mutation and normal-sized alleles, although uncommon, has been described in several cases of male with FXS (19, 21-27). Nolin et al. (1994) studied 61 FXS mosaic males and found 82% of mosaic full mutation/premutation, 3% of mosaic full mutation/normal size allele, and 1.7% of mosaic full mutation/deletion. The remaining 13% were methylation mosaics. It has been suggested that mosaicism for the presence of a full and a normal allele may originate during the early stages of development, preceding the germline segregation (28), when CGG repeat size modifications of the maternally transmitted premutation allele can originate resulting in expansions to the full mutation as well as less frequently in size reduction, thus, elucidating their somatic instability.
In addition to “size” mosaicism, some individuals exhibit “methylation” mosaicism, which occurs when some cells carry methylated alleles and some cells carry unmethylated alleles (with a CGG repeat number ranging from the premutation to the full mutation range) are present. The degree of methylation varies within or between different tissues in the same individual, giving rise to intra or inter-tissue mosaicism (17, 29). Such unmethylated alleles are transcriptionally active, and in many instances over expressed (30), and, therefore, potentially translated to produce FMRP (31). However, FMRP expression levels are negatively correlated with the CGG repeat number; and FMR1 alleles, particularly in the upper premutation range, show decreased expression likely due to a deficit in translational efficiency (32-34).
The spectrum of mosaicism can be further complicated by the presence of deletions in the FMR1 gene ranging from 1 bp to several Mbp (19, 35-47). If the smaller deletions result in the abrogation of the promoter region or if they affect the translational start site of FMR1, then no FMRP is made and the individual will develop FXS. In some instances, deletions are limited to the region between the transcription start site of FMR1 and the translation initiation codon and these alleles can still produce FMR1 mRNA (35, 37, 39), suggesting that the region encompassing the CGG repeats and the immediate surrounding sequences may not be essential for FMR1 expression.
Here, we report on the molecular and clinical measures in twelve cases of size- and methylation- mosaicism presenting with a clinical picture typical of FXS. Eight males had size mosaicism for the presence of a full mutation allele and normal/intermediate alleles (ranging from 16 to 54 CGG repeats). Of the remaining four males, two displayed size mosaicism and two methylation mosaicism; all had deletions, ranging in size from 158 to 396 bp, encompassing the entire CGG repeat and some of the flanking regions. Seven, out of eleven patients, approximately 63% of the total, were diagnosed with autism spectrum disorder (ASD), which is consistent with previous findings (48).
MATERIALS AND METHODS
Subjects
Twelve males, aging from 1 to 28 years, were included in the current study; they comprise all male patients seen clinically at our center who possess a normal allele together with an expanded allele or a deletion involving the CGG expansion. Specimens from five age-matched normal male controls were used for both measurement of FMR1 mRNA and FMRP expression levels. Genomic DNA derived from a normal female and from a male with a full mutation was used as negative and positive controls, respectively, in the Southern blot analysis. Informed consent was obtained from all participants who were evaluated at the Fragile X Research and Treatment Center at the UC Davis MIND Institute, Sacramento, California. The protocol was approved by the Institutional review board and ethics committee, at the University of California, Davis.
Molecular measures
CGG sizing and methylation status
Genomic DNA was isolated from peripheral blood leukocytes using standard methods (Qiagen, Valencia, CA). CGG repeat size was determined using a combination of both Southern blot and PCR analysis as previously described [49,50]. Southern blot was performed by digestion of 7–10 μg of genomic DNA with the restriction enzymes Eco RI and Nru I and the fragments were separated on 0.8% agarose gel containing Trisacetate- EDTA buffer, which did not contain ethidium bromide. DNA was then transferred onto a nylon membrane and hybridized with the FMR1-specific genomic probe StB12.3. Details of the method are as previously described [50]. The percent of methylation (expressing the percent of cell carrying a methylated allele) was calculated from the Southern blot autoradiographs as described in Tassone et al [51].
FMR1 mRNA analysis
Tot RNA was isolated from 3ml of whole blood, collected in Tempus tubes, according to the manufacturer’s instructions (Applied Biosystems, Foster City, CA). All quantifications of FMR1 mRNA were performed using a 7900 Sequence detector (Applied Biosystems, Foster City, CA) using specific primers and probes as previously detailed in (52).
Sequencing
PCR products of the deleted allele observed in cases 9 to 12 were purified using the MiniElute Gel Extraction Kit (Qiagen Inc., Valencia, CA) following a standard protocol. PCR products were sequenced by direct Sanger sequencing using the BigDye® Terminator v. 3.1 Cycle Sequencing Kit and the ABI Prism® 3730 Genetic to confirm the presence of the small deletions and to define deletions size and breakpoints. Primers used to amplify alleles containing the deleted region around the CGG repeat were as follows; C3 forward -primer 5′ TGT TTA CAC CCG CAG CGG GCC GGG GGT TC 3″, C5 forward -primer 5′ATT TCC CAC GCC ACT GAG TG, reverse- primer F 5′ TGG AGG AGC TGG TGG TGG AAG TGC GGG GCT 3′ and Free2 reverse- primer 5′AGA GGG GCT TCC AAC AGG CCCC 3′. Primer location of the primers with respect to the CGG repeat is shown in Figure 2.
Figure 2. Sequence analysis of the FMR1 allele containing a deletion including the CGG repeat in four subjects with FXS.
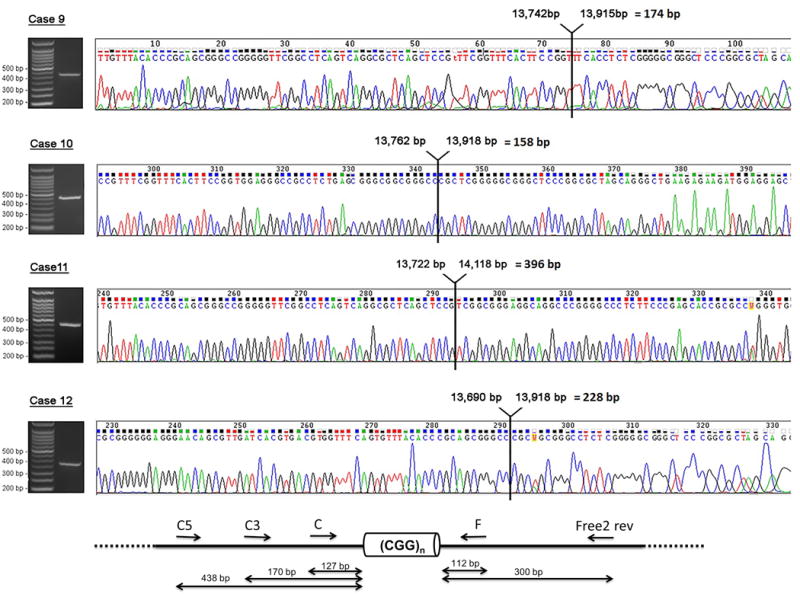
Sequence analysis of the FMR1 allele containing a deletion including the CGG repeat in four subjects with FXS. The presence of a deleted allele in four mosaic males was demonstrated by PCR and visualized by gel electrophoresis. Sequencing analysis showed the presence of a deletion in the 5ʹUTR of the FMR1 gene spanning the entire CGG repeat region as well as part of the upstream and downstream regions. The arrow specified the exact locations, in bp, of deletion within the FMR1 gene (L29074.1).
FMRP expression
The Cisbio Human FMRP assay (63ADK038PEC0) was utilized for detection of FMRP. The assay uses HTRF (homogeneous time resolved fluorescence) technology in which fluorescence resonance energy transfer occurs between antibody bound donor and acceptor groups located on the same FMRP molecule via the two fluorophores-conjugated antibodies [53]. Operation in a time-resolved fashion eliminates any background fluorescence. The donor is labeled with europium cryptate (Eu3+-Cryptate) and the acceptor is ‘d2.’ Two separate positive controls for FMRP were included on each plate: FMRP-positive control supplied by the assay as well as FMRP (TP322699) recombinant protein from Origene (Rockville, MD). Negative controls included reconstitution buffer only negative control, lysis buffer only negative control, and donor only Cryptate control, which resulted in baseline fluorescence readings. Total protein concentration was determined using the Thermo Fisher MicroBCA Assay (23235). For each sample, 6 and 3 μg of total protein were run for each experiment. FMRP levels were determined by comparing each sample to a standard curve of our fiducial control sample (RPM 7666; normal lymphoblasts [ATCC CCL114]) using 20, 10, 5, 2.5, and 1.25 μg. Protein levelswere determined in triplicate in a 384-well format following the instructions on the assay protocol. Samples were read on the PerkinElmer VictorX5.
Clinical Measures
Clinical measurement of participants included the domains of FSIQ, ASD, ADHD, perseveration, tantrums, and anxiety. Cognitive testing was carried out with standardized IQ measures depending on participants’ age and included Mullen Scales of Early Learning (MSEL) [54], WASI (Weschler abbreviated Scale of Intelligence [55], WISC-III (Wechsler Intelligence Scale for Children, 3rd edition) [56], Kaufman Assessment Battery for Children, and Stanford Binet V (Stanford–Binet Intelligence Scales [SB5], Fifth Edition) [57] as shown in Table1. Social and communication skills of participants were measured using Vineland Adaptive Behavior Scale [58] and Peabody Picture vocabulary test (PPVT) [59]. ASD was assessed by the Autism Diagnostic Observation Scale (ADOS) using a module that was developmentally appropriate [60]. ADHD was determined by clinical assessment [61].
Table 1.
Molecular and clinical measures data for the 12 cases
| Subject info. | Molecular Data | Clinical Data | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Age | Category | CGG repeat size | % Meth. | % Normal | % Pre/ Unmethylated | %deletion | FMRP | mRNA | Mullen | WAIS 3/Wasi | Stanford Binet | ASD |
| Case 1 | 23 | Size Mosaic | 606, 782, 875, 1130 (16) | 68% | 32% | 0.34 | 0.59 | N/A | YES | ||||
| Case 2 | 14 | Size Mosaic | 524, 685, 892, 1347 (24, 43) | 90% | 10% | 0.08 | 0.13 | Full scale IQ = 40, Non verbal IQ = 42, Verbal IQ = 44 | YES | ||||
| Case 3 | 28 | Size Mosaic | 303, 445, 642 (44) | 57% | 43% | 0.18 | 1.66 | Full scale IQ = 64, Verbal IQ = 67, Performance IQ = 67 | YES | ||||
| Case 4 | 6 | Size Mosaic | 700, 981 (52) | 87% | 13% | 0.21 | 0.23 | Full scale IQ = 42, Non Verbal IQ = 47, Verbal IQ = 43 | YES | ||||
| Case 5 | 2 | Size Mosaic | 493, 613, 927, 1215 (17) | 73% | 27% | 0.64 | 0.37 | ELC = 66 | NO | ||||
| Case 6 | 23 | Size Mosaic | 306, 455, 568, 692 (30) | 68% | 32% | 0.30 | 0.62 | Full scale IQ = 61, Verbal IQ = 67, Performance IQ = 59 | YES | ||||
| Case 7 | 10 | Size Mosaic | 174, 470, 670, 920 (13) | 83% | 17% | 0.18 | 0.42 | Full scale IQ = 40, Non verbal IQ = 42, Verbal IQ = 43 | YES | ||||
| Case 8 | 2 | Size Mosaic | 270-1000 (43) | 94% | 6% | 0.12 | 0.07 | ELC = 56 | N/A | ||||
| Case 9 | 18 | Meth mosaic | 353, 511, 632, 1068 (159-488 + del) | 59% | 28% | 13% | 0.32 | 0.64 | Full scale IQ = 47, Non verbal IQ = 53, Verbal IQ= 46 | YES | |||
| Case 10 | 9 | Size Mosaic | 455, 543, 626, 931, 1080 (118 + del) | 82% | 14% | 4% | 0.05 | 0.14 | Full scale IQ = 69, Verbal IQ = 73, Performance IQ = 69 | NO | |||
| Case 11 | 4 | Size mosaic | 420, 479, 1047 (84 + del) | 67% | 13% | 20% | 0.06 | 0.17 | ELC = 65 | Full Scale IQ = 66, Non verbal IQ = 82, Verbal IQ = 53 | NO | ||
| Case 12 | 1 | Meth mosaic | 250, 510, 710 (170-320+ del) | 68.5% | 22.5% | 9% | 0.11 | 0.72 | ELC = 90 | NO | |||
RESULTS
Fragile X diagnosis and gene expression analysis
Cognitive evaluation established that all 12 male participants were in the intellectual disabilities (ID) range with six males having an IQ close to borderline and five males with a low IQ. Seven out of eleven were diagnosed with ASD. For one patient testing of ASD was not available (Table 1).
DNA testing of 12 male participants revealed that they were all mosaics with a percent of cells, ranging from 57% to 94%, carrying methylated full mutation alleles. Eight probands were size mosaics for a full methylated and a normal/intermediate unmethylated alleles, two were size mosaics for the presence of a full methylated and of a premutation unmethylated allele, and two were methylation mosaics for the presence of full methylated and unmethylated alleles. Four probands with size and methylation mosaicism also exhibited an allele with a deletion of the CGG repeat element and of portions of the flanking region (Figure 1, case 9-12) in a percent of cells ranging from 4% to 28% (Table 1). Southern Blot results, showing the different mutation patterns, are illustrated in Figure 1.
Figure 1. Southern blot analysis of 12 cases.
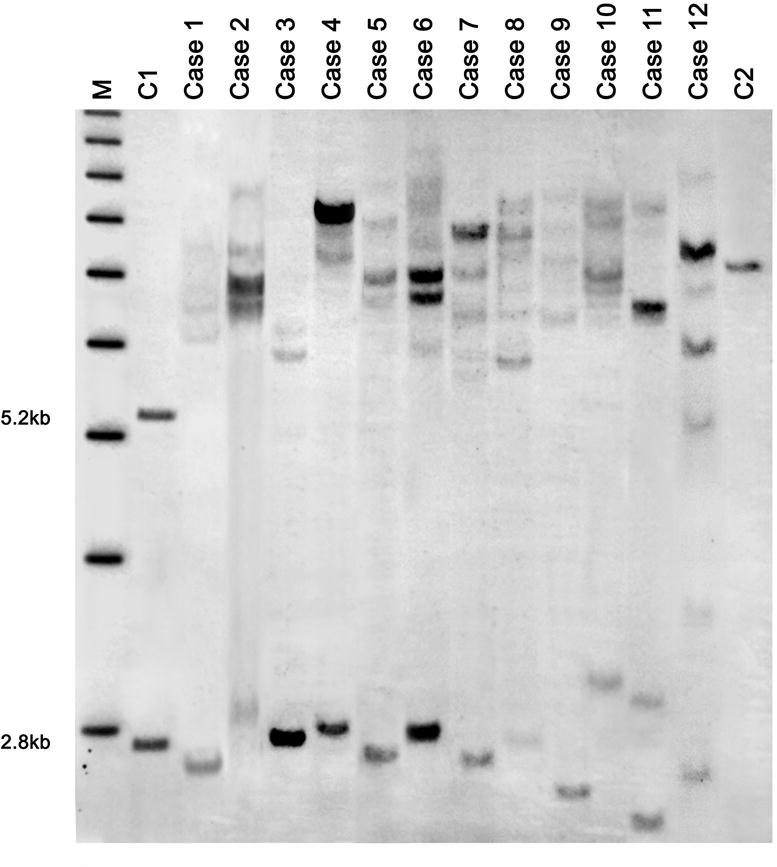
Southern blot analysis of genomic DNA isolated from normal control female. The normal allele on the active X chromosome (2.8 Kb) and the normal allele on the inactive X chromosome (5.2 Kb) are shown on the left (C1) and a positive male with a full mutation allele is shown on the right (C2). Results in Case 1 through Case 8 show mosaicism for the presence of full mutation alleles in addition to a normal/intermediate allele. Case 9 -12 show the four males with size or methylation mosaicism.
In order to determine the exact size and sequence of the deleted region in the proband’s smaller allele, direct sequencing of the DNA amplicon was carried out using PCR-amplified patient DNA patient DNA. Sequencing revealed that in all four cases, the fragment carried a deletion in the 5′UTR, spanning the entire CGG repeat region as well part of the upstream and downstream regions (Figure 1 and 2; cases 9-12) in a percent of cells ranging from 4% to 28% (Table 1). Southern Blot results, showing the different mutation patterns, are illustrated in Figure 1.
Gene expression analysis was carried out using qRT-PCR assays for the measurement of FMR1 mRNA and by HTRF analysis for the measurement of FMRP expression levels (Table 1). As expected, most of the probands showed reduced expression of both FMR1 mRNA and FMRP due to the presence of methylation in the majority of the cells. Among the 12 cases, in the ones who presented with higher IQ FMRP expression levels were not consistently higher than in those with lower IQ. Only in one case (case 3) the FMR1 mRNA expression level was within the normal range (30), likely due to the presence of a normal allele in over 40% of the cells; however, FMRP levels remained significantly lower than controls (Table 1).
Clinical history of selected cases
Case 1
This 37 year old man was diagnosed with FXS and ASD at 2-1/2 years of age. He has been severely affected by FXS, unable to tolerate clothes or leaving the house for the past few years, preferring to sit on a pad in his room. He has been on many medications in the past, including a variety of antipsychotic medications (risperidone, aripiprazole and quetiapine), but experienced side effects with all of medications, including severe muscle spasms, abnormal movements and/or weight gain. In particular, risperidone caused abnormal movements, significant regression in his behavior and bedwetting. Other medications that were marginally helpful at different times included SSRIs, SNRIs, lithium, clonidine and lorazepam, but no medication significantly helped his ability to tolerate clothes and move away from the pad in his room where he would play with his favorite toys.
He made the greatest improvement in behavioral at age 31 yr when he was put on a course of minocycline (200 mg/day). With minocycline he was able to tolerate clothes, leave the house and he began to use an iPad. He mumbles most of the time during the day, but on occasion he makes a clear statement; for instance, he was able to read the ingredients on a shampoo bottle and he said "this has cucumber in it", which was true.
He has intermittent tantrums and aggression when he is agitated, particularly with a change in his routine. When he is agitated he will perseverate on a behavior over- and- over again. He is tactilely defensive, with poor eye contact, stereotypic movements and he is very autistic. He likes to put books and papers into a box or basket and carry them around. He wets his bed regularly at night.
On examination, his height was 190 cm and his weight was 88.45 kg. Because of his agitation and pushing behavior it was not possible to get a blood pressure. HEENT examination demonstrates a long face, his ears are prominent with some ear-cupping bilaterally and his palate is high arched. He has scoliosis, in addition to kyphosis. His finger joints are not hyperextensible but his feet are flat with significant pronation. He has macroorchidism bilaterally. He did not have a tremor but he is unable to tandem walk or to cooperate with the examination. His deep tendon reflexes (DTRs) are 1+ and symmetrical and his Babinski reflex is negative.
He has severe autism and some cognitive strengths including limited reading ability but he has not tolerated cognitive testing.
DNA molecular testing indicated the presence of size mosaicism (full/normal) with a normal allele of 16 CGG repeats present in 32% of the cells; FMR1 mRNA and FMRP expression levels were 0.59 and 0.34 (Table 1), lower than controls (mean in controls was 1.26 ± 0.22 and 1.1 ± 0.23, respectively).
Case 5
Case 5 was diagnosed with FXS just prior to 2 years of age because his sibling also had FXS. His mother’s pregnancy was normal, birth weight was 7 lbs. 12 oz. and he did well in the newborn period. He had mild difficulties in coordinating his suck in the first day and was hypotonic but his developmental milestones were normal, including sitting at 6.5 months, crawling at 10 months, walking at 14 months, and saying 20 words by the age of 2. His eye contact was excellent and he was a joyful baby with frequent smiles, although he demonstrated separation anxiety on a daily basis. He was social and he did not present with ASD.
At age 2 his examination showed a head circumference of 48.5 cm, his weight was 12.4 kg and his height was 83.6 cm. HEENT exam was normal; his ear pinnae were not prominent, his joints were hyperextensible with metacarpal phalangeal (MP) extension to 90°, his thumbs were double-jointed, and his feet were flat.
On developmental testing at chronological age of 24 months using the Mullen Scales of Early Learning (MSEL), his gross motor was 16 months, visual reception 18 months, fine motor 20 months, receptive language 14 months, and expressive language 16 months, with an overall Early Learning Composite of 66 (normal mean is 100). On the Vineland Adaptive Behavior Scale his communication was 86, daily living skills 93, socialization 82, motor 82, and his overall Adaptive Behavior Composite (ABC) was 83 (normal mean is 100). In summary, Case 5 is a high functioning toddler with FXS and without ASD. His current medications include folic acid, 5 mg a day, and fish oil, 1 teaspoon a day. A trial of sertraline to improve overall language and development was recommended.
DNA molecular testing indicated the presence of size mosaicism (full/normal) with a normal allele of 17 CGG repeats present in 27% of the cells (Table 1). FMR1 mRNA and FMRP expression levels were 0.37 and 0.64, respectively (Table1). Surprisingly, FMRP expression levels were higher that the FMR1 mRNA levels observed in this subject. FMRP levels decrease with increasing CGG repeat throughout the permutation range; however, if the observed transcript levels derive completely from the normal allele, no deficit in translation efficiency should take place. However, it has also been shown that a broad variation in FMRP levels for any givenCGG-repeat size from the normal to the higher CGG expanded range may exists (including sampling differences and methodology) [31,62–64].
Case 9
The proband was born after a normal pregnancy with a birth weight of 7 lbs. 6 oz. His developmental milestones included walking at 13 months, saying words in the first year and phrases by 4 years of age. His early behavior included ADHD for which he was started on stimulant medication a long acting methylphenidate at 7 years of age. The proband also exhibited finger biting, poor eye contact, and hand flapping when excited. He did not exhibit tantrums or aggressive behavior; rather, he was described as shy.
He was “mainstreamed” in his educational program and his overall cognitive testing at age 4 on the Kaufman Assessment Battery demonstrated a mental processing composite of 83. Upon subsequent testing at age 12, cognitive evaluation by WASI (Weschler abbreviated Scale of Intelligence), demonstrated a verbal IQ (VIQ) of 70, performance IQ (PIQ) of 70, and full scale IQ (FSIQ) of 67. Subsequent cognitive testing at age 14, with the WISC-III, demonstrated a VIQ of 64, PIQ of 59 and a FS IQ of 58. His Peabody Picture vocabulary test at age 14 was 80 and his expressive one-word vocabulary test was 85. The mean for the general population is 100.
Evaluation with the Autism Diagnostic Observation Scale revealed that his overall score was in the Autism Spectrum Disorder (ASD) range. He was placed on sertaline (Zoloft) because his anxiety interfered with his social interactions, in addition to mixed amphetamine salts (Adderall) for ADHD.
Physical examination at age 14 revealed a head circumference in the 90th percentile for age, height in the 50th percentile and weight in the 60th percentile. Other characteristics included a long face, prominent ears with ear cupping bilaterally, and a normal palate. His finger joints were not hyperextensible, showing metacarpal phalangeal (MP) extension to 50° and his thumbs were not double-jointed. His feet were not flat, deep tendon reflexes were 2+ and symmetrical; and his genitalia had significant macroorchidism with a testicular volume of 50 ml.
In follow-up testing by WASI at age 18, he demonstrated a VIQ of 55, PIQ of 57 and a full scale IQ of 53. He was also given the Stanford Binet V, which yielded a VIQ of 46, NVIQ (non-verbal IQ) of 53 and FSIQ of 47. On his Vineland Adaptive Behavior Scales he scored higher, with Communication at 80; Daily living skills, 72; Socialization, 73; and an Overall Adaptive Behavior Composite of 73. ADOS testing at this time demonstrated a communication score of 5, Socialization score of 8 for a total of 13, which is in the ASD range. At this age he was placed on a trial of minocycline to assess its benefit as a targeted treatment for FXS (62).
DNA molecular testing indicated the presence of methylationnmosaicism (methylated and unmethylated full mutation alleles) with an allele encompassing a deletion across the CGG repeat present in 13% of the cells (Table 1, Figure 2). The total percent of cells carrying hypermethylated full mutation alleles was approximately 60%, FMR1 mRNA and FMRP expression levels were 0.64 and 0.32, respectively (Table1).
DISCUSSION
The mechanism leading to CGG instability is complex and still remains to be clarified. One of the proposed mechanisms is a DNA polymerase slippage or slipped strand mis-pairing (SSM) which can lead to reduction of the CGG length, either from a premutation to a normally sized allele or the contraction of a full mutation allele to a normal or deleted sized alleles [22,66]. This phenomenon may occur early in embryogenesis in a percentage of cells where the presence of long stretches of CGG repeats can induce mis-pairing of newly synthesized strand during DNA replication potentially leading to small deletions [67,68]. It has been demonstrated that CpG methylation and SSM can facilitate the formation of deletion events. Consequently, CpG-rich region may be prone to rearrangements, which results in deletion that can extend into the promoter sequence upstream of the CGG repeat [69]. Furthermore, post-zygotic deletion may result in unmethylated and normal-size alleles as seen in individuals with FXS presenting with size and methylation mosaicism [23,70]. Interestingly, a recent study on four mosaic males carrying both a full mutation and a normal pure FMR1 allele suggested the presence of a risk lineage-specific FMR1 haplotype predisposing to large contractions [70]. In this study, we report on 12 mosaic males with FXS including 8 males with size mosaicism (for a full and a normal/ intermediate size alleles) and 4 males who exhibit size and methylation mosaicism with alleles unmethylated in both the premutation and full mutation range, as well as alleles carrying a deletion spanning the CGG repeat. The presence of unmethylated normal or expanded alleles is what likely reflects gene activity of the FMR1 gene demonstrated by the expression, even at lower levels, of detectable FMR1 mRNA and FMRP. FMR1 mRNA expression measured by qRT-PCR was observed in all cases although at different levels, likely been produced from normal alleles, or the premutation alleles and/ or from the unmethylated full mutation allele [30].
Intriguingly, FMRP levels did not significantly correlate with the FMR1 mRNA levels, likely due to the decreased translation efficiency for longer expanded alleles [32–36]. This was also observed in previous studies [18,33].
Among the size mosaics, 50% (case 2, 4, 7, and 8) presented with a high percentage of methylation (>80%) accompanied by decreased FMR1 mRNA and FMRP expression levels, which appeared to correlate with more severe phenotype including lower IQ and ASD (Table 1). Males with lower percentage of methylation (<80%) appear to have higher cognitive abilities, although this does not appear to be always the case suggesting that other factors are likely to influence cognition; these observations are consistent with previous studies [18,37,40,71].
In one of these previous studies, it was observed that of 18 individuals with FXS, including 7 size mosaics and 6 methylation mosaics, the percent of methylation of the FMR1 gene was directly correlated with FMR1 mRNA and FMRP, in both blood and fibroblast tissues, and was negatively correlated with FSIQ scores. Further, higher IQ scores were found in subjects with higher levels of FMRP. The authors suggested that FXS individuals with low methylation and detectable FMRP levels could yield a better clinical outcome though they exhibited elevated FMR1 mRNA levels [18]. However, individuals in our study presented with impaired cognitive function likely due to the percent of cell carrying normal alleles, which were not sufficient to produce adequate FMRP levels particularly when methylation of the full mutation was present. For example, case 3 and case 7 both have very low levels of FMRP at 0.18 but case 3 with lower percentage of methylation (57%) presented with higher IQ (64) while case 7 with higher level of methylation (83%) had a much lower IQ (40). Similarly, McConkie-Rosell et al. [71] studied a fragile X family with six brothers and found that the degree of severity of the phenotypic expression of FXS correlates with the degree of FMR1 methylation. Additional cases of methylation mosaicism have also been reported supporting the notion that cognitive functions negatively correlate with the methylation status [72,73]. Case 9–12 demonstrated higher repeat instability due to the presence of methylated and unmethylated alleles spanning the entire CGG size range, in addition to a deletion of the CGG repeat tract and of a variable region upstream and downstream flanking regions (Figure 2). The role of these deletions relative to FMR1 gene activity (transcription and translation) is not clear. For example, case 11, a size mosaic male presenting with 67% of the cells methylated and a NVIQ in the low normal range, has approximately 20% of the cells harboring an allele bearing a deleted region of 396 base pairs (the largest of the four deletion cases here presented) upstream of the ATG start site and containing the entire CGG tract. In contrast, case 9 presented low methylation, high expression levels of both FMRP and FMR1 mRNA but showed lowest IQ among the deleted group. However, how the deletion may affect transcription is still need to be elucidated. It is possible that the binding domain regions important for the transcription of the gene are removed in the deleted alleles resulting in non-functional or defective alleles. It has been reported that deletion of the entire CGG repeat region does not abolish expression [39] as long as the transcription and translation start sites are unaffected by the deletion. These authors reported an unaffected female who had a small deletion at the 5′ end of the FMR1 gene on one of her X chromosomes, extending from about 65 bp upstream of the CGG repeat region to about 32 bp downstream which including all CGG repeats and 97 bp of flanking sequences but both transcription and translation start sites remained intact. Interestingly, this female had a large deletion on the other X chromosome, which abrogated the entire FMR1 gene. However, this X chromosome was silenced due to X-inactivation and the girl produced normal levels of FMRP from the other allele carrying the deletion, reflecting her normal phenotype. Thus, there has been a number of reports on deletions within the FMR1 gene; however, most of these studies did not report FMRP levels [20,24,39–47,74]. Only two deletion cases reported FMRP levels in mosaic males with FXS. de Graaf et al. [37] reported a male patient with FXS who exhibited a full mutation and a small deletion in 28% of the lymphocytes. The deletion included part of the CGG repeat and extended 30 bp 3′ of the repeat but did not seem to influence the expression of FMR1 gene as FMRP expression was observed in approximately the same percent of cells. Han et al. [40] reported on the presence of a deleted allele encompassing the CGG repeat region and 42 bp upstream of the repeats, with FMRP production in 22% of lymphocytes. In our study, our four probands with the deleted allele demonstrated lower FMRP production, ranging from 5% to 32% compared to controls, which resulted in their cognitive impairment. In all cases, the deletion occurred in the 5′ UTR upstream of the ATG start site likely not affecting the translation of FMRP. Interestingly, inter- and intra-tissue FMR1 gene expression differences have been shown, particularly in mosaic individuals [18]; therefore, the FMRP expression detected in the case’s lymphocytes of this study may be different from the expression in the brain.
In conclusion, our cases demonstrated the presence of size and methylation mosaicism in male patients with FXS who have a normal allele in eight subjects and a deletion in four subjects in addition to full mutation alleles. The presence of a normal allele particularly in routine PCR or in screening studies where the presence of the additional full mutation allele may not be easily detected by PCR can lead to a wrong diagnosis. This highlights the importance of being cautious particularly when the clinical picture is strongly indicative of FXS. Thus, in order to better characterize these atypical FXS mutational profiles, additional approaches, such as more robust PCR, i.e. using the CGG repeat-targeted primer [75,76] or a combination of different approaches as reported in Goncalves et al. [77], should be carried out to ensure that these types of mosaics do not go underdiagnosed and potentially missed by routine FXS testing. Genetic counseling and molecular genetic analysis are essential not only for the patients but also of the families for their future rehabilitation and family planning.
Key issues.
Atypical size- or methylation-mosaicism with both, full mutation and smaller (normal to premutation) alleles, as well as a combination of methylated and unmethylated alleles were observed in males with FXS.
Half of the size mosaic males exhibited a highly methylation with reduced FMR1 mRNA and FMRP expression levels which correlate with lower IQ and with the presence of ASD.
sMosaic males with a full and a normal allele should be investigated by an advanced PCR approach and detection rather a routine FXS testing (i.e. conventional PCR) to ensure that a correct and precise diagnosis does not go under-detected.
References
- 1.Hagerman RJ. Physical and behavioral phenotype In: Hagerman RJ, Hagerman PJ, editors Fragile X syndrome: diagnosis, treatment and research. 3rd. Baltimore: The Johns Hopkins University Press; 2002. pp. 3–109. [Google Scholar]
- 2.Verkerk AJ, Pieretti M, Sutcliffe JS, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65(5):905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 3.Pieretti M, Zhang FP, Fu YH, et al. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991;66(4):817–822. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- 4.Bagni C, Oostra BA. Fragile X syndrome: from protein function to therapy. Am J Med Genet A. 2013;161A(11):2809–2821. doi: 10.1002/ajmg.a.36241. Epub 2013/10/12. [DOI] [PubMed] [Google Scholar]
- 5.Maddalena A, Richards CS, McGinniss MJ, et al. Technical standards and guidelines for fragile X: the first of a series of disease-specific supplements to the standards and guidelines for clinical genetics laboratories of the American College of Medical Genetics. Quality Assurance Subcommittee of the Laboratory Practice Committee. Genet Med. 2001;3(3):200–205. doi: 10.1097/00125817-200105000-00010. Epub 2001/06/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devys D, Biancalana V, Rousseau F, et al. Analysis of full fragile X mutations in fetal tissues and monozygotic twins indicate that abnormal and somatic heterogeneity are established early in development. Am J Med Genet. 1992;43(1–2):208–216. doi: 10.1002/ajmg.1320430134. Epub 1992/04/01. [DOI] [PubMed] [Google Scholar]
- 7.Willemsen R, Bontekoe CJ, Severijnen LA, et al. Timing of the absence of FMR1 expression in full mutation chorionic villi. Hum Genet. 2002;110 doi: 10.1007/s00439-002-0723-5. [DOI] [PubMed] [Google Scholar]
- 8.Kumari D, Usdin K. The distribution of repressive histone modifications on silenced FMR1 alleles provides clues to the mechanism of gene silencing in fragile X syndrome. Hum Mol Genet. 2010;19 doi: 10.1093/hmg/ddq394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allingham-Hawkins DJ, Brown CA, Babul R, et al. Tissue-specific methylation differences and cognitive function in fragile X premutation females. Am J Med Genet. 1996;64(2):329–333. doi: 10.1002/(SICI)1096-8628(19960809)64:2<329::AID-AJMG19>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 10.Tassone F, Longshore J, Zunich J, et al. Tissue-specific methylation differences in a fragile X premutation carrier [Internet] Clin Genet. 1999;55(5):346–351. doi: 10.1034/j.1399-0004.1999.550508.x. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10422805. [DOI] [PubMed] [Google Scholar]
- 11.Pretto DI, Mendoza-Morales G, Lo J, et al. CGG allele size somatic mosaicism and methylation in FMR1 premutation alleles. J Med Genet. 2014;51(5):309–318. doi: 10.1136/jmedgenet-2013-102021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pietrobono R, Tabolacci E, Zalfa F, et al. Molecular dissection of the events leading to inactivation of the FMR1 gene. Hum Mol Genet. 2005;14 doi: 10.1093/hmg/ddi024. [DOI] [PubMed] [Google Scholar]
- 13.Tabolacci E, Moscato U, Zalfa F, et al. Epigenetic analysis reveals a euchromatic configuration in the FMR1 unmethylated full mutations. Eur J Hum Genet. 2008;16 doi: 10.1038/ejhg.2008.130. [DOI] [PubMed] [Google Scholar]
- 14.Lanni S, Goracci M, Borrelli L, et al. Role of CTCF protein in regulating FMR1 locus transcription. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alisch RS, Wang T, Chopra P, et al. Genome-wide analysis validates aberrant methylation in fragile X syndrome is specific to the FMR1 locus. BMC Med Genet. 2013;14 doi: 10.1186/1471-2350-14-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naumann A, Hochstein N, Weber S, et al. A distinct DNA-methylation boundary in the 5′-upstream sequence of the FMR1 promoter binds nuclear proteins and is lost in fragile X syndrome. Am J Hum Genet. 2009;85 doi: 10.1016/j.ajhg.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagerman RJ, Hull CE, Safanda JF, et al. High functioning fragile X males: demonstration of an unmethylated fully expanded FMR-1 mutation associated with protein expression. Am J Med Genet. 1994;51 doi: 10.1002/ajmg.1320510404. [DOI] [PubMed] [Google Scholar]
- 18.Pretto D, Yrigollen CM, Tang HT, et al. Clinical and molecular implications of mosaicism in FMR1 full mutations. Front Genet. 2014;5 doi: 10.3389/fgene.2014.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nolin SL, Glicksman A, Houck GE, Jr, et al. Mosaicism in fragile X affected males. Am J Med Genet. 1994;51(4):509–512. doi: 10.1002/ajmg.1320510444. Epub 1994/ 07/15. [DOI] [PubMed] [Google Scholar]
- 20.Hammond LS, Macias MM, Tarleton JC, et al. Fragile X syndrome and deletions in FMR1: new case and review of the literature. Am J Med Genet. 1997;72(4):430–434. Epub 1997/12/31 23: 38. [PubMed] [Google Scholar]
- 21.Fan H, Booker JK, McCandless SE, et al. Mosaicism for an FMR1 gene deletion in a fragile X female. Am J Med Genet. 2005;136A(2):214–217. doi: 10.1002/ajmg.a.30807. [DOI] [PubMed] [Google Scholar]
- 22.Schmucker B, Seidel J. Mosaicism for a full mutation and a normal size allele in two fragile X males. Am J Med Genet. 1999;84(3):221–225. [PubMed] [Google Scholar]
- 23.Orrico A, Galli L, Dotti MT, et al. Mosaicism for full mutation and normal-sized allele of the FMR1 gene: a new case. Am J Med Genet. 1998;78(4):341–344. Epub 1998/08/26. [PubMed] [Google Scholar]
- 24.Grasso M, Faravelli F, Lo Nigro C, et al. Mosaicism for the full mutation and a microdeletion involving the CGG repeat and flanking sequences in the FMR1 gene in eight fragile X patients. Am J Med Genet. 1999;85(3):311–316. doi: 10.1002/(sici)1096-8628(19990730)85:3<311::aid-ajmg24>3.0.co;2-a. Epub 1999/07/09. [DOI] [PubMed] [Google Scholar]
- 25.Mingroni-Netto RC, Haddad LA, Vianna-Morgante AM. The number of CGG repeats of the FMR1 locus in premutated and fully mutated heterozygotes and their offspring: implications for the origin of mosaicism. Am J Med Genet. 1996;64(2):270–273. doi: 10.1002/(SICI)1096-8628(19960809)64:2<270::AID-AJMG7>3.0.CO;2-Y. Epub 1996/08/ 09. [DOI] [PubMed] [Google Scholar]
- 26.Bonarrigo FA, Russo S, Vizziello P, et al. Think about it: FMR1 gene mosaicism. J Child Neurol. 2014;29(9):NP74–NP77. doi: 10.1177/0883073813503187. Epub 2013/09/ 26. [DOI] [PubMed] [Google Scholar]
- 27.Todorov T, Todorova A, Kirov A, et al. Fragile X mosaic male full mutation/normal allele detected by PCR/MS-MLPA. BMJ Case Rep. 2009;2009 doi: 10.1136/bcr.06.2008.0139. Epub 2009/01/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao XN, Usdin K. UPS and downs: mechanisms of repeat instability in the fragile X-related disorders. Genes (Basel) 2016;7(9):1–14. doi: 10.3390/genes7090070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Genc B, Muller-Hartmann H, Zeschnigk M, et al. Methylation mosaicism of 5ʹ-(CGG)(n)-3ʹ repeats in fragile X, premutation and normal individuals. Nucleic Acids Res. 2000;28(10):2141–2152. doi: 10.1093/nar/28.10.2141. Epub 2000/ 04/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tassone F, Hagerman RJ, Loesch DZ, et al. Fragile X males with unmethylated, full mutation trinucleotide repeat expansions have elevated levels of FMR1 messenger RNA. Am J Med Genet. 2000;94(3):232–236. doi: 10.1002/1096-8628(20000918)94:3<232::aid-ajmg9>3.0.co;2-h. Epub 2000/09/20. [DOI] [PubMed] [Google Scholar]
- 31.Ludwig AL, Espinal GM, Pretto DI, et al. CNS expression of murine fragile X protein (FMRP) as a function of CGG-repeat size. Hum Mol Genet. 2014;23(12):3228–3238. doi: 10.1093/hmg/ddu032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Primerano B, Tassone F, Hagerman RJ, et al. Reduced FMR1 mRNA translation efficiency in fragile X patients with premutations. RNA (New York, NY) 2002;8(12):1482–1488. Epub 2003/01/08. [PMC free article] [PubMed] [Google Scholar]
- 33.Peprah E, He W, Allen E, et al. Examination of FMR1 transcript and protein levels among 74 premutation carriers [Internet] J Hum Genet. 2010;55(1):66–68. doi: 10.1038/jhg.2009.121. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4122982&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allen EG, Sherman S, Abramowitz A, et al. Examination of the effect of the polymorphic CGG repeat in the FMR1 gene on cognitive performance. Behav Genet. 2005;35(4):435–445. doi: 10.1007/s10519-005-2792-4. Epub 2005/06/23. [DOI] [PubMed] [Google Scholar]
- 35.Kenneson A, Zhang F, Hagedorn CH, et al. Reduced FMRP and increased FMR1 transcription is proportionally associated with CGG repeat number in intermediate-length and premutation carriers. Hum Mol Genet. 2001;10(14):1449–1454. doi: 10.1093/hmg/10.14.1449. Epub 2001/07/13. [DOI] [PubMed] [Google Scholar]
- 36.Feng Y, Zhang F, Lokey L, et al. Translational suppression by trinucleotide repeat expansion at FMR1. Science (80-) 1995;268(5211):731–734. doi: 10.1126/science.7732383. [DOI] [PubMed] [Google Scholar]
- 37.de Graaff E, de Vries BB, Willemsen R, et al. The fragile X phenotype in a mosaic male with a deletion showing expression of the FMR1 protein in 28% of the cells. Am J Med Genet. 1996;64(2):302–308. doi: 10.1002/(SICI)1096-8628(19960809)64:2<302::AID-AJMG14>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 38.Coffee B, Ikeda M, Budimirovic DB, et al. Mosaic FMR1 deletion causes fragile X syndrome and can lead to molecular misdiagnosis. Am J Med Genet A. 2008 doi: 10.1002/ajmg.a.32261. epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grønskov K, Hjalgrim H, Bjerager MO, et al. Deletion of all CGG repeats plus flanking sequences in FMR1 does not abolish gene expression. Am J Hum Genet. 1997;61(4):961–967. doi: 10.1086/514872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han XD, Powell B, Phalin JL, et al. Mosaicism for a full mutation, premutation, and deletion of the CGG repeats results in 22% FMRP and elevated FMR1 mRNA levels in a high-functioning fragile X male. Am J Med Genet. 2006 Jul 1;140(13):1463–1471. doi: 10.1002/ajmg.a.31291. [DOI] [PubMed] [Google Scholar]
- 41.de Vries BB, Wiegers AM, de Graaff E, et al. Mental status and fragile X expression in relation to FMR-1 gene mutation. Eur J Hum Genet. 1993;1(1):72–79. doi: 10.1159/000472389. [DOI] [PubMed] [Google Scholar]
- 42.Mannermaa A, Pulkkinen L, Kajanoja E, et al. Deletion in the FMR1 gene in a fragile-X male. Am J Med Genet. 1996;64(2):293–295. doi: 10.1002/(SICI)1096-8628(19960809)64:2<293::AID-AJMG12>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 43.Mila M, Castellvi Bel S, Sanchez A, et al. Mosaicism for the fragile X syndrome full mutation and deletions within the CGG repeat of the FMR1 gene. J Med Genet. 1996;33(4):338–340. doi: 10.1136/jmg.33.4.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petek E, Kroisel PM, Schuster M, et al. Mosaicism in a fragile X male including a de novo deletion in the FMR1 gene. Am J Med Genet. 1999;84(3):229–232. Epub 1999/05/20. [PubMed] [Google Scholar]
- 45.Quan F, Zonana J, Gunter K, et al. An atypical case of fragile X syndrome caused by a deletion that includes the FMR1 gene. Am J Hum Genet. 1995;56(5):1042–1051. Epub 1995/05/01. [PMC free article] [PubMed] [Google Scholar]
- 46.Schmucker B, Ballhausen WG, Pfeiffer RA. Mosaicism of a microdeletion of 486 bp involving the CGG repeat of the FMR1 gene due to misalignment of GTT tandem repeats at chi-like elements flanking both breakpoints and a full mutation. Hum Genet. 1996;98(4):409–414. doi: 10.1007/s004390050230. Epub 1996/10/01. [DOI] [PubMed] [Google Scholar]
- 47.Snow K, Tester DJ, Kruckeberg KE, et al. Sequence analysis of the fragile X trinucleotide repeat: implications for the origin of the fragile X mutation. Hum Mol Genet. 1994;3(9):1543–1551. doi: 10.1093/hmg/3.9.1543. [DOI] [PubMed] [Google Scholar]
- 48.Harris SW, Hessl D, Goodlin-Jones B, et al. Autism profiles of males with fragile X syndrome. Am J Ment Retard. 2008;113(6):427–438. doi: 10.1352/2008.113:427-438. Epub 2009/01/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Filipovic-Sadic S, Sah S, Chen L, et al. A novel FMR1 PCR method for the routine detection of low abundance expanded alleles and full mutations in fragile X syndrome. Clin Chem. 2010;56(3):399–408. doi: 10.1373/clinchem.2009.136101. Epub 2010/01/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tassone F, Pan R, Amiri K, et al. A rapid polymerase chain reaction based screening method for identification of all expanded alleles of the fragile X (FMR1) gene in newborn and high-risk populations. J Mol Diagn. 2008;10(1):43–49. doi: 10.2353/jmoldx.2008.070073. Epub 2008/01/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tassone F, Hagerman RJ, Ikle DN, et al. FMRP expression as a potential prognostic indicator in fragile X syndrome. Am J Med Genet. 1999;84(3):250–261. Epub 1999/05/20. [PubMed] [Google Scholar]
- 52.Tassone F, Hagerman RJ, Taylor AK, et al. Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am J Hum Genet. 2000;66(1):6–15. doi: 10.1086/302720. Epub 2000/ 01/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schutzius G, Bleckmann D, Kapps-Fouthier S, et al. A quantitative homogeneous assay for fragile X mental retardation 1 protein [Internet] J Neurodev Disord. 2013;5(8) doi: 10.1186/1866-1955-5-8. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2710703&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mullen EM. Mullen scales of early learning. Circle Pines, MN: AGS. 1995:58–64. [Google Scholar]
- 55.Wechsler D. Wechsler abbreviated scale of intelligence WASI. San Antonio (TX): Physiological Corporation, Harcourt Brace; 1999. [Google Scholar]
- 56.Wechsler D. The Wechsler intelligence scale for children. 3rd. San Antonio (TX): The Psychological Corporation, Harcourt Brace; 1991. [Google Scholar]
- 57.Roid GH. Stanford-Binet Intelligence Scales. 5th. Itasca (IL): Riverside; 2003. [Google Scholar]
- 58.Sparrow S, Cicchetti D, Balla D. Vineland Adaptive Behavior Scales, Second Edition (VABS-II): Interview edition survey form. Circle, MN: American Guidance Service Publishing; 2005. [Google Scholar]
- 59.Dunn LM, Dunn LM, Bulheller S, Häcker H. Peabody picture vocabulary test. Circle Pines, MN: American Guidance Service; 1965. [Google Scholar]
- 60.Lord C, Rutter M, DiLavore PC, et al. Autism diagnostic observation schedule. Los Angeles: Western Psychological Services; 2002. [Google Scholar]
- 61.Swanson JM, Kraemer HC, Hinshaw SP, et al. Clinical relevance of the primary findings of the MTA: success rates based on severity of ADHD and ODD symptoms at the end of treatment. J Am Acad Child Adolesc Psychiatry. 2001;40(2):168–179. doi: 10.1097/00004583-200102000-00011. Epub 2001/02/24. [DOI] [PubMed] [Google Scholar]
- 62.Fauci La, et al. Fragile X screening by Quantification of FMRP in Dried Blood Spots by a Luminex Immunoassay. J Mol Diagn. 2013 Jul;15(4):508–517. doi: 10.1016/j.jmoldx.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 63.Brouwer J, Mientjes E, Bakker C, et al. Elevated Fmr1 mRNA levels and reduced protein expression in a mouse model with an unmethylated Fragile X full mutation. Exp Cell Res. 2007;313(2):244–253. doi: 10.1016/j.yexcr.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Entezam A, Biacsi R, Orrison B, et al. Regional FMRP deficits and large repeat expansions into the full mutation range in a new Fragile X premutation mouse model. Gene. 2007;395(1–2):125–134. doi: 10.1016/j.gene.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leigh MJ, Nguyen DV, Mu Y, et al. A randomized double-blind, placebo-controlled trial of minocycline in children and adolescents with fragile x syndrome. J Dev Behav Pediatr. 2013;34(3):147–155. doi: 10.1097/DBP.0b013e318287cd17. Epub 2013/04/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tabolacci E, Pomponi MG, Pietrobono R, et al. A unique case of reversion to normal size of a maternal premutation FMR1 allele in a normal boy. Eur J Hum Genet. 2008;16(2):209–214. doi: 10.1038/sj.ejhg.5201949. Epub 2007/11/ 01. [DOI] [PubMed] [Google Scholar]
- 67.Chiurazzi P, Kozak L, Neri G. Unstable triplets and their mutational mechanism: size reduction of the CGG repeat vs. germline mosaicism in the fragile X syndrome. Am J Med Genet. 1994;51(4):517–521. doi: 10.1002/ajmg.1320510446. Epub 1994/07/15. [DOI] [PubMed] [Google Scholar]
- 68.Garcia Arocena D, de Diego Y, Oostra BA, et al. A fragile X case with an amplification/deletion mosaic pattern. Hum Genet. 2000;106(3):366–369. doi: 10.1007/s004390051052. Epub 2000/05/08. [DOI] [PubMed] [Google Scholar]
- 69.Nichol Edamura K, Pearson CE. DNA methylation and replication: implications for the “deletion hotspot” region of FMR1. Hum Genet. 2005;118(2):301–304. doi: 10.1007/s00439-005-0037-5. Epub 2005/09/01. [DOI] [PubMed] [Google Scholar]
- 70.Maia N, Loureiro JR, Oliveira B, et al. Contraction of fully expanded FMR1 alleles to the normal range: predisposing haplotype or rare events? J Hum Genet. 2016 doi: 10.1038/jhg.2016.122. Epub 2016/10/28. [DOI] [PubMed] [Google Scholar]
- 71.McConkie-Rosell A, Lachiewicz AM, Spiridigliozzi GA, et al. Evidence that methylation of the FMR-I locus is responsible for variable phenotypic expression of the fragile X syndrome. Am J Hum Genet. 1993;53(4):800–809. [PMC free article] [PubMed] [Google Scholar]
- 72.Helderman-van den Enden AT, Maaswinkel-Mooij PD, Hoogendoorn E, et al. Monozygotic twin brothers with the fragile X syndrome: different CGG repeats and different mental capacities. J Med Genet. 1999;36(3):253–257. [PMC free article] [PubMed] [Google Scholar]
- 73.Wohrle D, Salat U, Glaser D, et al. Unusual mutations in high functioning fragile X males: apparent instability of expanded unmethylated CGG repeats. J Med Genet. 1998;35(2):103–111. doi: 10.1136/jmg.35.2.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Quan F, Grompe M, Jakobs P, et al. Spontaneous deletion in the FMR1 gene in a patient with fragile X syndrome and cherubism. Hum Mol Genet. 1995;4(9):1681–1684. doi: 10.1093/hmg/4.9.1681. [DOI] [PubMed] [Google Scholar]
- 75.Chen L, Hadd A, Sah S, et al. An information-rich CGG repeat primed PCR that detects the full range of fragile X expanded alleles and minimizes the need for southern blot analysis. J Mol Diagn. 2010;12(5):589–600. doi: 10.2353/jmoldx.2010.090227. Epub 2010/07/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hayward BE, Zhou Y, Kumari D, et al. Set of assays for the comprehensive analysis of FMR1 alleles in the fragile X–related disorders [Internet] J Mol Diagnostics. 2016;18(5):762–774. doi: 10.1016/j.jmoldx.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goncalves TF, Dos Santos JM, Goncalves AP, et al. Finding FMR1 mosaicism in Fragile X syndrome. Expert Rev Mol Diagn. 2016;16(4):501–507. doi: 10.1586/14737159.2016.1135739. Epub 2015/12/31. [DOI] [PMC free article] [PubMed] [Google Scholar]


