Abstract
A cultivation-dependent approach revealed that highly diverse populations of Streptomyces were present in atmospheric precipitations from a hailstorm event sampled in February 2016 in the Cantabrian Sea coast, North of Spain. A total of 29 bioactive Streptomyces strains isolated from small samples of hailstone and rainwater, collected from this hailstorm event, were studied here. Taxonomic identification by 16S rRNA sequencing revealed more than 20 different Streptomyces species, with their closest homologs displaying mainly oceanic but also terrestrial origins. Backward trajectory analysis revealed that the air-mass sources of the hailstorm event, with North Western winds, were originated in the Arctic Ocean (West Greenland and North Iceland) and Canada (Labrador), depending on the altitude. After traveling across the North Atlantic Ocean during 4 days the air mass reached Europe and precipitated as hailstone and rain water at the sampling place in Spain. The finding of Streptomyces species able to survive and disperse through the atmosphere increases our knowledge of the biogeography of genus Streptomyces on Earth, and reinforces our previous dispersion model, suggesting a generalized feature for the genus which could have been essential in his evolution. This unique atmospheric-derived Streptomyces collection was screened for production of bioactive secondary metabolites. Analyses of isolates ethyl acetate extracts by LC-UV-MS and further database comparison revealed an extraordinary diversity of bioactive natural products. One hundred molecules were identified, mostly displaying contrasted antibiotic and antitumor/cytotoxic activities, but also antiparasitic, antiviral, anti-inflammatory, neuroprotector, and insecticide properties. More interestingly, 38 molecules not identified in natural products databases might represent new natural products. Our results revealed for the first time an extraordinary diversity of Streptomyces species in the atmosphere able to produce an extraordinary repertoire of bioactive molecules, thus providing a very promising source for the discovery of novel pharmaceutical natural products.
Keywords: hailstorm, bioaerosols, Streptomyces, antibiotic, antimicrobial, antitumor
Introduction
Natural products are essential to human health and constitute a primary resource in biomedicine and biotechnology. Streptomyces species (Phylum Actinobacteria) are the most prolific source of bioactive natural products with pharmaceutical activities. New trends in the discovery of novel drugs, such antibiotics and antitumor compounds, are focused on the search of producing microorganisms from unexplored habitats (Subramani and Aalbersberg, 2013; Behie et al., 2016; Maciejewska et al., 2016; Law et al., 2017).
Although Streptomyces species have been traditionally considered as soil bacteria, in the last decades became evident their presence and wide distribution in oceanic ecosystems and associated to diverse marine organisms. Previous work in the North Atlantic region, Cantabrian Sea (Bay of Biscay), Northern Spain, revealed the presence of a great number of Streptomyces strains in intertidal seaweeds, and deep-sea coral reef invertebrates at the Aviles Canyon. New natural products with antimicrobial and cytotoxic activities against tumor cell lines were recently discovered in this submarine Canyon (Braña et al., 2015, 2017a,b; Sarmiento-Vizcaíno et al., 2015, 2016, 2017a,b).
Besides Earth and oceans, there is increasing evidence of the presence of Streptomyces strains in the atmosphere. In culture-dependent approaches, Streptomyces strains were isolated from cloud water at Puy de Dôme, Southern France (Amato et al., 2007) and repeatedly isolated from atmospheric precipitations such as rainwater, hailstone and snow in the Cantabrian region, during 2013–2014 (Braña et al., 2015; Sarmiento-Vizcaíno et al., 2016). These included three ubiquitous species, Streptomyces albidoflavus, Streptomyces cyaneofuscatus, and Streptomyces carnosus, previously isolated from terrestrial and oceanic environments (Braña et al., 2015; Sarmiento-Vizcaíno et al., 2016).
Following this line of evidence, we have previously proposed an atmospheric dispersion model, which follows the Earth hydrological cycle, to explain the biogeography and distribution among terrestrial, marine and atmospheric environments of Streptomyces species (Sarmiento-Vizcaíno et al., 2016). According to this hypothetical cycle, oceanic bioaerosols-forming clouds contribute to streptomycetes dissemination from marine ecosystems to the atmosphere, where they undergo long distances transport by winds and finally fall down to inland and oceanic ecosystems by precipitation.
Clouds have been defined as atmospheric air masses with water condensed in ice crystals or liquid state (Amato et al., 2007). They are considered as low-temperature “aquatic” environments which contribute to transport and aerial connection between Earth ecosystems (Amato et al., 2007), having been considered as possible atmospheric oases for microorganisms (Amato et al., 2017). Recent studies at the Puy de Dôme Mountain revealed that some microorganisms are even metabolically active in clouds (Amato et al., 2017). Rainwater droplets can coalesce into hailstones which circulate inside storm clouds following unpredictable pathways (Amato et al., 2017). Biogeochemical studies on hailstones indicate that storm clouds can be considered among the most extreme habitats for microbial life on Earth. (Šantl-Temkiv et al., 2013).
Here is reported the exploration of the phylogenetic and biosynthetic diversity of atmospheric-derived Streptomyces strains collected from a storm cloud in Northern Spain, using hailstone and rainwater precipitations as natural sampling sources. This work constitutes the first large insight into the Streptomyces diversity existing in hailstone and rainwater and the natural compounds produced. Meteorological analyses addressed to estimate the air mass sources and trajectories support our previous Streptomyces dispersion cycle model.
Materials and methods
Sampling of atmospheric precipitations (hailstone and rain water)
Cloud precipitations samples, such as hailstone and rainwater, were taken during a thunderstorm discharge over the coastal location of Gijón (Asturias) in the afternoon of 14th February 2016 at 16.00 h. Gijón (43°32′ N, 5°39′ W) is located in the North of Spain (Bay of Biscay, Figure 1). The prevailing wind direction during this storm event in this area was Northwestern.
Figure 1.
Sampling location. Overview of the Western European Seas (Atlantic Ocean). Star indicates the sampling location at Gijón, North of Spain (Iberian Peninsula).
Hailstone and rainwater samples from this storm event were collected in sterile recipients, while they were falling on the ground, in a terrace in front of the sea at about 30 m above sea level. Samples were stored at −20°C (hailstone) and 4°C (rain water) until immediately processing as has been reported (Braña et al., 2015).
Air mass backward trajectories analysis
In order to investigate the long-range transport journey of air masses that originated the precipitation event, backward trajectories were obtained using the HYSPLIT model (Hybrid Single Particle Lagrangian Integrated Trajectory) obtained from the Global Data Assimilation System of National Oceanic and Atmospheric Administration, USA (Stein et al., 2015). To track the transport pathways of air masses, 5-day backward trajectories (commonly used in bioaerosol studies) were generated using the NOAA model (http://ready.arl.noaa.gov/hypub-bin/trajtype.pl?runtype=archive) to determine the origin of a given air parcel. The sampling location for this study was used as the backward trajectory start point with altitudes of 30, 1,000, and 3,000 m, respectively above the ground level to estimate the accurate trajectories of atmospheric air masses.
Isolation of Streptomyces strains and culture media
Atmospheric samples were inoculated on selective agar media prepared with cycloheximide (80 μg ml−1) as antifungal and nalidixic acid (20 μg ml−1) as anti-Gram negative bacteria, using MOPS BLEB 1/6 (Oxoid) basal medium as previously reported (Sarmiento-Vizcaíno et al., 2016). For selection, two different media either prepared with distilled water or with a supplement of 3.5% NaCl were used. After 2–3 weeks of incubation at 28°C, growing colonies were selected based on different morphological features and pigment production on R5A agar plates. Isolates obtained in pure cultures were conserved in 20% glycerol at −20°C, and at −70°C. For addressing halotolerance studies, MOPS BLEB 1/6 (Oxoid) was used as the basal medium, adding NaCl at of 0, 3.5, 7.0, and 10.5% (w/v) final concentrations. For secondary metabolite production streptomycetes isolates were cultured on R5A medium as previously described (Braña et al., 2015).
Antimicrobial bioassays
Agar diffusion methods were used to determine antimicrobial activities. Antibiotic production was assessed using the following indicator microorganisms: the Gram-positive bacteria Micrococcus luteus ATCC 14452 and Streptomyces 85E ATCC 55824 (Shanbhag et al., 2015), the Gram-negative Escherichia coli ESS, and the yeast Saccharomyces cerevisiae var. carlsbergensis. Analyses were performed in TSA1/2 (Merck) against bacteria and in Sabouraud 1/2 (Pronadisa) against yeast. Bioassays were carried out both with agar plugs (7 mm diameter) and in parallel with different ethyl acetate extracts obtained from solid cultures of the isolates.
16S RNA phylogenetic analysis
For phylogenetic analysis of the strains based on 16S rRNA sequences, DNA was extracted with a microbial isolation kit (Ultra Clean, MoBio Laboratories, Inc.) and standard methods were used for checking the purity (Russell and Sambrook, 2001). Partial 16S rRNA gene sequences of the bacterial strains were obtained by using the 616V (forward) and 699R (reverse) primers (Arahal et al., 2008) in PCR amplification as previously described (Braña et al., 2015). Once obtained the nucleotide sequences were compared to sequences in databases using the BLAST program (Basic Local Alignment Search Tool) against the NCBI (National Centre for Biotechnology Information). The nucleotide sequences were submitted and deposited in the EMBL sequence database. Phylogenetic analysis of the strains based on 16S rRNA sequences was carried out as previously reported (Sarmiento-Vizcaíno et al., 2017a).
Chromatographic analysis
Plugs of R5A plates (about 7 ml) were extracted using ethyl acetate in neutral and acidic (with 1% formic acid) conditions. After evaporation, the organic fraction residue was redissolved in 100 μL of a mixture of DMSO and methanol (50:50). The analysis of the samples were performed by reversed phase liquid chromatography as has been described (Braña et al., 2014; Sarmiento-Vizcaíno et al., 2016).
Identification of compounds by LC-UV-Vis and LC-UV-HRMS analyses
Samples were first analyzed and evaluated using an in-house HPLC-UV-Vis database. LC-UV-HRMS analyses were carried out as has been described (Pérez-Victoria et al., 2016) and major peaks in each chromatogram were searched against the MEDINA's internal database and also against the Dictionary of Natural Products (DNP) (Chapman and Hall/CRC, 2015).
Results
Backward transport trajectories
To estimate the sources of the air masses that caused the precipitation event in the Cantabrian Sea coast, 120 h backward trajectories were determined at three different arriving heights (30, 1,000, and 3,000 m). As shown in Figure 2, the results of the NOAA meteorological analysis indicated three different routes followed by the air layers depending on the altitude. The air-mass at 30 m altitude originate from Newfoundland and Labrador (Canada); the one at 1,000 m came from the Davies Strait in the Arctic Ocean, between North Canada and West Greenland; the air layer at 3,000 m originated at Northwest Iceland. All air-masses at different altitudes crossed the Atlantic Ocean, and after 4 days reached the Iberian Peninsula precipitating as hailstones and rainwater in the North of Spain, were samples were collected. In addition, possible mixing events were detected between different air layers at different altitudes meanwhile traveling across the Atlantic Ocean (Figure 2). Thus, the estimated backward trajectories mainly revealed an oceanic route, involving both the Arctic and Atlantic Oceans, but there was also a terrestrial route from continental America.
Figure 2.

Five-day backward trajectories of air masses generating the storm that arrived at Gijón (North Spain) on February 14, 2016, at 16.00, calculated with the NOAA Hysplit Model with three different transects with different arriving height: red = 30 m; blue = 1,000 m; green = 3,000 m. Sampling time and place are indicated by the black asterisk.
Isolation of bioactive Streptomyces from a hailstorm event
A cultivation-dependent approach revealed in atmospheric precipitations the presence of highly diverse Streptomyces populations by using a sample of hailstone (300 ml of unfrozen hailstones) and a sample of 25 ml of rainwater. Only strains cultivable at 28°C and atmospheric pressure were recovered on selective agar plates, prepared either with 3.5% NaCl (to simulate the salt content of the Cantabrian Sea water) or without salt. The percentage of streptomycete colonies recovered on selective medium without salt was higher, approximately 66% from hailstone and 56% from rainwater, than on saline medium.
Among a total of 136 streptomycete colonies isolated on selection plates (92 colonies from hailstone and 44 from rainwater), 45 morphologically different isolates from hailstone and 14 from rainwater samples were selected after the dereplication process. None of these isolates required NaCl for growth. An important feature observed in all isolates, with a single exception, was their halotolerance. Most isolates tolerate around 7% NaCl, which doubles the salt concentration in the Cantabrian Sea (3.5% average). This is in agreement with previous studies of NaCl tolerance for the genus Streptomyces in which it was estimated that around 50% of the species tolerate up to 7% NaCl (Tresner et al., 1968).
All isolates were initially tested for antimicrobial activity using agar diffusion assays. Further 29 different bioactive strains, displaying diverse antimicrobial activities, were selected for this study (Table 1). Most of the isolates displayed antibiotic activities against M. luteus and Streptomyces 85E (Gram-positive bacteria), and also against the yeast S. cerevisiae; whereas only three strains were active against the E. coli ESS (Gram-negative).
Table 1.
Antibiotic activities of Streptomyces cultures (agar plugs) against Gram-positive, Gram-negative bacteria, and yeasts.
| Strain | M. luteus | Streptomyces 85E | E. coli | S. cerevisiae |
|---|---|---|---|---|
| A-185 | 20 | – | – | 20 |
| A-186 | 22 | – | – | – |
| A-189 | – | – | – | 15 |
| A-191 | 11 | 26 | – | 10 |
| A-192 | – | – | 18* | – |
| A-193 | – | 13 | – | – |
| A-196 | – | 12 | – | – |
| A-197 | 20 | 21 | 18 | – |
| A-198 | – | – | – | 19 |
| A-201 | 12 | 10 | – | – |
| A-202 | 11 | 12 | – | – |
| A-203 | 13 | 17 | – | 10 |
| A-204 | 27 | 28 | – | – |
| A-206 | 14 | 22 | 19 | 13 |
| A-208 | 16 | – | – | – |
| A-209 | 19 | – | – | – |
| A-210 | 13 | 11 | – | – |
| A-211 | 13 | 13 | – | – |
| A-214 | 11 | 36 | – | – |
| A-215 | 16 | – | – | – |
| A-217 | 26 | 33 | – | – |
| A-221 | 16 | 33 | 14 | 14 |
| A-222 | – | – | – | – |
| A-225 | 19 | 15 | – | – |
| A-226 | 14 | – | – | – |
| A-227 | – | 11 | – | – |
| A-228 | – | 10 | – | 16 |
| A-229 | 13 | – | – | 11 |
| A-230 | – | – | – | 15 |
| A-231 | 11 | – | – | 11 |
Activities were measured as the zones of complete inhibition (diameters in mm). Bioassays with ethyl acetate extracts in acid and neutral conditions were also performed in parallel. The asterisk indicates that the activity was detected in the extract.
Taxonomic identification of isolates
For taxonomic identifications of the atmospheric-derived bioactive strains, we sequenced fragments of their 16S rDNA and deposited the nucleotide sequences in the EMBL database. Table 2 displays the accession numbers of the strains. Based on 16S rRNA gene alignments, phylogenetic analyses clearly demonstrated that all 29 isolates belonged to the Streptomyces genus, since all of them shared 99–100% identity with previously known Streptomyces species. The relationship between the atmospheric isolates and their closest phylogenetic relatives with indication of their isolation site is shown in Table 2. A phylogenetic tree was built to display the diversity among atmospheric isolates and assess their phylogenetic relationship (Figure 3).
Table 2.
Phylogenetic diversity of atmospheric-derived bioactive Streptomyces isolates.
| Strain | Source | EMBL A. N. | NaCl % | Closest homolog | A. N. | % Homology (bp) | Isolation source (reference) |
|---|---|---|---|---|---|---|---|
| Streptomyces geldanamycininus A-185 | Rain water | LT899923 | <3.5 | Streptomyces geldanamycininus NRRL B-3602 | NR_043722 | 99.9 (727/728) | Soil (Goodfellow et al., 2007) |
| Streptomyces sp. A-186 | Rain water | LT907817 | 7 | Streptomyces chilikensis RC 1830* | NR_118246 | 100 (684/684) | Brackish water sediment Chilika Lake (India) (Ray et al., 2013) |
| Streptomyces sp. A-189 | Rain water | LT907818 | 3.5 | Streptomyces chartreusis ISP 5085* | NR_114825 | 100 (769/769) | African soil (Leach et al., 1953) |
| Streptomyces sp. A-191 | Rain water | LT907819 | 7 | Streptomyces litmocidini NRRL B-3635* | NR_116096 | 99.7 (768/770) | Soil (Dodzin et al., 1998) |
| Streptomyces sp. A-192 | Rain water | LT907820 | 7 | Streptomyces albus NRRL B-1811* | NR_118467 | 99.6 (665/668) | Soil; marine sediment (westsouthern coast Iberian Peninsula) (Schleissner et al., 2011; Labeda et al., 2014) |
| Streptomyces sp. A-193 | Rain water | LT907821 | 7 | Streptomyces thinghirensis S10* | NR_116901 | 99.5 (663/666) | Rhizosphere soil of Vitis vinifer (Morocco) (Loqman et al., 2009) |
| Streptomyces fradiae A-196 | Rain water | LT899924 | 7 | Streptomyces fradiae NBRC 12773 | AB184134 | 100 (724/724) | Soil samples (Egypt and Saudi Arabia) (El-Naggar et al., 2016) |
| Streptomyces flavofuscus A-197 | Rain water | LT899925 | 7 | Streptomyces flavofuscus NRRL B-2594 | NR_115965 | 99.5 (823/827) | Colliery spoil heaps (Czech Republic) (Chronáková et al., 2010) |
| Streptomyces chumphonensis A-198 | Rain water | LT899926 | 7 | Streptomyces chumphonensis KK1-2 | AB738400 | 98.5 (746/757) | Marine sediment (Tailand) (Phongsopitanun et al., 2014) |
| Streptomyces sp. A-201 | Hailstone | LT907822 | 7 | Streptomyces lunaelactis MM109* | NR_134822 | 99.9 (728/729) | Moonmilk deposit from a cave (Belgium) (Maciejewska et al., 2015) |
| Streptomyces sp. A-202 | Hailstone | LT907823 | 7 | Streptomyces pratensis ch24* | JQ806215 | 99.4 (855/860) | Grassy fields (Rong et al., 2013) |
| Streptomyces thermospinosisporus A-203 | Hailstone | LT899927 | 7 | Streptomyces thermospinosisporus AT10 | AF333113 | 99.5 (745/749) | Soil (UK) (Kim and Goodfellow, 2002) |
| Streptomyces olivaceus A-204 | Hailstone | LT899928 | 7 | Streptomyces olivaceus NBRC 3200 | AB184743 | 99.6 (781/784) | Marine (Yue et al., 2016) |
| Streptomyces sp. A-206 | Hailstone | LT907824 | 7 | Streptomyces chilikensis RC 1830* | NR_118246 | 100 (717/717) | Brackish water sediment Chilika Lake (India) (Ray et al., 2013) |
| Streptomyces sulphureus A-208 | Hailstone | LT899929 | 10 | Streptomyces sulphureus NRRL B-1627 | DQ442546 | 99.7 (621/623) | Marine sediment (China); Submarine Canyon (Spain) (Zhao et al., 2012; Sarmiento-Vizcaíno et al., 2017b) |
| Streptomyces sp. A-209 | Hailstone | LT907825 | 7 | Streptomyces lunaelactis MM109* | NR_134822 | 100 (733/733) | Moonmilk deposit from a cave (Belgium) (Maciejewska et al., 2015) |
| Streptomyces cyaneofuscatus A-211 | Hailstone | LT899930 | 7 | Streptomyces cyaneofuscatus NBRC 13190 | AB184860 | 99.9 (788/789) | Marine, terrestrial and atmospheric (Spain) (Braña et al., 2015) |
| Streptomyces californicus A-214 | Hailstone | LT899931 | 7 | Streptomyces californicus NBRC 3386 | AB184755 | 100 (789/789) | Saline Soil (USA) (Killham and Firestone, 1984) |
| Streptomyces cyaneofuscatus A-215 | Hailstone | LT899932 | 7 | Streptomyces cyaneofuscatus NBRC 13190 | AB184860 | 99.2 (763/769) | Marine, terrestrial and atmospheric (Spain) (Braña et al., 2015) |
| Streptomyces carnosus A-217 | Hailstone | ND | 7 | Similar to Streptomyces carnosus M-40 | HG965214 | ND | Marine, terrestrial and atmospheric (Spain) (Braña et al., 2015) |
| Streptomyces sp. A-221 | Hailstone | LT907826 | 7 | Streptomyces chilikensis RC 1830* | NR_118246 | 99.6 (746/749) | Brackish water sediment Chilika Lake (India) (Ray et al., 2013) |
| Streptomyces rishiriensis A-222 | Hailstone | LT899933 | 3.5 | Streptomyces rishiriensis NRRL B-3239 | NR_044141 | 98.7 (602/610) | Soil (Japan) (Matsumoto et al., 1996) |
| Streptomyces sp. A-225 | Hailstone | LT907827 | 3.5 | Streptomyces avidinii NBRC 13429* | NR_041132 | 99.5 (605/608) | Marine sediment (India) (Sudha and Masilamani, 2012) |
| Streptomyces coelicolor A-226 | Hailstone | ND | 7 | Streptomyces coelicolor A3(2) | AB184196 | ND | Soil (UK) (Bentley et al., 2002) |
| Streptomyces sp. A-227 | Hailstone | LT907828 | 7 | Streptomyces thermocarboxydus NBRC 16323* | NR_112585 | 99.7 (749/751) | Soil (Kim et al., 1998) |
| Streptomyces sp. A-228 | Hailstone | LT907829 | 3.5 | Streptomyces lunaelactis MM109* | NR_134822 | 100 (656/656) | Moonmilk deposit from a cave (Belgium) (Maciejewska et al., 2015) |
| Streptomyces hygroscopicus A-229 | Hailstone | LT899934 | 3.5 | Streptomyces hygroscopicus NRRL 2387 | AJ391820 | 99.8 (803/805) | Soil (Australia) (Jensen, 1931) |
| Streptomyces sp. A-230 | Hailstone | LT907830 | 7 | Streptomyces iconiensis BNT558* | NR_134198 | 98.9 (730/738) | Salt lake and saltern (Turkey) (Tatar et al., 2014) |
| Streptomyces albidoflavus A-231 | Hailstone | ND | 7 | Similar to Streptomyces albidoflavus T-199 | LN626360 | ND | Marine, terrestrial and atmospheric (Spain) (Sarmiento-Vizcaíno et al., 2016) |
The asterisk indicates that only one species is shown when more than one closest homolog was found. ND, not determined.
Figure 3.
Neighbor-joining phylogenetic tree generated by distance matrix analysis of 16S rDNA sequences from atmospheric Streptomyces strains (highlighted) and nearest phylogenetic relatives. The numbers on branch nodes indicate bootstrap values (1,000 resamplings; only values >70% are shown). Bar represents 0.5% sequence divergence.
As shown by their identification, strains related to more than 20 different Streptomyces species have been isolated from a small sample of hailstone and rainwater (Table 2). This constitutes a significant proportion of the global number of Streptomyces species described in our planet so far, estimated in 550–823 (http://www.bacterio.net/streptomyces.html). Among 29 isolates, 15 were identified at species level and were designated with the species name (see Table 2), whereas the rest displayed 16S rDNA gene similarity to more than one species. Unfortunately, just based on these 16S rRNA sequences it is not enough to discriminate among closely related species and further assays are needed to complement the results of 16S rRNA sequence analysis for Streptomyces identification at specific level. Among identified isolates, S. albidoflavus, S. cyaneofuscatus, and S. carnosus, were repeatedly isolated in our geographical area from atmospheric precipitations, marine ecosystems (intertidal seaweeds and deep-sea corals) and terrestrial lichens. Interestingly the model species Streptomyces coelicolor, the genetically best known representative of the genus, was here isolated from hailstone; and Streptomyces albus, used as heterologous host in many laboratories, was isolated from rain water. Rare or infrequent Streptomyces species, here obtained from atmospheric precipitations, were previously isolated from highly diverse environments, mainly from the North Hemisphere (Table 2).
The habitats of known Streptomyces species closely related to the atmospheric-derived strains include soils and aquatic environments, such as oceans, lakes, and groundwater. This information was used together with the backward trajectory analysis (see above) to estimate the possible sources of airborne Streptomyces reported here.
Identification of secondary metabolites by metabolite profiling analysis
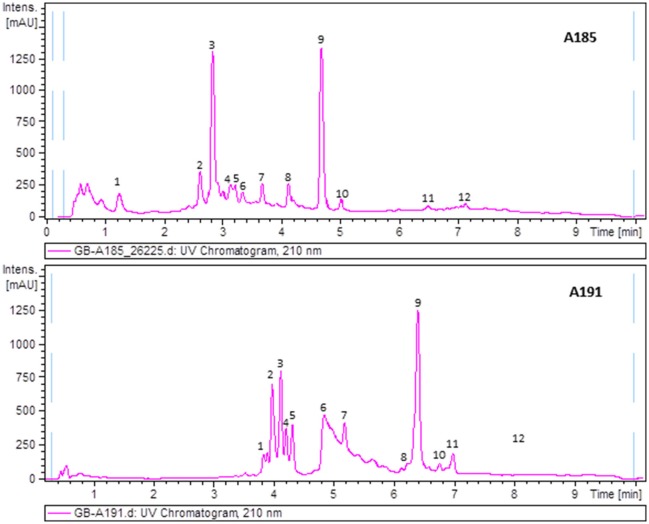
To uncover the biosynthetic abilities of the airborne Streptomyces strains, ethyl acetate extracts were screened for secondary metabolites production by LC/UV and LC/HRMS analyses in combination with searches in UV and MS databases or the DNP after generation of a molecular formula of each peak based on HRMS results. Most of the strains displayed complex metabolic profiles revealing their high potential as a source of novel natural products (Supplementary Material 1). As an example, Figure 4 displays UV210nm chromatograms corresponding to samples A-185 and A-191 showing identified compounds.
Figure 4.
UV210nm chromatogram of samples A185 and A191 with peaks annotated showing components identified by HRMS. Dereplicated components in sample A185: (1) Ketomycin, (2) Thiazinogeldanamycin, (3) 4,5-Dihydrogeldanamycin, (4) Reblastatin, (5) Herbimycin E, (6) 15R-Hydroxygeldanamycin, (7) 6-Demethoxy-6-methylgeldanamycin, (8) 8-Demethylgeldanamycin or 17-O-Demethylgeldanamycin, (9) Geldanamycin, (10) C30H40N2O10 (geldanamycin related but with a molecular formula not found in the Dictionary of Natural Products), (11) Nigericin, and (12) 30-O-Acetylnigericin. Dereplicated components in sample A191: (1) Antibiotic RK 397, (2) Flavofungin I, (3 and 4) – C36H56O9 (related to flavofungin but with a molecular formula not included in the Dictionary of Natural Products), (5) Flavofungin II, (6) Prodigiosin 25b, (7) C18H30O (molecular formula not present in the Dictionary of Natural Products as produced by prokaryotes), (8) Undecylprodigiosin, (9) Spectinabilin or Arabilin, (10) Lyngbyamide D, (11) C28H33NO5 (molecular formula not found in the Dictionary of Natural Products as produced by prokaryotes), and (12) Pamamycins.
Among a total of 138 metabolites detected by HPLC/MS in the ethyl acetate extracts of the studied strains (Supplementary Material 1), 100 were identified after metabolite profiling analysis and comparison with natural products databases (some of them only at the family level). Among identified products, 45 were reported to display biological activities as antibiotics (17 antifungal and 13 antibacterial), 29 as cytotoxic/ antitumor agents, 5 as antiviral, 3 as antiparasitic, 3 as immunosupresors, 2 as anti-inflammatory, 1 as neuroprotector, 1 as insecticide, and other compounds with physiological roles in the Streptomyces life cycle (Table 3).
Table 3.
Bioactive compounds produced by atmospheric-derived Streptomyces strains and their biological activities.
| Compound | Strain | Biological activities | Identification |
|---|---|---|---|
| 2-Acetamidobenzamide | A-196 | Antifungal against phytopathogenic filamentous fungi (Phay et al., 1996) | MS |
| 4,5-Dihydrogeldanamycin | A-185, A-229 | Anticancer (Schnur et al., 1995; Wu et al., 2012) | MS |
| 6-Prenyltryptophol | A-206 | Cytotoxic (Sánchez López et al., 2003) | MS |
| 8-Demethylgeldanamycin/17-O-Demethylgeldanamycin* | A-185, A-229 | Moderate cytotoxicity against the human breast cancer cell line (Buchanan et al., 2005)/Unknown | MS |
| Abierixin | A-229 | Antibiotic (David et al., 1985), weak cytotoxicity, antimalarial activity (Supong et al., 2016) | MS |
| Actinorhodin | A-226 | Antibiotic (Wright and Hopwood, 1976) | UV |
| Aggreceride B | A-203, A-206, A-221 | Platelet aggregation inhibitor (Omura et al., 1986) | MS |
| Albonoursin | A-192 | Antibacterial, antitumor in mice (Fukushima et al., 1973) | MS |
| Alpha-lipomycin | A-186 | Antibiotic (Bihlmaier et al., 2006) | MS |
| Alteramide-derivative | A-201, A-209, A-231 | Unknown | UV |
| Alteramide A | A-211, A-214, A-228 | Cytotoxic (Shigemori et al., 1992); antifungal (Moree et al., 2014) | MS |
| Alteramide B | A-211, A-214, A-228 | Antifungal (Moree et al., 2014) | MS |
| Antibiotic RK 397 | A-191 | Antibiotic, cytotoxic (Kobinata et al., 1993) | MS |
| Antibiotic TMC (1A/B or TMC 1F) | A-241 | Antibiotic, moderate cytotoxicity (Kohno et al., 1996) | MS |
| Antimycins (A4; A5a/A5b; A6a/A6b/A18 and A11) | A-206, A-222 | Antifungal (Seipke et al., 2012); antiviral (Raveh et al., 2013); cytotoxic (Takimoto et al., 1999); apoptosis inducer (Seipke and Hutchings, 2013) | UV, MS |
| Bafilomycin C1 | A-228 | Antibiotic, cytotoxic (Moon et al., 2003) | MS |
| Blastmycin | A-222 | Fungicide (Endo and Yonehara, 1970), cytotoxic (Fujita et al., 2004) | MS |
| Caboxamycin | A-228 | Anti-Gram-positive, antitumor (Hohmann et al., 2009) | UV |
| Cyclo(4-hydroxyprolylleucyl) | A-193 | Moderate toxicity toward brine shrimp larvae (Gao et al., 2014) | MS |
| Cyclo(leucylprolyl) | Several strainsA | Antibiotic, cytotoxic activity (Santos et al., 2015) | MS |
| Cyclo(prolylvalyl) | A-197, A-203, A-221, A-225, A-241 | Antifungal (Kumar et al., 2014) | MS |
| Deisovalerylblastmycin | A-222 | Antifungal (Ishiyama et al., 1976) | MS |
| Dihydromaltophilin | A-209 | Antifungal (Fiedler et al., 2005) | MS |
| Feigrisolide C | A-214 | Moderate activity on Coxsackie virus B3 (Tang et al., 2000), lysis of Plasmopara viticola, Phytophthora capsici, and Aphanomyces cochlioides zoospores (Islam et al., 2016) | MS |
| Flavofungin I and II | A-191 | Antifungal antibiotic (Uri and Bekesi, 1958), anti-glioma and antifungal activities (Wang et al., 2017a) | MS |
| Fogacin | A-226 | Antimicrobial activities against C. albicans (Lu et al., 2014) | MS |
| Geldanamycin | A-185; A-229 | Antifungal, anticancer, neurotrophic and neuroprotective (Tadtong et al., 2007) | MS |
| Germicidin A | Several strainsB | Spore germination, hypha elongation (Aoki et al., 2011) | MS |
| Germicidin B | A-193, A-203, A-217, A-221, A-226 | Spore germination, hypha elongation (Aoki et al., 2011) | MS |
| Germicidin D | A-193 | Spore germination, hypha elongation (Aoki et al., 2011) | MS |
| Grecocycline A | A-202 | Cytotoxic (Paululat et al., 2010) | MS |
| Griseorhodins | A-214 | Antibiotics, cytotoxic (Stroshane et al., 1979); inhibition of HIV reverse transcriptase and human telomerase (Lin et al., 2014) | UV |
| Herbimycin E | A-185 | Hsp90α affinity (Alzheimer's disease pathogenesis) (Shaaban et al., 2013) | MS |
| Ikarugamycin epoxide | A-214 | Moderate activities against Gram-positive bacteria and fungi, strongly cytotoxic (HMO2 and MCF 7) (Bertasso et al., 2003) | MS |
| Ilamycin A, C1 or C2 | A-215 | Cytotoxic (Ma et al., 2017) | MS |
| Indanomycin | A-222 | Antibacterial, insecticidal (Zhang et al., 1997) | MS |
| Izumiphenazine C | A-196 | Synergistic activity in sensitizing TRAIL-resistant AGS cells (Abdelfattah et al., 2010) | MS |
| Juglomycin A | A-215 | Antibiotic (Fiedler et al., 1994) | MS |
| Kandenol C | A-228 | Moderate antimicrobial activity againts Mycobacterium vaccae (Ding et al., 2012) | MS |
| Ketomycin | A-185 | Antibiotic (Takeda et al., 1984) | MS |
| Lobophorin A | A-204, A-217 | Anti-inflammatory, antituberculosis, anti-BCG (Jiang et al., 1999; Chen et al., 2013) | UV, MS |
| Lobophorin B | A-204, A-218 | Anti-inflammatory, antituberculosis, anti-BCG (Jiang et al., 1999; Chen et al., 2013) | UV, MS |
| Lobophorin K | A-204, A-219 | Cytotoxic, moderate antibiotic activity againts Staphylococcus aureus (Braña et al., 2017a) | UV, MS |
| Maltophilins | A-214, A-228 | Antifungal (Fiedler et al., 2005) | UV, MS |
| Methylsulfomycin I | A-209 | Antibiotic (Vijaya Kumar et al., 1999) | MS |
| N-Butanoylhomoserine lactone | A-228 | Quorum-sensing signal molecule in Gram-negative bacteria (Chan et al., 2011) | MS |
| Neoenactin B1 or B2 | A-221 | Antifungal (Roy et al., 1987) | MS |
| Nigericin | A-185, A-229 | Antibiotic, strong cytotoxicity (A2780 and SKOV3) (Wang et al., 2017b) | MS |
| Nonactins | A-209, A-214 | Ammonium ionophore, antibacterial, antiviral, antitumor (Zhan and Zheng, 2016) | MS |
| Okicenone | A-189 | Cytotoxic activity (Komiyama et al., 1991) | MS |
| Oxostaurosporine | A-198 | Protein kinase C inhibitor (Osada et al., 1992) | MS |
| Pamamycins (607, 621 A/B/C/D, 635 A/B/C/D/E/F and 663) | A-191 | Aerial mycelium and secondary metabolite production inducing (Hashimoto et al., 2011), anti-Gram-positive and antifungal (Hanquet et al., 2016) | MS |
| Paulomycin B | A-231 | Anti-Gram-positive, gonococcal and Chlamydia infections (Argoudelis et al., 1982; Novak, 1988) | UV |
| Phenazinoline or Izumiphenazine derivative | A-196 | Unknown | MS |
| Phenelfamycin | A-189, A-210 | Anti-Gram-positive (Brötz et al., 2011) | UV |
| Radamycin | A-209 | tipA promoter inducer (González Holgado et al., 2002) | MS |
| Reblastatin | A-185 | Cell cycle inhibitor (Takatsu et al., 2000), Hsp90 ATPase inhibitor (Wu et al., 2012) | MS |
| Spectinabilin/Arabilin* | A-191 | Antimalarial and cytotoxic (Isaka et al., 2002)/Androgen receptor antagonist in prostate cancer LNCaP cells (Kawamura et al., 2010) | MS |
| Staurosporine | A-198, A-230 | Protein kinase C inhibitor (Mori et al., 1994), Antifungal, pan-kinase inhibitor (Tamaoki et al., 1986; Song et al., 2017) | MS |
| Tetranactin | A-214 | Antibiotic, immunosuppressive and anti-proliferative (Tanouchi and Shichi, 1988) | MS |
| Thiazinogeldanamycin | A-185 | Cytotoxic (Ni et al., 2011) | MS |
| Trinactin | A-214 | Antibiotic, immunosuppressive (Tanouchi and Shichi, 1987) | MS |
| Trioxacarcin A | A-186 | Antitumor, antibiotic (Tomita et al., 1981) | MS |
| Undecylprodigiosin | A-191, A-193, A-203, A-226, A-241 | Antibiotic, cytotoxic (Petrović et al., 2017), immunosuppressor (Songia et al., 1997; Williamson et al., 2006) | UV, MS |
| Urauchimycin A/B and C | A-222 | Antibiotic (Imamura et al., 1993) | MS |
| Valinomycin | A-211 | Antibiotic, antiparasitary, antiviral (Perkins et al., 1990; Cheng, 2006; Pimentel-Elardo et al., 2010) | MS |
| WS 9326A | A-198 | Tachykinin receptor antagonist (Hayashi et al., 1992); quorum sensing inhibitor in Gram-positive bacteria (Desouky et al., 2015) | MS |
| β-Indomycinone/Saptomycin A/Rubimycinone A* | A-197 | Antibiotic, cytotoxic (Tsukahara et al., 2014)/Antimicrobial (Abe et al., 1993)/Unknown | MS |
The asterisk means that more than one compound was identified.
A: A-189, A-197, A-201, A-203, A-206, A-214, A-221, A-222, A-225, A-228, A-230.
B: A-186, A-193, A-196, A-203, A-204, A-217, A-221, A-222, A-226.
Remarkably, there are 38 metabolites whose molecular formulae determined by HRMS does not correspond to any compound included in Natural Products Databases and remained unidentified. These molecules might be new natural products, and therefore constitute an excellent starting point for the discovery of new bioactive molecules with pharmaceutical interest.
Discussion
We provide here the first insight ever made into the Streptomyces species diversity within a small storm cloud sample, which was collected after a hailstone and rainwater precipitation event during the afternoon of the 14th of February 2016 in the Cantabrian Sea coast (North of Spain). Our results revealed a striking richness of Streptomyces species present in the atmosphere. A generalized feature observed in the airborne strains here isolated (with a single exception) is their high halotolerance, since they mostly grew very well in culture media containing 7% NaCl, thus suggesting a marine origin. This has been previously proposed for Streptomyces species isolated from atmospheric precipitations in Northern Spain (Braña et al., 2015; Sarmiento-Vizcaíno et al., 2016).
Air mass backward trajectories analysis of this precipitation event revealed two main sources, oceanic (mainly from the Arctic Ocean, Greenland and Iceland) and continental (Canada) depending on the altitude. Mixing of the different air layers during the travel was observed. All air masses crossed the Atlantic Ocean and arrived to continental Europe after 4 days, reaching the North of Spain where samples were collected by atmospheric precipitation. Consistent with estimated air mass backward trajectories all isolated strains are similar to previously isolated species from highly diverse environments, either marine or terrestrial, mainly from the North hemisphere. The results of Blast search from 16S rRNA partial sequences herein provided revealed the presence of 29 strains belonging to 20–25 different Streptomyces species. Bearing in mind that the currently estimated number of Streptomyces species is of 550–823, the number of species isolated during this hail event represents a non-negligible 3–4% of all Streptomyces species known so far in our planet. This fact suggests that the presence in the atmosphere could be a generalized phenomenon within the Streptomyces genus and is in agreement with our previously stablished atmospheric dispersion model (Sarmiento-Vizcaíno et al., 2016). This model has recently received further support from culture-independent report from precipitations in Japan. That work also shows seasonal variations of microbial communities in the atmosphere in correlation with estimated air mass trajectories (Hiraoka et al., 2017).
Overall, the most relevant feature of the atmospheric-derived Streptomyces strains here studied is that they represent a striking great reservoir of structurally diverse bioactive compounds (Table 3). One hundred molecules have been identified, and for 60 of them different biological activities, mainly antimicrobial (antibacterial, antifungal, and antiviral) and antitumor properties, have been previously described. Interestingly 38 potentially bioactive natural products have not been identified and their possible novelty is the subject of current active research. During the time of writing this manuscript a new natural product was identified, after purification and NMR structure elucidation, in one of the strains here isolated (unpublished results). The number of produced secondary metabolites for these strains is estimated to be much higher than the one presented here, since only diffusible apolar molecules produced in a unique culture condition (R5A medium, 28°C) were analyzed so far, and possible diffusible polar or volatile products were not analyzed. Even more, most of the Streptomyces metabolic abilities are mainly hidden, not expressed under standard culture conditions. This silent (or cryptic) potential represents most of the metabolome (Reen et al., 2015).
Conclusion
Our findings highlight the relevance of the atmosphere as a novel source of highly diverse Streptomyces species able to produce an incredible reservoir of natural products, which has been overlooked so far. Atmospheric precipitations might represent a relevant unexplored environment for discovering bioactive natural products with pharmacological and biotechnological interest.
Author contributions
GB isolated the strains and analyzed the air masses backward trajectories. AS-V performed the taxonomic identification and phylogenetic analyses of the strains. AS-V and JE conducted the bioactivity assays. AS-V and AB analyzed the compounds by LC-UV. JM and FR performed the metabolite profiling analysis and identified the compounds produced by LC-MS. GB wrote the manuscript which has been revised and approved by all the authors. LG and GB conceived and coordinated the project.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors want to thank the TBR group from the Universidad de Oviedo (GRUPIN14-140) for financial support and to Julian Davies for sending us the Streptomyces 85E strain. We also thank Victor González and Manuel Mora from the Oviedo Meteorological Observatory (AEMET), for their assistance with the NOAA analysis. We are also grateful to José L. Martínez and Daniel Serna (Servicios científico-técnicos, edificio Severo Ochoa, Universidad de Oviedo) for their great help in strains identification. This is a contribution of the Asturias Marine Observatory.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00773/full#supplementary-material
UV 210 nm chromatograms corresponding to all samples.
References
- Abdelfattah M. S., Kazufumi T., Ishibashi M. (2010). Izumiphenazines A-C: isolation and structure elucidation of phenazine derivatives from Streptomyces sp. IFM 11204. J. Nat. Prod. 73, 1999–2002. 10.1021/np100400t [DOI] [PubMed] [Google Scholar]
- Abe N., Nakakita Y., Nakamura T., Enoki N., Uchida H., Munekata M. (1993). Novel antitumor antibiotics, saptomycins. I. Taxonomy of the producing organism, fermentation, HPLC analysis and biological activities. J. Antibiot. 46, 1530–1535. 10.7164/antibiotics.46.1530 [DOI] [PubMed] [Google Scholar]
- Amato P., Joly M., Besaury L., Oudart A., Taib N., Moné A. I., et al. (2017). Active microorganisms thrive among extremely diverse communities in cloud water. PLoS ONE 12:e0182869. 10.1371/journal.pone.0182869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato P., Parazols M., Sancelme M., Laj P., Mailhot G., Delort A. M. (2007). Microorganisms isolated from the water phase of tropospheric clouds at the Puy de Dôme: major groups and growth abilities at low temperatures. FEMS Microbiol. Ecol. 59, 242–254. 10.1111/j.1574-6941.2006.00199.x [DOI] [PubMed] [Google Scholar]
- Aoki Y., Matsumoto D., Kawaide H., Natsume M. (2011). Physiological role of germicidins in spore germination and hyphal elongation in Streptomyces coelicolor A3(2). J. Antibiot. 64, 607–611. 10.1038/ja.2011.59 [DOI] [PubMed] [Google Scholar]
- Arahal D. R., Sánchez E., Macián M. C., Garay E. (2008). Value of recN sequences for species identification and as a phylogenetic marker within the family “Leuconostocaceae.” Int. Microbiol. Off. J. Span. Soc. Microbiol. 11, 33–39. 10.2436/20.1501.01.42 [DOI] [PubMed] [Google Scholar]
- Argoudelis A. D., Brinkley T. A., Brodasky T. F., Buege J. A., Meyer H. F., Mizsak S. A. (1982). Paulomycins A and B. Isolation and characterization. J. Antibiot. 35, 285–294. 10.7164/antibiotics.35.285 [DOI] [PubMed] [Google Scholar]
- Behie S. W., Bonet B., Zacharia V. M., McClung D. J., Traxler M. F. (2016). Molecules to ecosystems: actinomycete natural products In situ. Front. Microbiol. 7:2149. 10.3389/fmicb.2016.02149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley S. D., Chater K. F., Cerdeño-Tárraga A. M., Challis G. L., Thomson N. R., James K. D., et al. (2002). Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147. 10.1038/417141a [DOI] [PubMed] [Google Scholar]
- Bertasso M., Holzenkämpfer M., Zeeck A., Stackebrandt E., Beil W., Fiedler H.-P. (2003). Ripromycin and other polycyclic macrolactams from Streptomyces sp. Tü 6239: taxonomy, fermentation, isolation and biological properties. J. Antibiot. 56, 364–371. 10.7164/antibiotics.56.364 [DOI] [PubMed] [Google Scholar]
- Bihlmaier C., Welle E., Hofmann C., Welzel K., Vente A., Breitling E., et al. (2006). Biosynthetic gene cluster for the polyenoyltetramic acid alpha-lipomycin. Antimicrob. Agents Chemother. 50, 2113–2121. 10.1128/AAC.00007-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braña A. F., Fiedler H.-P., Nava H., González V., Sarmiento-Vizcaíno A., et al. (2015). Two Streptomyces species producing antibiotic, antitumor, and anti-inflammatory compounds are widespread among intertidal macroalgae and deep-sea coral reef invertebrates from the central Cantabrian Sea. Microb. Ecol. 69, 512–524. 10.1007/s00248-014-0508-0 [DOI] [PubMed] [Google Scholar]
- Braña A. F., Rodríguez M., Pahari P., Rohr J., García L. A., Blanco G. (2014). Activation and silencing of secondary metabolites in Streptomyces albus and Streptomyces lividans after transformation with cosmids containing the thienamycin gene cluster from Streptomyces cattleya. Arch. Microbiol. 196, 345–355. 10.1007/s00203-014-0977-z [DOI] [PubMed] [Google Scholar]
- Braña A. F., Sarmiento-Vizcaíno A., Osset M., Pérez-Victoria I., Martín J., de Pedro N., et al. (2017a). Lobophorin K, a new natural product with cytotoxic activity produced by Streptomyces sp. M-207 associated with the deep-sea coral Lophelia pertusa. Mar. Drugs 15:144. 10.3390/md15050144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braña A. F., Sarmiento-Vizcaíno A., Pérez-Victoria I., Otero L., Fernández J., Palacios J. J., et al. (2017b). Branimycins B and C, antibiotics produced by the abyssal actinobacterium Pseudonocardia carboxydivorans M-227. J. Nat. Prod. 80, 569–573. 10.1021/acs.jnatprod.6b01107 [DOI] [PubMed] [Google Scholar]
- Brötz E., Kulik A., Vikineswary S., Lim C. T., Tan G. Y. A., Zinecker H., et al. (2011). Phenelfamycins G and H, new elfamycin-type antibiotics produced by Streptomyces albospinus Acta 3619. J. Antibiot. 64, 257–266. 10.1038/ja.2010.170 [DOI] [PubMed] [Google Scholar]
- Buchanan G. O., Regentin R., Piagentini M., Rascher A., McDaniel R., Galazzo J. L., et al. (2005). Production of 8-demethylgeldanamycin and 4,5-epoxy-8-demethylgeldanamycin from a recombinant strain of Streptomyces hygroscopicus. J. Nat. Prod. 68, 607–610. 10.1021/np0496744 [DOI] [PubMed] [Google Scholar]
- Chan K.-G., Puthucheary S. D., Chan X. Y., Yin W. F., Wong C. S., Too W. S. S., et al. (2011). Quorum sensing in Aeromonas species isolated from patients in Malaysia. Curr. Microbiol. 62, 167–172. 10.1007/s00284-010-9689-z [DOI] [PubMed] [Google Scholar]
- Chen C., Wang J., Guo H., Hou W., Yang N., Ren B., et al. (2013). Three antimycobacterial metabolites identified from a marine-derived Streptomyces sp. MS100061. Appl. Microbiol. Biotechnol. 97, 3885–3892. 10.1007/s00253-012-4681-0 [DOI] [PubMed] [Google Scholar]
- Cheng Y. Q. (2006). Deciphering the biosynthetic codes for the potent anti-SARS-CoV cyclodepsipeptide valinomycin in Streptomyces tsusimaensis ATCC 15141. Chembiochem Eur. J. Chem. Biol. 7, 471–477. 10.1002/cbic.200500425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chronáková A., Kristufek V., Tichý M., Elhottová D. (2010). Biodiversity of streptomycetes isolated from a succession sequence at a post-mining site and their evidence in Miocene lacustrine sediment. Microbiol. Res. 165, 594–608. 10.1016/j.micres.2009.10.002 [DOI] [PubMed] [Google Scholar]
- David L., Leal Ayala H., Tabet J. C. (1985). Abierixin, a new polyether antibiotic. Production, structural determination and biological activities. J. Antibiot. 38, 1655–1663. 10.7164/antibiotics.38.1655 [DOI] [PubMed] [Google Scholar]
- Desouky S. E., Shojima A., Singh R. P., Matsufuji T., Igarashi Y., Suzuki T., et al. (2015). Cyclodepsipeptides produced by actinomycetes inhibit cyclic-peptide-mediated quorum sensing in Gram-positive bacteria. FEMS Microbiol. Lett. 362:fnv109. 10.1093/femsle/fnv109 [DOI] [PubMed] [Google Scholar]
- Ding L., Maier A., Fiebig H.-H., Lin W.-H., Peschel G., Hertweck C. (2012). Kandenols A-E, eudesmenes from an endophytic Streptomyces sp. of the mangrove tree Kandelia candel. J. Nat. Prod. 75, 2223–2227. 10.1021/np300387n [DOI] [PubMed] [Google Scholar]
- Chapman and Hall/CRC (2015). Dictionary of Marine Natural Products. Available online at: http://dnp.chemnetbase.com
- Dodzin M. E., Vinogradova K. A., Kotova I. B. (1998). [Novel L-glutamate oxidase producing organisms: Streptomyces litmocidini and Streptomyces cremeus]. Antibiot. Khimioterapiia Antibiot. Chemoterapy Sic. 43, 7–13. [PubMed] [Google Scholar]
- El-Naggar Nel-A., Deraz S. F., Soliman H. M., El-Deeb N. M., El-Ewasy S. M. (2016). Purification, characterization, cytotoxicity and anticancer activities of L-asparaginase, anti-colon cancer protein, from the newly isolated alkaliphilic Streptomyces fradiae NEAE-82. Sci. Rep. 6:32926 10.1038/srep32926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T., Yonehara H. (1970). Chemical studies on blastmycin. 3. Gas-liquid chromatography of antimycin A-blastmycin antibiotics. J. Antibiot. 23, 91–95. 10.7164/antibiotics.23.91 [DOI] [PubMed] [Google Scholar]
- Fiedler H. P., Bruntner C., Bull A. T., Ward A. C., Goodfellow M., Potterat O., et al. (2005). Marine actinomycetes as a source of novel secondary metabolites. Antonie Van Leeuwenhoek 87, 37–42. 10.1007/s10482-004-6538-8 [DOI] [PubMed] [Google Scholar]
- Fiedler H. P., Kulik A., Schüz T. C., Volkmann C., Zeeck A. (1994). Biosynthetic capacities of actinomycetes. 2. Juglomycin Z, a new naphthoquinone antibiotic from Streptomyces tendae. J. Antibiot. 47, 1116–1122. 10.7164/antibiotics.47.1116 [DOI] [PubMed] [Google Scholar]
- Fujita K. I., Kiso T., Usuki Y., Tanaka T., Taniguchi M. (2004). UK-2A, B, C and D, novel antifungal antibiotics from streptomyces sp. 517-02 VI (3). Role of substituents on dilactone ring of UK-2A and antimycin A3 against generation of reactive oxygen species in porcine renal proximal tubule LLC-PK1 cells. J. Antibiot. 57, 687–690. 10.7164/antibiotics.57.687 [DOI] [PubMed] [Google Scholar]
- Fukushima K., Yazawa K., Arai T. (1973). Biological activities of albonoursin. J. Antibiot. 26, 175–176. 10.7164/antibiotics.26.175 [DOI] [PubMed] [Google Scholar]
- Gao C., Lin L., Long B., Chen Y., He B., Sun H., et al. (2014). A new diketopiperazine from the gorgonian coral Menella kanisa. Nat. Prod. Res. 28, 473–476. 10.1080/14786419.2013.879134 [DOI] [PubMed] [Google Scholar]
- González Holgado G., Castro Rodríguez J., Cañedo Hernández L. M., Díaz M., Fernández-Abalos J. M., Trujillano I., et al. (2002). Radamycin, a novel thiopeptide produced by streptomyces sp. RSP9. I. Taxonomy, fermentation, isolation and biological activities. J. Antibiot. 55, 383–390. 10.7164/antibiotics.55.383 [DOI] [PubMed] [Google Scholar]
- Goodfellow M., Kumar Y., Labeda D. P., Sembiring L. (2007). The Streptomyces violaceusniger clade: a home for Streptomycetes with rugose ornamented spores. Antonie Van Leeuwenhoek 92, 173–199. 10.1007/s10482-007-9146-6 [DOI] [PubMed] [Google Scholar]
- Hanquet G., Salom-Roig X., Lanners S. (2016). New insights into the synthesis and biological activity of the pamamycin macrodiolides. Chimia 70, 20–28. 10.2533/chimia.2016.20 [DOI] [PubMed] [Google Scholar]
- Hashimoto M., Katsura H., Kato R., Kawaide H., Natsume M. (2011). Effect of pamamycin-607 on secondary metabolite production by Streptomyces spp. Biosci. Biotechnol. Biochem. 75, 1722–1726. 10.1271/bbb.110251 [DOI] [PubMed] [Google Scholar]
- Hayashi K., Hashimoto M., Shigematsu N., Nishikawa M., Ezaki M., Yamashita M., et al. (1992). WS9326A, a novel tachykinin antagonist isolated from Streptomyces violaceusniger no. 9326. I. Taxonomy, fermentation, isolation, physico-chemical properties and biological activities. J. Antibiot. 45, 1055–1063. 10.7164/antibiotics.45.1055 [DOI] [PubMed] [Google Scholar]
- Hiraoka S., Miyahara M., Fujii K., Machiyama A., Iwasaki W. (2017). Seasonal analysis of microbial communities in precipitation in the greater Tokyo area, Japan. Front. Microbiol. 8:1506. 10.3389/fmicb.2017.01506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann C., Schneider K., Bruntner C., Irran E., Nicholson G., Bull A. T., et al. (2009). Caboxamycin, a new antibiotic of the benzoxazole family produced by the deep-sea strain Streptomyces sp. NTK 937. J. Antibiot. 62, 99–104. 10.1038/ja.2008.24 [DOI] [PubMed] [Google Scholar]
- Imamura N., Nishijima M., Adachi K., Sano H. (1993). Novel antimycin antibiotics, urauchimycins A and B, produced by marine actinomycete. J. Antibiot. 46, 241–246. 10.7164/antibiotics.46.241 [DOI] [PubMed] [Google Scholar]
- Isaka M., Jaturapat A., Kramyu J., Tanticharoen M., Thebtaranonth Y. (2002). Potent in vitro antimalarial activity of metacycloprodigiosin isolated from Streptomyces spectabilis BCC 4785. Antimicrob. Agents Chemother. 46, 1112–1113. 10.1128/AAC.46.4.1112-1113.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiyama T., Endo T., Otake N., Yonehara H. (1976). Deisovalerylblastmycin produced by Streptomyces sp. J. Antibiot. 29, 804–808. 10.7164/antibiotics.29.804 [DOI] [PubMed] [Google Scholar]
- Islam M. T., Laatsch H., von Tiedemann A. (2016). Inhibitory effects of macrotetrolides from Streptomyces spp. on Zoosporogenesis and motility of Peronosporomycete zoospores are likely linked with enhanced ATPase activity in mitochondria. Front. Microbiol. 7:1824. 10.3389/fmicb.2016.01824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen H. L. (1931). Contributions to our knowledge of the Actinomycetales. II. The definition and subdivision of the genus Actinomyces, with a preliminary account of Australian soil Actinomycetes. Proc. Linn. Soc. New South Wales 56, 345–370. [Google Scholar]
- Jiang Z. D., Jensen P. R., Fenical W. (1999). Lobophorins A and B, new antiinflammatory macrolides produced by a tropical marine bacterium. Bioorg. Med. Chem. Lett. 9, 2003–2006. 10.1016/S0960-894X(99)00337-6 [DOI] [PubMed] [Google Scholar]
- Kawamura T., Fujimaki T., Hamanaka N., Torii K., Kobayashi H., Takahashi Y., et al. (2010). Isolation and structure elucidation of a novel androgen antagonist, arabilin, produced by Streptomyces sp. MK756-CF1. J. Antibiot. 63, 601–605. 10.1038/ja.2010.98 [DOI] [PubMed] [Google Scholar]
- Killham K., Firestone M. K. (1984). Salt stress control of intracellular solutes in Streptomycetes indigenous to saline soils. Appl. Environ. Microbiol. 47, 301–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. B., Falconer C., Williams E., Goodfellow M. (1998). Streptomyces thermocarboxydovorans sp. nov. and Streptomyces thermocarboxydus sp. nov., two moderately thermophilic carboxydotrophic species from soil. Int. J. Syst. Bacteriol. 48(Pt 1), 59–68. 10.1099/00207713-48-1-59 [DOI] [PubMed] [Google Scholar]
- Kim S. B., Goodfellow M. (2002). Streptomyces thermospinisporus sp. nov., a moderately thermophilic carboxydotrophic streptomycete isolated from soil. Int. J. Syst. Evol. Microbiol. 52, 1225–1228. 10.1099/00207713-52-4-1225 [DOI] [PubMed] [Google Scholar]
- Kobinata K., Koshino H., Kudo T., Isono K., Osada H. (1993). RK-397, a new oxo pentaene antibiotic. J. Antibiot. 46, 1616–1618. 10.7164/antibiotics.46.1616 [DOI] [PubMed] [Google Scholar]
- Kohno J., Nishio M., Kawano K., Nakanishi N., Suzuki S., Uchida T., et al. (1996). TMC-1 A, B, C and D, new antibiotics of the manumycin group produced by Streptomyces sp. taxonomy, production, isolation, physico-chemical properties, structure elucidation and biological properties. J. Antibiot. 49, 1212–1220. 10.7164/antibiotics.49.1212 [DOI] [PubMed] [Google Scholar]
- Komiyama K., Funayama S., Anraku Y., Ishibashi M., Takahashi Y., Kawakami T., et al. (1991). A new antibiotic, okicenone. I. taxonomy, fermentation, isolation and biological characteristics. J. Antibiot. 44, 814–818. 10.7164/antibiotics.44.814 [DOI] [PubMed] [Google Scholar]
- Kumar S. N., Nambisan B., Sundaresan A., Mohandas C., Anto R. J. (2014). Isolation and identification of antimicrobial secondary metabolites from Bacillus cereus associated with a rhabditid entomopathogenic nematode. Ann. Microbiol. 64, 209–218. 10.1007/s13213-013-0653-6 [DOI] [Google Scholar]
- Labeda D. P., Doroghazi J. R., Ju K. S., Metcalf W. W. (2014). Taxonomic evaluation of Streptomyces albus and related species using multilocus sequence analysis and proposals to emend the description of Streptomyces albus and describe Streptomyces pathocidini sp. nov. Int. J. Syst. Evol. Microbiol. 64, 894–900. 10.1099/ijs.0.058107-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J. W. F., Ser H. L., Duangjai A., Saokaew S., Bukhari S. I., Khan T. M., et al. (2017). Streptomyces colonosanans sp. nov., A novel Actinobacterium isolated from malaysia mangrove soil exhibiting antioxidative activity and cytotoxic potential against human colon cancer cell lines. Front. Microbiol. 8:877. 10.3389/fmicb.2017.00877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach B. E., Calhoun K. M., Johnson L. E., Teeters C. M., Jackson W. G. (1953). Chartreusin, a new antibiotic produced by Streptomyces chartreusis, a new species. J. Am. Chem. Soc. 75, 4011–4012. 10.1021/ja01112a040 [DOI] [Google Scholar]
- Lin Z., Zachariah M. M., Marett L., Hughen R. W., Teichert R. W., Concepcion G. P., et al. (2014). Griseorhodins D-F, neuroactive intermediates and end products of post-PKS tailoring modification in Griseorhodin biosynthesis. J. Nat. Prod. 77, 1224–1230. 10.1021/np500155d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loqman S., Bouizgarne B., Barka E. A., Clément C., von Jan M., Spröer C., et al. (2009). Streptomyces thinghirensis sp. nov., isolated from rhizosphere soil of Vitis vinifera. Int. J. Syst. Evol. Microbiol. 59, 3063–3067. 10.1099/ijs.0.008946-0 [DOI] [PubMed] [Google Scholar]
- Lu Y., Li S., Zhou D., Zhang Y. (2014). [Isolation and identification of termitarium antagonistic actinomycetes BYC 01 and its active metabolites]. Wei Sheng Wu Xue Bao 54, 754–759. [PubMed] [Google Scholar]
- Ma J., Huang H., Xie Y., Liu Z., Zhao J., Zhang C., et al. (2017). Biosynthesis of ilamycins featuring unusual building blocks and engineered production of enhanced anti-tuberculosis agents. Nat. Commun. 8:391. 10.1038/s41467-017-00419-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejewska M., Adam D., Martinet L., Naômé A., Całusinska M., Delfosse P., et al. (2016). A phenotypic and genotypic analysis of the antimicrobial potential of cultivable streptomyces isolated from cave moonmilk deposits. Front. Microbiol. 7:1455. 10.3389/fmicb.2016.01455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejewska M., Pessi I. S., Arguelles-Arias A., Noirfalise P., Luis G., Ongena M., et al. (2015). Streptomyces lunaelactis sp. nov., a novel ferroverdin A-producing Streptomyces species isolated from a moonmilk speleothem. Antonie Van Leeuwenhoek 107, 519–531. 10.1007/s10482-014-0348-4 [DOI] [PubMed] [Google Scholar]
- Matsumoto N., Tsuchida T., Maruyama M., Sawa R., Kinoshita N., Homma Y., et al. (1996). Lactonamycin, a new antimicrobial antibiotic produced by Streptomyces rishiriensis. J. Antibiot. 49, 953–954. 10.7164/antibiotics.49.953 [DOI] [PubMed] [Google Scholar]
- Moon S. S., Hwang W. H., Chung Y. R., Shin J. (2003). New cytotoxic bafilomycin C1-amide produced by Kitasatospora cheerisanensis. J. Antibiot. 56, 856–861. 10.7164/antibiotics.56.856 [DOI] [PubMed] [Google Scholar]
- Moree W. J., McConnell O. J., Nguyen D. D., Sanchez L. M., Yang Y. L., Zhao X., et al. (2014). Microbiota of healthy corals are active against fungi in a light-dependent manner. ACS Chem. Biol. 9, 2300–2308. 10.1021/cb500432j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T., Ando M., Takagi K. (1994). Staurosporine and its derivatives enhance f-Met-Leu-Phe-induced superoxide production via phospholipase D activation in human polymorphonuclear leukocytes. Int. J. Clin. Pharmacol. Ther. 32, 422–428. [PubMed] [Google Scholar]
- Ni S., Wu L., Wang H., Gan M., Wang Y., He W., et al. (2011). Thiazinogeldanamycin, a new geldanamycin derivative produced by Streptomyces hygroscopicus 17997. J. Microbiol. Biotechnol. 21, 599–603. [PubMed] [Google Scholar]
- Novak E. (1988). Treating Chlamydia Infections With Paulomycin. Patent PCT/US1987/002420: The Upjohn Company.
- Omura S., Nakagawa A., Fukamachi N., Otoguro K., Kobayashi B. (1986). Aggreceride, a new platelet aggregation inhibitor from Streptomyces. J. Antibiot. 39, 1180–1181. 10.7164/antibiotics.39.1180 [DOI] [PubMed] [Google Scholar]
- Osada H., Koshino H., Kudo T., Onose R., Isono K. (1992). A new inhibitor of protein kinase C, RK-1409 (7-oxostaurosporine). I. Taxonomy and biological activity. J. Antibiot. 45, 189–194. 10.7164/antibiotics.45.189 [DOI] [PubMed] [Google Scholar]
- Paululat T., Kulik A., Hausmann H., Karagouni A. D., Zinecker H., Imhoff J. F., et al. (2010). Grecocyclines: new angucyclines from Streptomyces sp. Acta 1362. Eur. J. Org. Chem. 2010, 2344–2350. 10.1002/ejoc.201000054 [DOI] [Google Scholar]
- Pérez-Victoria I., Martín J., Reyes F. (2016). Combined LC/UV/MS and NMR strategies for the dereplication of marine natural products. Planta Med. 82, 857–871. 10.1055/s-0042-101763 [DOI] [PubMed] [Google Scholar]
- Perkins J. B., Guterman S. K., Howitt C. L., Williams V. E., Pero J. (1990). Streptomyces genes involved in biosynthesis of the peptide antibiotic valinomycin. J. Bacteriol. 172, 3108–3116. 10.1128/jb.172.6.3108-3116.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrović S., Vasić V., Mitrović T., Lazović S., Leskovac A. (2017). The impact of concentration and administration time on the radiomodulating properties of undecylprodigiosin in vitro. Arh. Hig. Rada Toksikol. 68, 1–8. 10.1515/aiht-2017-68-2897 [DOI] [PubMed] [Google Scholar]
- Phay N., Yada H., Higashiyama T., Yokota A., Ichihara A., Tomita F. (1996). NP-101A, antifungal antibiotic from Streptomyces aurantiogriseus NPO-101. J. Antibiot. 49, 703–705. 10.7164/antibiotics.49.703 [DOI] [PubMed] [Google Scholar]
- Phongsopitanun W., Thawai C., Suwanborirux K., Kudo T., Ohkuma M., Tanasupawat S. (2014). Streptomyces chumphonensis sp. nov., isolated from marine sediments. Int. J. Syst. Evol. Microbiol. 64, 2605–2610. 10.1099/ijs.0.062992-0 [DOI] [PubMed] [Google Scholar]
- Pimentel-Elardo S. M., Kozytska S., Bugni T. S., Ireland C. M., Moll H., Hentschel U. (2010). Anti-parasitic compounds from Streptomyces sp. strains isolated from Mediterranean sponges. Mar. Drugs 8, 373–380. 10.3390/md8020373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raveh A., Delekta P. C., Dobry C. J., Peng W., Schultz P. J., Blakely P. K., et al. (2013). Discovery of potent broad spectrum antivirals derived from marine actinobacteria. PLoS ONE 8:e82318. 10.1371/journal.pone.0082318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray L., Suar M., Pattnaik A. K., Raina V. (2013). Streptomyces chilikensis sp. nov., a halophilic streptomycete isolated from brackish water sediment. Int. J. Syst. Evol. Microbiol. 63, 2757–2764. 10.1099/ijs.0.046284-0 [DOI] [PubMed] [Google Scholar]
- Reen F. J., Romano S., Dobson A. D. W., O'Gara F. (2015). The sound of silence: activating silent biosynthetic gene clusters in marine microorganisms. Mar. Drugs 13, 4754–4783. 10.3390/md13084754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong X., Doroghazi J. R., Cheng K., Zhang L., Buckley D. H., Huang Y. (2013). Classification of Streptomyces phylogroup pratensis (Doroghazi and Buckley, 2010) based on genetic and phenotypic evidence, and proposal of Streptomyces pratensis sp. nov. Syst. Appl. Microbiol. 36, 401–407. 10.1016/j.syapm.2013.03.010 [DOI] [PubMed] [Google Scholar]
- Roy S. K., Inouye Y., Nakamura S., Furukawa J., Okuda S. (1987). Isolation, structural elucidation and biological properties of neoenactins B1, B2, M1 and M2, neoenactin congeners. J. Antibiot. 40, 266–274. 10.7164/antibiotics.40.266 [DOI] [PubMed] [Google Scholar]
- Russell D. W., Sambrook J. F. (2001). Molecular Cloning: A Laboratory Manual, 3rd Edn. New York, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Sánchez López J. M., Martínez Insua M., Pérez Baz J., Fernández Puentes J. L., Cañedo Hernández L. M. (2003). New cytotoxic indolic metabolites from a marine Streptomyces. J. Nat. Prod. 66, 863–864. 10.1021/np0204444 [DOI] [PubMed] [Google Scholar]
- Santos O. C., Soares A. R., Machado F. L. S., Romanos M. T. V., Muricy G., Giambiagi-deMarval M., et al. (2015). Investigation of biotechnological potential of sponge-associated bacteria collected in Brazilian coast. Lett. Appl. Microbiol. 60, 140–147. 10.1111/lam.12347 [DOI] [PubMed] [Google Scholar]
- Sarmiento-Vizcaíno A., Braña A. F., González V., Nava H., Molina A., Llera E., et al. (2016). Atmospheric dispersal of bioactive Streptomyces albidoflavus strains among terrestrial and marine environments. Microb. Ecol. 71, 375–386. 10.1007/s00248-015-0654-z [DOI] [PubMed] [Google Scholar]
- Sarmiento-Vizcaíno A., Braña A. F., Pérez-Victoria I., Martín J., de Pedro N., de la Cruz M., et al. (2017a). Paulomycin G, a new natural product with cytotoxic activity against tumor cell lines produced by deep-sea sediment derived Micromonospora matsumotoense M-412 from the Avilés Canyon in the Cantabrian Sea. Mar. Drugs 15:271. 10.3390/md15090271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmiento-Vizcaíno A., González V., Braña A. F., Molina A., Acuña J. L., García L. A., et al. (2015). Myceligenerans cantabricum sp. nov., a barotolerant actinobacterium isolated from a deep cold-water coral. Int. J. Syst. Evol. Microbiol. 65, 1328–1334. 10.1099/ijs.0.000107 [DOI] [PubMed] [Google Scholar]
- Sarmiento-Vizcaíno A., González V., Braña A. F., Palacios J. J., Otero L., Fernández J., et al. (2017b). Pharmacological potential of phylogenetically diverse actinobacteria isolated from deep-sea coral ecosystems of the submarine Avilés Canyon in the Cantabrian Sea. Microb. Ecol. 73, 338–352. 10.1007/s00248-016-0845-2 [DOI] [PubMed] [Google Scholar]
- Šantl-Temkiv T., Finster K., Dittmar T., Hansen B. M., Thyrhaug R., Nielsen N. W., et al. (2013). Hailstones: a window into the microbial and chemical inventory of a storm cloud. PLoS ONE 8:e53550. 10.1371/journal.pone.0053550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleissner C., Pérez M., Losada A., Rodríguez P., Crespo C., Zúñiga P., et al. (2011). Antitumor actinopyranones produced by Streptomyces albus POR-04-15-053 isolated from a marine sediment. J. Nat. Prod. 74, 1590–1596. 10.1021/np200196j [DOI] [PubMed] [Google Scholar]
- Schnur R. C., Corman M. L., Gallaschun R. J., Cooper B. A., Dee M. F., Doty J. L., et al. (1995). Inhibition of the oncogene product p185erbB-2 in vitro and in vivo by geldanamycin and dihydrogeldanamycin derivatives. J. Med. Chem. 38, 3806–3812. 10.1021/jm00019a010 [DOI] [PubMed] [Google Scholar]
- Seipke R. F., Hutchings M. I. (2013). The regulation and biosynthesis of antimycins. Beilstein J. Org. Chem. 9, 2556–2563. 10.3762/bjoc.9.290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seipke R. F., Kaltenpoth M., Hutchings M. I. (2012). Streptomyces as symbionts: an emerging and widespread theme? FEMS Microbiol. Rev. 36, 862–876. 10.1111/j.1574-6976.2011.00313.x [DOI] [PubMed] [Google Scholar]
- Shaaban K. A., Wang X., Elshahawi S. I., Ponomareva L. V., Sunkara M., Copley G. C., et al. (2013). Herbimycins D-F, ansamycin analogues from Streptomyces sp. RM-7-15. J. Nat. Prod. 76, 1619–1626. 10.1021/np400308w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag P., Bhave S., Vartak A., Kulkarni-Almeida A., Mahajan G., Villanueva I., et al. (2015). Screening of microbial extracts for anticancer compounds using Streptomyces kinase inhibitor assay. Nat. Prod. Commun. 10, 1287–1291. [PubMed] [Google Scholar]
- Shigemori H., Bae M. A., Yazawa K., Sasaki T., Kobayashi J. (1992). Alteramide A, a new tetracyclic alkaloid from a bacterium Alteromonas sp. associated with the marine sponge Halichondria okadai. J. Org. Chem. 57, 4317–4320. 10.1021/jo00041a053 [DOI] [Google Scholar]
- Song X., Liu X., Ding X. (2017). Staurosporine scaffold-based rational discovery of the wild-type sparing reversible inhibitors of EGFR T790M gatekeeper mutant in lung cancer with analog-sensitive kinase technology. J. Mol. Recognit. 30:e2590. 10.1002/jmr.2590 [DOI] [PubMed] [Google Scholar]
- Songia S., Mortellaro A., Taverna S., Fornasiero C., Scheiber E. A., Erba E., et al. (1997). Characterization of the new immunosuppressive drug undecylprodigiosin in human lymphocytes: retinoblastoma protein, cyclin-dependent kinase-2, and cyclin-dependent kinase-4 as molecular targets. J. Immunol. Baltim. Md 158, 3987–3995. [PubMed] [Google Scholar]
- Stein A. F., Draxler R. R., Rolph G. D., Stunder B. J. B., Cohen M. D., Ngan F. (2015). NOAA's HYSPLIT atmospheric transport and dispersion modeling system. Bull. Am. Meteorol. Soc. 96, 2059–2077. 10.1175/BAMS-D-14-00110.1 [DOI] [Google Scholar]
- Stroshane R. M., Chan J. A., Rubalcaba E. A., Garretson A. L., Aszalos A. A., Roller P. P. (1979). Isolation and structure elucidation of a novel griseorhodin. J. Antibiot. 32, 197–204. 10.7164/antibiotics.32.197 [DOI] [PubMed] [Google Scholar]
- Subramani R., Aalbersberg W. (2013). Culturable rare Actinomycetes: diversity, isolation and marine natural product discovery. Appl. Microbiol. Biotechnol. 97, 9291–9321. 10.1007/s00253-013-5229-7 [DOI] [PubMed] [Google Scholar]
- Sudha S., Masilamani S. M. (2012). Characterization of cytotoxic compound from marine sediment derived actinomycete Streptomyces avidinii strain SU4. Asian Pac. J. Trop. Biomed. 2, 770–773. 10.1016/S2221-1691(12)60227-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supong K., Thawai C., Choowong W., Kittiwongwattana C., Thanaboripat D., Laosinwattana C., et al. (2016). Antimicrobial compounds from endophytic Streptomyces sp. BCC72023 isolated from rice (Oryza sativa L.). Res. Microbiol. 167, 290–298. 10.1016/j.resmic.2016.01.004 [DOI] [PubMed] [Google Scholar]
- Tadtong S., Meksuriyen D., Tanasupawat S., Isobe M., Suwanborirux K. (2007). Geldanamycin derivatives and neuroprotective effect on cultured P19-derived neurons. Bioorg. Med. Chem. Lett. 17, 2939–2943. 10.1016/j.bmcl.2006.12.041 [DOI] [PubMed] [Google Scholar]
- Takatsu T., Ohtsuki M., Muramatsu A., Enokita R., Kurakata S. I. (2000). Reblastatin, a novel benzenoid ansamycin-type cell cycle inhibitor. J. Antibiot. 53, 1310–1312. 10.7164/antibiotics.53.1310 [DOI] [PubMed] [Google Scholar]
- Takeda Y., Mak V., Chang C. C., Chang C. J., Floss H. G. (1984). Biosynthesis of ketomycin. J. Antibiot. 37, 868–875. 10.7164/antibiotics.37.868 [DOI] [PubMed] [Google Scholar]
- Takimoto H., Machida K., Ueki M., Tanaka T., Taniguchi M. (1999). UK-2A, B, C and D, novel antifungal antibiotics from Streptomyces sp. 517-02. IV. Comparative studies of UK-2A with antimycin A3 on cytotoxic activity and reactive oxygen species generation in LLC-PK1 cells. J. Antibiot. 52, 480–484. 10.7164/antibiotics.52.480 [DOI] [PubMed] [Google Scholar]
- Tamaoki T., Nomoto H., Takahashi I., Kato Y., Morimoto M., Tomita F. (1986). Staurosporine, a potent inhibitor of phospholipid/Ca++dependent protein kinase. Biochem. Biophys. Res. Commun. 135, 397–402. 10.1016/0006-291X(86)90008-2 [DOI] [PubMed] [Google Scholar]
- Tang Y. Q., Sattler I., Thiericke R., Grabley S., Feng X. Z. (2000). Feigrisolides A, B, C and D, new lactones with antibacterial activities from Streptomyces griseus. J. Antibiot. 53, 934–943. 10.7164/antibiotics.53.934 [DOI] [PubMed] [Google Scholar]
- Tanouchi Y., Shichi H. (1987). Immunosuppressive effects of polynactins (tetranactin, trinactin and dinactin) on experimental autoimmune uveoretinitis in rats. Jpn. J. Ophthalmol. 31, 218–229. [PubMed] [Google Scholar]
- Tanouchi Y., Shichi H. (1988). Immunosuppressive and anti-proliferative effects of a macrotetrolide antibiotic, tetranactin. Immunology 63, 471–475. [PMC free article] [PubMed] [Google Scholar]
- Tatar D., Guven K., Spröer C., Klenk H.-P., Sahin N. (2014). Streptomyces iconiensis sp. nov. and Streptomyces smyrnaeus sp. nov., two halotolerant actinomycetes isolated from a salt lake and saltern. Int. J. Syst. Evol. Microbiol. 64, 3126–3133. 10.1099/ijs.0.062216-0 [DOI] [PubMed] [Google Scholar]
- Tomita F., Tamaoki T., Morimoto M., Fujimoto K. (1981). Trioxacarcins, novel antitumor antibiotics. I. Producing organism, fermentation and biological activities. J. Antibiot. 34, 1519–1524. 10.7164/antibiotics.34.1519 [DOI] [PubMed] [Google Scholar]
- Tresner H. D., Hayes J. A., Backus E. J. (1968). Differential tolerance of streptomycetes to sodium chloride as a taxonomic aid. Appl. Microbiol. 16, 1134–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukahara K., Toume K., Ito H., Ishikawa N., Ishibashi M. (2014). Isolation of β-indomycinone guided by cytotoxicity tests from Streptomyces sp. IFM11607 and revision of its double bond geometry. Nat. Prod. Commun. 9, 1327–1328. [PubMed] [Google Scholar]
- Uri J., Bekesi I. (1958). Flavofungin, a new crystalline antifungal antibiotic: origin and biological properties. Nature 181:908. 10.1038/181908a0 [DOI] [PubMed] [Google Scholar]
- Vijaya Kumar E. K., Kenia J., Mukhopadhyay T., Nadkarni S. R. (1999). Methylsulfomycin I, a new cyclic peptide antibiotic from a Streptomyces sp. HIL Y-9420704. J. Nat. Prod. 62, 1562–1564. 10.1021/np990088y [DOI] [PubMed] [Google Scholar]
- Wang W., Song T., Chai W., Chen L., Chen L., Lian X. Y., et al. (2017a). Rare polyene-polyol macrolides from mangrove-derived Streptomyces sp. ZQ4BG. Sci. Rep. 7:1703. 10.1038/s41598-017-01912-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Zhao Y., Yao S., Cui X., Pan W., Huang W., et al. (2017b). Nigericin inhibits epithelial ovarian cancer metastasis by suppressing the cell cycle and epithelial-mesenchymal transition. Biochem. Biokhimiia 82, 933–941. 10.1134/S0006297917080089 [DOI] [PubMed] [Google Scholar]
- Williamson N. R., Fineran P. C., Leeper F. J., Salmond G. P. (2006). The biosynthesis and regulation of bacterial prodiginines. Nat. Rev. Microbiol. 4, 887–899. 10.1038/nrmicro1531 [DOI] [PubMed] [Google Scholar]
- Wright L. F., Hopwood D. A. (1976). Actinorhodin is a chromosomally-determined antibiotic in Streptomyces coelicolar A3(2). J. Gen. Microbiol. 96, 289–297. 10.1099/00221287-96-2-289 [DOI] [PubMed] [Google Scholar]
- Wu C. Z., Jang J. H., Ahn J. S., Hong Y. S. (2012). New geldanamycin analogs from Streptomyces hygroscopicus. J. Microbiol. Biotechnol. 22, 1478–1481. 10.4014/jmb.1206.06026 [DOI] [PubMed] [Google Scholar]
- Yue C., Niu J., Liu N., Lü Y., Liu M., Li Y. (2016). Cloning and identification of the lobophorin biosynthetic gene cluster from marine Streptomyces olivaceus strain FXJ7.023. Pak. J. Pharm. Sci. 29, 287–293. [PubMed] [Google Scholar]
- Zhan Y., Zheng S. (2016). Efficient production of nonactin by Streptomyces griseus subsp. griseus. Can. J. Microbiol. 62, 711–714. 10.1139/cjm-2016-0248 [DOI] [PubMed] [Google Scholar]
- Zhang D., Nair M. G., Murry M., Zhang Z. (1997). Insecticidal activity of indanomycin. J. Antibiot. 50, 617–620. 10.7164/antibiotics.50.617 [DOI] [PubMed] [Google Scholar]
- Zhao X., Geng X., Chen C., Chen L., Jiao W., Yang C. (2012). Draft genome sequence of the marine actinomycete Streptomyces sulphureus L180, isolated from marine sediment. J. Bacteriol. 194:4482. 10.1128/JB.00900-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
UV 210 nm chromatograms corresponding to all samples.