Abstract
The phytohormone ethylene regulates many aspects of plant growth, development, and environmental responses. Much of the developmental regulation of ethylene responses in tomato (Lycopersicon esculentum) occurs at the level of hormone sensitivity. In an effort to understand the regulation of ethylene responses, we isolated and characterized tomato genes with sequence similarity to the Arabidopsis ETR1 (ethylene response 1) ethylene receptor. Previously, we isolated three genes that exhibit high similarity to ETR1 and to each other. Here we report the isolation of two additional genes, LeETR4 and LeETR5, that are only 42% and 40% identical to ETR1, respectively. Although the amino acids known to be involved in ethylene binding are conserved, LeETR5 lacks the histidine within the kinase domain that is predicted to be phosphorylated. This suggests that histidine kinase activity is not necessary for an ethylene response, because mutated forms of both LeETR4 and LeETR5 confer dominant ethylene insensitivity in transgenic Arabidopsis plants. Expression analysis indicates that LeETR4 accounts for most of the putative ethylene-receptor mRNA present in reproductive tissues, but, like LeETR5, it is less abundant in vegetative tissues. Taken together, ethylene perception in tomato is potentially quite complex, with at least five structurally divergent, putative receptor family members exhibiting significant variation in expression levels throughout development.
Ethylene, a gaseous phytohormone, plays an important regulatory role in such diverse plant developmental processes as fruit ripening, abscission, seed germination, stem elongation, and leaf and flower senescence (Abeles et al., 1992). Ethylene action can be regulated at the level of both hormone synthesis and sensitivity. The regulation of ethylene biosynthesis is especially well characterized (McKeon and Yang, 1987). Ethylene is synthesized from S-adenosyl-l-Met, which is converted to ACC by ACC synthase. This is followed by the conversion of ACC to ethylene by ACC oxidase. The biosynthetic pathway appears to be regulated at the level of ACC synthase gene transcription (Olson et al., 1991; Rottmann et al., 1991; Oetiker et al., 1997), and ACC synthase genes are differentially regulated in response to various developmental and environmental stimuli (Rottmann et al., 1991; Kieber and Ecker, 1993; Theologis, 1993). ACC oxidase activity, although present constitutively, is also regulated and may be responsible for the fine regulation of ethylene levels present in plant tissues (Kende, 1993).
Although the regulation of ethylene biosynthesis has been studied extensively, much less is known about the regulation of ethylene perception. Because ethylene is diffusible from the site of synthesis, ethylene biosynthesized in one part of the plant can affect other tissues as well. The ability of plants to regulate the diverse developmental and environmental responses to ethylene in a tissue-specific manner indicates that the perception of ethylene is a complex process and that ethylene perception must be regulated. For example, tomato (Lycopersicon esculentum) fruit undergoes a well-defined transition in ethylene sensitivity at the onset of ripening (Liu et al., 1985). Recent research has focused on the isolation of genes involved in the ethylene signal transduction pathway in both Arabidopsis and tomato. Several Arabidopsis mutants deficient in the classic triple response of etiolated seedlings to ethylene have been identified (Bleecker et al., 1988; Guzman and Ecker, 1990; Ecker, 1995; Roman et al., 1995; Kieber, 1997). Isolation of the mutant genes responsible for these phenotypes has enhanced our understanding of the mechanisms of ethylene perception and signal transduction (Chang et al., 1993; Kieber and Ecker, 1993; Lehman et al., 1996; Chao et al., 1997). Multiple mutations in one gene of Arabidopsis, ETR1 (ethylene response 1), result in dominant ethylene insensitivity (Bleecker et al., 1988). The ETR1 protein exhibits saturable, high-affinity binding of ethylene, and mutations in the hydrophobic amino-terminal domain reduce or abolish that binding (Schaller and Bleecker, 1995). Based on these results, it has been concluded that ETR1 acts as an ethylene receptor in Arabidopsis. Recently, it was shown that ETR1 is a member of a gene family consisting of five members: ETR1, ERS1, ETR2, EIN4, and ERS2 (Hua et al., 1995, 1998; Sakai et al., 1998). Mutations in the membrane-spanning domains of each of these genes result in dominant ethylene insensitivity (Hua et al., 1998; Sakai et al., 1998), indicating a likely role for all of these proteins in ethylene signal transduction. Loss-of-function mutants of the Arabidopsis ETR1, ERS1, ETR2, EIN4, and ERS2 genes did not have defects in ethylene responses; however, mutants in three or four of these genes have constitutive ethylene phenotypes in the absence of ethylene, indicating that these genes negatively regulate ethylene responses (Hua and Meyerowitz, 1998).
ETR1 exhibits significant sequence homology to a class of His kinases known as two-component regulators (Chang et al., 1993). In bacteria, these signal transduction systems consist of two proteins, a sensor and a response regulator, and mediate responses to a range of environmental stimuli (Parkinson, 1993). The Arabidopsis ETR1 protein exists as a membrane-associated dimer and can be divided into three domains (Chang et al., 1993; Schaller et al., 1995). The amino-terminal sensor domain contains three putative transmembrane segments within which all known mutations resulting in loss of ethylene sensitivity are located. This domain has been shown to bind ethylene when expressed in yeast, and the etr1-1 mutation abolishes ethylene binding (Schaller and Bleecker, 1995). The second domain exhibits homology to His kinases that, in bacterial two-component sensing systems, are autophosphorylated. This portion of the ETR1 protein has been shown to exhibit His kinase activity in vitro (Gamble et al., 1998). The third domain, the response regulator, may receive the phosphate from the His of the His kinase domain at an Asp residue (Chang et al., 1993).
Nr (Never ripe) is a semidominant mutant of tomato originally identified by the inability of its fruit to undergo ripening (Rick and Butler, 1956). Nr is now known to be ethylene insensitive in all tissues (Lanahan et al., 1994). For example, dark-grown seedlings do not show the “triple response” of shortened, thickened hypocotyls and roots and a pronounced apical hook when grown in the presence of ethylene. The mutant also exhibits defects in ethylene-regulated processes, such as greatly impaired pedicel abscission and significant delays in the onset of leaf and flower petal senescence. The mutation responsible for the Nr phenotype is a single-base change resulting in a Pro-to-Leu substitution in a protein, designated NR, that exhibits significant sequence similarity to ETR1 of Arabidopsis (Wilkinson et al., 1995). As in Arabidopsis, tomato contains a family of putative ethylene-receptor genes (Yen et al., 1995). Previously, we and others characterized three tomato gene family members, LeETR1, LeETR2, and NR, the first two of which contain response-regulator domains (Zhou et al., 1996a, 1996b; Lashbrook et al., 1998). In this paper, we describe the isolation of two additional tomato genes, designated LeETR4 and LeETR5, with sequence similarity to Arabidopsis ETR1 and tomato NR, bringing the number of tomato ETR1 homologs to five. LeETR4 and LeETR5 are divergent from the other tomato ETR1 homologs and exhibit structural features not seen in these proteins. The presence of multiple structurally divergent, putative ethylene receptors suggests that the regulation of ethylene responses may be more complex than previously recognized.
MATERIALS AND METHODS
Plant Material
Flowers from field-grown tomato (Lycopersicon esculentum cv Pearson and the Nr mutant) plants were tagged at anthesis, and the fruit was harvested at the reported days after anthesis. Wild-type fruit at 30, 40, 50, and 58 d after anthesis were immature, mature green, turning, and red ripe, respectively. Leaf, epicotyl, and hypocotyl tissues were harvested from greenhouse-grown cv Pearson and Nr seedlings. Flowers were harvested from greenhouse-grown plants. Arabidopsis cv Columbia plants were grown in a growth chamber under a 16-h daylength at 22°C.
Nucleic Acid Analysis
The Arabidopsis ETR2 (ethylene response 2) cDNA (Sakai et al., 1998) was kindly provided by Dr. Tony Bleecker (University of Wisconsin, Madison). The ETR2 insert was used to screen under low stringency a phosphate-stressed tomato root cDNA library in λ ZAPII (Stratagene) (kindly provided by Dr. K.G. Raghothama, Purdue University, West Lafayette, IN). The resulting clones were sequenced using synthetic oligonucleotide primers on a sequence analyzer (ABI, Columbia, MD). Sequences of LeETR4 (accession no. AF118843) and LeETR5 (accession no. AF118844) have been deposited in GenBank. Amino acid identities and similarities were calculated using the Gap program, and sequence alignments were performed using the PileUp program with sequence-analysis software (Genetics Computer Group, Madison, WI). Predictions of membrane-spanning domains were performed using the EMBL PredictProtein e-mail server (Rost et al., 1995).
RNase Protection Assays
Total RNAs were extracted from tissues frozen in liquid N2 and stored at −80°C, as described previously (Lashbrook et al., 1994). RNase protection assays were performed using the RPA II kit (Ambion, Austin, TX) with modifications (Lashbrook et al., 1998) using pBluescript vectors harboring a PstI fragment containing nucleotides 2266 to 2565 of the LeETR4 cDNA and a HindIII/ApaI fragment containing nucleotides 2718 to 3106 of the LeETR5 cDNA as the probes. Levels of mRNA were quantified using sense RNAs generated from a construct containing the 3′ 1697 bp or the 3′ 2010 bp of the LeETR4 and LeETR5 cDNAs, respectively.
DNA Mutagenesis
Mutations in each of the tomato genes were introduced with the QuikChange site-directed mutagenesis kit from Stratagene using oligonucleotides with the Cys TGT codon analogous to Cys-65 of ETR1 mutated to Ser TCT (NR, Cys-65; LeETR4, Cys-68; and LeETR5, Cys-90). Mutations were confirmed by sequencing. The full-length mutated or wild-type versions of LeETR4 and LeETR5 were cloned into a vector under the control of the figwort mosaic virus promoter (Richins et al., 1987) and followed by the Agrobacterium tumefaciens nopaline synthase (nos) 3′ terminator. The promoter/mutated or wild-type ETR1 homolog/nos 3′ cassettes were cloned into a vector containing the neomycin phosphotransferase II (nptII) gene under the control of the A. tumefaciens nos promoter and followed by the A. tumefaciens nos 3′ terminator for selection of transgenic plants by kanamycin resistance. This plasmid was introduced into A. tumefaciens, and Arabidopsis plants were transformed according to the method of Bechtold et al. (1993). Introduction of the transgene was confirmed by PCR using oligonucleotides specific for the nptII gene and by seedling germination on kanamycin-containing plates.
Ethylene-Insensitivity Assay
Seeds from transgenic Arabidopsis plants were plated on medium containing Murashige and Skoog salts, 1% Suc, 0.5 mm ACC, 1 mm GA3, and 1% agar and then stored at 4°C for 3 d and at room temperature for 6 d in the dark.
RESULTS
Cloning of Tomato Ethylene-Receptor Homologs
To identify genes that may be involved in ethylene perception in tomato, the Arabidopsis ETR2 cDNA (Sakai et al., 1998) was used to screen a tomato-root cDNA library at low stringency. Sequencing of four ETR2-hybridizing clones resulted in the identification of two classes of tomato cDNAs. The longest clones within each class were designated LeETR4 and LeETR5. LeETR4 is a cDNA of 3337 bp that contains a single open reading frame encoding a protein of 761 amino acids with a predicted molecular mass of 85.1 kD. The LeETR5 cDNA consists of 3370 bp and encodes a deduced protein of 767 amino acids with a predicted molecular mass of 85.9 kD. The predicted LeETR4 and LeETR5 proteins are somewhat larger than the other members of the tomato ETR1 family (LeETR1, 84.2 kD; LeETR2, 81.7 kD; and NR, 71.0 kD). Each of these cDNAs has an unusually long 5′-untranslated sequence: 576 nucleotides in LeETR4 and 680 nucleotides in LeETR5. The 3′-untranslated region consists of 389 nucleotides in LeETR5 and 477 nucleotides in LeETR4. LeETR4 is 78% similar and 60% identical to the Arabidopsis ETR2 protein, whereas LeETR5 is 76% similar and 54% identical.
Structural Analysis of the Tomato Ethylene-Receptor Homologs
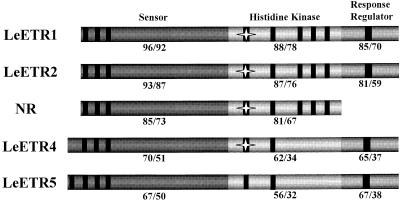
The Arabidopsis ETR1 protein has been divided into three domains: the sensor, the His kinase, and the response regulator (Chang et al., 1993). Figure 1 shows the amino acid identities and similarities of the tomato ETR1 homologs compared with the Arabidopsis ETR1 ethylene receptor in each of the three domains. The similarity among these genes is most evident in the sensor domain, whereas the His kinase and response-regulator domains are more divergent. NR is the only member of the tomato putative ethylene-receptor family that lacks the response-regulator domain. An alignment of the tomato and Arabidopsis ETR1 and ETR2 deduced amino acid sequences is shown in Figure 2. The three membrane-spanning regions critical for ethylene binding are well conserved; however, LeETR5 is predicted to have a fourth membrane-spanning domain within an amino-terminal extension of the protein (Fig. 2). An alternative prediction of membrane-spanning domains using the method of Hofman and Stoffel (1993) posits a fourth membrane-spanning domain in both LeETR4 and LeETR5.
Figure 1.
Schematic representation of the structure of the tomato LeETR proteins. By analogy to the Arabidopsis ETR1 protein, the three domains of the proteins include the sensor domain, with three or four hydrophobic regions (represented by black boxes) capable of spanning a membrane (amino acids 1–325 of ETR1); the signaling domain (amino acids 326–609 of ETR1), with the conserved domains of bacterial His kinases represented by black boxes and the conserved His represented by a star; and the response-regulator domain (amino acids 610–738 of ETR1), with the conserved domain containing an Asp that is capable of receiving the phosphate from the His kinase represented by a black box (Chang et al., 1993). The percentage of amino acid identity and similarity between the Arabidopsis ETR1 protein and each of the tomato ETR1 homologs within each of the three domains is shown below the schematic of each protein.
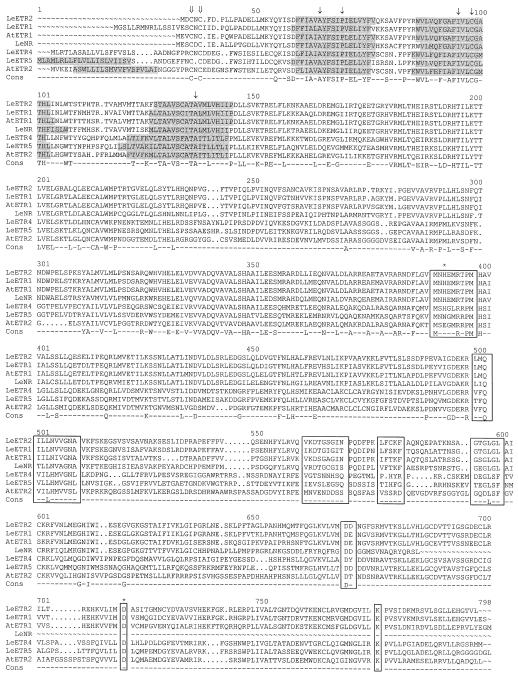
Figure 2.
Amino acid sequence alignments of the five tomato ETR1 homologs and Arabidopsis ETR1 and ETR2. Tomato sequences are designated LeETR1, LeETR2 (Lashbrook et al., 1998), LeNR (Wilkinson et al., 1995), LeETR4, and LeETR5 (this paper). Arabidopsis ETR1 and ETR2 are designated AtETR1 and AtETR2, respectively (Chang et al.,1993; Sakai et al., 1998). Shaded areas represent putative membrane-spanning domains. Cons, Consensus sequence; ⇓ , Cys residues involved in dimerization (Schaller et al., 1995); ↓, amino acids in which mutations result in dominant ethylene insensitivity in Arabidopsis (Ala-31 to Val, Ile-62 to Phe, Cys-65 to Tyr, and Ala-102 to Thr) or tomato (Pro-36 to Leu in NR) (Wilkinson et al., 1995); * at nucleotide 391, the autophosphorylated His (Gamble et al., 1998); * at nucleotide 718, Asp suggested to act as a receiver of the phosphate; boxed areas are conserved regions in bacterial His kinases and response regulators (Parkinson and Kofoid, 1992).
The amino acids mutated in the known dominant ethylene-insensitive mutants of Arabidopsis and tomato are conserved in the tomato ETR1 homologs (Fig. 2, arrows). Other invariant amino acids include the two Cys residues (at positions 4 and 6 of ETR1), which were demonstrated to be involved in covalent dimerization of the ETR1 protein, although the intervening amino acid is absent in LeETR5 (Schaller et al., 1995). The five sequence motifs characteristic of bacterial His kinases (Fig. 2, boxed regions) are well conserved in LeETR1, LeETR2, and NR; however, the third and fourth motifs are highly divergent in LeETR4 and LeETR5. The putative autophosphorylated His (Fig. 2, asterisk at nucleotide 391) in the His kinase domain is present in all of the tomato homologs except LeETR5. This His is also missing in the Arabidopsis ETR2 and ERS2 proteins (Hua et al., 1998; Sakai et al., 1998), and when it is mutated in the ETR1 protein, phosphorylation activity is abolished (Gamble et al., 1998). The three domains conserved in bacterial response regulators are also conserved in the tomato proteins containing a response-regulator domain (Fig. 2, boxed regions). The Asp that has been suggested to act as a phosphate receiver is present in all of the proteins containing the response-regulator domain.
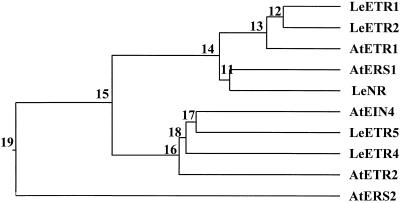
Phylogenetic analysis of the Arabidopsis and tomato ETR1 homologs (Fig. 3) indicates that LeETR1 and LeETR2 are most closely related to ETR1 of Arabidopsis, whereas NR is most closely related to the ERS protein of Arabidopsis. LeETR4 and LeETR5 are more closely related to Arabidopsis ETR2 and EIN4. Arabidopsis ERS2 is divergent from all other known tomato and Arabidopsis ETR1 homologs.
Figure 3.
Phylogenetic analysis of the Arabidopsis and tomato ETR1 homologs aligned by the Clustal program. Tomato sequences are designated LeETR1, LeETR2 (Lashbrook et al., 1998), LeNR (Wilkinson et al., 1995), LeETR4, and LeETR5 (this paper). Arabidopsis sequences are designated AtETR1 (Chang et al., 1993), AtERS1 (Hua et al., 1995), AtEIN4 (Hua et al., 1998), AtETR2 (Sakai et al., 1998), and AtERS2 (Hua et al., 1998). Numbers represent the percentages of amino acids that differ between two proteins (Saitou and Nei, 1987).
Mutagenesis of the Tomato Ethylene-Receptor Homologs
Introduction of mutations into the membrane-spanning domains of the Arabidopsis ETR1, ERS, EIN4, ETR2, and ERS2 proteins results in ethylene insensitivity in Arabidopsis (Chang et al., 1993; Hua et al., 1995, 1998; Sakai et al., 1998). To assess the potential role of LeETR4 and LeETR5 in ethylene perception, we introduced mutations into the membrane-spanning domain of these proteins. We chose to mutate the Cys aligning with Cys-65 of ETR1 to Ser because this mutation has been shown to be effective in conferring ethylene insensitivity in Arabidopsis and resulted in a loss of ethylene binding when the mutated etr1-1 was expressed in yeast (Chang et al., 1993; Schaller and Bleecker, 1995). The mutated or wild-type tomato cDNAs under control of the figwort mosaic virus 35S promoter (Richins et al., 1987) were introduced into Arabidopsis. This cross-species approach has been shown to confer ethylene insensitivity in petunia and tomato plants using the Arabidopsis etr1-1 gene under the control of a constitutively expressed promoter (Wilkinson et al., 1997). Progeny from transgenic plants for each of the gene constructs were tested for ethylene insensitivity, and the results are shown in Table I and Figure 4. Introduction of each of the mutated tomato genes (Cys-65 to Ser) conferred ethylene insensitivity, as judged by elongation of the hypocotyl and the absence of an apical hook, comparable to that of ein3-1 in approximately two-thirds of the transgenic lines (Fig. 4). Introduction of the wild-type LeETR4 gene did not result in increased ethylene insensitivity of transgenic Arabidopsis; however, 36% of the transgenic lines were significantly shorter than wild-type Columbia when grown in the presence of ACC (data not shown). These results may indicate an increased sensitivity to ethylene or an effect of the transgene on seedling growth. Introduction of the wild-type LeETR5 into Arabidopsis resulted in a somewhat longer phenotype in 23% of transformed lines; however, the frequency of the longer hypocotyl and root phenotype was much less than that exhibited by plants transformed with the mutant LeETR5 gene construct (data not shown). As shown in Figure 4, the phenotype of both LeETR4 and LeETR5 wild-type overexpressers is consistent with ethylene sensitivity, as indicated by the shorter, thicker hypocotyl and the exaggerated apical hook.
Table I.
Ethylene insensitivity of Arabidopsis seedlings from independent transgenic lines transformed with the mutated (mut) or wild-type (WT) tomato ethylene-receptor homolog genes, LeETR4 and LeETR5, and control, wild-type Columbia and ethylene-insensitive ein3-1 seedlings
| Line | Length |
|---|---|
| mm | |
| Columbia | 4.9 ± 0.2 |
| ein3-1 | 7.6 ± 0.3 |
| LeETR4 mut-1 | 7.2 ± 0.2 |
| LeETR4 mut-2 | 7.2 ± 0.2 |
| LeETR4 mut-3 | 7.4 ± 0.6 |
| LeETR4 WT-1 | 3.8 ± 0.2 |
| LeETR4 WT-2 | 4.0 ± 0.2 |
| LeETR4 WT-3 | 4.1 ± 0.2 |
| LeETR5 mut-1 | 7.3 ± 0.3 |
| LeETR5 mut-2 | 7.3 ± 0.2 |
| LeETR5 mut-3 | 7.5 ± 0.2 |
| LeETR5 WT-1 | 4.3 ± 0.2 |
| LeETR5 WT-2 | 3.9 ± 0.3 |
| LeETR5 WT-3 | 4.4 ± 0.2 |
Data shown indicate lengths (±se) of etiolated seedlings grown in the presence of ACC, as described in Methods.
Figure 4.
Phenotype of etiolated Arabidopsis seedlings transformed with the mutated or wild-type tomato putative ethylene-receptor genes, LeETR4 or LeETR5, under the control of a constitutive promoter grown in the presence of ACC, as described in Methods. A, Wild-type Columbia; B, ethylene-insensitive mutant ein3-1; C, transgenic Arabidopsis expressing the mutated LeETR4 gene; D, transgenic Arabidopsis expressing the wild-type LeETR4 gene; E, transgenic Arabidopsis expressing the mutated LeETR5 gene; and F, transgenic Arabidopsis expressing the wild-type LeETR5 gene.
Expression Patterns of the Tomato Ethylene-Receptor Homologs
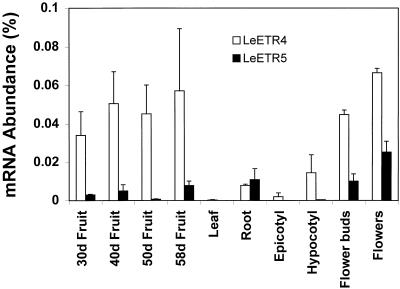
RNA expression patterns of LeETR4 and LeETR5 in tomato tissues were determined by RNase protection assays (Fig. 5). Expression levels of LeETR4 and LeETR5 mRNAs were highly regulated among plant tissues, with high levels of expression in reproductive tissues but low levels in vegetative tissues. High levels of LeETR4 and LeETR5 RNA expression were found in flower buds and increased in mature flowers. LeETR5 constitutes approximately 0.025% of the total RNA in mature flowers, the highest level of this mRNA in all tomato plant tissues examined. Low levels of LeETR4 and LeETR5 RNA expression were found in leaves, epicotyls, and hypocotyls. LeETR5 RNA expression in roots was similar to that in ripe fruit and flower buds, whereas low levels of LeETR4 were found in roots. In wild-type cv Pearson fruit, expression of LeETR4 and LeETR5 RNA did not change significantly from 30 to 58 d after flowering. In ripening fruit, the levels of LeETR4 were the highest among all of the tomato ETR1 homologs, indicating that this gene, if encoding a functional ethylene receptor, should play an important role in ethylene perception in tomato. LeETR5 mRNA levels in ripening fruit were somewhat lower than NR mRNA levels. LeETR4 was calculated to constitute approximately 0.06% of total RNA in 58-d-old fruit, whereas LeETR1, LeETR2, and NR mRNAs constituted approximately 0.01%, 0.002%, and 0.03%, respectively, of total RNA in ripening fruit (Lashbrook et al., 1998). In wild-type fruit, NR mRNA levels increased rapidly at the onset of ripening and continued to increase until fruit reached the pink stage. In comparison, the levels of NR mRNA expression in ethylene-insensitive Nr fruit were greatly reduced. These results suggest that the levels of ethylene sensitivity and ethylene evolution are both regulated during fruit ripening (Wilkinson et al., 1995). We examined LeETR4 and LeETR5 mRNA levels in tissues of Nr plants to determine the effects of diminished ethylene sensitivity on RNA expression. Overall, LeETR4 and LeETR5 mRNA expression patterns were similar in Nr and wild-type fruit, flowers, and vegetative tissues (data not shown), suggesting that the regulation of these genes is not affected by ethylene insensitivity of the Nr plants.
Figure 5.
LeETR4 and LeETR5 mRNA expression in fruit, flowers, and vegetative tissues of cv Pearson tomato plants. Fruit at 30, 40, 50, and 58 d after anthesis are approximately immature, mature green, turning, and red ripe, respectively. RNase protection assays were performed using 5 and 10 μg of total RNA for LeETR4 and LeETR5, respectively.
DISCUSSION
Tomato is an excellent system in which to study the biochemistry and physiology of ethylene perception because hormone sensitivity is developmentally regulated in many processes, such as fruit ripening, petal senescence, and leaf and flower abscission (Liu et al., 1985). The roles of the ethylene biosynthetic enzymes ACC synthase and ACC oxidase in fruit ripening are well established. Elimination of either ACC synthase or ACC oxidase gene expression results in fruit that ripen only after exogenous ethylene application. However, ethylene perception is less well understood. Because ethylene is readily diffusible within the plant, the regulation of differential tissue and developmental responses to ethylene is essential to the plant's survival. An understanding of the tomato putative ethylene-receptor family's functions, expression patterns, and interactions is essential to understanding responses to ethylene. Previously, we examined the expression patterns of LeETR1, LeETR2, and NR mRNAs in tomato plant development (Lashbrook et al., 1998). In the experiments reported here, we cloned two novel tomato ethylene-receptor candidates using the Arabidopsis ETR2 cDNA as a hybridization probe. These genes bring the total number of cloned tomato ethylene-receptor homologs to five.
Structurally, the known tomato and Arabidopsis ethylene-receptor homologs can be divided into three classes (Figs. 1–3). The first class, incorporating two subclasses, includes the tomato proteins most closely related to ETR1. The first subclass consists of AtETR1, LeETR1, and LeETR2. The second subclass consists of proteins that, although similar to ETR1, do not have a response-regulator domain (LeNR and AtERS1). The proteins of the first class are highly homologous in the five conserved regions of residues characteristic of bacterial His kinases, each of which includes the autophosphorylated His (Fig. 2) (Parkinson and Kofoid, 1992). The Asp suggested to act as a phosphate receiver is also conserved in the proteins that have the response-regulator domain (Fig. 2). The second class consists of AtETR2, AtEIN4, LeETR4, and LeETR5, proteins that are structurally divergent from those in the first class. These proteins are also less similar in the conserved regions of His kinases, especially the third and fourth conserved regions. The autophosphorylated His is absent in LeETR5, indicating that it may not be an active His kinase. This His is also absent from AtETR2 and AtERS2 (Hua et al., 1998; Sakai et al., 1998). Although a high level of sequence similarity is evident in the membrane-spanning region, an additional predicted membrane-spanning domain is present in LeETR5, AtETR2, AtERS2, AtEIN4, and possibly LeETR4. The sole member of the third class of receptors, AtERS2, is highly divergent from all other tomato and Arabidopsis ethylene-receptor homologs, with only 59% similarity and 38% identity to AtETR1. The role of this protein in ethylene signal perception has not been determined.
Of the known tomato proteins, only NR lacks the response-regulator domain. The significance of this domain for signal transduction has yet to be determined. In bacteria, many two-component sensors lack a contiguous response-regulator domain. Rather, the phosphate is transferred to an Asp on a separate protein responsible for signal transduction (Parkinson and Kofoid, 1992). Several response-regulator proteins capable of accepting a phosphate have been identified in Arabidopsis (Imamura et al., 1998). The yeast osmosensing pathway contains a multistep phosphorelay system consisting of a two-component His kinase/receiver protein (SLN1), a protein (YPD1) that accepts the phosphate from the receiver of SLN1, and a second receiver protein (SSK1) that accepts the phosphate from YPD1 (Posas et al., 1996). A family of proteins with significant homology to YPD1 has been identified in the Arabidopsis expressed sequence tag database (H. Klee, unpublished data). The absence of a response-regulator domain in a subset of putative ethylene receptors, the possibility of heterodimer formation, and the presence of multiple separate response regulators and YPD1-like proteins suggest that the phosphorelay mechanism may be quite complex and vary with different ethylene receptors.
The lack of the autophosphorylated His and the presence of a fourth membrane-spanning region in LeETR5 may indicate a divergent function for this protein. How the presence of a fourth membrane-spanning domain might affect dimerization is not known; however, the Cys residues involved in dimerization are present in a region predicted to be outside of the membrane, between the first and second membrane-spanning domains of LeETR5 (Fig. 2). The missing autophosphorylated His in LeETR5 also presents the interesting possibility of an ethylene receptor that cannot interact with the next step in the signal transduction pathway. This protein does have the Asp that is essential for response-regulator activity; therefore, conceivably it could accept a phosphate from an active His kinase. This possibility would depend on formation of heterodimers, something that has not yet been formally demonstrated. Mutations in the membrane-spanning regions of LeETR4 and LeETR5 result in ethylene insensitivity in transgenic Arabidopsis, suggesting involvement in ethylene signal perception (Table I; Fig. 4). Similar results have been seen with mutated versions of the Arabidopsis ETR2 and ERS2 genes that are missing the autophosphorylated His (Hua et al., 1998; Sakai et al., 1998). These results suggest that His kinase activity is not essential to the function of this protein. Alternatively, the missing His in LeETR5 is not the His that is autophosphorylated; however, mutation of the analogous His in the Arabidopsis ETR1 protein resulted in the loss of autophosphorylation in vitro (Gamble et al., 1998). There is a His conserved in all of the tomato and Arabidopsis proteins seven amino acids from the autophosphorylated His.
Although the expression of the mutant LeETR4 and LeETR5 genes in Arabidopsis resulted in a phenotype consistent with ethylene insensitivity, the introduction of the wild-type versions of LeETR4 and LeETR5 into Arabidopsis also resulted in a change in phenotype in some transgenic lines (data not shown). The shorter phenotype of Arabidopsis plants expressing LeETR4 and the longer phenotype in some lines expressing LeETR5 may be the result of an ethylene-independent role of the putative receptors, as suggested by the etr1 loss-of-function mutants of Arabidopsis that have a shorter phenotype in the presence of ethylene (Hua and Meyerowitz, 1998). The altered phenotype may also be the result of cosuppression of gene expression by the introduction of the transgene under the control of a strong promoter. A constitutive ethylene phenotype has been seen in Arabidopsis loss-of-function mutants of multiple ethylene-receptor genes (Hua and Meyerowitz, 1998).
The presence of structurally divergent ethylene receptors in plants may be one mechanism whereby plants coordinate and distinguish their many responses to a single phytohormone. The differential expression patterns of members of the tomato putative ethylene-receptor gene family suggest another mechanism for regulating ethylene insensitivity. For example, tomato fruit undergo a fundamental change in ethylene responsiveness just before the onset of ripening (Liu et al., 1985). NR expression is known to increase concomitantly with the developmental shift in responsiveness (Wilkinson et al., 1995) and may contribute to the shift in ethylene sensitivity. In contrast, published expression data indicate that LeETR1 and LeETR2 are expressed at similar levels in most tissues, including fruit (Zhou et al., 1996a; Lashbrook et al., 1998). LeETR4 and LeETR5 are also present at relatively constant levels during fruit development. However, LeETR4 is expressed at a very high level, accounting for more than 90% of the putative receptor expression in green fruit and approximately 50% of the putative receptor expression in ripening fruit. LeETR4 and LeETR5 are also the predominantly expressed putative receptors in flowers, but they are expressed at relatively low levels in vegetative tissues.
In conclusion, we have shown that the tomato ethylene-receptor homolog gene family consists of at least five members that exhibit overlapping expression in virtually all tissues. However, relative levels of expression of the individual genes vary significantly during development. The individual putative receptor proteins exhibit significant structural divergence. This divergence within the functional domains of the proteins could greatly affect the mechanisms and efficiencies with which they transduce the ethylene signal. The differential responses to ethylene in response to developmental and environmental stimuli may be a result of several factors working singly or in combination, including the differential regulation of gene expression, the differential binding affinities of the receptors for ethylene, the formation of receptor heterodimers or homodimers from the complement of receptors found in a tissue, the absence of the response-regulator domain in NR, and the differential interactions with downstream components of the ethylene signal transduction pathway. If the members of the ethylene-receptor gene family act as negative regulators of the ethylene signal transduction pathway (Hua and Meyerowitz, 1998), high levels of receptors in fruit tissues may act to modulate the fruit response to the high levels of ethylene produced during fruit ripening. Although the actual mechanisms for modulating ethylene sensitivity have yet to be elucidated, it is clear that there is ample opportunity for plants to adjust ethylene responses at the receptor level.
The accession numbers for the sequences reported in this article are AF118843 (LeETR4) and AF118844 (LeETR5).
ACKNOWLEDGMENTS
We thank Dr. K.G. Raghothama for the phosphate-stressed root cDNA library and Dr. A. Bleecker for the ETR2 gene. We also thank Dr. Elliot Meyerowitz for sharing the unpublished sequences of EIN4 and ETR2, Dr. Mark Taylor for excellent care of the plants, and Dr. David Clark for assistance with the figures.
Footnotes
This work was supported in part by the U.S. Department of Agriculture (grant no. 95-37304-2326 to H.J.K.). This is Florida Agricultural Experiment Station journal series no. R-06316.
LITERATURE CITED
- Abeles FB, Morgan PW, Saltveit ME (1992). Ethylene in Plant Biology, Ed 2. Academic Press, San Diego, CA, pp 264–296
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis plants. CR Acad Sci Paris. 1993;316:1194–1199. [Google Scholar]
- Bleecker AB, Estelle MA, Somerville C, Kende H. Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis. Science. 1988;241:1086–1089. doi: 10.1126/science.241.4869.1086. [DOI] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM. Arabidopsis ethylene-response gene ETR1: similarity of products to two-component regulators. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker J. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell. 1997;89:1133–1144. doi: 10.1016/s0092-8674(00)80300-1. [DOI] [PubMed] [Google Scholar]
- Ecker JR. The ethylene signal transduction pathway in plants. Science. 1995;268:667–675. doi: 10.1126/science.7732375. [DOI] [PubMed] [Google Scholar]
- Gamble RL, Coonfield ML, Schaller GE. Histidine kinase activity of the ETR1 ethylene receptor from Arabidopsis. Proc Natl Acad Sci USA. 1998;95:7825–7829. doi: 10.1073/pnas.95.13.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman P, Ecker J. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell. 1990;2:513–523. doi: 10.1105/tpc.2.6.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofman K, Stoffel W. Tmbase-A database of membrane spanning protein segments. Biol Chem. 1993;347:166. [Google Scholar]
- Hua J, Chang C, Sun Q, Meyerowitz EM. Ethylene insensitivity conferred by Arabidopsis ERS gene. Science. 1995;269:1712–1714. doi: 10.1126/science.7569898. [DOI] [PubMed] [Google Scholar]
- Hua J, Meyerowitz EM. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis. Cell. 1998;94:261–271. doi: 10.1016/s0092-8674(00)81425-7. [DOI] [PubMed] [Google Scholar]
- Hua J, Sakai H, Nourizadeh S, Chen QG, Bleecker AB, Ecker JR, Meyerowitz EM. EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell. 1998;10:1321–1332. doi: 10.1105/tpc.10.8.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura A, Hanaki N, Umeda H, Nakamura A, Suzuki T, Ueguchi C, Mizuno T. Response regulators implicated in His-to-Asp phosphotransfer signaling in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:2691–2696. doi: 10.1073/pnas.95.5.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kende H. Ethylene biosynthesis. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:283–307. [Google Scholar]
- Kieber JJ. The ethylene response pathway in Arabidopsis. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:277–296. doi: 10.1146/annurev.arplant.48.1.277. [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Ecker JR. Ethylene gas: it's not just for ripening any more! Trends Genet. 1993;9:356–362. doi: 10.1016/0168-9525(93)90041-f. [DOI] [PubMed] [Google Scholar]
- Lanahan MB, Yen H-C, Giovannoni JJ, Klee HJ. The Never ripe mutation blocks ethylene perception in tomato. Plant Cell. 1994;6:521–530. doi: 10.1105/tpc.6.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashbrook CC, Gonzalez-Bosch C, Bennett AB. Two divergent endo-β-1,4-glucanase genes exhibit overlapping expression in ripening fruit and abscising flowers. Plant Cell. 1994;6:1485–1493. doi: 10.1105/tpc.6.10.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashbrook CC, Tieman DM, Klee HJ. Differential regulation of the tomato ETR gene family throughout plant development. Plant J. 1998;15:243–252. doi: 10.1046/j.1365-313x.1998.00202.x. [DOI] [PubMed] [Google Scholar]
- Lehman A, Black R, Ecker JR. HOOKLESS1, an ethylene response gene is required for differential cell elongation in Arabidopsis. Cell. 1996;85:183–194. doi: 10.1016/s0092-8674(00)81095-8. [DOI] [PubMed] [Google Scholar]
- Liu Y, Hoffman N, Yang SF. Promotion by ethylene of the capability to convert 1-aminocyclopropane-1-carboxylic acid to ethylene in preclimacteric tomato and cantaloupe fruits. Plant Physiol. 1985;77:407–411. doi: 10.1104/pp.77.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon T, Yang SF. Biosynthesis and metabolism of ethylene. In: Davies P, editor. Plant Hormones and Their Role in Plant Growth and Development. Boston, MA: Martinus Nijhoff; 1987. pp. 94–112. [Google Scholar]
- Oetiker J, Olson D, Shiu O, Yang SF. Differential induction of seven 1-aminocyclopropane-1-carboxylate synthase genes by elicitor in suspension cultures of tomato (Lycopersicon esculentum) Plant Mol Biol. 1997;34:275–286. doi: 10.1023/a:1005800511372. [DOI] [PubMed] [Google Scholar]
- Olson DC, White JA, Edelman L, Harkins RN, Kende H. Differential expression of two genes for 1-aminocyclopropane-1-carboxylate synthase in tomato fruits. Proc Natl Acad Sci USA. 1991;88:5340–5344. doi: 10.1073/pnas.88.12.5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. Signal transduction schemes of bacteria. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- Parkinson JS, Kofoid EC. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- Posas F, Wurgler-Murphy S, Maeda T, Witten E, Thai TC, Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two component” osmosensor. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- Richins RD, Scholthof HB, Shepard RJ. Sequence of the figwort mosaic virus DNA (caulimovirus group) Nucleic Acids Res. 1987;15:8451–8466. doi: 10.1093/nar/15.20.8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rick C, Butler L. Phytogenetics of the tomato. Adv Genet. 1956;8:267–382. [Google Scholar]
- Roman G, Lubarsky B, Kieber J, Rothenberg M, Ecker J. Genetic analysis of ethylene signal transduction in Arabidopsis: five novel mutant loci integrated into a stress response pathway. Genetics. 1995;139:1393–1409. doi: 10.1093/genetics/139.3.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B, Casadio R, Fariselli P, Sander C. Prediction of helical transmembrane segments at 95% accuracy. Protein Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottmann WH, Peter GF, Oeller PW, Keller JA, Shen NF, Nagy BP, Taylor LP, Campbell AD, Theologis A. 1-Aminocyclopropane-1-carboxylate synthase in tomato is encoded by a multigene family whose transcription is induced during fruit and floral senescence. J Mol Biol. 1991;222:937–961. doi: 10.1016/0022-2836(91)90587-v. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sakai H, Hua J, Chen QG, Chang C, Medrano LJ, Bleecker AB, Meyerowitz EM. ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:5812–5817. doi: 10.1073/pnas.95.10.5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Bleecker AB. Ethylene binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science. 1995;270:1809–1811. doi: 10.1126/science.270.5243.1809. [DOI] [PubMed] [Google Scholar]
- Schaller GE, Ladd AN, Lanahan MB, Spanbauer JM, Bleecker AB. The ethylene response mediator ETR1 from Arabidopsis forms a disulfide-linked dimer. J Biol Chem. 1995;270:12526. doi: 10.1074/jbc.270.21.12526. 12530. [DOI] [PubMed] [Google Scholar]
- Theologis A. One rotten apple spoils the whole bushel: the role of ethylene in fruit ripening. Cell. 1993;70:181–184. doi: 10.1016/0092-8674(92)90093-r. [DOI] [PubMed] [Google Scholar]
- Wilkinson JQ, Lanahan MB, Clark DG, Bleecker AB, Chang C, Meyerowitz EM, Klee HJ. A dominant mutant receptor from Arabidopsis confers ethylene insensitivity in heterologous plants. Nat Biotechnol. 1997;15:444–447. doi: 10.1038/nbt0597-444. [DOI] [PubMed] [Google Scholar]
- Wilkinson JQ, Lanahan MB, Yen H-C, Giovannoni JJ, Klee HJ. An ethylene-inducible component of signal transduction encoded by Never-ripe. Science. 1995;270:1807–1809. doi: 10.1126/science.270.5243.1807. [DOI] [PubMed] [Google Scholar]
- Yen H-C, Lee S, Tanksley S, Lanahan M, Klee H, Giovannoni J. The tomato Never-ripe locus regulates ethylene-inducible gene expression and is linked to a homologue of the Arabidopsis ETR1 gene. Plant Physiol. 1995;107:1343–1353. doi: 10.1104/pp.107.4.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Kalaitzis P, Mattoo A, Tucker M. The mRNA for an ETR1 homologue in tomato is constitutively expressed in vegetative and reproductive tissues. Plant Mol Biol. 1996a;30:1331–1338. doi: 10.1007/BF00019564. [DOI] [PubMed] [Google Scholar]
- Zhou D, Mattoo A, Tucker M. Molecular cloning of a tomato cDNA encoding an ethylene receptor. Plant Physiol. 1996b;110:1435–1436. [Google Scholar]