Abstract
A massive effort is underway to map the structure of the Drosophila nervous system and to genetically dissect its function. Virtual Fly Brain (VFB; http://www.virtualflybrain.org) is a popular, OWL-based resource providing neuroinformatics support for this work. It provides: curated descriptions of brain regions and neurons; queries for neurons based on their relationship to gross neuroanatomy; and queries for reagents based on their expression patterns. Query results are enriched by OWL axiomatisation allowing basic mereological reasoning. To keep reasoning fast and scalable, VFB confines expressiveness to the EL profile of OWL. As a result, VFB does not provide queries involving negation, despite there being both demand and sufficient information to support them. Recent developments in reasoning technology may make more expressive queries practical. Here we present design patterns to support queries with negation that are compatible with the mereological reasoning used in VFB.
Keywords: OWL, neurobiology, neuron, DL reasoning, negation, closure axioms, ontology design pattern
1. Introduction
1.1. Mapping and genetically dissecting the Drosophila nervous system
A massive effort is underway to map the neural circuitry of the Drosophila nervous system and to genetically dissect its function. New microscopy and image analysis techniques are facilitating the collection and integration of the large 3D image data sets required to map the structure and connectivity of the nervous system down to the single neuron level [1, 8]. New genetic techniques allow researchers to precisely inhibit or activate elements of the neural circuitry in order to assess the effects on function and behaviour [3]. The scale of this effort, and the huge volumes of data involved, mean that its success depends on suitable informatics support. Virtual Fly Brain (VFB) [9, 10] is an OWL-based, open source resource dedicated to this role. Usage is growing rapidly among the community it serves. The site currently gets 15-20,000 page views per month.
The adult Drosophila nervous system contains an estimated 200,000 neurons. These can be grouped into classes that share characteristics such as similar lineage, morphology and location. Each brain includes multiple members of most classes, so the number of such classes is likely to be much lower than the number of neurons - proabably by at least an order of magnitude.
Mapping the neural circuitry of Drosophila requires ways to track the classification of these neurons and their properties, including their relationships to each other and to the gross anatomy of the nervous system, musculature, sense organs and neuro-endocrine system. This work requires the synthesis of many qualitative assertions from the literature and their integration with information from bulk data sources, much of it quantitative. OWL is an ideal technology for building and maintaining these queryable classifications. Although there will always be a need for direct mathematical access to quantitative data, if suitable cutoffs can be chosen to make qualitative assertions from quantitative data, OWL provides a means to integrate qualitative and quantitative data into a queryable whole.
Modulating the activity of particular neuron classes requires finding reagents whose expression is sufficiently specific. Finding such reagents frequently requires mining 3D image data of expression patterns. Integrating the phenotypic results of modulating neuronal activity into the bigger picture of nervous system function requires ways to keep track of the phenotypes associated with modulating the neuronal activity of connected neurons. Annotation with OWL ontology terms - either semi-formalised in a database or fully formalised in an OWL knowledgebase provides a means of storing this information in queryable form.
1.2. Virtual Fly Brain
The Drosophila anatomy ontology
Virtual Fly Brain is built around the Drosophila anatomy ontology (DAO) [2], an OWL ontology of Drosophila anatomy, over 45% of which (3875/8576 classes) is devoted to representing neuroanatomy. The DAO is largely manually curated from the literature and includes a large textual component in the form of referenced synonym lists and definitions/descriptions - making it searchable by and accessible to biologists. These synonyms are used to drive auto-suggestion based searching on VFB and to populate term information pages for specific neuron classes and nervous system regions. The DAO is also richly formalised, using 44 object properties in >17000 Subclassing axioms and >2000 Equivalent Class axioms. This axiomatisation infers almost 50% of >10,000 classifications and allows a rich variety of biologically interesting queries. In order to keep reasoning tractable, expressiveness is kept almost entirely within the EL profile of OWL1, allowing us to use the fast reasoner ELK [7]. Classification of the ontology is complete in under 500ms. Query answering time, taking advantage of incremental reasoning via ELK, is in the 10s of milliseconds range.
Annotation queries
One major usage of VFB is as a means to query for expression of genes, transgenes and phenotypes in specified anatomical classes. These queries use information curated from the literature and bulk data sets by VFB and FlyBase curators using a semi-formalised tagging system. All queries of these annotations start with a query for subclasses, parts and overlapping cells. The resulting list is then used to query the FlyBase SQL database of annotations. 10’s of thousands of annotations are available from these queries.
OWL queries and design patterns for neuroanatomy
The DAO uses an integrated set of relations and design patterns to classify neurons according to their location, connectivity, lineage and function [9, 10]. The neuronal connectivity relations (defined in detail [10]) drive the query system on VFB (see figure 1). VFB takes advantage of term classification in the DAO to serve only queries that are appropriate to the term displayed. So, for example, the queries available for neurons are different to those available for brain regions.
Fig. 1.
VFB query menus (left) with the DL queries they run (right). The top panel shows nested queries for classes of neurons based on overlaps and its subproperties. Each of these queries returns classes based on assertions further down the partonomy than the query term. The bottom panel show queries for individual neurons with and without clustering. A 3D rendering of a cluster is shown on the bottom right.
The typical mereological relationship between a neuron and gross neuroanatomy is overlap: most neurons have parts in many parts of the brain. In an insect brain, each neuron has a cell body (soma) in the cortex and many have long, branching projections that extend to multiple brain regions. Projections bundle (fasciculate) together to form tracts. On exiting a tract, the projection enters a region called neuropil where it typically branches extensively and connects to other neuron projections via synapses.
The Drosophila brain contains many neuron classes that can be defined via some combination of: soma location, tracts fasciculated with; neuropils in which they form input or output synaptic connections with other neurons; neuron classes synapsed with; the developmental origin of the neuron. The DAO takes advantage of this to automate classification of neurons based on these properties via EquivalentClass expressions.
Central to the basic mereological reasoning on VFB is an overlaps relation defined using part_of and its inverse has_part
X overlaps Y iff: exists some Z and X part_of Z and Y has_part Z2
part_of subPropertyOf overlaps
has_part subPropertyOf overlaps
has_part o part_of subPropertyOf overlaps
overlaps o part_of subPropertyOf overlaps
has_part o overlaps subPropertyOf overlaps
The property chains allow inference over partonomy. This is central to the function of the query system on VFB - allowing queries for overlap from any level of granularity in the partonomy.
Typically, overlaps is too abstract to be directly useful in class restrictions. Instead we use a range of subproperties of overlaps that record something useful about the nature of the overlap, such as which tract(s) a neuron fasciculates with and which neuropils it forms synapses in. Like overlaps, relations recording synaptic terminal location also propagate over partonomy via property chains, allowing queries from any level of the partonomy. For example:
has_synaptic_terminal_in o part_of subPropertyOf has_synaptic_terminal_in
has_part o has_synaptic_terminal in subPropertyOf has_synaptic_terminal_in
VFB also provides combinatorial query functionality, via its query builder tool3, allowing users to query for neurons based on their pattern of synapsing. This functionality is currently limited to query legs combined with ‘and’, and does not support negation. (See [10] for details.)
2. Integration of images into VFB using OWL
Neurobiology is a very visual subject. While it is useful to read both informal and formal descriptions of neuron classes and brain regions, there is no substitute for being able to see images of them. VFB is built around a standard 3D adult brain template. Major brain regions are defined as 3D painted regions on this template according to an expert-defined standard [5]. These regions are modelled as individual members of the relevant ontology classes, but are also related to brain region classes via an axiom of the form:
has_exemplar value ‘individual region’
This indicates that the individual provides a standard reference for the boundaries of a brain region. A simple DL query is used to find images to illustrate term pages for these brain regions.
VFB also incorporates large datasets of 3D images of single neurons (>16,000), neuron clones (>200) and expression patterns (>3500). As for painted brain regions, structures depicted in these images are modelled as OWL individuals. Importantly, all of these images are registered (morphed) onto the standard brain. This allows direct comparison - both automated and manual - of registered images.
From image analysis, we can determine which gross brain regions a neuron, clone or expression pattern overlaps, recording this using a Type statement on the individual. These axioms drive queries for single neuron images by location (figure 1). A more sophisticated form of image analysis, developed by G Jefferis [unpublished], compares pairs of neurons, giving each pair a similarity score based on morphology and location. A clustering algorithm is then used to group neurons with similar morphology and location and to assign an exemplar neuron for each cluster.
We treat clusters as individuals, with single neurons standing in a member_of relationship to a cluster. A subproperty of member_of, exemplar_of, is used to relate exemplars to clusters. This simple formalism allows VFB to group the very large numbers of images that often result from queries of brain regions for overlapping neurons into a much smaller number of clusters of similar neurons (see figure 1).
In many cases, the resulting clusters correspond largely or completely to well characterized neurons from the literature, for which the DAO has classes defined by lineage, tract and location of synaptic connections. Where this is the case, we add manual typing statements. In other cases, manual annotation of neurons with Type statements provides sufficient information for automated classification in the ontology.
2.1. Modelling expression patterns in OWL
A key aim of VFB is to provide a means for biologists to find candidate transgenes with expression patterns suitable for targeting expression to specific neurons or brain regions.
To formalise what we mean by expression pattern, we first define a relation, expresses:
expresses: This relation holds between an anatomical entity (a) and a gene or transgene (g) where the anatomical entity is either a cell or has cells as a part and in all of those cells, some instance of GO:gene expression that has input g is occuring.
We use this relation to record expression in any type of cell or multicellular anatomical entity including single neurons, neuron clones and complete expression patterns. Images of single neurons and neuron clones typically depict only fragments of expression patterns. To keep these separable from images of complete expression patterns, we define an expression pattern as an anatomical entity consisting of mereological sum of all cells that express a particular gene or transgene. This is axiomatised using the following pattern:
‘gene B expression pattern’ EquivalentTo: ‘expression pattern’ that expresses some ‘gene B’
GCI : expresses some ‘gene B’ EquivalentTo part_of some ‘B expression pattern’
Classification time for the full ontology combined with the knowledgebase (¿ 21,000 expression patterns, clones and neurons) is 1500ms. DL query answering time, taking advantage of incremental reasoning with ELK, is under 100ms.
Representation of expression patterns in the knowledgebase is currently used on VFB to provide images of transgene expression patterns found via SQL queries. The above formalisation provides an obvious way to convert the semi-formalised annotations in SQL to OWL.
For brain regions:
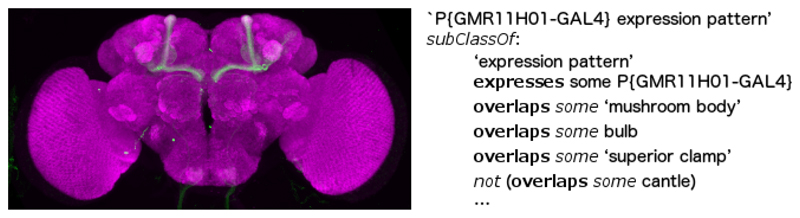
‘expression pattern of X’ overlaps some ‘brain region Y’ (see figure 3 for an example)
Fig. 3.
The left panel shows the expression pattern of the PGMR11H01-GAL4 transgene in the adult brain. The right panel shows its representation in OWL. Only one explicit negation is shown, but the full OWL representation includes negative expression assertions for 30 brain regions. These explicit negations are necessary in the absence of sufficient information to add closure axioms.
For cells, we can make a stronger assertion:
‘expression pattern of X’ has_part4 some ‘cell Y’
We can then find anatomical structures in which there is some expression via “overlaps some X”
As discussed in the next section, this formalisation can be used to as part of a pattern that allows safe queries for expression patterns involving negation.
3. Beyond EL: supporting queries with negation
In order to remain computationally tractable and scalable, VFB restricts expressiveness to the EL profile of OWL and uses the ELK reasoner [7] during development and to drive live OWL queries on the site. Recent advances in reasoning technology may make scaling with more expressive forms of OWL practical. For example, Zhou and colleagues have recently published impressive results for fast query answering by combining triple store based RL reasoning with a HermiT DL reasoner [12].
Some types of queries that would be extremely useful to our users require more expressiveness. In particular, there are a number of cases where queries involving negation would be useful. For example, for some neurons, we know all of the brain regions overlapped, all of the tracts fasciculated with and the location of all synaptic terminals. It would be useful, in such cases, to allow users to add negative legs to the compound queries for neuron classes that VFB already supports. For some transgene expression patterns in the adult brain, we have both negative and positive assertions about where a transgene is expressed. It would be very useful for researchers to be able to add negative clauses to queries for expression as this can be critical for choosing specific reagents that can be used to modulate the activity of particular neurons to assess their function.
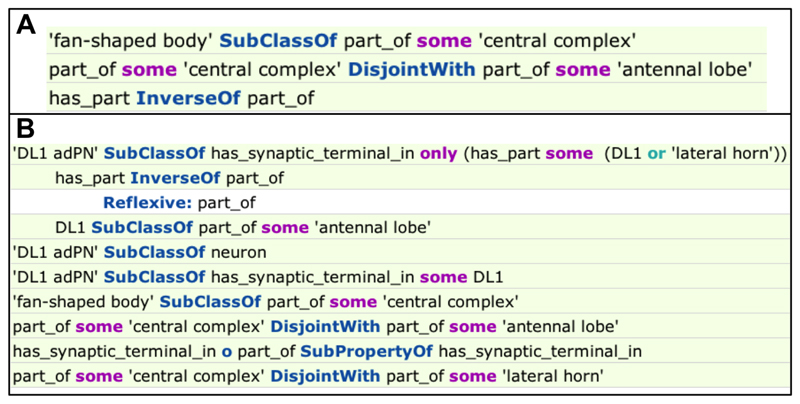
The most efficient way to support queries involving negation is to combine closure axioms and disjointness declarations. For example, the neuron DL1 adPN fasciculates with only one tract, the mALT. We currently record this as:
‘DL1 adPN’ subClassOf fasciculates_with some mALT
But if we also have the axioms:
‘DL1 adPN’ subClassOf fasciculates_with only mALT ‘great commissure’ disjointWith mALT
Then we can find ’DL1 adPN’ with the query:
neuron and not (fasciculates_with some ‘great commissure’)
For cases where a neuron fasciculates_with multiple tracts, the closure axioms can simply combine multiple classes using or. Unfortunately, our use of inference over partonomy rules out this pattern of closure axioms for many important relations used in querying. For example:
overlaps o part_of subPropertyOf overlaps
X overlaps some Y
X overlaps only Y
Y part_of some Z
Z disjointWith X
=>inconsistency: X overlaps some Z, X not (overlaps some Z)
We can get around this by using closure axioms of the form “rel only (has_part some X)” and declaring spatial disjointness between brain regions (which also provides a useful integrity check). Spatial disjointness can be declared using a simple GCI:
part_of some X disjointWith part_of some Y
For example, we can represent that the neuron DL1 adPN only has synaptic terminals in DL1 (part of the antennal lobe) and the lateral horn5 with:
‘DL1 adPN’
subClassOf : has_synaptic_terminals_in some DL1
subClassOf : has_synaptic_terminals_in some ‘lateral horn’
subClassOf : has_synaptic_terminals_in only (has_part only (DL1 or ‘lateral horn’))
‘fan-shaped body’ subClassOf : part_of some ‘central complex’
DL1 subClassOf : part_of some ‘antennal lobe’
With ‘antennal lobe’, ‘lateral horn’ and ‘central complex’ declared spatially disjoint, DL1 adPN is returned by the query:
neuron that (has_synaptic_terminal_in some ‘antennal lobe’) and not (has_synaptic_terminal_in some ‘fan-shaped body’)
An explanation is shown in figure 2B. There is no need to assert has_part relationships. The inverseOf axiom between has_part and part_of is sufficient to infer not has_part from spatial disjointness axioms (figure 2A).
Fig. 2.
A. Explanation for why the query “not has_part some ‘fan-shaped body’ ” returns ‘antennal lobe’. Note that direct assertion of has_part restriction axioms is not necessary. B. Explanation for why the query “neuron that (has_synaptic_terminal_in some ‘antennal lobe’) and not (has_synaptic_terminal_in some ‘fan-shaped body’)” returns the neuron ‘DL1 adPN’.
This pattern is also dependent on the reflexive nature of part_of and has_part (figure 2B).
Negative query legs in compound queries for expression patterns would be especially useful to our users. Our ability to provide these is limited by the extent to which it is possible to specify which regions lack expression. It is generally not possible to provide an exhaustive list of all regions lacking expression to use to define a closure axiom. However some datasets come with explicit assertions about regions not overlapped. For example, the largest transgene expression dataset that VFB currently hosts [6] was provided with annotations recording the presence or absence of expression in every major neuropil in the adult brain. These can easily be translated programmatically into restriction axioms asserting overlaps and not overlaps on expression pattern classes (figure 3).
4. Discussion and future directions
Virtual Fly Brain uses OWL to provide a unique service to the Drosophila neurobiology community, integrating a wealth of information from the literature and bulk datasets into an easily queryable resource. Much of this would be difficult or impossible to provide using a conventional relational database. OWL provides a sustainable way to develop and maintain a queryable classification of anatomical structures and neurons. OWL axiomatisation allowing inference over partonomy drives queries that return complete information about neuronal overlap and synaptic terminal location from any level of the partonomy. OWL reasoning also provides a way to group annotations of expression and phenotypes based on classification, partonomy and cell overlap. This massively enriches the results of annotation queries.
VFB has so far avoided taking advantage of the full expressiveness of OWL. Restricting expressiveness to the EL profile allows us to use the ELK reasoner, which gives classification and query answering times suitable for live use on the web. Reasoners such as HermiT[4] and FaCT++ [11] are many orders of magnitude slower at classifying the DAO and answering queries and, in our tests, are unable to completely classify the combined DAO and knowledgebase. However, we have one use case for which DL expressiveness would be extremely useful: compound queries for neurons or expression patterns involving negation.
There are two major barriers to achieving this. The most serious barrier is the ability to query across an ontology or combined ontology and knowledgeBase with DL expressiveness. Zhou and colleagues have recently published impressive results for fast query answering by combining triple store based RL reasoning with a HermiT DL reasoner [12]. We are working with the authors to test query speed for compound queries with negation for test datasets using the design patterns outlined in this paper.
A more clearly surmountable barrier is the lack of tooling support for some of the axiomatisation required in the design patterns we propose. In particular, adding GCIs to record spatial disjointness is currently very tedious to do by hand in Protege 5. This may be accomplished by scripting, but in order for the approach to be accessible for any ontology builder this would ideally be achieved via a plugin for a popular editor such as Protege. By analogy with support for the addition of class disjointness axioms in Protege, this could work by allowing users to navigate down a partonomy tree, adding disjointness axioms to whole sets of sibling terms at once.
5. Methods
For details of construction and maintenance of the Drosophila anatomy ontology please see Costa et al., 2013 [2]. The ontology is available from http://purl.obolibrary.org/obo/fbbt
VFB is an open source project. All code is available from https://github.com/VirtualFlyBrain OWL individuals files used on VFB are available from https://github.com/VirtualFlyBrain/VFB_owl/tree/master/src/owl. A test ontology illustrating implementation of the DL patterns for negative queries can be found at: http://purl.obolibrary.org/obo/fbbt/vfb/demo/owled2014_demo.owl.
5.1. VFB architecture
All queries for anatomical classes or individuals on VFB are live DL queries via the elk OWL reasoner. All queries of annotation begin with a DL query for subclasses, parts and overlapping cells. The resulting list is then used to query annotations store in the FlyBase Postgresql database. More details of the overall architecture of the project cen be found at https://github.com/VirtualFlyBrain/VFB#overall-architecture-of-project
5.2. Database representation of OWL individuals
Details of individuals are maintained in a SQL database (https://github.com/VirtualFlyBrain/VFB_owl/wiki/Individuals-DB) and programmatically converted to OWL using the OWL-API (https://github.com/VirtualFlyBrain/VFB_owl/). A standard DB representation of OWL ontologies/individuals would be preferable to our bespoke solution, which limits axiom expressiveness in order to keep the DB structure simple. We are currently unaware of any viable, non-proprietary alternatives.
Acknowledgments
We thank all those who have contributed to the development of VFB who are not authors on this paper: FlyBase, Michael Ashburner, J. Douglas Armstrong, Nestor Milyaev, Simon Reeve and Gregory S.X.E Jefferis.
Funding This work was largely supported by:‘Standardising the representation of Drosophila anatomy and development for databases’ BBBSRC:BB/G02233X/1 awarded 2009 to J.Douglas Armstrong, Michael Ashburner, Cahir O’Kane and David Osumi-Sutherland; An Isaac Newton Trust grant to Cahir O’Kane to fund the work of Marta Costa, awarded 2012:‘Neuroinformatic identification of new types of neuron in the Drosophila brain.’ VFB is currently supported by a Wellcome Trust grant to Cahir O’Kane, J. Douglas Armstrong, Gregory S.X.E Jefferis, Helen Parkinson and David Osumi-Sutherland: ‘Virtual Fly Brain: a global informatics hub for Drosophila neurobiology’ WT105023MA.
Footnotes
We stray outside the EL profile with inverse objectProperty declarations. To our knowledge, and based on extensive testing, these have no effect on classification and query answering with our current axiomatisation.
part_of and has_part are both transitive and reflexive; part_of inverseOf has_part
has_part entails overlaps
There is actually one additional region, but we simplify here in order to provide a more compact example
Author’s contributions
The OWL design patterns and queries presented in this paper were designed and tested by DOS He also designed the database representation of OWL individuals and wrote the code the translates this representation into OWL. DOS, MC, and RC all contributed to the representation of neuroanatomy in the DAO. MC was also responsible for annotation of individual neurons in the VFB knowledgeBase.
References
- 1.Chiang AS, Lin CY, Chuang CC, Chang HM, Hsieh CH, Yeh CW, Shih CT, Wu JJ, Wang GT, Chen YC, Wu CC, et al. Three-dimensional reconstruction of brain-wide wiring networks in Drosophila at single-cell resolution. Curr Biol. 2011 Jan;21(1):1–11. doi: 10.1016/j.cub.2010.11.056. [DOI] [PubMed] [Google Scholar]
- 2.Costa Marta, Reeve Simon, Grumbling Gary, Osumi-Sutherland David. The Drosophila anatomy ontology. Journal of Biomedical Semantics. 2013 Jan;4(1):32. doi: 10.1186/2041-1480-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.del Valle Rodriguez A, Didiano D, Desplan C. Power tools for gene expression and clonal analysis in Drosophila. Nat Methods. 2012 Jan;9(1):47–55. doi: 10.1038/nmeth.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glimm Birte, Horrocks Ian, Motik Boris, Shearer Rob, Stoilos Giorgos. A novel approach to ontology classification. Web Semantics: Science, Services and Agents on the World Wide Web. 2012;14(0) [Google Scholar]
- 5.Ito K, Shinomiya K, Ito M, Armstrong JD, Boyan G, Hartenstein V, Harzsch S, Heisenberg M, Homberg U, Jenett A, Keshishian H, et al. A systematic nomenclature for the insect brain. Neuron. 2014 Feb;81(4):755–765. doi: 10.1016/j.neuron.2013.12.017. [DOI] [PubMed] [Google Scholar]
- 6.Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, Iyer N, et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2012 Oct;2(4):991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kazakov Yevgeny, Krötzsch Markus, Simančík František. The incredible elk. Journal of Automated Reasoning. 2014;53(0):1–61. [Google Scholar]
- 8.Manton James D, Ostrovsky Aaron D, Goetz Lea, Costa Marta, Rohlfing Torsten, Jefferis Gregory SXE. Combining genome-scale drosophila 3d neuroanatomical data by bridging template brains. bioRxiv. 2014 [Google Scholar]
- 9.Milyaev N, Osumi-Sutherland D, Reeve S, Burton N, Baldock RA, Armstrong JD. The Virtual Fly Brain browser and query interface. Bioinformatics. 2012 Feb;28(3):411–415. doi: 10.1093/bioinformatics/btr677. [DOI] [PubMed] [Google Scholar]
- 10.Osumi-Sutherland D, Reeve S, Mungall CJ, Neuhaus F, Ruttenberg A, Jefferis GS, Armstrong JD. A strategy for building neuroanatomy ontologies. Bioinformatics. 2012 May;28(9):1262–1269. doi: 10.1093/bioinformatics/bts113. [DOI] [PubMed] [Google Scholar]
- 11.Tsarkov Dmitry, Horrocks Ian. FaCT++ description logic reasoner: System description. Lecture Notes in Artificial Intelligence; Proc of the Int Joint Conf on Automated Reasoning (IJCAR 2006); Springer; 2006. pp. 292–297. [Google Scholar]
- 12.Zhou Yujiao, Nenov Yavor, Grau Bernardo Cuenca, Horrocks Ian. Pay-as-you-go OWL query answering using a triple store. Proc of the 28th Nat Conf on Artificial Intelligence (AAAI 14); 2014. [Google Scholar]