Abstract
Two β-galactosidases from Lactobacillus, including a heterodimeric LacLM type enzyme from Lactobacillus reuteri L103 and a homodimeric LacZ type β-galactosidase from Lactobacillus bulgaricus DSM 20081, were studied for immobilization on chitin using a carbohydrate-binding domain (chitin-binding domain, ChBD) from a chitinolytic enzyme. Three recombinant enzymes, namely, LacLM-ChBD, ChBD-LacLM, and LacZ-ChBD, were constructed and successfully expressed in Lactobacillus plantarum WCFS1. Depending on the structure of the enzymes, either homodimeric or heterodimeric, as well as the positioning of the chitin-binding domain in relation to the catalytic domains, that is, upstream or downstream of the main protein, the expression in the host strain and the immobilization on chitin beads were different. Most constructs showed a high specificity for the chitin in immobilization studies; thus, a one-step immobilizing procedure could be performed to achieve up to 100% yield of immobilization without the requirement of prior purification of the enzyme. The immobilized-on-chitin enzymes were shown to be more stable than the corresponding native enzymes; especially the immobilized LacZ from L. bulgaricus DSM20081 could retain 50% of its activity when incubated at 37 °C for 48 days. Furthermore, the immobilized enzymes could be recycled for conversion up to eight times with the converting ability maintained at 80%. These results show the high potential for application of these immobilized enzymes in lactose conversion on an industrial scale.
Keywords: β-galactosidase, Lactobacillus, immobilization, chitin-binding domain
Introduction
β-Galactosidases (β-gals) (lactases, EC 3.2.1.23) are known as important enzymes for applications in the dairy industry,1 where they are used for lactose hydrolysis to produce low-lactose or lactose-free products as a response to lactose intolerance of consumers, which affects approximately 70% of the world population.2 Another useful property of β-gals is their transgalactosylation activity, by which health-promoting prebiotic galacto-oligosaccharides (GOS) can be formed from lactose.1,3 Many studies have demonstrated the indirect health benefits of GOS, where GOS promote growth and activity of beneficial intestinal microbes in the host.4–6
It is conceivable that β-gals from probiotic strains produce GOS that are also specific for these probiotics,7 and therefore β-gals from several probiotic Bifidobacterium spp.8,9 and prominently Lactobacillus spp. such as Lactobacillus reuteri, Lactobacillus plantarum, Lactobacillus sakei, Lactobacillus pentosus, or Lactobacillus bulgaricus have been studied in relation to GOS production during the past decade.10–14 β-Galactosidases from Lactobacillus species are reported to be of two main types, consisting of either two different subunits (heterodimeric or LacLM type) or two identical subunits (homodimeric or LacZ type).15,16 Two well-studied examples are the LacLM β-galactosidase from L. reuteri L10314 and the LacZ β-galactosidase from Lactobacillus delbrueckii subsp. bulgaricus (L. bulgaricus) DSM 20081,16 both belonging to glycoside hydrolase family GH2.
The recombinant β-gals from L. reuteri L103 and L. bulgaricus DSM20081 were successfully developed and overexpressed in food grade Lactobacillus hosts,16–18 yielding remarkably high levels of activity, for example, ca. 23−53 kU per liter of fermentation broth, respectively.16,17 The lactose conversion toward GOS of both native and recombinant β-gals from L. reuteri L103 and L. bulgaricus DSM 20081 were studied in detail in both batch and continuous bioreactors.14,16,19–21 Maximum GOS yields achieved were approximately 38% of total sugars (at a lactose conversion rate of 80% and an initial lactose concentration of ∼200 g/L) using LacLM from L. reuteri L103,20 or 50% GOS for recombinant LacZ from L. bulgaricus DSM 20081 (at 90–95% of lactose conversion and an initial lactose concentration of ∼200 g/L).16 These GOS mixtures contained mainly (nonlactose) disaccharides, trisaccharides, and tetrasaccharides, in which the transferred galactosyl moiety is attached via β-1,3- and β-1,6-linkages. These structures are important for potential prebiotics.20
To study the application of these enzymes in more detail, we aimed at immobilization of these two β-gals. To date, there have been no efforts on immobilization of β-gal from L. reuteri L103, even though β-gals are well-studied enzymes in terms of immobilization.22 Immobilization will enable the reuse of these biocatalysts, and it might also contribute to stabilization of the two β-gals, because their thermostability could be a limiting factor for their application.23
The chitin-binding domain (ChBD) is part of some chitin- or chitosan-degrading enzymes, chitinases or chitosanases, where it is responsible for the tight binding of the enzyme onto the substrate, thus increasing the hydrolytic activity.24,25 The ChBD of Bacillus circulans WL-12 chitinase A1 is one of the most well studied carbohydrate-binding domains. It belongs to the carbohydrate-binding module family 12 of the CAZY (carbohydrate-active enzyme) database. The three-dimensional structure of this ChBD has been determined by NMR.26 It consists of 45 amino acids, Ala655–Gln699, including several hydrophobic and aromatic residues with low solvent accessibility, thus forming a rigid and compact structure.26 Two β-sheets are composed of five strands including Thr660–Tyr662, Gln666–Tyr670, Lys673–Cys677, His681–Ser683, and Trp696–Leu698 residues.26 This ChBD has been reported to bind to only insoluble or crystalline chitin but not to the chitooligosaccharide, soluble derivatives of chitin, or other polysaccharides.27 On the basis of the high affinity to chitin, ChBD from WL-12 has been successfully applied for immobilization of several enzymes such as d-hydantoinase28 or levansucrase29 on chitin material. The procedure for the simultaneous purification and immobilization was a simple mixing of the enzyme solution with the insoluble chitin without the requirement of purified enzyme.28,29 Moreover, this method can overcome the disadvantages of other immobilization methods. For example, the covalent binding method immobilization required the harsh condition to create the bond between the enzyme molecule to the support material.23 The absorption method is based on weak forces such as van der Waals, hydrophobic interaction, or hydrogen interaction; thus, the immobilized enzymes are loosely bound to support materials.30
This study focuses on the immobilization of heterodimeric L. reuteri L103 LacLM and homodimeric L. bulgaricus DSM20081 LacZ β-gal on chitin using ChBD of B. circulans WL-12 chitinase A1. The recombinant enzymes were fused to the ChBD via DNA-based molecular methods. The biochemical characteristics of these fusion enzymes in comparison to the native enzymes are shown; thus, the effects of the ChBD and the immobilization on the properties of these enzymes are elucidated.
Materials and Methods
Chemicals and Reagents
All chemicals were purchased from Sigma (St. Louis, MO, USA) unless otherwise stated and were of the highest quality available. The endonucleases were purchased from New England BioLabs (Ipswich, MA, USA) and used as recommended by the supplier. T4 DNA ligase was from Fermentas (Vilnius, Lithuania).
Bacterial Strains and Media
The bacterial strains, plasmids, and primers used in this study are listed in Tables 1 and 2. L. plantarum WCFS131 was grown in MRS medium (Oxoid, Basingstoke, UK) at 37 °C without agitation. Escherichia coli NEB5 α (New England BioLabs, Ipswich, MA, USA) as cloning host was cultivated in Luria-Bertani (LB) medium at 37 °C with shaking at 200 rpm. Agar media were prepared by adding 1.5% agar to the respective media. Unless otherwise stated, the antibiotic concentrations were 5 or 200 μg/mL of erythromycin (Erm) for Lactobacillus or E. coli, respectively, and 100 μg/mL of ampicillin for E. coli.
Table 1.
Strains and Plasmids Used in This Study
| strain or plasmid | relevant characteristic | reference |
|---|---|---|
| strains | ||
| L. plantarum WCFS1 | wild type | 31 |
| E. coli NEB5α | cloning host | NEB |
| plasmids | ||
| pJETlacZ | source for lacZ of L. bulgaricus DSM20081 | 16 |
| pSIP403 | Emr, pSIP401 derivative, gusA controlled by PsppA | 32 |
| pTH103.3 | source for lacLM from L. reuteri | 15 |
| pTxB1 | source for chbd sequence | NEB |
| pSCBD | derivative of pSIP403, carrying chbd | this study |
| pSCBDlac1 | pSCBD, gusA replaced by lacLM | this study |
| pSCBDlac2 | pSIP403, gusA replaced by fused chbd_lacLM | this study |
| pSCBDlac3 | pSCBD, gusA replaced by lacZ from L. bulgaricus | this study |
Table 2.
Primers Used in This Study
| target gene | primer | sequencea (5′−3′) |
|---|---|---|
| chbd from pTxB1 | F1 (XhoI) | C TCACTC GAGACGACAAATCCTGGTGTATCCGC |
| R1 (Acc65I) | C TTCGGTACCTCATTGAAGCTGCCACAAGGC | |
| F2 (BsaI) | GCAGGTCTC CCATG GCG ACA AAT CCT GGT GTA TCC | |
| R2 | CCA TTT TAT ATT TGC TTG TTGAAGCTGCCAC | |
| lacLM of L. reuteri L103 | F3 | G TGG CAG CTT CAA CAA GCA AAT ATA AAA TGG |
| R3 (XhoI) | CG CTCGAG TTA TTT TGC ATT CAA TAC AAA CG | |
| lacZ of L. bulgaricus DSM20081 | F4 (BsmbI) | GCTGCGTCTCCC ATG AGC AAT AAG TTA GTA AAA G |
| R4 (XhoI) | GAAGCTCGAG TGA TTT TAG TAA AAG GGG |
The italic sequences in R2 and F3 are complementary. The restriction enzyme recognizing sites are underlined.
Oligonucleotide primers for PCR amplification were supplied by VBC-Biotech Service GmbH (Vienna, Austria). The appropriate endonuclease restriction sites were introduced in the forward and reverse primers (Table 2).
Plasmid Construction and Transformation
The DNA amplification was performed with Phusion High-Fidelity DNA polymerase (Finnzymes, Espoo, Finland). Plasmid DNA from E. coli was purified using the Gene Elute plasmid Miniprep kit (Sigma, St. Louis, MO, USA). DNA was purified with the WizardRSV Gel and PCR Clean-up system kit (Promega, Madison, WI, USA). The pJET1.2 plasmid (CloneJET PCR cloning kit, Fermentas) was used for subcloning when necessary.
Three recombinant fusion proteins were constructed. Two are based on LacLM from L. reuteri L103, and the chitin-binding domain ChBD was attached upstream of LacL (termed ChBD-LacLM) or downstream of LacM (termed LacLM-ChBD). The third fusion protein was based on LacZ of L. bulgaricus DSM20081 with ChBD linked downstream of LacZ (termed LacZ-ChBD).
For construction of the expression plasmids of the fusion protein LacLM-ChBD or LacZ-ChBD, the fragment of chbd of B. circulans WL-12 chitinase A1 was amplified from the plasmid pTxB1 (New England BioLabs) with the primer pair F1 and R1 (Table 2). After double digestion with XhoI and Acc65I, this fragment was ligated to the fragment of pSIP403,32 which contained gusA as the original reporter gene and was treated by the same endonucleases, resulting in plasmid pSCBD-gusA (Table 1; Figure 1). The gusA gene was then excised by double digestion with NcoI and XhoI to obtain the empty vector pSCBD. This empty vector was ligated to the NcoI–XhoI fragment of lacLM (encoding LacLM from L. reuteri L103) from pHA103.1,15 resulting in pSCBDlac1 (Table 1; Figure 1). Similarly, lacZ was amplified from pJETlacZ16 by PCR with the two primers F4 and R4 (Table 2) and then digested by BsmBI and XhoI prior to ligation into empty pSCBD, resulting in pSCBDlac3 (Table 1; Figure 1).
Figure 1.
Construction of expression plasmids: pSCBDlac1 (for LacLM-ChBD) and pSCBDlac3 (for LacZ-ChBD) (A) and pSCBDlac2 (for ChBD-LacLM) (B).
For construction of the expression plasmid for ChBD-LacLM, chbd from pTxB1 was amplified with the two primers F2 and R2, resulting in a fragment of F2-chbd-R2. The lacLM gene was PCR-amplified from the lacLM template in pTH103.1 using the F3 and R3 primers, resulting in the fragment F3-lacLM-R3. Because of the 18 nt complementary sequence between F3 and R2 (Table 2), the fragment F2-chbd-R2 could be used as a mega forward primer in combination with R3 as reverse primer to amplify the F3-lacLM-R3 template, resulting in the fragment F2-chbd-lacLM-R3. The PCR product from this amplification was digested by BsaI and XhoI and ligated to NcoI–XhoI-digested pSIP403, resulting in pSCBDlac2 (Table 1; Figure 1).
This strategy resulted in expression vectors, in which the target gene, lacLM or lacZ linked to the chbd sequence, is controlled by the inducible promoter PsppA, similar to gusA in the original pSIP403 vector.32 These plasmids were then electroporated into competent cells of L. plantarum WCFS1 as previously described.33 The transformants carrying the plasmids were determined and screened by using a colony PCR amplification of the target fusion genes.
Evaluation of the Expression of Recombinant β-Galactosidases
This experiment was performed following a previous method.18
Fermentation
To obtain sufficient amounts of the enzymes for characterization, immobilization, and application, L. plantarum WCFS1 harboring different plasmids was cultivated in 1 L of medium to obtain larger amounts of the recombinant enzymes. The bacterial cells were induced at OD600 nm ∼ 0.3 and harvested at OD600 nm ∼ 6. The cell pellets were disrupted by homogenization with a French press (Aminco, Silver Spring, MD, USA). The cell-free extract, clarified by ultracentrifugation (Beckman, USA) at 30000g and 4 °C for 20 min, was used either directly for immobilization on chitin beads (New England BioLabs) or for a single-step affinity purification using the p-aminobenzyl 1-thio-β-d-galactopyranoside (ABTG) resin (Sigma) as described in a previous study.14
Immobilization on Chitin Beads
Before use, the chitin beads were washed three times with sodium phosphate buffer (50 mM, pH 6.5) and resuspended in the same buffer. Five hundred microliters of chitin bead suspension was mixed with the same volume of diluted cell-free crude extracts of 1000 U/mL of oNPG activity. The immobilization experiments were carried out at 4 °C with gentle agitation for 1–18 h. Chitin beads were separated from the supernatant by filtration and rinsed with sodium phosphate buffer. The supernatant and wash solutions were collected and pooled for SDS-PAGE analysis and protein measurement. Chitin beads were resuspended in buffer for further studies.
The immobilization yield (IY) and the activity retention (AR) were calculated according to the methods of Klein et al.34 IY (%) is defined as the ratio of immobilized activity in relation to total applied activity, in which the immobilized activity was determined by subtraction of the residual activity in the supernatant after immobilization from the total applied activity. AR (%) is the percentage of activity measured on chitin beads in relation to the theoretical immobilized activity.
The amount of chitin-bound protein was indirectly estimated by subtraction of the protein concentration in the supernatant after immobilization from that in the crude extract (prior to the immobilization).
Influence of pH and Temperature on the Activity and Stability of Immobilized Enzymes
The pH optimum of β-gal activity was evaluated in Britton–Robinson buffer17 with the pH ranging from 4 and 9. To determine the pH stability, the immobilized enzymes were incubated at 30 or 37 °C in Britton-Robinson buffer of different pH values. At various time intervals, the residual activities were measured with the oNPG assay. The inactivation constants kin were obtained by linear regression of ln(residual activity) versus time. The half-life values τ1/2 were calculated using τ1/2 = ln(2)/kin.35
The optimum temperature for hydrolysis activity of β-gals with both substrates lactose and oNPG was determined in the range of 20–80 °C. For estimation of the kinetic thermostability, enzymes were incubated in 50 mM sodium phosphate buffer, pH 6.5, at different temperatures ranging from 20 to 80 °C. At various time intervals, residual activities were measured with oNPG as substrate and plotted versus the incubation time. The half-life values of thermal inactivation (τ1/2) were similarly calculated as above.
Steady-State Kinetic Measurements
To estimate the kinetic parameters of recombinant and fusion β-gals for lactose and oNPG, substrate concentrations were varied from 1 to 25 mM for oNPG and from 10 to 600 mM for lactose.14 The enzyme assays were performed for 10 min at 30 °C in 50 mM sodium phosphate buffer, pH 6.5. The kinetic parameters were calculated by using the Henri–Michaelis–Menten model and nonlinear least-squares regression.
Reusability of the Immobilized Enzymes
To assess the possibility of recycling the immobilized enzyme preparations for conversion, immobilized β-gal LacLM L. reuteri L103 was added to sodium phosphate buffer (50 mM, pH 6.5) containing 600 mM lactose and maintained at 30 °C and 600 rpm agitation for the conversion of lactose. Every 24 h, the chitin beads carrying the enzyme were filtered off, rinsed with buffer, and reused in another batch conversion experiment under identical conditions. The filtrates from every conversion cycle were collected to determine the glucose concentration by using a commercial d-glucose assay kit (GOPOD Format, Megazyme, Wicklow, Ireland). The glucose concentration in the filtrate from the first conversion was taken as 100% of converting ability of the immobilized enzyme preparation.
Immobilized LacZ from L. bulgaricus DSM 20081 was also tested for its reusability but using the substrate oNPG. After 5 min of incubation with o-nitrophenyl-β-d-galactopyranoside (oNPG) at 30 °C, chitin beads carrying LacZ were separated from the liquid phase and reused in the next conversion experiments. The concentration of oNPG in the liquid phases after the conversion was measured with a spectrophotometer at 420 nm and used to calculate the residual converting ability of enzyme in each repeating conversion.
Lactose Hydrolysis and Transgalactosylation Experiments
The transformation of lactose was carried out in batch mode. Both chitin-immobilized I-LacLM-ChBD and purified soluble LacLM-ChBD were added at equal concentrations of 1.5 Ulactose/mL to sodium phosphate buffer (50 mM, pH 6.5) containing 10 mM MgCl2 and 205 g/L lactose. Lactose conversion experiments were performed at 30 °C for 24 h at 600 rpm of agitation.
The chitin-immobilized β-gal of L. bulgaricus was applied in different amounts ranging from 1.7 to 9.7 Ulactose/mL to sodium phosphate buffer (50 mM, pH 6.5) containing 10 mM MgCl2 and 50 or 205 g/L lactose. This enzyme preparation was also used for conversion of lactose in ultrahigh-temperature-treated whole cow’s milk. The incubation temperature was varied from 37 to 60 °C.
In all conversion experiments, samples were periodically withdrawn, heated at 99 °C for 5 min, and further analyzed for lactose, galactose, glucose, and GOS present in the samples. Qualitative analysis of sugar was performed by thin-layer chromatography (TLC) as described previously by Nguyen and co-workers.14 Furthermore, samples from conversion experiments were quantitatively analyzed by high-performance liquid chromatography (HPLC) (Dionex, Germany) equipped with a refractive index detector and an Aminex HPX-87K (300 mm × 7.8 mm) carbohydrate analysis column (Bio-Rad, Hercules, CA, USA). The chromatographic separation was performed at 80 °C with ultrapure water used as eluting solvent at a flow rate of 0.5 mL/min. The concentration of saccharides was calculated by interpolation from external standards. Total GOS concentration was calculated by subtraction of the quantified saccharides (lactose, glucose, galactose) from the initial lactose concentration. The GOS yield (%) was defined as the percentage of GOS produced in the samples compared to initial lactose.
β-Galactosidase Assay
β-Galactosidase activity was determined using oNPG or lactose as substrate following the method of Nguyen et al.14 In brief, the activity assays were performed in 50 mM sodium phosphate buffer of pH 6.5 at 30 °C, and the final substrate concentrations in the 10 min assays were 22 mM for oNPG and 600 mM for lactose. One unit of oNPG activity was defined as the amount of enzyme releasing 1 μmol of oNP per minute, whereas 1 unit of lactase activity was defined as the amount of enzyme releasing 1 μmol of d-glucose per minute under the given conditions.
Protein Measurement
Protein concentration was determined by using the method of Bradford36 with bovine serum albumin (BSA; Sigma) as standard.
Sodium Dodecyl Sulfate–Polyacrylamide Gel Electrophoresis (SDS-PAGE) Analysis
For visual observation of the expression level of the recombinant β-gals in L. plantarum WCFS1 and the effectiveness of the immobilization, cell-free extracts, liquid phases, and chitin beads (after immobilization) were analyzed by SDS-PAGE using 3-(N-morpholino)propanesulfonic acid (MOPS) as running buffer.16
Statistical Analysis
All experiments and measurements were performed at least in duplicate, and the data are given as the mean ± standard deviation when appropriate. Student’s t test was used for the comparison of data with significance value α = 0.05.
Results and Discussion
Plasmid Construction and Expressions of Recombinant β-Galactosidases in L. plantarum WCFS1
In this work, the three expression plasmids pSCBDlac1, pSCBDlac2, and pSCBDlac3, containing the sequences of the three recombinant fusion proteins LacLM-ChBD, ChBD-LacLM, and LacZ-ChBD, respectively, were constructed and successfully electroporated into L. plantarum WCFS1. The transcription of the encoding sequences lacLM-chbd, chbd-lacLM, and lacZ-chbd is regulated by the inducible promoter PsppA in these plasmids (Figure 1). The expression levels of the different constructs and the recombinant β-galactosidases in L. plantarum WCFS1 were investigated.
Induced cells formed intracellular LacLM-ChBD at around 32 U/mL of fermentation broth with a specific activity of ca. 179 U/mg protein (Table 3). ChBD-LacLM was expressed in 3-fold lower yields (p < 0.05), giving ca. 11 U of β-galactosidase activities per milliliter of fermentation broth with a specific activity of 54 U/mg. The basal expression from the expression plasmids in noninduced cells was unexpectedly high (7.35 and 4.06 U/mg for LacLM-ChBD and ChBD-LacLM, respectively (Table 3)). The induction ratios (ratio of the β-galactosidase activities obtained under induced conditions divided by the activity under noninduced conditions in cells harvested at similar OD600 values of 1.8) were 24 and 14, respectively, which is lower than previously reported for different target genes expressed with the same expression vector pSIP403.16,17 It should be noted that the activity level produced by the wildtype host strain under identical growth conditions is very low (0.07 U/mg),15 and hence the activities measured can be attributed solely to the recombinant enzymes.
Table 3.
β-Galactosidase Activity in Cell-free Lysate of the Induced and Noninduced Cells of L. plantarum WCFS1 Carrying Various Plasmidsa
| volumetric activity (U/mL fermentation broth) |
specific activity (U/mg protein) |
||||
|---|---|---|---|---|---|
| plasmid | noninduced | induced | noninduced | induced | induction factor |
| pSCBDlac1 | 1.45 ± 0.48 | 32.86 ± 3.55a | 7.35 ± 2.44 | 178.90 ± 10.05e | 24 |
| pSCBDlac2 | 0.77 ± 0.19 | 11.31 ± 1.20b | 4.06 ± 1.09 | 57.29 ± 6.24f | 14 |
| pSCBDlac3 | 16.77 ± 1.02 | 65.83 ± 1.53c | 9.43 ± 0.14 | 40.27 ± 2.36g | 4 |
The data are expressed as the mean ± standard deviation from three independent cultivations. The induction factors are calculated by dividing the specific activity (U/mg) of induced and noninduced cells. The letters indicate significant difference (p < 0.05).
The expression of LacZ-ChBD in L. plantarum WCFS1 gave a higher volumetric activity (around 65.83 U/mL) but lower specific activity (40.27 U/mg) when compared to the expression of LacLM-ChBD (p < 0.05). Again, a rather high basal activity was observed in noninduced cultivation (16.77 U/mL and 9.43 U/mg), even though PsppA is known as a tightly controlled promoter.32,37 As a consequence, a low induction factor of around 4 was obtained with this system (Table 3).
The fusion of ChBD had significant effects on expression levels of the recombinant enzymes. This is obvious from, for example, LacZ, as expression of the lacZ gene without fusion to the chbd fragment gave much higher expression (i.e., 180-190 U/mg) with the same host, expression system, and induction conditions.16 Moreover, the expression levels of the recombinant proteins are remarkably different depending on the position of ChBD when fused to LacLM. The lower expression levels were observed when chbd was fused upstream of lacL (ChBD binding N-terminally to the large subunit LacL) (Table 3). It is shown that the nucleotide sequences at the beginning of a gene can change the stability of the secondary structure of the transcribed mRNA and thus affect the translation process.38 The secondary structures of the chbd-lacLM mRNA and lacLM-chbd mRNA are predicted by the Mfold tool (http://unafold.rna.albany.edu/?q=mfold) (data not shown). With chbd-lacLM, a hairpin loop including the ribosome-binding site RBS (AGGAG) is predicted in the region of the 5'UTR and the initial nucleotides of the chbd. This hairpin loop might prevent the binding of ribosome to the mRNA to initiate the translation process. Meanwhile, with lacLM-chbd, the RBS is not involved in the hairpin loop; hence, it might be an explanation for the higher expression level of the LacLM-ChBD fusion protein mentioned above.
SDS-PAGE analysis of cell-free extracts of L. plantarum WCFS1 harboring the different plasmids clearly indicates strong bands for the recombinant proteins at approximately 72 and 41 kDa (lane 2, Figure 2A) for LacLM-ChBD, 78 and 35 kDa (lane 4, Figure 2A) for ChBD-LacLM, and ∼120 kDa (lane 2, Figure 2C) for LacZ-ChBD. These sizes are in agreement with those of LacL (72 kDa) and LacM (35 kDa) from L. reuteri L10314,15 and lacZ from L. bulgaricus DSM20081 (115 kDa),16 and ChBD (∼6 kDa).
Figure 2.
SDS-PAGE analysis of expression and immobilization of recombinant enzymes. (A) Cell-free extracts of noninduced cells (lanes 1, 3) and induced cells (lanes 2, 4) of L. plantarum WCFS1 harboring pSCBlac1 (containing LacLM-ChBD) and pSCBlac2 (containing ChBD-LacLM), respectively. (B) Cell-free crude extracts (lanes 1, 4), flow through during immobilization (lanes 2, 5), and chitin-bound β-gals (lanes 3, 6) of L. plantarum harboring plasmids pSCBDlac1 (containing LacLM-ChBD) and pSCBDlac2 (containing ChBD-LacLM), respectively. (C) Cell-free crude extracts of L. plantarum WCFS1 harboring pSCBDlac3 (containing LacZ-ChBD) at OD ~ 0.3 (before induction) (lane 1) and at 16 h of induction (lane 2); flow through during immobilization (lane 3) and rinsing fractions (lanes 4, 5), chitin-bound LacZ (lane 6); and chitin beads (lane 7). Lane M shows the Precision Plus Protein standard (Bio-Rad). The arrows indicate subunits of the recombinant β-galactosidases.
The deduced amino acid sequences of the three fusion proteins were used to predict their 3D structures using the RaptorX tool (http://raptorx.uchicago.edu/). The prediction shows that the ChBD domain arranges well separated from the main protein LacM (in the case of LacLM-ChBD) and LacZ (in the case of LacZ-ChBD), but not with LacL (for ChBD-LacLM), where it interacts more tightly with the LacL subunit (see Figure 10). It should be noted that with LacZ-ChBD, the ChBD fold was not predicted accurately, as it was not in agreement with the structure of ChBD alone (see Figure 10D) or in LacM-ChBD (see Figure 10A); nevertheless, it was predicted to be separately positioned from the main protein (Figure 10C). It should be noted that the chbd from pTxB1 encodes for ChBD from chitinase A1 of B. circulans WL-12 with 45 amino acids, from Ala655 to Gln699, as previously reported26 and 7 extra residues at the N-terminus. Moreover, 5 residues upstream of Thr660 are not involved in the essential structural region of ChBD.26 The sequence of 12 residues might be a flexible linkage between LacM or LacZ to ChBD. Meanwhile, for ChBD-LacLM, the linkage is only the last Gln699 of ChBD, which is not involved in forming the β-sheet.26 These predicted structures indicate that in both LacLM-ChBD and LacZ-ChBD, the interaction of ChBD with chitin beads will affect the main protein to a lesser extent than in ChBD-LacLM. Even though these structures need to be confirmed by crystallization/structure elucidation, they are in agreement with our further results in this study (see below).
Figure 10.
Predicted 3D structures of LacM-ChBD (A), ChBD-LacL (B), LacZ-ChBD (C), and ChBD (D). The 3D structures were predicted by the RaptorX tool (http://raptorx.uchicago.edu/) on the basis of the deduced amino acid sequences.
Immobilization of β-Galactosidases on Chitin Beads
The three different fusion proteins were used at equal activities of 500 UoNPG to study their immobilization on chitin beads (11 mg of dry weight), and the residual activities in the liquid phase after the immobilization reaction as well as the bound activities were subsequently analyzed for all three constructs (Table 4). An important parameter in immobilization is the immobilization yield (IY) as it is an indication of how much of the applied protein is actually bound to the carrier. Immobilization yields for both LacLM-ChBD and LacZ-ChBD were very high, at >91%, and especially the IY of LacLM- ChBD, which was close to 100% (0.67% of residual activity detected in the supernatant after immobilization), is very promising. In contrast, the IY of ChBD-LacLM was much lower, at ca. 52% (Table 4). These values are further corroborated by SDS-PAGE analysis using samples of the liquid phases and chitin beads after immobilization (Figures 2B and 2C). The bands of ChBD-LacL and lacM or LacZ-ChBD can be seen in the lanes of liquid phase samples (lane 5 in Figure 2B; lane 3, 4, and 5 in Figure 2C). The patterns of chitin bead samples, with only two bands corresponding to the two subunits of immobilized LacLM (LacL and LacM-ChBD in lane 3 and ChBD-LacL and LacM in lane 6, Figure 2B) or the single strong band of LacZ-ChBD (Figure 2C, lane 6) with only a minor very faint band indicating an impurity, indicate the high specific affinity of ChBD to chitin.25 This observation is in accordance with many other studies using ChBD for enzyme immobilization.28,29,39 LacLM from L. reuteri L103 is a heterodimer consisting of two subunits. Fusing the ChBD fragment N-terminally to LacL or C-terminally to LacM clearly had different effects on the immobilization of the recombinant fusion enzymes on chitin beads. A possible explanation for these results could be that in the ChBD-LacLM fusion the ChBD is positioned in such a way that it cannot interact with the chitin beads unperturbed. This is in agreement with the predicted structures of these fusion proteins (see Figure 10). LacZ from L. bulgaricus DSM20081 is a homodimer,16 and therefore each of the identical subunits will carry its own ChBD. These two chitin-binding domains per fusion protein might interfere with each other when binding on chitin, which could be the reason for the slightly lower efficiency in immobilization compared to LacLM-ChBD (Table 4).
Table 4.
Immobilization of Recombinant Enzymes on Chitin Beadsa
| enzyme | residual activity in liquid phase (%) | IYb (%) | residual activity on beadc (%) | ARd (%) | residue activity (kU/g chitin bead) |
|---|---|---|---|---|---|
| LacLM-ChBD | 0.67 ± 0.15e | 99.33 | 25.89 ± 2.49 | 26.06 | 11.77 |
| ChBD-lacLM | 48.36 ± 4.50f | 51.64 | 9.19 ± 0.50 | 19.00 | 4.17 |
| lacZ-ChBD | 8.75 ± 0.47g | 91.25 | 12.50 ± 0.61 | 13.70 | 6.25 |
The applied enzyme activity was 500 U, and 11 mg (dry weight) of chitin beads were used. The immobilization was used for 18 h at 4 °C with gentle agitation. Letters e, f, and g indicate significant difference (p < 0.05).
IY (%) was calculated by subtraction of the residual enzyme activity (%) in the liquid phase from the total activity applied (100%).
Residual activity (%) on beads is the percentage of total activity on chitin beads to the applied activity. To determine activity on a bead, the bead suspension was used in enzyme assay.
Activity retention, AR (%) is the ratio of residual activity to IY.
Even though the immobilization yields were very high for two of the constructs (Table 4), the activity retention (AR) on chitin beads was much lower than expected. The residual activities on chitin beads for ChBD-LacLM and LacLM-ChBD were ∼9.19 and 25.89% in comparison with the initially applied activity, respectively. This amounted to AR values of 19–25%. A low AR value of ca. 13.7% was also observed for LacZ-ChBD. However, AR depends on the applied activity of our enzymes, because applying lower activities resulted in higher AR values (data not shown). To date, various methods have been used for immobilization of fungal or bacterial β-gals, yielding different ARs.22,40 For instance, a range of 2.5–9.5% of ARs of β-galactosidase from E. coli, which was immobilized on poly(2-hydroxyethyl methacrylate) membrane using entrapment, was reported by Baran and co-workers.41 However, most of these immobilization methods are based on purified enzymes, whereas in this study a single-step procedure was performed for the immobilization.
Partial Characterization of Immobilized β-Galactosidases
The effect of ChBD and of the immobilization on the characteristics of the β-gals was further assessed. Three chitin-immobilized enzymes of LacLM-ChBD, ChBD-LacLM, and LacZ-ChBD are termed I-LacLM-ChBD, I-ChBD-LacLM, and I-LacZ-ChBD, respectively, hereafter. Free LacLM-ChBD was also purified and partially characterized to compare with the immobilized I-LacLM-ChBD.
Steady-State Kinetic Constants
Table 5 presents the kinetic constants of the recombinant fusion β-gals for their two substrates, lactose and oNPG. For I-LacLM-ChBD, the vmax values for both substrates are higher than those of I-ChBD-LacLM (p < 0.05). With 1.74 and 0.72 μmol min−1 mg−1 these vmax values for the natural substrate lactose are much lower than the corresponding value of the free fusion protein (17.8 μmol min−1 mg−1), as well as LacLM isolated from its natural source L. reuteri L103,14 or expressed recombinantly either in E. coli15 or in L. plantarum WCFS17 with vmax values ranging from 34 to 43 μmol min−1 mg−1 (Table 5). The Michaelis constant Km was also negatively affected by the immobilization, but judging from the Km values determined, this effect is much less dramatic than for vmax. This obviously indicates that the hydrolysis activity of the chitin-bound fusion enzymes is negatively influenced by immobilization. The effect of immobilization on kinetic parameters of I-LacZ-ChBD was also negative, but here the decrease in vmax and hence kcat was less drastic (Table 5).
Table 5.
Kinetic Parameters of the Immobilized β-d-Galactosidases for the Hydrolysis of Lactose and o-Nitrophenyl-β-d-galactopyranoside (oNPG) in Comparison to the Respective Free Enzymes without ChBDa
| substrate | method for determination of enzyme activity | kinetic parameter | LacLM-ChBDb | I-LacLM-ChBD | I-ChBD-LacLM | L103c | EcoL103d | LpL103e | I-LacZ-ChBD | LacZf |
|---|---|---|---|---|---|---|---|---|---|---|
| lactose | release of d-glucose | vmax,Glu (μmol min−1 mg−1) | 17.83 ± 0.14 | 1.74 ± 0.07 | 0.72 ± 0.01 | 34 | 38 | 43 | 39.9 ± 0.9 | 123 ± 5 |
| Km,Lac (mM) | 51.74 ± 1.80 | 36.82 ± 0.33 | 12.25 ± 0.39 | 13 | 12 | 12 | 45.5 ± 4.2 | 19.2 ± 3.8 | ||
| Kcat (s−1) | 9.00 ± 0.07 | 0.88 ± 0.03 | 0.36 ± 0.01 | 18 | 20 | 23 | 79.8 ± 1.8 | 234 ± 13 | ||
| Kcat/Km (s−1 M−1) | 174 | 24 | 29 | 1389 | 1681 | 1902 | 1754 | 12300 | ||
| oNPG | release of oNP | vmax,oNP (μmol min−1 mg−1) | 103.9 ± 1.82 | 65.40 ± 1.78 | 32.09 ± 0.14 | nd | nd | nd | 87.1 ± 2.8 | 317 ± 6 |
| Km,oNPG (mM) | 0.91 ± 0.05 | 2.18 ± 0.45 | 1.34 ± 0.03 | nd | nd | nd | 13.7 ± 1.1 | 0.92 ± 0.09 | ||
| Kcat (s−1) | 52.39 ± 0.92 | 32.98 ± 0.90 | 16.18 ± 0.07 | nd | nd | nd | 174.2 ± 5.6 | 603 ± 15 | ||
| Kcat/Km (s−1 M−1) | 54970 | 15096 | 12032 | nd | nd | nd | 12715 | 655000 | ||
The three chitin-immobilized enzymes of LacLM-ChBD, ChBD-LacLM, and LacZ-ChBD are designated I-LacLM-ChBD, I-ChBD-LacLM, and I-LacZ-ChBD, respectively. The bound amount of protein on the chitin beads was calculated by subtraction of the protein content in the cell crude extract (before immobilization) and in the liquid phase (after immobilization). The difference of vmax and Km values among the immobilized enzymes and the free-soluble enzyme is statistically significant (p < 0.05). nd, not determined.
Free soluble LacLM-ChBD.
Native enzyme from L. reuteri L103.14
Recombinant enzyme expressed and purified from E. coli.15
Recombinant enzyme expressed and purified from L. plantarum WCFS1.17
Purified recombinant enzyme LacZ from L. bulgaricus expressed in L. plantarum.16
Effect of Temperature and pH on Enzyme Activity
The immobilized fusion enzymes typically showed their highest activity at 50–55 °C with both oNPG and lactose as substrate for the 10 min assays (Figure 3); this is also the optimal range of temperature observed with one of the counterparts, the soluble enzyme LacLM-ChBD (data not shown). At higher temperatures (>70 °C) the enzyme activity was rapidly lost. The range of optimal temperature for the activity of the immobilized enzymes is comparable to that of the free enzyme without ChBD from the native strain L. reuteri L103 or recombinant LacLM expressed in different hosts.14,15,17 Interestingly, the immobilized preparation I-LacZ-ChBD showed a slightly increased temperature optimum of 65 °C (Figure 3) compared to the free enzyme LacZ without the ChBD with an optimum of 60 °C.16 We did not observe a similar increase for I-LacLM-ChBD and I-ChBD-LacLM, the reason for which might be that only one subunit is involved in binding to chitin when LacLM is used.
Figure 3.
Temperature optimum of the immobilized enzymes. I-LacLM-ChBD (●) and I-ChBD-LacLM (▽) indicate the recombinant enzymes with the ChBD fused to the C-terminal of LacM and the N-terminal of LacL, respectively. I-LacZ-ChBD (◆) indicates the immobilized LacZ-ChBD. The solid and dotted line indicate the substrate oNPG and lactose, respectively.
The thermostability of the different immobilized β-galactosidase preparations was tested at various temperatures in 50 mM sodium phosphate buffer, pH 6.5, by determination of the half-life times τ1/2 (Table 6). I-LacZ-ChBD proved to be the most stable preparation at all tested temperatures. For example, τ1/2 of this enzyme was 203 h at 37 °C, whereas it was <170 h for I-LacLM-ChBD and I-ChBD-LacLM at 30 °C. Free β-gal of L. bulgaricus was previously shown to be relatively thermostable (τ1/2 for 145 and 345 h without and with, respectively, the presence of 10 mM Mg2+ at 37 °C).16 Furthermore, the stability of the immobilized preparation I-LacZ-ChBD was further increased in the presence of 10 mM Mg2+ to a half-life time τ1/2 of almost 48 days (1155 h) at 37 °C (Table 7). The soluble fusion protein LacLM-ChBD was considerably less thermostable than its immobilized counterpart I-LacLM-ChBD (τ1/2 of 19 h versus 169 h at 30 °C; Table 6); thus, immobilization onto chitin beads via the chitin-binding domain can improve the kinetic stability of an enzyme significantly.
Table 6.
Thermostability of Recombinant Immobilized β-Galactosidases from Lactobacillusa
| temperature (°C) | LacLM-ChBD |
I-LacLM-ChBD |
I-ChBD-LacLM |
I-LacZ-ChBD |
||||
|---|---|---|---|---|---|---|---|---|
| kin (h−1) | τ1/2 (h) | kin (h−1) | τ1/2 (h) | kin (h−1) | τ1/2 (h) | kin (h−1) | τ1/2 (h) | |
| 30 | 0.036 | 19.254 | 0.0041 | 169.060 | 0.006 | 115.524 | ndb | nd |
| 37 | nd | nd | nd | nd | nd | nd | 0.0034 | 203.867 |
| 40 | 0.33 | 2.100 | 0.095 | 7.296 | 0.188 | 3.687 | nd | nd |
| 50 | 26.84 | 0.026 | 1.92 | 0.361 | 3.64 | 0.190 | nd | nd |
| 55 | nd | nd | nd | nd | nd | nd | 0.9543 | 0.726 |
| 60 | 115.78 | 0.006 | 10.924 | 0.063 | 10.82 | 0.064 | nd | nd |
| 65 | nd | nd | nd | nd | nd | nd | 8.3593 | 0.083 |
The immobilized enzymes were incubated in 50 mM sodium phosphate buffer. pH 6.5. at various temperatures.
nd, not determined.
Table 7.
Effect of pH on Enzyme Stabilitya
| I-LacLM-ChBD |
I-ChBD-LacLM |
I-LacZ-ChBD |
||||
|---|---|---|---|---|---|---|
| pH | kin (h−1) | τ1/2 (h) | kin (h−1) | τ1/2 (h) | kin (h−1) | τ1/2 (h) |
| 4 | 0.5318 | 1.303 | 2.161 | 0.321 | 0.0984 | 7.0 |
| 5 | 0.0373 | 18.583 | 0.0685 | 10.119 | 0.0035 | 198.0 |
| 6 | 0.0078 | 88.865 | 0.0106 | 65.391 | 0.0006 | 1155.2 |
| 6.5 | 0.0038 | 182.407 | 0.0061 | 113.630 | 0.0006 | 1155.2 |
| 7 | 0.077 | 9.002 | 0.0432 | 16.045 | 0.0008 | 866.4 |
| 8 | 0.718 | 0.965 | 1.0722 | 0.646 | 0.0087 | 79.7 |
Half-life times of the immobilized β-galactosidases were determined by incubating the enzymes at 30 °C (for I-LacLM-ChBD and I-ChBD-LacLM) or at 37 °C (for I-LacZ-ChBD in the presence of 10 mM MgCl2) at different pH values.
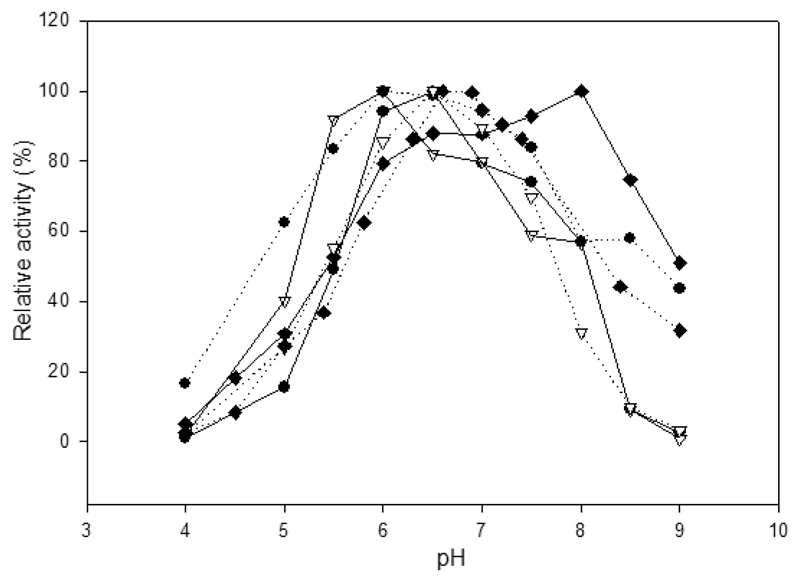
The immobilized enzyme preparations showed their optimum activity in the pH range of 6–6.5 for both substrates lactose and oNPG, with the exception of I-LacZ-ChBD and lactose having an optimal pH of 8 without clear reason (Figure 4). It is likely related to the isoelectric point (pH 9) of ChBD.27 When stored at different pH values, the enzymes were most stable in the pH range that also coincides with the pH optima, that is, 6–6.5 (Table 7). The binding of ChBD from B. circulans WL-12 chitinase to chitin was shown to be best at pH 9 (binding capacity of 90%) and less at pH 4 (binding capacity of 80%).27 However, LacLM from L. reuteri L103 and LacZ from L. bulgaricus were reported to have very low activity at pH 4,14,16 which was also observed with the immobilized enzymes in this study (Figure 4). This suggests that the effect of pH on the fusion enzymes is mainly on the catalytic activity rather than the binding of ChBD to chitin.
Figure 4.
Optimum pH values of β-galactosidase activity of the immobilized enzymes. I-LacLM-ChBD (●) and I-ChBD-LacLM (▽) indicate the recombinant enzymes with the ChBD fused to C-terminal of the LacM and the N-terminal of the LacL, respectively. I-LacZ-ChBD (◆) indicates the immobilized LacZ-ChBD. The solid and dotted lines indicate substrates oNPG and lactose, respectively.
Reuse of Immobilized Enzymes
A study on the reusability of the immobilized β-gals, using subsequent hydrolysis steps with oNPG or lactose as substrate, was carried out. The residual converting ability of immobilized enzyme preparations was plotted versus the number of cycles of application (Figure 5). In this study I-LacLM-ChBD and I-LacZ-ChBD exhibited better stability, because these immobilized enzymes retained approximately 80% of their initial ability of substrate conversion after four to eight cycles. I-ChBD-LacLM showed a more rapid decrease of relative activity (Figure 5).
Figure 5.
Reusability of the chitin-immobilized enzymes I-LacLM-ChBD (●), I-ChBD-LacLM (○), and I-LacZ-ChBD (▲). The converting ability (%) was calculated from the ratio of the product released (glucose or oNP released from lactose or oNPG, respectively) in different reaction cycles to the product obtained at the first batch (assumed as 100%).
These results are comparable with previously reported data. Chiang et al. immobilized levansucrase on chitin using the B. circulans WL-12 ChBD, which retained approximately half of its initial activity at the end of the seventh cycle.29
Lactose Transformation
Figures 6 and 7 show a qualitative analysis by TLC of products formed during lactose conversion using different immobilized β-gal preparations under various conditions. In general, the profile of products shown is comparable to the profile of the corresponding free enzymes previously reported14,16 as well as to the profile of Vivinal GOS (Amersfoort, The Netherlands) used as “reference” (Figure 6). This indicates that similar GOS mixtures are formed by the free and immobilized enzymes.
Figure 6.
TLC analysis of different batches of lactose conversion by the immobilized β-galactosidase from L. bulgaricus, I-LacZ-ChBD. The batch conversions were carried out in 50 mM sodium phosphate buffer, pH 6.5, 10 mM MgCl2 with various initial lactose concentrations, activities, and temperatures: 50 g/L initial lactose at 55 °C using 3.2 Ulactose/mL (A); 205 g/L initial lactose concentration at 55 °C using 1.7 Ulactose/mL (B); 50 g/L initial lactose concentration at 60 °C using 9.7 Ulactose/mL (C); commercial whole milk, 1 h at 60 °C using 9.7 Ulactose/mL (D).
Figure 7.
TLC of products from the batch lactose conversions by the immobilized enzymes I-LacLM-ChBD, I-ChBD-LacLM, and LacLM-ChBD (free soluble enzyme). The conversions were performed in 50 mM sodium phosphate buffer, pH 6.5, 10 mM MgCl2, with an initial concentration of 205 g/L lactose at 30 °C, using 1.5 Ulactose/ml. Glc, glucose; Gal, galactose; Lac, lactose; GOS, galacto-oligosaccharides. The standards include a mixture of glucose, galactose, and lactose (1); Vivinal GOS (2); and GOS from conversion of free LacLM from L. reuteri L103 (3).
As expected, a higher enzyme activity or higher temperatures led to faster conversion (Figure 6). The β-gal LacZ from L. bulgaricus was shown to be a thermostable enzyme,16 and therefore it catalyzed lactose conversion efficiently at the higher temperature of 50 °C. In this study, the chitin-immobilized LacZ again showed good thermostability, thus resulting in a comparable profile of products at 60 °C compared to 37 °C. Regardless of the conditions of the conversion, the maximal GOS yield was around 23–24% (Table 8; Figure 8), which is lower than the yield obtained with the free enzyme LacZ (approximately 50%) as reported by Nguyen and co-workers.16 A possible explanation could be that the immobilization reduces the contact between enzyme and lactose as well as the monosaccharide sugars for transgalactosylation, thus resulting in reduced GOS products. This may be also the reason for the rather high residual lactose concentration at the end of the conversion run (Table 8; Figure 6). This effect of immobilization was also observed by Sheu et al. when using chitosan-immobilized β-gal from Aspergillus oryzae for lactose conversion.42 The conversion of lactose in the samples of ∼5% w/v, which is the concentration of lactose in milk, using immobilized recombinant β-gal I-LacZ-ChBD with a relatively high activity (9.7 Ulactose/mL) at a high temperature of 60 °C was fast, as 81% of lactose was converted after only 1 h (Table 8).
Table 8.
Lactose Conversions Using the Immobilized β-Galactosidase from L. bulgaricus under Various Conditions
| initial lactose concentrationa |
|||||
|---|---|---|---|---|---|
| 50 g/L |
205 g/L |
||||
| temperature (°C) | 37 | 55 | 60 | 37 | 55 |
| Ulactose/mL | 3.2 | 3.2 | 9.7 | 1.7 | 1.7 |
| run time (h) | 3 | 3 | 1 | 24 | 24 |
| glucose (%) | 26.0 | 41.7 | 39.3 | 24.8 | 30.6 |
| galactose (%) | 18.5 | 33.6 | 32.8 | 11.6 | 15.7 |
| total GOS (%) | 10.4 | 2.5 | 10.4 | 23.5 | 23.9 |
| lactose conversion rate (%) | 54.2 | 77.2 | 81.2 | 59.8 | 70.2 |
In 50 mM sodium phosphate buffer, pH 6.5, 10 mM MgCl2.
Figure 8.
Carbohydrate composition during lactose transformation by the immobilized β-galactosidase from L. bulgaricus (I-LacZ-ChBD) determined by HPLC. The batch conversions were carried out in sodium phosphate buffer 50 mM, pH 6.5, in the presence of 10 mM MgCl2 at various temperatures, enzyme activities, and initial lactose concentrations: (A) 50 g/L initial lactose concentration, at 55 °C using 3.2 Ulactose/mL; (B) 50 g/L initial lactose concentration at 37 °C using 3.2 Ulactose/mL; (C), 205 g/L initial lactose concentration at 37 °C using 1.7 Ulactose/mL; (D) 205 g/L initial lactose concentration at 55 °C using 1.7 Ulactose/mL. The concentrations of lactose (◆), glucose (□), galactose (▲), and total galacto-oligosaccharides (○) were analyzed during the conversion.
When used for batch conversion experiments of lactose, I-LacLM-ChBD and I-ChBD-LacLM also resulted in a profile of GOS products comparable to that of the native β-gal L103 (Figure 7). Lactose conversion was faster with the free enzyme of LacLM-ChBD compared to I-LacLM-ChBD at equal activity loading (Figure 9), which might be explained by limitations in diffusion of the substrate to the active site. A maximum GOS level of ca. 39–40% of total sugars was reached by both enzyme preparations, soluble and immobilized (Figure 9), yet the time for reaching this maximum was significantly longer for I-LacLM-ChBD at 12 h (Figure 9B) compared to 8 h for LacLM-ChBD (Figure 9A). These maximum yields were obtained at a similar lactose conversion of ca. 85%. This indicates that the conversion of lactose and the yield of GOS obtained by the recombinant enzymes are comparable to the conversion catalyzed by their counterpart, the native enzyme LacLM L. reuteri L103.14
Figure 9.
Carbohydrate composition during lactose transformation by the recombinant β-galactosidase LacLM-ChBD from L. reuteri L103 as determined by HPLC. The batch conversions were carried out in 50 mM sodium phosphate buffer, pH 6.5, in the presence of 10 mM MgCl2 at 30 °C; 1.5 Ulactose/mL, and an initial concentration of lactose of 205 g/L with the free soluble LacLM-ChBD (A) or with the immobilized enzyme I-LacLM-ChBD (B). The concentrations of lactose (◆), glucose (□), galactose (▲), and total galacto-oligosaccharides (○) were analyzed during the conversion.
In conclusion, this work describes the immobilization of two lactobacillal β-galactosidases, a homodimeric β-galactosidase of the LacZ type and a heterodimeric β-galactosidase of the LacLM type, onto chitin via a chitin-binding domain. This could provide a promising and efficient approach for lactose hydrolysis and production of prebiotic galacto-oligosaccharides because the enzyme can be purified from the crude cell extract and immobilized in one simple step. The immobilized fusion enzyme can be stably reused for several cycles for lactose hydrolysis and transformation. Preliminary results from an ongoing investigation of lactose conversion in continuous mode using these immobilization enzymes also show a high potential for an industrial application of these immobilized enzymes.
Funding
This work was supported by the National Foundation for Science and Technology Development (NAFOSTED) Vietnam under Grant 106.16-2011.60. M.-L.P. thanks the European Commission for the Erasmus Mundus scholarship under the ALFABET project. T.-H.N. acknowledges support from the Austrian Science Fund (FWF Project P24868-B22).
Footnotes
ORCID
Mai-Lan Pham: 0000-0003-0352-9906
Tien-Thanh Nguyen: 0000-0002-3203-8228
Notes
The authors declare no competing financial interest.
References
- (1).Nakayama T, Amachi T. Beta-galactosidase, enzymology. In: Flickinger MC, Drew SW, editors. Encyclopedia of Bioprocess Technology: Fermentation, Biocatalysis, and Bioseparation. Wiley; New York: 1999. pp. 1291–1305. [Google Scholar]
- (2).Lomer MC, Parkes GC, Sanderson JD. Review article: Lactose intolerance in clinical practice – myths and realities. Aliment Pharmacol Ther. 2008;27:93–103. doi: 10.1111/j.1365-2036.2007.03557.x. [DOI] [PubMed] [Google Scholar]
- (3).Park AR, Oh DK. Galacto-oligosaccharide production using microbial beta-galactosidase: current state and perspectives. Appl Microbiol Biotechnol. 2010;85:1279–1286. doi: 10.1007/s00253-009-2356-2. [DOI] [PubMed] [Google Scholar]
- (4).Gosling A, Stevens GW, Barber AR, Kentish SE, Gras SL. Recent advances refining galactooligosaccharide production from lactose. Food Chem. 2010;121:307–318. [Google Scholar]
- (5).Macfarlane GT, Steed H, Macfarlane S. Bacterial metabolism and health-related effects of galacto-oligosaccharides and other prebiotics. J Appl Microbiol. 2008;104:305–344. doi: 10.1111/j.1365-2672.2007.03520.x. [DOI] [PubMed] [Google Scholar]
- (6).Macfarlane S, Macfarlane GT, Cummings JH. Review article: prebiotics in the gastrointestinal tract. Aliment Pharmacol Ther. 2006;24:701–714. doi: 10.1111/j.1365-2036.2006.03042.x. [DOI] [PubMed] [Google Scholar]
- (7).Rabiu BA, Jay AJ, Gibson GR, Rastall RA. Synthesis and fermentation properties of novel galacto-oligosaccharides by beta-galactosidases from Bifidobacterium species. Appl Environ Microbiol. 2001;67:2526–2530. doi: 10.1128/AEM.67.6.2526-2530.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Goulas T, Goulas A, Tzortzis G, Gibson GR. Comparative analysis of four beta-galactosidases from Bifidobacterium bifidum NCIMB41171: purification and biochemical characterisation. Appl Microbiol Biotechnol. 2009;82:1079–1088. doi: 10.1007/s00253-008-1795-5. [DOI] [PubMed] [Google Scholar]
- (9).Yi SH, Alli I, Park KH, Lee B. Overexpression and characterization of a novel transgalactosylic and hydrolytic beta-galactosidase from a human isolate Bifidobacterium breve B24. New Biotechnol. 2011;28:806–813. doi: 10.1016/j.nbt.2011.07.006. [DOI] [PubMed] [Google Scholar]
- (10).Iqbal S, Nguyen TH, Nguyen HA, Nguyen TT, Maischberger T, Kittl R, Haltrich D. Characterization of a heterodimeric GH2 beta-galactosidase from Lactobacillus sakei Lb790 and formation of prebiotic galacto-oligosaccharides. J Agric Food Chem. 2011;59:3803–3811. doi: 10.1021/jf103832q. [DOI] [PubMed] [Google Scholar]
- (11).Iqbal S, Nguyen TH, Nguyen TT, Maischberger T, Haltrich D. Beta-galactosidase from Lactobacillus plantarum WCFS1: biochemical characterization and formation of prebiotic galacto-oligosaccharides. Carbohydr Res. 2010;345:1408–1416. doi: 10.1016/j.carres.2010.03.028. [DOI] [PubMed] [Google Scholar]
- (12).Maischberger T, Leitner E, Nitisinprasert S, Juajun O, Yamabhai M, Nguyen TH, Haltrich D. Beta-galactosidase from Lactobacillus pentosus: purification, characterization and formation of galacto-oligosaccharides. Biotechnol J. 2010;5:838–847. doi: 10.1002/biot.201000126. [DOI] [PubMed] [Google Scholar]
- (13).Nguyen TH, Splechtna B, Krasteva S, Kneifel W, Kulbe KD, Divne C, Haltrich D. Characterization and molecular cloning of a heterodimeric beta-galactosidase from the probiotic strain Lactobacillus acidophilus R22. FEMS Microbiol Lett. 2007;269:136–144. doi: 10.1111/j.1574-6968.2006.00614.x. [DOI] [PubMed] [Google Scholar]
- (14).Nguyen TH, Splechtna B, Steinbock M, Kneifel W, Lettner HP, Kulbe KD, Haltrich D. Purification and characterization of two novel beta-galactosidases from Lactobacillus reuteri. J Agric Food Chem. 2006;54:4989–4998. doi: 10.1021/jf053126u. [DOI] [PubMed] [Google Scholar]
- (15).Nguyen TH, Splechtna B, Yamabhai M, Haltrich D, Peterbauer C. Cloning and expression of the beta-galactosidase genes from Lactobacillus reuteri in Escherichia coli. J Biotechnol. 2007;129:581–591. doi: 10.1016/j.jbiotec.2007.01.034. [DOI] [PubMed] [Google Scholar]
- (16).Nguyen TT, Nguyen HA, Arreola SL, Mlynek G, Djinovic-Carugo K, Mathiesen G, Nguyen TH, Haltrich D. Homodimeric beta-galactosidase from Lactobacillus delbrueckii subsp. bulgaricus DSM 20081: expression in Lactobacillus plantarum and biochemical characterization. J Agric Food Chem. 2012;60:1713–1721. doi: 10.1021/jf203909e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Halbmayr E, Mathiesen G, Nguyen TH, Maischberger T, Peterbauer CK, Eijsink VG, Haltrich D. High-level expression of recombinant beta-galactosidases in Lactobacillus plantarum and Lactobacillus sakei using a Sakacin P-based expression system. J Agric Food Chem. 2008;56:4710–4719. doi: 10.1021/jf073260+. [DOI] [PubMed] [Google Scholar]
- (18).Nguyen TT, Mathiesen G, Fredriksen L, Kittl R, Nguyen TH, Eijsink VG, Haltrich D, Peterbauer CK. A food-grade system for inducible gene expression in Lactobacillus plantarum using an alanine racemase-encoding selection marker. J Agric Food Chem. 2011;59:5617–5624. doi: 10.1021/jf104755r. [DOI] [PubMed] [Google Scholar]
- (19).Splechtna B, Nguyen TH, Haltrich D. Comparison between discontinuous and continuous lactose conversion processes for the production of prebiotic galacto-oligosaccharides using beta-galactosidase from Lactobacillus reuteri. J Agric Food Chem. 2007;55:6772–6777. doi: 10.1021/jf070643z. [DOI] [PubMed] [Google Scholar]
- (20).Splechtna B, Nguyen TH, Steinbock M, Kulbe KD, Lorenz W, Haltrich D. Production of prebiotic galacto-oligosaccharides from lactose using β-galactosidases from Lactobacillus reuteri. J Agric Food Chem. 2006;54:4999–5006. doi: 10.1021/jf053127m. [DOI] [PubMed] [Google Scholar]
- (21).Splechtna B, Nguyen TH, Zehetner R, Lettner HP, Lorenz W, Haltrich D. Process development for the production of prebiotic galacto-oligosaccharides from lactose using beta-galactosidase from Lactobacillus sp. Biotechnol J. 2007;2:480–485. doi: 10.1002/biot.200600230. [DOI] [PubMed] [Google Scholar]
- (22).Grosova Z, Rosenberg M, Rebros M. Perspectives and applications of immobilised beta-galactosidase in food industry – a review. Czech J Food Sci. 2008;26:1–14. [Google Scholar]
- (23).Panesar PS, Kumari S, Panesar R. Potential applications of immobilized beta-galactosidase in food processing industries. Enzyme Res. 2010;2010:473137. doi: 10.4061/2010/473137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Eijsink V, Hoell I, Vaaje-Kolstad G. Structure and function of enzymes acting on chitin and chitosan. Biotechnol Genet Eng Rev. 2010;27:331–366. doi: 10.1080/02648725.2010.10648156. [DOI] [PubMed] [Google Scholar]
- (25).Kurek DV, Lopatin SA, Varlamov VP. Prospects of application of the chitin-binding domains to isolation and purification of recombinant proteins by affinity chromatography. Appl Biochem Microbiol. 2009;45:1–8. [PubMed] [Google Scholar]
- (26).Ikegami T, Okada T, Hashimoto M, Seino S, Watanabe T, Shirakawa M. Solution structure of the chitin-binding domain of Bacillus circulans WL-12 chitinase A1. J Biol Chem. 2000;275:13654–13661. doi: 10.1074/jbc.275.18.13654. [DOI] [PubMed] [Google Scholar]
- (27).Hashimoto M, Ikegami T, Seino S, Ohuchi N, Fukada H, Sugiyama J, Shirakawa M, Watanabe T. Expression and characterization of the chitin-binding domain of chitinase A1 from Bacillus circulans WL-12. J Bacteriol. 2000;182:3045–3054. doi: 10.1128/jb.182.11.3045-3054.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Chern JT, Chao YP. Chitin-binding domain based immobilization of d-hydantoinase. J Biotechnol. 2005;117:267–275. doi: 10.1016/j.jbiotec.2005.02.001. [DOI] [PubMed] [Google Scholar]
- (29).Chiang CJ, Wang JY, Chen PT, Chao YP. Enhanced levan production using chitin-binding domain fused levansucrase immobilized on chitin beads. Appl Microbiol Biotechnol. 2009;82:445–451. doi: 10.1007/s00253-008-1772-z. [DOI] [PubMed] [Google Scholar]
- (30).Tanaka K, Kawamoto T. Cell and enzyme immoblization. In: Demain AL, Davies JE, Atlas RM, Cohen G, Hershberger CL, Hu WS, Sherman DH, Wilson RC, Wu JHD, editors. Manual of Industrial Microbiology and Biotechnology. ASM Press; Washington, DC, USA: 1999. pp. 94–102. [Google Scholar]
- (31).Kleerebezem M, Boekhorst J, van Kranenburg R, Molenaar D, Kuipers OP, Leer R, Tarchini R, Peters SA, Sandbrink HM, Fiers MW, Stiekema W, et al. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc Natl Acad Sci U S A. 2003;100:1990–1995. doi: 10.1073/pnas.0337704100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Sorvig E, Mathiesen G, Naterstad K, Eijsink VG, Axelsson L. High-level, inducible gene expression in Lactobacillus sakei and Lactobacillus plantarum using versatile expression vectors. Microbiology. 2005;151:2439–2449. doi: 10.1099/mic.0.28084-0. [DOI] [PubMed] [Google Scholar]
- (33).Aukrust TW, Brurberg MB, Nes IF. Transformation of Lactobacillus by electroporation. Methods Mol Biol. 1995;47:201–208. doi: 10.1385/0-89603-310-4:201. [DOI] [PubMed] [Google Scholar]
- (34).Klein MP, Nunes MR, Rodrigues RC, Benvenutti EV, Costa TM, Hertz PF, Ninow JL. Effect of the support size on the properties of beta-galactosidase immobilized on chitosan: advantages and disadvantages of macro and nanoparticles. Biomacromolecules. 2012;13:2456–2464. doi: 10.1021/bm3006984. [DOI] [PubMed] [Google Scholar]
- (35).Polizzi KM, Bommarius AS, Broering JM, Chaparro-Riggers JF. Stability of biocatalysts. Curr Opin Chem Biol. 2007;11:220–225. doi: 10.1016/j.cbpa.2007.01.685. [DOI] [PubMed] [Google Scholar]
- (36).Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- (37).Sorvig E, Gronqvist S, Naterstad K, Mathiesen G, Eijsink VG, Axelsson L. Construction of vectors for inducible gene expression in Lactobacillus sakei and L. plantarum. FEMS Microbiol Lett. 2003;229:119–126. doi: 10.1016/S0378-1097(03)00798-5. [DOI] [PubMed] [Google Scholar]
- (38).Mathiesen G, Huehne K, Kroeckel L, Axelsson L, Eijsink VG. Characterization of a new bacteriocin operon in sakacin P-producing Lactobacillus sakei, showing strong translational coupling between the bacteriocin and immunity genes. Appl Environ Microbiol. 2005;71:3565–3574. doi: 10.1128/AEM.71.7.3565-3574.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Harris JM, Epting KL, Kelly RM. N-terminal fusion of a hyperthermophilic chitin-binding domain to xylose isomerase from Thermotoga neapolitana enhances kinetics and thermostability of both free and immobilized enzymes. Biotechnol Prog. 2010;26:993–1000. doi: 10.1002/btpr.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Husain Q. Beta galactosidases and their potential applications: a review. Crit Rev Biotechnol. 2010;30:41–62. doi: 10.3109/07388550903330497. [DOI] [PubMed] [Google Scholar]
- (41).Baran T, Arica MY, Denizli A, Hasirci V. Comparison of beta-galactosidase immobilization by entrapment in and adsorption on poly(2-hydroxyethylmethacrylate) membranes. Polym Int. 1997;44:530–536. [Google Scholar]
- (42).Sheu D-C, Li S-Y, Duan K-J, Chen CW. Production of galactooligosaccharides by beta-galactosidase immobilized on glutaraldehyde-treated chitosan beads. Biotechnol Tech. 1998;12:273–276. [Google Scholar]