Significance
Biological specimens, such as whole blood, contain many different types of cells that have important applications for monitoring health, including tracking vaccine efficacy and HIV progression, and for treating complex diseases, like cancer. However, current cell-sorting technologies require expensive and large pieces of equipment and can only isolate a few types of cells at one time. Here, we develop a cell-sorting platform by engineering dynamic DNA devices that can label, sort, and recover different cell populations at high purity en masse. This DNA-based nanotechnology can potentially be expanded to exceed the capacity of current methods for sorting multiple types of cells, improve biomedical diagnostics, and provide new insights into cell biology.
Keywords: cell sorting, DNA nanotechnology, strand displacement, protein conjugates, lymphocytic choriomeningitis virus
Abstract
The ability to analyze and isolate cells based on the expression of specific surface markers has increased our understanding of cell biology and produced numerous applications for biomedicine. However, established cell-sorting platforms rely on labels that are limited in number due to biophysical constraints, such as overlapping emission spectra of fluorophores in FACS. Here, we establish a framework built on a system of orthogonal and extensible DNA gates for multiplexed cell sorting. These DNA gates label target cell populations by antibodies to allow magnetic bead isolation en masse and then selectively unlock by strand displacement to sort cells. We show that DNA gated sorting (DGS) is triggered to completion within minutes on the surface of cells and achieves target cell purity, viability, and yield equivalent to that of commercial magnetic sorting kits. We demonstrate multiplexed sorting of three distinct immune cell populations (CD8+, CD4+, and CD19+) from mouse splenocytes to high purity and show that recovered CD8+ T cells retain proliferative potential and target cell-killing activity. To broaden the utility of this platform, we implement a double positive sorting scheme using DNA gates on peptide-MHC tetramers to isolate antigen-specific CD8+ T cells from mice infected with lymphocytic choriomeningitis virus (LCMV). DGS can potentially be expanded with fewer biophysical constraints to large families of DNA gates for applications that require analysis of complex cell populations, such as host immune responses to disease.
Advances in cytometry and cell-sorting technologies have led to fundamental insights in cell biology and important applications in biomedicine (1–3). For example, in fluorescent-based platforms such as fluorescence-activated cell sorting (FACS), each different cell type is labeled with fluorophore-tagged antibodies and sorted along fluorescent gates; this well-established approach has been used to define new types of stem cells (4–7), discover biomarkers for cancer (8, 9), and isolate rare cells for cell-based therapies (10, 11). However, for complex systems involving many different types of cells, sorting by fluorescent gates is challenging because only a limited number of fluorophores can be used simultaneously due to overlapping emission spectra. This has led to the development of methods, such as combinatorial staining (12, 13) and fluorescent cell barcoding (FCB) (14), to improve the multiplexing capacity of flow cytometry, enabling applications such as high-content single-cell drug screening (15). The emergence of cytometry by time of flight (CyTOF) takes an entirely different approach by staining surface markers with heavy metal tags in lieu of fluorophores and then analyzing cells by mass spectrometry. Because the number of unique heavy metals exceeds the number of spectrally distinct fluorophores that can be combined in a single staining panel, CyTOF allows analysis of systems that would otherwise be challenging by flow, such as mapping cellular differentiation pathways (e.g., hematopoiesis) (16–18) and analyzing single cells at a systems level (19–21). However, in CyTOF, cells are ionized during detection, limiting throughput and preventing recovery of cells for downstream functional assays. Ideally, a high-throughput cell-sorting technology would be built from a multiplexed system of sorting gates that are orthogonal and highly extensible while enabling recovery of target cells for additional studies.
Here, we designed a multiplexed cell-sorting platform called DNA-gated sorting (DGS) by engineering DNA gates that capture, release, and recover target cells from a complex biological specimen (Fig. 1A). We developed DGS to leverage the ability to assemble large, orthogonal libraries of DNA gates by sequence design that could potentially be multiplexed at a scale that exceeds fluorophores or heavy metal tags (i.e., the pool of all possible DNA sequences from which gates can be designed increases exponentially with the number of bases). Moreover, synthetic DNA approaches have been used previously for cell-based applications, including patterning cells onto DNA-functionalized surfaces (22–26), multiplexed imaging of RNA (27) and proteins (28, 29), sorting beads (30), and analyzing cell surfaces using autonomous cascades (31, 32). Our DNA gates consist of different strand displacement reactions that are uniquely mapped to cell surface markers via affinity agents, such as antibodies. These DNA gates are analogous to fluorescence-based gates that are used in FACS to identify and then sort target cells from a general population; however, while FACS sorting occurs for cells above a threshold fluorescent intensity, cell sorting by DGS triggers when a DNA gate unlocks by DNA strand displacement (Fig. 1B). We show that DNA gates are triggered to completion within minutes on the surface of cells and, when combined with magnetic beads, enable target cells to be sorted with purity, viability, and yield equivalent to commercial magnetic activated cell sorting (MACS) reagents. We apply DGS for sorting multiple immune cell subsets from murine splenocytes and show that CD8+ T cells retain key cell functions, such as proliferation and cell-killing activity. We then extend DGS to peptide-MHC (pMHC) tetramers and use antibody and tetramer DGS in combination to isolate antigen-specific CD8+ T cells from mice undergoing an acute response to lymphocytic choriomeningitis virus (LCMV) infection. This work establishes a scalable framework for multiplexed sorting of complex biological samples based on DNA gates.
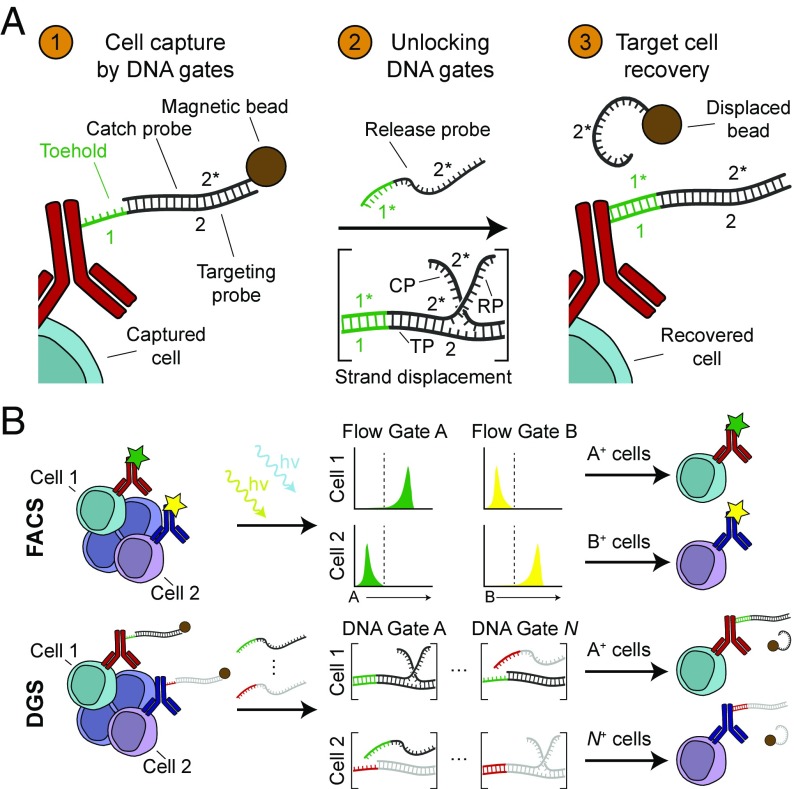
Fig. 1.
Multiplexed cell sorting by orthogonal antibody DNA gates. (A) In step 1, antibodies encoded with a targeting probe bind to cell surface markers on target cells. Annealing partially complementary catch probe-coated magnetic beads permits magnetic capture of target cells but leaves a toehold (1) for subsequent displacement. In step 2, release probes displace the catch probe through toehold-mediated strand displacement, removing the magnetic label from target cells. In step 3, target cells are recovered, due to the absence of a magnetic label. (B) DNA gates are analogous to fluorescent gates used in FACS. Labeled target cells are interrogated with different lasers (denoted hν; FACS) or release probes (DGS). In FACS, cells are sorted if a threshold fluorescence in a particular channel is exceeded while, in DGS, cells are sorted if strand displacement in a particular DNA gate occurs. DNA gates can be extended to N gates through sequence design.
Results
Engineering Dynamic Antibody DNA Gates.
An ideal system of high performance antibody DNA gates should be extensible, independently addressable, and kinetically tunable. We designed our DNA gates based on strand displacement reactions because the thermodynamic parameters that govern displacement kinetics [e.g., toehold length, guanine and cytosine (GC) content, temperature] are rigorously characterized (33–36) and therefore allow rapid sequence design and prediction of strand dynamics in silico (37–39). Also, orthogonal sets of DNA displacement reactions have previously been used for multiplexed applications in biological systems, such as imaging mRNA (27) and protein targets in situ (28, 29). Here, we engineered antibody DNA gates comprised of a system of three single stranded DNA sequences—a targeting probe (TP), a catch probe (CP), and a release probe (RP)—that mediate magnetic target cell capture, cell release via unlocking of DNA gates, and target cell recovery (Fig. 1A). The targeting probe consists of two domains in tandem (a toehold domain [1] and a hybridization domain [2]) and is conjugated to an antibody to label target cells for annealing to a catch probe [2*]. The catch probe is coupled to a magnetic bead to isolate target cells from a complex mixture but is unlocked after addition of a release probe [1*2*] via DNA strand displacement to recover target cells. By designing a system of individually addressable DNA gates coupled to different antibodies, individual cell populations can be isolated from a complex biological specimen by multiplexed cell sorting (Fig. 1B).
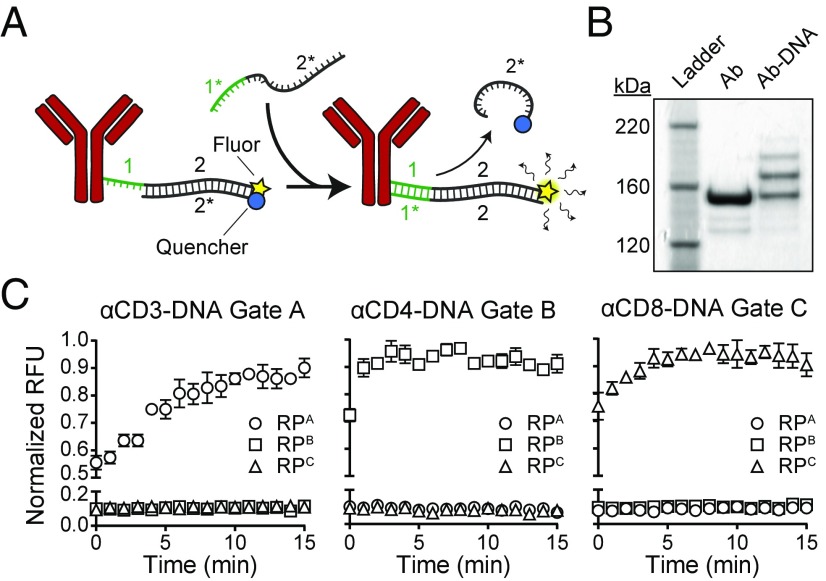
We first designed sequences in silico for TPs, CPs, and RPs for orthogonal DNA gates using a domain-based approach (37), where 6-base toehold and 20-base hybridization domains are independently optimized (50% GC content, 5′ G/C on the toehold domain). These domains were then concatenated to form candidate TP sequences that were screened in silico using NUPACK (40) to minimize secondary structures and cross-hybridization under relevant conditions for sorting (4 to 25 °C, 150 mM NaCl). Using this approach, we designed libraries of 3 (A–C) (Table S1) and 24 gates (α–ω) (Table S2). To validate orthogonality and the kinetics of DNA displacement from gates A–C, we annealed each Cy5-labeled TP with its complementary Iowa Black-labeled CP and then incubated complexes with an equimolar concentration of RP from each gate in separate wells. In samples where RP was reacted with TP:CP duplexes from the same gate, we observed complete displacement within 5 min (Fig. S1). Next, we examined whether strand displacement reactions occur when DNA complexes are covalently linked to antibodies (Ab), as the proximity of a conjugated protein could sterically shield the RP from binding to the toehold region. We conjugated quenched TP:CP complexes to antibodies using hydrazone chemistry, labeling antibodies against canonical lymphocyte cell surface markers CD3, CD4, and CD8 with Gate A, B, and C sequences, respectively (Fig. 2A). To determine conjugation efficiency, we purified Ab-TP:CP conjugates by size-exclusion chromatography and performed a mobility shift assay (Fig. 2B). We observed a distribution of higher molecular weight bands in the Ab-DNA lane compared with the free Ab lane, which was expected under nonsaturating molar ratios that we selected to preserve antibody affinity (22). To validate strand displacement kinetics and confirm DNA gate orthogonality on Ab-DNA complexes, we arrayed quenched Ab-TP:CP conjugates from each gate with one of the three RP strands. Consistent with our free strand results, only Ab-TP:CP complexes incubated with RP strands from the same gate displaced, achieving complete displacement within 5 to 10 min (Fig. 2C). Taken together, these results demonstrate that the specificity and rate of strand displacement reactions are negligibly altered when coupled to the surface of antibodies.
Fig. 2.
DNA gates conjugated to antibodies exhibit rapid displacement kinetics. (A) Schematic of Ab-DNA complexes used in kinetic studies. TP:CP duplexes containing a quenched fluorophore and possessing a toehold (1) were conjugated to antibodies via free amines. Addition of RP from a matching gate initiated strand displacement, releasing the CP strand and permitting fluorescence. (B) SDS/PAGE of Ab-TP:CP complexes. Multiple DNA complexes were conjugated per antibody. (C) Kinetics of DNA strand displacement on antibodies. Complete displacement occurred within 10 min when RP from a matching gate was added (n = 3). Data shown as mean ± SD.
High Performance Cell Isolation by Strand Displacement.
We next set out to integrate DNA-gated antibodies with magnetic beads to allow selective cell isolation en masse. To do so, we first examined the kinetic efficiency of DNA strand displacement on the surface of cells. We stained three human T cell lines—Jurkat, CCRF-CEM, and TALL-104—as representative CD3+, CD4+, and CD8+ cells with one of the quenched Ab-TP:CP conjugates (αCD3-TPA:CPA, αCD4-TPB:CPB, and αCD8-TPC:CPC, respectively) and measured baseline fluorescence on cells by flow cytometry (Fig. 3A). After incubating cells with RP strands from the matching gate (RPA, RPB, or RPC) to trigger strand displacement, we observed an increase in fluorescent intensity on all three cell types, indicating that strand displacement was preserved on the surface of cells. We then tested the use of Ab-DNA gates to label target cells with magnetic beads, followed by the reversible release of beads for target cell recovery. We stained mouse CD8+ T cells with anti-mouse CD8 conjugated with TPA, incubated stained T cells with CPA-labeled magnetic beads, and then looked for bead-bound cells by microscopy. We found that annealing of TPA and CPA strands bound beads to cells, resulting in an average of 5.8 beads per cell (Fig. 3 B and C). After incubating bead-bound cells with RPA, the magnetic beads were released from the surface of cells with high efficiency (0.1 beads per cell) (****P < 0.0001 by unpaired t test, Fig. 3C). These results show that cells expressing a target cell surface marker can be magnetically captured via DNA hybridization and released by DNA gates.
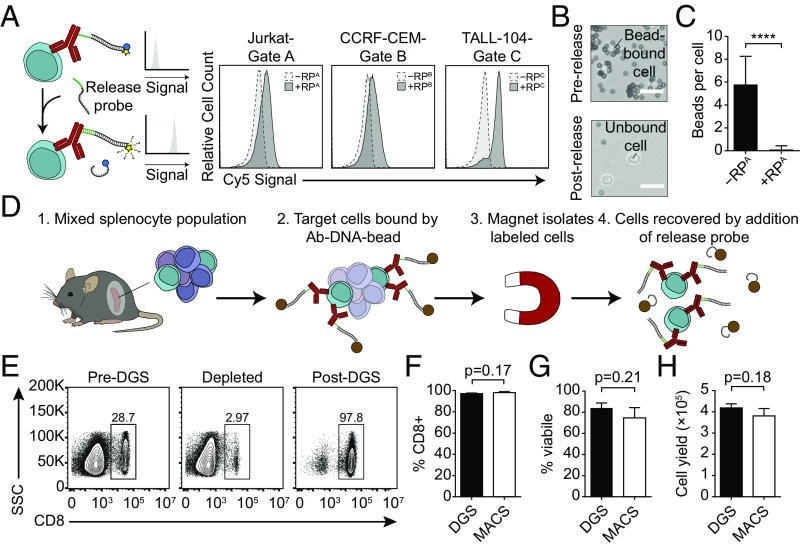
Fig. 3.
Magnetic capture and release of target cells by DNA gates. (A) CD3+ Jurkat, CD4+ CCRF-CEM, and CD8+ TALL-104 cells stained with quenched anti–CD3-TPA:CPA, anti–CD4-TPB:CPB, and anti–CD8-TPC:CPC complexes, respectively, exhibited increased fluorescence when incubated with RP from a matching gate. (B) Brightfield images of target cells magnetically labeled by annealing CP-coated magnetic beads (Upper). After addition of RP strands, beads were released from the cell surface (Lower). (Scale bars: 15 μm.) (C) Quantification of beads per cell before and after addition of release probe (P < 0.0001, Student’s t test, n = 29 for −RPA, n = 18 for +RPA). Data shown as mean ± SD. (D) CD8+ target cells were isolated from C57BL/6J mice by DGS. (E) Flow plots showing frequency of CD8+ cells in the native tissue (Left) and in the unlabeled (Middle) and labeled (Right) fractions after DGS. Comparison of CD8+ cell sorting (F) purity, (G) viability, and (H) yield by DGS and MACS (Student’s t test, n = 3 for all experiments). Data shown as mean ± SD.
We next applied DGS to isolate cell populations from a complex biological sample and compared DGS sorting efficiency to magnetic activated cell sorting (MACS), a commercial platform that is routinely used for cell enrichment. As a test bed, we chose to isolate CD8+ T cells from a mouse spleen because splenocytes are comprised of multiple immune cell populations, including CD8+ and CD4+ T cells, B cells, monocytes, and dendritic cells (Fig. 3D). We stained splenocytes from C57BL/6J mice with anti-mouse CD8-TPA and then annealed CPA-coated magnetic beads to labeled CD8+ cells. After capturing target cells in a magnetic column to separate the unbound CD8-depleted fraction, we added RPA strands to displace beads and isolate CD8+ cells. We then stained both the unbound cell fraction and recovered target cells to check for CD8+ cell frequency, finding that DGS enriched CD8+ target cells to a purity >97% with ∼90% recovery (CD8+ frequency decreased from 28.7% in the unsorted sample to 2.97% in the unbound fraction) (Fig. 3E). To measure the performance of sorting with our DNA-gated antibodies, we benchmarked DGS against sorting with a commercial CD8+ positive selection MACS kit and found no significant difference in several key parameters, including CD8+ purity (DGS: 97.1% vs. MACS: 98.1%, P = 0.17 by unpaired t test, n = 3), cell viability (DGS: 84.0% vs. MACS: 74.6%, P = 0.21 by unpaired t test, n = 3), and yield (DGS: 4.19 × 105 cells vs. MACS: 3.81 × 105 cells, P = 0.18 by unpaired t test, n = 3) (Fig. 3 F–H and Fig. S2). These results demonstrate that the cell-sorting efficiency of DGS from complex cell samples is equivalent to that of commercial platforms.
Multiplexed DGS of Murine Splenocytes Preserves Key Cell Functions.
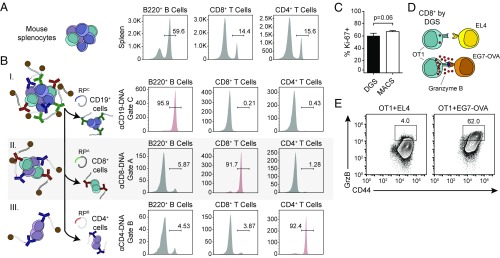
In contrast to standard bead-based sorting, which lacks the ability to isolate multiple cell populations simultaneously, DGS is theoretically not limited in the number of cell types it can isolate from a sample because cell sorting is based on the orthogonality of DNA gates and the number of all possible DNA sequences from which we can build orthogonal DNA strand displacement reactions scales exponentially with sequence length (4N). To demonstrate the potential for parallel sorting by DGS, we used our panel of orthogonal DNA-gated antibodies for multiplexed sorting of primary CD19+ B cells, CD8+ T cells, and CD4+ T cells from mouse splenocytes. We harvested splenocytes from C57BL/6J mice and measured initial CD19+, CD8+, and CD4+ immune cell frequencies (Fig. 4A) before simultaneously staining cells with anti-mouse CD8-TPA, anti-mouse CD4-TPB, and anti-mouse CD19-TPC. After annealing CP-coated magnetic beads to target cells for magnetic capture, the sequential addition of RP strands from each DNA gate resulted in recovery of specific target cell populations. We verified the purity of each cell fraction by staining for B220, CD8, and CD4 cell surface markers, finding that target cell purity after each displacement was >90% (Fig. 4B). B220 was used as a proxy for CD19 expression because binding of the DNA-gated antibody blocked staining of an alternate CD19 antibody clone used to measure frequency; we confirmed that CD19 and B220 are coexpressed on B cells by costaining (Fig. S3). To confirm that cells remain functional after sorting by DNA-gated antibodies, we examined both T cell proliferation and killing markers. First, we plate activated DGS- and MACS-sorted CD8+ T cells using anti-mouse CD3/CD28 and stained for the proliferation marker Ki-67 5 d after sorting. We observed that 60.2% of DGS-sorted cells exhibited proliferative capacity, equivalent to that of MACS-sorted cells (DGS: 60.2% vs. MACS: 67.0%, P = 0.06 by unpaired t test, n = 3) (Fig. 4C). Additionally, we performed an in vitro killing assay where we coincubated DGS-sorted CD8+ T cells from transgenic OT1 mice, which express a T cell receptor specific for OVA257–264 antigen, with either EL4 (non–OVA-expressing) controls or EG7-OVA (OVA-expressing) target cells and measured effector Granzyme B expression (Fig. 4D). We observed a 15-fold increase in the number of CD8+ T cells expressing Granzyme B when incubated with EG7-OVA cells compared with EL4 cells, consistent with the Granzyme B elevation seen in MACS-sorted cells cultured with EG7-OVA cells (Fig. 4E and Fig. S4). These results show that multiplexed DGS enriches populations of several cell types from a single biological tissue to a high purity while preserving cellular function.
Fig. 4.
Multiplexed sorting of major immune cell types from murine splenocytes by DGS retains cell function. (A) Initial B220+, CD8+, and CD4+ cell frequencies in splenocytes harvested from C57BL/6J mice. (B) Splenocytes were magnetically labeled with DNA-gated antibodies targeting CD19, CD8, and CD4. Individual populations were sequentially recovered by addition of appropriate RP strands (Top, B220+ cells; Middle, CD8+ cells; Bottom, CD4+ cells). (C) Released CD8+ cells retain proliferative potential compared with MACS-sorted CD8+ cells (Student’s t test, n = 3). Data shown as mean ± SD. (D and E) CD8+ cells purified by DGS from OT1 mice produce elevated Granzyme B when coincubated with EG7-OVA target cells compared with EL4 control cells.
Dual Gated DGS Isolates Antigen-Specific CD8+ T Cells.
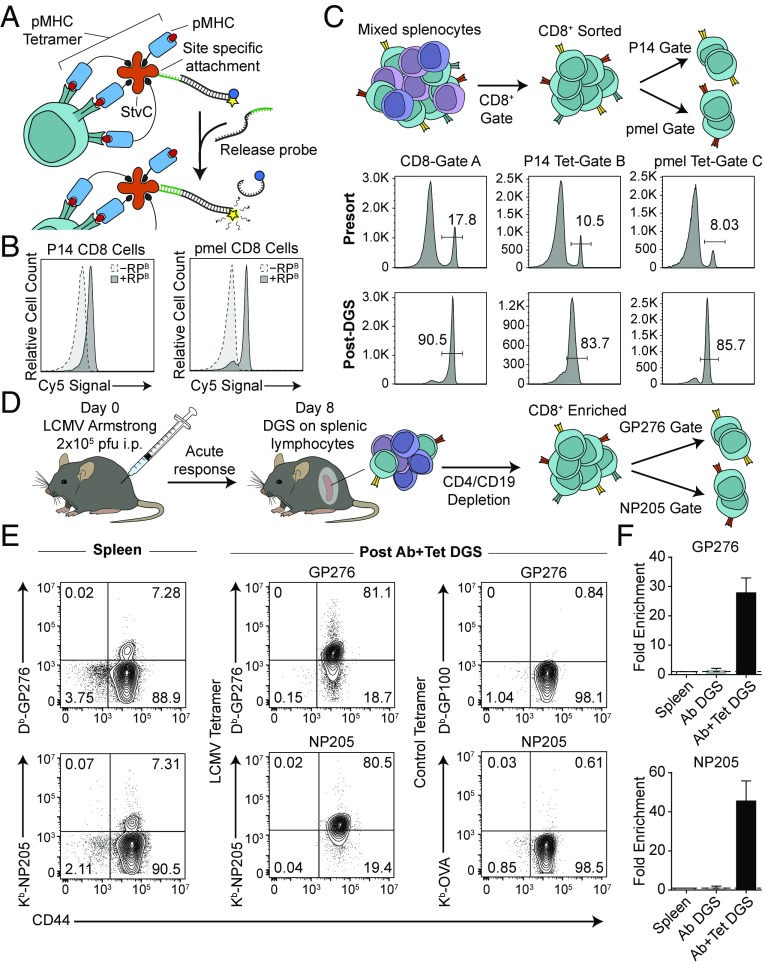
To expand DGS to dual gated sorting of cells expressing multiple markers, we extended the use of DNA gates to peptide-MHC tetramers for isolation of antigen-specific CD8+ T cells. We first synthesized DNA-gated pMHC tetramers by expressing recombinant streptavidin engineered with a C-terminal cysteine residue (StvC) (23, 41) to site-specifically attach DNA at a location that would not impair binding of biotinylated pMHC monomers. We then confirmed efficient DNA strand displacement on TCR transgenic P14 or pmel T cells within whole splenocytes using their respective pMHC tetramers (Db-GP33 or Db-GP100) (Fig. 5 A and B). In a dual gated system designed to sort CD8+Tet+ cells, we combined anti-mouse CD8-Gate A, Tet- Db-GP33-Gate B, and Tet-Db-GP100-Gate C to a mixture of P14 and pmel splenocytes. CD8+ T cells were isolated in bulk to similar purities as in our single marker sort by addition of RPA. From the CD8+ enriched subpopulation, P14 and pmel CD8+ T cells were eluted into two distinct samples by unlocking gates B and C with RPB and RPC (Fig. 5C). To validate the composition of recovered cells, we stained isolated cells with fluorescent pMHC tetramer and observed enrichment to high purity for both P14 and pmel splenocytes (Fig. S5).
Fig. 5.
Dual gated DGS with antibodies and pMHC tetramers isolates viral-specific CD8+ T cells from an endogenous polyclonal immune response. (A) Quenched DNA gates were site-specifically conjugated to pMHC tetramers. Upon addition of the corresponding RP, the quencher-labeled CP was displaced by strand displacement, permitting fluorescence. (B) Splenocytes from P14 and pmel TCR transgenic mice stained with corresponding quenched tetramer-TPB:CPB complexes showed increased fluorescence after addition of RPB strands. (C) CD8+ T cells from P14 and pmel mice were sorted by antibody and tetramer DGS from a mixture of B6, P14, and pmel splenocytes using a double positive gating scheme. (D) C57BL/6J mice were injected i.p. with 2 × 105 pfu of LCMV Armstrong, and LCMV-specific CD8+ T cells were isolated at day 8 postinfection using antibody DGS to deplete CD4+ and CD19+ cells, followed by tetramer DGS to isolate LCMV-derived GP276- or NP205-specific populations. (E) Frequencies of GP276- or NP205-specific T cells in the spleen and after sorting by DGS. Specificity of isolated cells was verified by staining with allele-matched control tetramer. Data shown are gated on CD8+ cells. (F) Fold enrichment in GP276- or NP205-specific T cell populations from the spleen after CD4 and CD19 depletion (Ab DGS) and after DGS with tetramers (Ab+Tet DGS). Data shown as mean ± SD, n = 2 for GP276, n = 3 for NP205.
We next sought to apply dual gated tetramer sorting to isolate antigen-specific T cells during an endogenous polyclonal immune response to infection using the model virus LCMV (Fig. 5D). During acute clearance [8 d post infection (p.i.)], we analyzed CD8+ T cells from the spleen specific for the known LCMV-derived antigens GP276 and NP205 by tetramer staining (Fig. 5E). To sort GP276- and NP205-specific CD8+ T cells, we enriched CD8+ T cells from the spleen in bulk using negative selection by depleting CD4+ and CD19+ cells with DGS; we did not observe any change in the frequency of tetramer-positive cells within the CD8+ population postdepletion (Fig. S6). Antigen-specific CD8+ T cells were isolated from the CD8-enriched sample by addition of RP strands to recover putative LCMV-specific populations. To validate the specificity of tetramer DGS, we stained sorted samples with allele-matched control tetramers (Db-GP100 for Db-GP33 and Kb-OVA for Kb-NP205) and observed no binding (Fig. 5E). By contrast, staining with fluorescent tetramers used for DGS showed that both GP276- and NP205-specific populations were sorted to high purity and enriched ∼30- to 50-fold from the spleen (Fig. 5 E and F). These results show that multiplexed DGS can be extended to complex targeting ligands and dual gating strategies to isolate cells based on expression of multiple surface markers.
Discussion
Here, we introduced the concept of DNA gates for multiplexed sorting of target cells from a biological specimen. We achieved DGS sorting of CD8+, CD4+, and CD19+ immune subsets from a mouse spleen with >90% purity, and, compared with MACS, we found no significant difference in sorting purity, viability, yield, or downstream cellular function. Unlike bead-based sorting methods which require serial labeling and purifying steps to isolate multiple populations of cells from a sample, DGS uses a single capture step followed by sequential release steps to sort cells. Furthermore, each gate can be uniquely attached to different cell capture agents to permit sorting of cells based on expression of multiple surface markers as we demonstrate with pMHC tetramers to isolate antigen-specific CD8+ T cells. Additionally, families of high performance DNA gates can be readily extended to sort and analyze additional types of cells by designing additional orthogonal gates using existing sequence design frameworks (33, 38, 39, 42, 43). By comparison, extending the number of channels in flow cytometry beyond the use of single fluorophores requires combinatorial staining or fluorescent cell barcoding (FCB), which scales with limitations and increases the complexity of analysis (12–14). Expansion of DGS by DNA sequence design may be particularly important in settings where a large number of orthogonal channels are required, such as mapping hematopoiesis and monitoring polyclonal patient responses to vaccines.
Materials and Methods
Additional details for materials and methods can be found in Supporting Information.
DNA Sequence Design.
TP, CP, and RP strand sequences were designed in silico using a domain-based approach. To generate TP strands, pools of 6-nt toehold domain and 20-nt hybridization domain sequences each containing 50% GC content were generated and checked for minimal secondary structure (i.e., little to no intramolecular binding) at 25 °C using NUPACK software. After filtering out oligos with significant structure, each remaining toehold domain was concatenated to the 5′ end of a hybridization domain, and lack of secondary structure was again verified in silico. Corresponding CP and RP strands were generated by taking the reverse complement of the hybridization domain or the entire TP strand, respectively. Orthogonality between gates was checked by inputting TP and CP sequences for all gates with and without all RP sequences and analyzing the resulting species at equilibrium as predicted by NUPACK.
DNA Conjugation to Antibodies and Streptavidin.
Recombinant streptavidin expressing a C-terminal cysteine residue (StvC) was expressed and purified as previously described (44). Antibodies were reacted with 50-fold excess succinimidyl-6-hydrazino-nicotinamide (S-HyNic) (Solulink), StvC was reduced with 10 mM Tris(2-carboxyethyl)phosphine (TCEP) and then reacted with 50-fold excess maleimido-6-hydraziniumpyridine (MHPH) (Solulink), and amine-terminated DNA with 20-fold excess succinimidyl-4-formylbenzamide (S-4FB) (Solulink) for 4 h. Excess linker was removed by buffer exchanging into citrate buffer (50 mM sodium citrate, 150 mM NaCl, pH 6) using Amicon spin filters (Millipore). Functionalized DNA was combined with antibodies or StvC at a 20:1 or 1:1 ratio, respectively, and reacted overnight. Ab-DNA gates were purified on a Superdex 200 Increase 10/300 GL column using an AKTA Pure FPLC (GE Healthcare). StvC-DNA gates were purified using Pierce spin columns (Thermo) packed with iminobiotin agarose (Thermo) according to the manufacturer’s instructions. Conjugation was verified by SDS/PAGE followed by Coomassie staining.
Antibody DNA-Gated Cell Sorting.
All animal work was approved by the Georgia Tech Institutional Animal Care and Use Committee. Each cell-sorting reaction started with 5 × 106 C57BL/6J splenocytes resuspended in 100 μL of sorting buffer (1× PBS + 0.1% BSA + 2 mM EDTA). Cells were blocked with anti-mouse CD16/CD32 (Mouse BD Fc Block) before staining with 1 μg of Ab-TP conjugate for 30 min on ice. Then, 250 pmol of biotinylated CP strands were reacted with 1 × 107 Dynabeads Biotin Binder (Invitrogen) for 15 min at room temperature before quenching with 50 to 125 μM d-biotin (Avidity). CP-coated beads were washed five times with sorting buffer using a MACSxpress Separator (Miltenyi Biotec) and then annealed to Ab-TP stained cells for 30 min at 4 °C. Cell samples were washed five times with sorting buffer using the MACSxpress Separator to remove unlabeled cells. Target cells were first released by adding RP strands (5 μM final concentration) to cells resuspended in 500 μL of sorting buffer and incubating in a tube rotator for 1 h at room temperature and then recovered by washing five times with sorting buffer in the MACSxpress Separator. For multiplexed sorts, subsequent RP strand incubation and magnetic washes were performed to recover additional target cell populations. Annealing and release of magnetic beads were visualized by imaging cells using an EVOS FL Auto Imaging System (40× objective; Life Technologies) before and after addition of RP strands. Target cell purity was measured by staining recovered cells with alternate antibody clones against the targeted cell surface marker and analyzing on a BD Accuri C6.
Dual Gated DNA-Gated Cell Sorting with pMHC Tetramers.
Splenocytes from pooled pmel and P14 mice or LCMV Arm-infected mice (day 8 p.i.) were CD8-purified by positive or negative selection, respectively, using Ab-DGS as described above. For DGS using pMHC tetramers, 250 pmol of biotinylated CP strands were reacted with 1 × 107 Dynabeads Biotin Binder (Invitrogen) for 15 min at room temperature and then washed eight times with sorting buffer using a DynaMag-2 magnet (Invitrogen). Then, 5 μg of corresponding StvC-TP was mixed with appropriate CP-coated beads for 30 min at 4 °C and then washed five times with sorting buffer. Next, 18 μg of biotinylated pMHC monomer (Db-GP33 for P14, Db-GP100 for pmel, or Db-GP276, Kb-NP205 for LCMV experiments) was added to StvC-coated beads for 30 min at 4 °C and then washed five times with sorting buffer. CD8+ purified cells were then incubated with pMHC tetramer-coated beads for 30 min at 4 °C, washed five times with sorting buffer to remove unlabeled cells, and then serially incubated with 5 μM appropriate RP strands for 1 h at room temperature to recover target antigen-specific cells. Purity was measured by staining released cells with fluorescent tetramer.
Software and Statistical Analysis.
Graphs were plotted and Student’s t tests were conducted using GraphPad Prism 6.0. Brightness/contrast of microscopy images were adjusted using ImageJ (NIH). Flow cytometry data were analyzed using FlowJo X.
Supplementary Material
Acknowledgments
We thank R. Ahmed (Emory) for insightful discussions. This work was funded by NIH Director’s New Innovator Award DP2HD091793 and by National Center for Advancing Translational Sciences of the NIH Award UL1TR000454. S.N.D. is supported by NSF Graduate Research Fellowships Program Grant DGE-1650044 and NSF Integrative Graduate Education and Research Traineeship Grant 0965945. Y.M.C. is supported by National Institute of General Medical Sciences of the NIH Award T32GM8169-30. G.A.K. holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund. This work was performed in part at the Georgia Tech Institute for Electronics and Nanotechnology, a member of the National Nanotechnology Coordinated Infrastructure, which is supported by NSF Grant ECCS-1542174.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.F. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714820115/-/DCSupplemental.
References
- 1.Bendall SC, Nolan GP, Roederer M, Chattopadhyay PK. A deep profiler’s guide to cytometry. Trends Immunol. 2012;33:323–332. doi: 10.1016/j.it.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaye DL, Bray RA, Gebel HM, Harris WA, Waller EK. Translational applications of flow cytometry in clinical practice. J Immunol. 2012;188:4715–4719. doi: 10.4049/jimmunol.1290017. [DOI] [PubMed] [Google Scholar]
- 3.Spitzer MH, Nolan GP. Mass cytometry: Single cells, many features. Cell. 2016;165:780–791. doi: 10.1016/j.cell.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 5.Morikawa S, et al. Prospective identification, isolation, and systemic transplantation of multipotent mesenchymal stem cells in murine bone marrow. J Exp Med. 2009;206:2483–2496. doi: 10.1084/jem.20091046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tani H, Morris RJ, Kaur P. Enrichment for murine keratinocyte stem cells based on cell surface phenotype. Proc Natl Acad Sci USA. 2000;97:10960–10965. doi: 10.1073/pnas.97.20.10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uchida N, et al. Direct isolation of human central nervous system stem cells. Proc Natl Acad Sci USA. 2000;97:14720–14725. doi: 10.1073/pnas.97.26.14720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang W, et al. Biomarkers on melanoma patient T cells associated with ipilimumab treatment. J Transl Med. 2012;10:146. doi: 10.1186/1479-5876-10-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daud AI, et al. Tumor immune profiling predicts response to anti-PD-1 therapy in human melanoma. J Clin Invest. 2016;126:3447–3452. doi: 10.1172/JCI87324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen CJ, et al. Isolation of neoantigen-specific T cells from tumor and peripheral lymphocytes. J Clin Invest. 2015;125:3981–3991. doi: 10.1172/JCI82416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Coppi P, et al. Isolation of amniotic stem cell lines with potential for therapy. Nat Biotechnol. 2007;25:100–106. doi: 10.1038/nbt1274. [DOI] [PubMed] [Google Scholar]
- 12.Newell EW, Klein LO, Yu W, Davis MM. Simultaneous detection of many T-cell specificities using combinatorial tetramer staining. Nat Methods. 2009;6:497–499. doi: 10.1038/nmeth.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hadrup SR, et al. Parallel detection of antigen-specific T-cell responses by multidimensional encoding of MHC multimers. Nat Methods. 2009;6:520–526. doi: 10.1038/nmeth.1345. [DOI] [PubMed] [Google Scholar]
- 14.Krutzik PO, Nolan GP. Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nat Methods. 2006;3:361–368. doi: 10.1038/nmeth872. [DOI] [PubMed] [Google Scholar]
- 15.Krutzik PO, Crane JM, Clutter MR, Nolan GP. High-content single-cell drug screening with phosphospecific flow cytometry. Nat Chem Biol. 2008;4:132–142. doi: 10.1038/nchembio.2007.59. [DOI] [PubMed] [Google Scholar]
- 16.Bendall SC, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu P, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011;29:886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Porpiglia E, et al. High-resolution myogenic lineage mapping by single-cell mass cytometry. Nat Cell Biol. 2017;19:558–567. doi: 10.1038/ncb3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bodenmiller B, et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat Biotechnol. 2012;30:858–867. doi: 10.1038/nbt.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newell EW, Sigal N, Bendall SC, Nolan GP, Davis MM. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity. 2012;36:142–152. doi: 10.1016/j.immuni.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spitzer MH, et al. Systemic immunity is required for effective cancer immunotherapy. Cell. 2017;168:487–502.e15. doi: 10.1016/j.cell.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bailey RC, Kwong GA, Radu CG, Witte ON, Heath JR. DNA-encoded antibody libraries: A unified platform for multiplexed cell sorting and detection of genes and proteins. J Am Chem Soc. 2007;129:1959–1967. doi: 10.1021/ja065930i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwong GA, et al. Modular nucleic acid assembled p/MHC microarrays for multiplexed sorting of antigen-specific T cells. J Am Chem Soc. 2009;131:9695–9703. doi: 10.1021/ja9006707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Onoe H, et al. Cellular microfabrication: Observing intercellular interactions using lithographically-defined DNA capture sequences. Langmuir. 2012;28:8120–8126. doi: 10.1021/la204863s. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Z, Chen N, Li S, Battig MR, Wang Y. Programmable hydrogels for controlled cell catch and release using hybridized aptamers and complementary sequences. J Am Chem Soc. 2012;134:15716–15719. doi: 10.1021/ja307717w. [DOI] [PubMed] [Google Scholar]
- 26.Todhunter ME, et al. Programmed synthesis of three-dimensional tissues. Nat Methods. 2015;12:975–981. doi: 10.1038/nmeth.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi HM, et al. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat Biotechnol. 2010;28:1208–1212. doi: 10.1038/nbt.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schweller RM, et al. Multiplexed in situ immunofluorescence using dynamic DNA complexes. Angew Chem Int Ed Engl. 2012;51:9292–9296. doi: 10.1002/anie.201204304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jungmann R, et al. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat Methods. 2014;11:313–318. doi: 10.1038/nmeth.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Probst CE, Zrazhevskiy P, Gao X. Rapid multitarget immunomagnetic separation through programmable DNA linker displacement. J Am Chem Soc. 2011;133:17126–17129. doi: 10.1021/ja2072324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rudchenko M, et al. Autonomous molecular cascades for evaluation of cell surfaces. Nat Nanotechnol. 2013;8:580–586. doi: 10.1038/nnano.2013.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.You M, et al. DNA “nano-claw”: Logic-based autonomous cancer targeting and therapy. J Am Chem Soc. 2014;136:1256–1259. doi: 10.1021/ja4114903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang DY, Winfree E. Control of DNA strand displacement kinetics using toehold exchange. J Am Chem Soc. 2009;131:17303–17314. doi: 10.1021/ja906987s. [DOI] [PubMed] [Google Scholar]
- 34.Panyutin IG, Hsieh P. The kinetics of spontaneous DNA branch migration. Proc Natl Acad Sci USA. 1994;91:2021–2025. doi: 10.1073/pnas.91.6.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Q, Luan G, Guo Q, Liang J. A new class of homogeneous nucleic acid probes based on specific displacement hybridization. Nucleic Acids Res. 2002;30:E5. doi: 10.1093/nar/30.2.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fern J, et al. DNA strand-displacement timer circuits. ACS Synth Biol. 2017;6:190–193. doi: 10.1021/acssynbio.6b00170. [DOI] [PubMed] [Google Scholar]
- 37.Zhang DY. Towards domain-based sequence design for DNA strand displacement reactions. Lect Notes Comput Sci. 2011;6518:162–175. [Google Scholar]
- 38.Lakin MR, Youssef S, Polo F, Emmott S, Phillips A. Visual DSD: A design and analysis tool for DNA strand displacement systems. Bioinformatics. 2011;27:3211–3213. doi: 10.1093/bioinformatics/btr543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dirks RM, Lin M, Winfree E, Pierce NA. Paradigms for computational nucleic acid design. Nucleic Acids Res. 2004;32:1392–1403. doi: 10.1093/nar/gkh291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zadeh JN, et al. NUPACK: Analysis and design of nucleic acid systems. J Comput Chem. 2011;32:170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- 41.Reznik GO, Vajda S, Cantor CR, Sano T. A streptavidin mutant useful for directed immobilization on solid surfaces. Bioconjug Chem. 2001;12:1000–1004. doi: 10.1021/bc015507t. [DOI] [PubMed] [Google Scholar]
- 42.Lakin MR, Youssef S, Cardelli L, Phillips A. Abstractions for DNA circuit design. J R Soc Interface. 2012;9:470–486. doi: 10.1098/rsif.2011.0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thubagere AJ, et al. Compiler-aided systematic construction of large-scale DNA strand displacement circuits using unpurified components. Nat Commun. 2017;8:14373. doi: 10.1038/ncomms14373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sano T, Cantor CR. Expression of a cloned streptavidin gene in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:142–146. doi: 10.1073/pnas.87.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.