Significance
Pentameric ligand-gated ion channels (pLGICs) mediate fast signal transduction in animal nerve cells through neurotransmitters. Mutation of some of these receptors in the brain causes severe nervous system diseases. The high sequence diversity of prokaryotic receptors makes them unique model systems to understand evolutionary conservation in gating and sensitivity to allosteric modulators. We present the 2.3 Å X-ray structure of a pLGIC (sTeLIC) from a gammaproteobacteria that is activated at alkaline pH. The structure at pH 8.0 displays an unusually open pore. It is unchanged, but less flexible, in the presence of a positive allosteric modulator that binds in a cavity where benzodiazepines are found in Erwinia chrysanthemi pLGIC. This cavity is also present (and druggable) in the 5HT3-receptor.
Keywords: ligand-gated ion channel, structural biology, crystallography, electrophysiology, druggable cavity
Abstract
Pentameric ligand-gated ion channels (pLGICs) constitute a widespread class of ion channels, present in archaea, bacteria, and eukaryotes. Upon binding of their agonists in the extracellular domain, the transmembrane pore opens, allowing ions to go through, via a gating mechanism that can be modulated by a number of drugs. Even though high-resolution structural information on pLGICs has increased in a spectacular way in recent years, both in bacterial and in eukaryotic systems, the structure of the open channel conformation of some intensively studied receptors whose structures are known in a nonactive (closed) form, such as Erwinia chrysanthemi pLGIC (ELIC), is still lacking. Here we describe a gammaproteobacterial pLGIC from an endo-symbiont of Tevnia jerichonana (sTeLIC), whose sequence is closely related to the pLGIC from ELIC with 28% identity. We provide an X-ray crystallographic structure at 2.3 Å in an active conformation, where the pore is found to be more open than any current conformation found for pLGICs. In addition, two charged restriction rings are present in the vestibule. Functional characterization shows sTeLIC to be a cationic channel activated at alkaline pH. It is inhibited by divalent cations, but not by quaternary ammonium ions, such as tetramethylammonium. Additionally, we found that sTeLIC is allosterically potentiated by aromatic amino acids Phe and Trp, as well as their derivatives, such as 4-bromo-cinnamate, whose cocrystal structure reveals a vestibular binding site equivalent to, but more deeply buried than, the one already described for benzodiazepines in ELIC.
Pentameric ligand-gated ion channels (pLGICs), also known as Cys-loop receptors (1, 2) or Pro-loop receptors (3), play an essential role in nerve impulse transmission in animals by mediating the transduction of a chemical signal (release of a neurotransmitter in the synaptic cleft) into an electrical signal (modification of the membrane potential) in the millisecond range. The receptors are divided into two different functional families: cationic channels, namely nicotinic acetylcholine (nACh-R) and serotonergic type 3 receptors (5HT3-R); and anionic channels, including glycinergic (Gly-R) and GABA-ergic type A receptors (GABAA-R), yet they retain a very similar folding topology and molecular architecture (1). Following the binding of agonists in the extracellular domain (ECD), the receptors undergo an allosteric conformational change, whereby the transmembrane domain (TMD) switches from a closed to an open state, letting select ions permeate through the pore down their electrochemical gradient.
Dysfunction of Cys-loop receptors in the brain causes severe neurological diseases, and these receptors are the subject of many functional, structural, and biophysical studies (4). Interestingly, these receptors are the targets of numerous natural and synthetic compounds of pharmacological interest, including nicotine (5), alcohols (6, 7), benzodiazepines (8), barbiturates (9), and general anesthetics (10–12). Therefore, understanding their molecular properties and the mechanisms of the gating-process during channel opening, at an atomic level, remains a central issue in structural neurobiology and pharmacology with potentially far-reaching consequences for human health.
There are also numerous members of the pLGIC family throughout the bacterial world and their physiological role is not known, although they are thought to be involved in bacterial cell-to-cell communication, such as chemotaxis and quorum sensing (3, 13). The first two full-length crystal structures of members of the pLGICs family were obtained with bacterial pLGICs, the Erwinia chrysanthemi and Gloeobacter violaceus pentameric ligand-gated ion channels (ELIC and GLIC) (14–16), followed by the first crystal structure of a eukaryotic pLGIC, GluClα, an anionic channel from Caenorhabditis elegans (17). Since then, all major eukaryotic classes of pLGICs had at least one member whose structure was solved either by crystallography or cryo-electron microscopy (cryo-EM), both anionic ones [a human homopentameric β3-GABAA-R (18), a human homopentameric α3-Gly-R (19), a zebrafish homopentameric α1-Gly-R (20)] and cationic ones [a mouse homopentameric 5HT3A-R (21), a human heteropentameric α4β2-nACh-R (5)]. Paradoxically, there has not been any new high-resolution structure of bacterial pLGICs from other species since 2009, even though they certainly display a wide range of pharmacologies and allosteric conformations. Thus, the wealth of information contained in the diversity of bacterial pLGICs has not been fully explored. Additionally, to understand their evolutionary history, it is important to understand all of the differences between eukaryotic and bacterial pLGICs at the molecular level (22).
GLIC was the first pLGIC to be solved both in an apparently open form, at a resolution of 2.4 Å (23), and in the resting form. The latter form reached only a resolution of 4.35 Å but contained four pentamers in the asymmetric unit, which allowed the unambiguous tracing of all of the chains after noncrystallographic symmetry averaging (24). In molecular dynamics simulations, the structure of GLIC obtained at low pH relaxes to a slightly more open form where some permeation events could be identified (23). Other crystal forms, called the locally closed forms (LC forms), were identified through mutations introducing disulfide bridges at key positions, or even single-point mutations in either the M2 α-helix TMD region or the M2–M3 loop (25–27). It is possible that these LC forms represent a preactivation state on the reaction pathway between the closed to open forms, as suggested by experimental (28) and computational studies (29). An LC form was also found in one crystal structure as coexisting with the open form at pH 4 in the same asymmetric unit (24). Recently, another crystal structure conformation was found that was suggested to represent a desensitized form (30). Finally, GLIC proved an excellent model system to study allosteric modulation at the structural level by general anesthetics (6, 7, 11, 31), alcohols (7), xenon (32), and barbiturates (9). Structural studies and determination of the binding sites of allosteric modulators correlate well with chemical affinity-labeling studies in solution (33). The precise mechanism by which protons activate the channel remains unknown, but a systematic mutational analysis identified a key cluster of residues, mainly located at the ECD–TMD interface, as critical for GLIC gating (34).
The other bacterial receptor (ELIC) for which structural information is available, also has many entries (17) in the Protein Data Bank (PDB), involving several mutants and some very informative complexes with divalent cations (35), benzodiazepines (8), and general anesthetics (36). Electronic-spin resonance studies have also been conducted on this channel (37). However, its crystal structure has only been obtained with the ion channel pore in the closed form. Indeed, all ELIC crystal structures are found in a closed-pore conformation, even in the presence of a bound agonist and for mutants favoring the active form (38); therefore, the functional state represented by the crystal structure is still not completely understood. It was even suggested that this closed form is actually an uncoupled form, reminiscent of the uncoupled form of the nAChR (39, 40). It was suggested that ELIC could be a representative member of a new subfamily of pLGICs due to its peculiar behavior in the presence of quaternary ammonium ions that are usually channel blockers, and that its gating properties should be extrapolated only with great care to other pLGICs (41). Analysis of the aromatic residues at the interface of M1/M3 and M4 helices also suggested that ELIC’s TMD is dissimilar to GLIC’s (40).
For these reasons, we decided to get more structural information from bacterial pLGICs closely related to ELIC, especially in the gammaproteobacteria genus, in the hope of crystallizing it in an open form or both its closed and open forms. In the following we describe our results for a gammaproteobacteria pLGIC (referred to as sTeLIC in the rest of the paper), having 28% sequence identity with ELIC, found in the endosymbiont of Tevnia jerichonana. This symbiont bacteria is found inside the giant tube-worm T. jerichonana, which lives close to deep-sea hydrothermal vents (42).
Results
sTeLIC Is Activated at Alkaline pH.
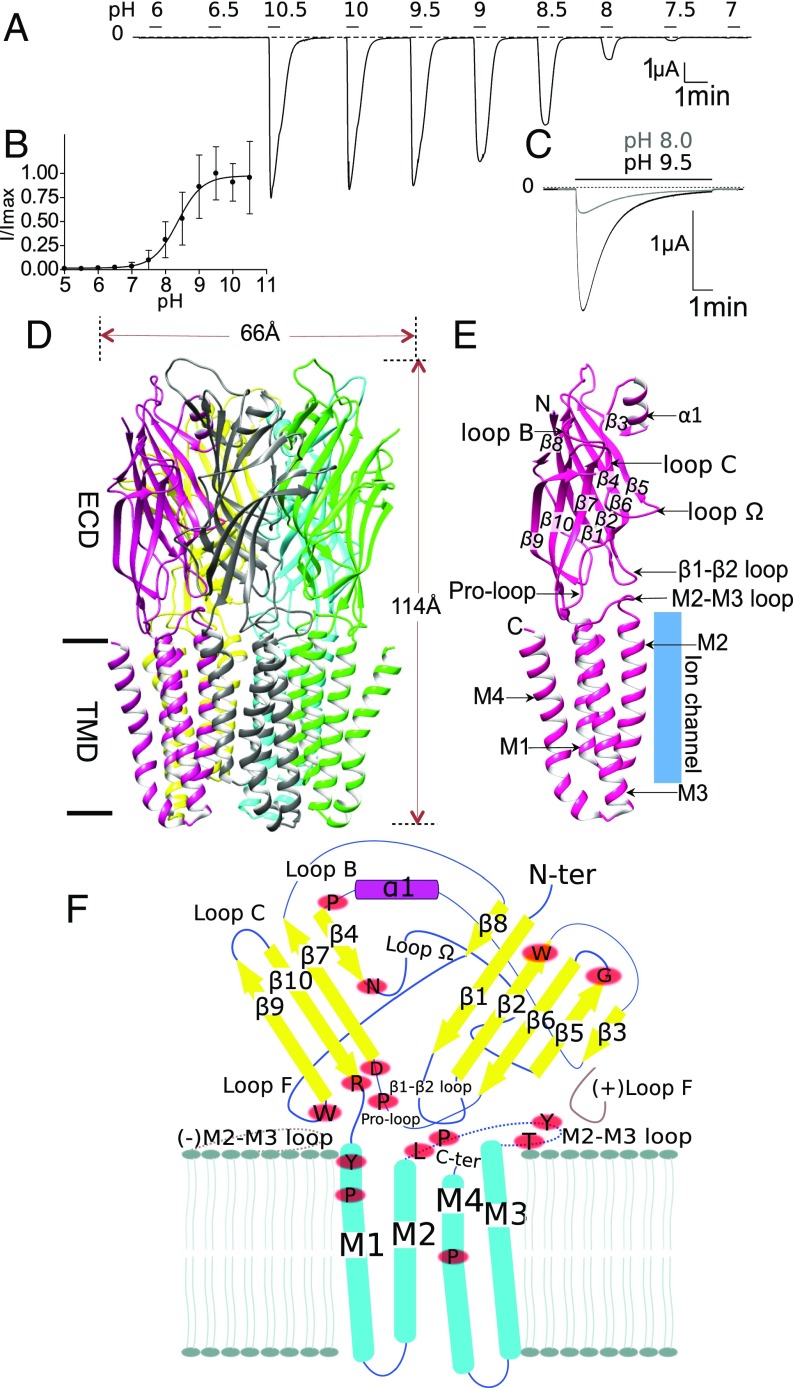
Initial electrophysiological characterization of sTeLIC expressed in Xenopus oocytes showed it is potentiated by MES buffer and inhibited by divalent cations. Therefore, Hepes and Tris buffers were used for all subsequent evaluations, after no effect was found for either buffer. Calcium was removed from the recording solution, whereas to maintain the stability of the membrane during recordings Mg2+, the weakest inhibitor, was maintained as a divalent cation. For experiments where long washes were needed, 1 mM CaCl2 was present in the wash solution only. Under these conditions, sTeLIC was found to be open at basic/neutral pHs and closed at acidic pHs (Fig. 1A). Using a perfusion buffer at pH 5, which keeps sTeLIC closed, pH-jumps to more basic conditions elicit robust currents characterized by a fast activation, followed by a rapid desensitization. Small currents start to be seen at pH 7.5 and maximal currents occur at pH 9.5. Fitting all pH activation data according to the Hill model yielded a pH50 of 8.6 ± 0.4 (Fig. 1B). Long duration applications at high pH resulted in a strong current decay (Fig. 1C), which was reversible upon return to pH 5, a property characteristic of desensitization.
Fig. 1.
Overall characterization of sTeLIC, a bacterial ion channel of the pLGIC family, structure, and activation at alkaline pH. (A) Activation at high pH. Voltage-clamp current trace obtained from an oocyte expressing sTeLIC, showing the effect of successive 30-s-long applications of an extracellular solution at various pH values. (B) pH dose–response curve. Graph of the average ± SD of normalized peak current values with a nonlinear regression fit. The pH50 of 8.6 ± 0.4 with a Hill-slope of 2 ± 1 was found by fitting individual dose–response curves from 13 oocytes. Values are reported as mean ± SD. (C) Superimposed current traces obtained from an oocyte at 19-min interval, showing the effect of 7-min applications of solution at pH 9.5 and then pH 8 (lower and upper traces, respectively) from pH 5. The strong high-pH–induced current decay observed is reversed at pH 5. (D) Side-view cartoon representation of sTeLIC X-ray crystal structure solved at pH 8.0, with subunits individually colored for clarity. (E) Schematic representation of an individual subunit, highlighting the secondary structure elements with labels. This includes the β-strands: β1–β10; α-helices: α1 in the ECD and M1–M4 in the TMD; as well as functionally important loops: loop Ω, loop B, loop C, Pro-loop, the M2–M3 loop, and the β1–β2 loop. (F) Schematic representation of the topology and secondary structure elements of sTeLIC, with neighboring complementary (−) and principal (+) subunit interactions showing the structural context of functionally important loops (in brown), and the strictly conserved residues (red ovals) among bacterial receptors (whose multialignment is shown in SI Appendix, Fig. S13).
Crystal Structure at pH 8.0 Displays a Widely Open Pore.
Initial crystallization screens yielded crystals of sTeLIC diffracting only up to 6 Å. Extensive optimization, including seeding and dehydration protocols, gave crystals diffracting to 3.5 Å. After further optimization with various additives, we found that adding 6.5 mM nonyl-β-d-glucose (NDG) gave the best crystals, at pH 8.0, diffracting up to 2.3 Å resolution. The structure of sTeLIC was solved by molecular replacement using the open structure of GLIC (PDB ID code 4HFI) (23) as the search model. The final model of sTeLIC, which leaves only six unresolved residues at the N terminus and four residues at the C terminus, has excellent refinement statistics (SI Appendix, Table S1) and electron-density maps of high quality (SI Appendix, Fig. S1). The crystal-packing molecular arrangement shows both parallel and antiparallel, head-to-tail, pentamer interactions, which consist essentially of ECD–ECD or TMD–TMD contacts (SI Appendix, Fig. S2), a phenomenon already seen in the GLIC (15) (PDB ID code 3EAM) and β3-GABAA-R (18) (PDB ID code 4COF) structures.
The overall architecture of the sTeLIC is the same as for other receptors from the pLGIC family. The five monomers are arranged along a fivefold symmetry axis perpendicular to the cell membrane with small deviations from C5 exact symmetry evaluated for pairs (1,2), (1,3), (1,4), and (1,5), respectively, as 0.17, 0.09, 0.14, and 0.14 Å. The receptor is 66 Å in the largest diameter and 114 Å tall (Fig. 1D). The TMD of one monomer is composed of a bundle of four transmembrane α-helices (M1–M4), with M2 facing the ion channel pore (Fig. 1E), flanked by M1 and M3 helices of the same subunit and the M1 π/α-helix from a neighboring (complementary) subunit. The ECD of the monomer contains 10 β-strands (β1–β10), which form a compact curled β-sandwich, where β4, β7, β9, and β10 compose the inner β-sandwich [in the principal (+) interface], and the rest form the outer β-sandwich [in the complementary (−) interface]. An amphiphilic α-helix α1 is inserted between the β3 and β4 strands, at the top ECD vestibule, with its hydrophobic part interacting with the top section of the β-sandwich and its hydrophilic part facing the vestibule (Fig. 1E). Other important regions, such as the β1–β2 loop, the M2–M3 loop, and the Pro-loop at the TMD–ECD interface are also indicated in Fig. 1E, as well as loops known to be important to bind the orthosteric-site agonist, such as loop C (β9–β10 loop) and loop B (β7–β8 loop). In Fig. 1F we have highlighted all strictly conserved residues resulting from a multiple alignment of 11 bacterial pLGICs, including sTeLIC, ELIC, and GLIC. In addition, we define here loop Ω (β4–β5 loop), which forms an almost continuous ring in the lumen because of C5 symmetry (Fig. 1 E and F).
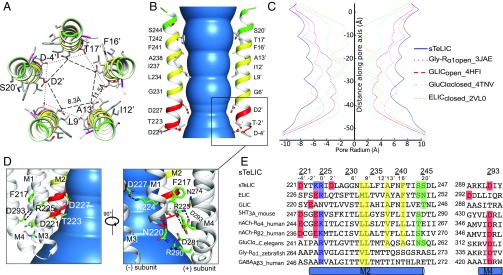
The columnar-shaped ion channel pore is constituted by the five M2 α-helices, the pore-facing residues of which shape the ion translocation pathway (Fig. 2 A and B). The diameter of the pore ranges from 11 to 15 Å (Fig. 2C), and the residues bordering the ion channel pore from the beginning to the end of M2 α-helix, are D-4′, D2′, G6′, L9′, I12′, A13′, F16′, T17′, and S20′. The distribution of the pore-lining residues is in accordance with those of other members of the Cys-loop receptor family (Fig. 2E), with the exception of G6′ (instead of S or T), D2′ (generally polar but not charged), and K-1′, whose charge is counter-balanced by a salt bridge with D2′ of the neighboring subunit (see below). The negatively charged residues, D-4′, whose side chains point to the center of the pore in its cytoplasmic end, create a strong binding site for a Na+/K+ cation (Fig. 3B) and strongly suggest that sTeLIC is a cationic channel. Compared with other members of the pLGIC family, the ion channel pore of sTeLIC has the widest diameter, with a minimum value of 11 Å at the level of D2′ (Fig. 2C). The closest comparable pore diameter is that found in the open-state structure of the α1-Gly-R, whose structure was determined by cryo-EM at 3.9 Å resolution (20). Superimposition of sTeLIC with the open state of α1-Gly-R reveals that in sTeLIC the top of the M3 α-helix and the M2–M3 loop move further outward and the pre-M1 region moves closer to the M2 and M3 α-helices to form a more compact helical bundle (SI Appendix, Fig. S3L). The widely open conformation of the pore is stabilized by two salt bridges, as illustrated in Fig. 2D, one between D293 in M4 and R225 in M2, and one between K224 (-1′) and D227 (2′). The profile of the pore is wider than other resolved pore profiles that have been assigned to the open-state (Fig. 2C). We therefore assign this conformation to an open/active state, as the transmembrane pore diameter is compatible with permeation of sodium and potassium (or chloride) ions even with their hydration shells (43). In the electron-density map of the pore itself, we found five strong residual densities in the Fo–Fc map on the surface of the ion channel pore during the model building, which were modeled by five NDG detergent molecules. NDG was used as an additive to get the high-resolution diffracting crystals; its curled aliphatic tail appears to be inserted into the crevice formed by two adjacent subunits, while its hydrophilic sugar head faces the channel pore (SI Appendix, Fig. S4 A–D). A structure obtained from crystals grown without NDG shows that the conformation of the ion channel pore is unchanged (SI Appendix, Fig. S4E), thereby excluding the influence of the NDG upon the widely open transmembrane pore conformation.
Fig. 2.
sTeLIC has a widely open ion-channel pore. (A) Top-down view of sTeLIC M2 α-helices. Amino acids facing the ion channel pore are shown as sticks with the helical path colored in green (polar), yellow (hydrophobic), and red (charged) according to the property of the corresponding side chain. The distance of negatively charged amino acids with their counterparts from the adjacent and symmetry-related subunits are shown in a dashed line. (B) Side view of M2 α-helices. The pore is colored in blue. Only two helices are shown for clarity, using the same color code as in A. Both absolute (Left) and relative (Right) numbering of amino acids facing the pore are shown. (C) Pore radius profile of sTeLIC (blue), Gly-Rα1open 3JAE (pink), GLICopen 4HFI (red), GluClαclosed 4TNV (cyan), and ELICclosed 2VL0 (light green) along their fivefold symmetry axis. (D) Two different views of the bottom part of TMD, highlighting two different salt bridges with a dashed line (D293-R225 and D227-K224). (E) Sequence alignment of the M2 α-helix and part of its neighbor M4 α-helix containing the residues involved in salt bridges between sTeLIC and other members of the pLGIC family with a known structure. The pair D227-K224 is clearly nonconserved, while the R225-D293 pair is conserved except in GLIC. negatively charged residues facing the pore at the beginning of M2 are in red, hydrophobic ones in yellow, and polar ones in green.
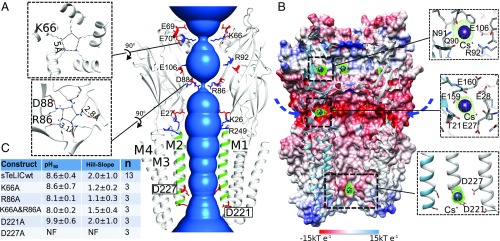
Fig. 3.
Monovalent ion binding sites and permeation pathway in sTeLIC. (A) The solvent-accessible region from the vestibule of the ECD to the pore of the TMD is shown in blue. Two of the front subunits are removed for clarity. M2 α-helices are colored in green. Charged amino acids along the putative ion permeation pathway are represented in sticks, with side chains of negatively charged amino acids colored in red and positively charged amino acids colored in blue. The two pore residues D221 and D227 are boxed for emphasis. The top-down view showing the two constriction rings for K66 and R86 are shown as Insets (Left). (B) Cutaway view of receptor inner surface colored by electrostatic potential from red (−15 kT e−1) to blue (15 kT e−1). The putative lateral ion permeation pathways are indicated with arrows. An anomalous Fourier difference electron-density map (contoured at 4.5 σ), with diffraction data collected at the peak of the anomalous signal of Cs+ (PDB ID 6FVR), is shown in a green mesh, with the Cs+ ions shown in purple spheres. (C) Summary of the electrophysiological data of five mutated constructs of sTeLIC: D221A, D227A, K66A, R86A, and the double-mutant K66A-R86A. The mean ± SD value of pH50 and Hill-slope are presented. The mutant-construct D227A was nonfunctional (NF) (see also SI Appendix, Fig. S5).
Monovalent Ion Binding Sites and Permeation Pathway.
To experimentally investigate the interaction of sTeLIC with monovalent cations or anions, we performed crystal-soaking experiments with Cs+ or Br− that have both a substantial anomalous signal at wavelengths attainable in synchrotron beamlines. Soaking the crystals with 150 mM KBr gave no signal in the anomalous map, indicating no strong binding site for bromide anions. On the other hand, crystals soaked at a final concentration of 150 mM CsCl gave anomalous difference Fourier maps that revealed two Cs+ binding sites per monomer in the vestibule of the ECD (Fig. 3B). There is one Cs+ binding site around E106, N91, and Q90, as well as another one close to loop F (with E28, E159, and E160). Additionally, one strong Cs+ anomalous peak is located at the bottom of the central ion channel pore on the C5 axis in between D-4′ and D2′. These residues were mutated into an alanine and the D2′A (D227A) mutant was found to be nonfunctional, while D-4′A (D221A) has a marked loss of function phenotype (Fig. 3C), namely a significantly higher pH50. The lack of currents seen with D2′A was not a result of expression problems, as an expression study shows the mutant and WT sTeLIC similarly localizing to the membrane (SI Appendix, Fig. S5). The removal of D2′ via an alanine mutation would remove the neutralization of K-1′, thereby potentially disrupting ion flow by inserting a positive charge into the pore.
Based on the structure, the electrostatic potential was calculated and mapped onto the protein surface (Fig. 3B). Clearly there is an intermediate chamber in the lumen of the ECD with a strong negative potential, mainly from residues E69, E70, E106, D88, and E27, explaining the binding sites of the bound cations in the vestibule (Fig. 3A). Taken together, these structural data strongly suggest that sTeLIC is a cationic ion channel. This prediction is consistent with the conclusion of the electrophysiological analysis presented below. Surprisingly, the surface representation reveals that there are two constriction rings in the ECD, one at the level of K66 in the α1 amphipathic helix, and another one at the level of D88–R86 in loop Ω (β4–β5 loop) (Fig. 3 A and B). The carboxylate group of D88 interacts with atom Nε of R86, which indicates a mesomeric form of the guanidinium group where the positive charge is carried by this atom. The distance of terminal Nε amino groups of symmetry-related mates is only 2.8–3.2 Å, showing potential hydrogen bonds between them. If these side chains were frozen in this position in solution, they would prevent ion flow through the vestibule.
The functional significance of the constriction rings was tested by site-directed mutagenesis and electrophysiological experiments in oocytes, which showed that suppression of the charged side chains by mutation to alanine of R86 or K66, or both, produced functional channels and did not significantly affect the pH50 (Fig. 3C). R86 is not conserved in either GLIC or ELIC, whereas D88 (as D86) is present in both. Strikingly, D86—along with D88—in GLIC binds soft divalent ions, such as Ni++ (PDB ID code 4NPP) (24), while hard divalent ions, such as Ba++, bind to D86 in ELIC (PDB ID code 2YN6) (SI Appendix, Fig. S6 B and C) (35).
Interestingly, similar constriction rings in the lumen at the level corresponding to loop Ω were also observed in eukaryotic members of the pLGIC family, such as: (i) in the 5HT3A-R structure (PDB ID code 4PIR) at the level of the five side chains of K108 (21) that collectively bind a central sulfate ion, and (ii) in the GluClα structure (PDB ID code 4TNV) at the level of the five side chains of Y99 that bind to a central citrate ion (44). However, 4TNV is believed to be in a nonconductive conformation and 4PIR has an uncertain functional annotation (SI Appendix, Fig. S6 D and E).
The crystal structure of the R86A mutant obtained in the same conditions as the WT-sTeLIC shows no change in the pore architecture and simply removes the constriction point seen in the WT structure’s vestibule (SI Appendix, Fig. S7).
sTeLIC Is a Cationic Channel.
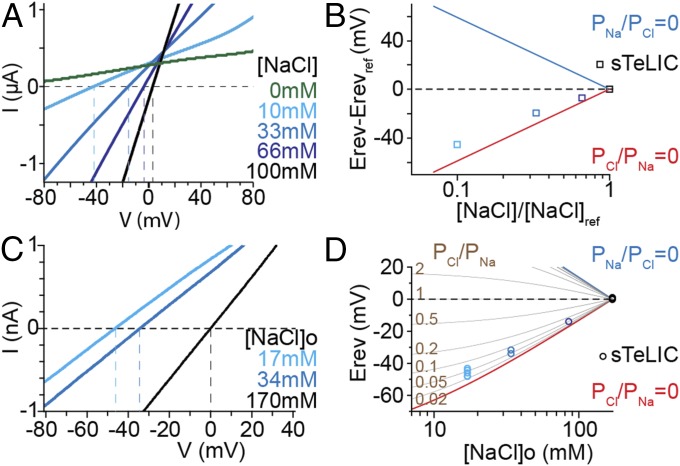
Ion charge selectivity of sTeLIC was examined in oocytes using a change in pH from 5 to 8. Reduction of the extracellular Na+ and Cl− concentrations (kept equal and referred to as the NaCl concentration) produced a left shift of the reversal potential (Fig. 4A), and the shift varied with the ratio of extracellular NaCl concentrations almost as predicted by the Goldman, Hodgkin, and Katz (GHK) voltage equation assuming PCl/PNa = 0 (Fig. 4B). This analysis, which only supposes constant intracellular ion concentrations, leads us to conclude that PCl/PNa is low for sTeLIC. Subsequent patch-clamp recordings from baby hamster kidney (BHK) cells provided recordings of current elicited by a potentiator at pH 7.5 and measurements of reversal potentials (Erev) at various extracellular NaCl concentrations, using a NaCl-based intracellular solution (Fig. 4C). A graphic plot of the Erev values measured at various extracellular NaCl concentrations showed PCl/PNa < 0.1 (Fig. 4D). From individual Erev values obtained at 17 mM [NaCl]o, calculation of the permeability ratio using the GHK equation indicates a PCl/PNa value of 0.05 ± 0.02 (n = 5). This charge selectivity analysis, based on a PCl/PNa evaluation, leads to the conclusion that sTeLIC is a cationic channel.
Fig. 4.
Cationic selectivity of sTeLIC. Ion charge selectivity was evaluated from measurements of the reversal potential of the sTeLIC current using voltage ramps, with both an oocyte two-electrode voltage-clamp set-up (A and B) and a BHK path-clamp set-up (C and D). Simultaneously and equally diminishing the extracellular concentrations of Na+ and Cl− (denoted [NaCl]) was counterbalanced with sorbitol. (A) Current-to-voltage plot of typical pH 8, from pH 5, elicited current, in 10 mM Hepes/NMDG, with 1 mM MgCl2 solution. For each [NaCl] value, currents recorded with the solution at pH 5 were subtracted from currents recorded at pH 8 on the same oocyte. (B) Plot of the shift in reversal potential (Erev-Erevref), measured in experiments as shown in A, as a function of the ratio [NaCl]/[NaCl]ref. Erevref denotes the reversal potential measured with the reference concentration, 100 mM NaCl ([NaCl]ref). Data, given as mean ± SD (error bar is smaller than the symbol), are from three oocytes. The theoretical values of the shift in Erev for anionic and cationic channels, assuming perfect selectivity for Cl− over Na+ (PNa/PCl = 0), or for Na+ over Cl− (PCl/PNa = 0) are also shown. (C) Current-to-voltage plot of sTeLIC current recorded from a BHK cell, with 17, 34, or 170 mM [NaCl] (306, 272, or 0 mM sorbitol) outside, and 170 mM [NaCl] inside, in addition to 1 mM MgCl2 and 10 mM Hepes/Na, at pH 7.5 in the presence of 30 µM 4-BrC. Average of three traces collected after desensitization was subtracted from the average of two traces near the maximal current, in each condition. (D) Plot of the reversal potential (Erev), measured in ion conditions as in C, as a function of [NaCl] outside. Erev values expected for various PCl/PNa ratios are also plotted.
Characterization of Allosteric Potentiators of sTeLIC.
To identify small molecules acting on sTeLIC, known pLGIC agonists (nicotine, acetylcholine, 5-HT, GABA, glycine, glutamate, and histamine) and other compounds were tested for the potentiation or inhibition of the pH 7.5-elicited currents, with or without potentiator (SI Appendix, Table S2). Most compounds were inactive, but some amino acids were found to have a small potentiating effect (l-Met, l-His, l-Leu), with the aromatic amino acids l-Phe and l-Trp having the most promising potentiating effect (l-Tyr could not be tested beyond 1 mM due to its low solubility). The effect of aromatic amino acids and their derivatives was systematically studied. It was found that they robustly potentiate the pH 7.5-elicited currents, and their dose–response curves at pH 7.5 were established (SI Appendix, Table S3). All compounds, including the nontitratable Nα-acetyl-l-tryptophan ethyl-ester, do not elicit currents at pH 5, suggesting that they cannot activate the receptor by themselves and that they rather act as allosteric potentiators of the pH-elicited currents (SI Appendix, Fig. S8).
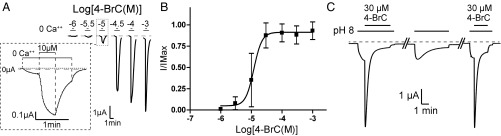
From the study presented in SI Appendix, Table S3, it appears that compounds with an aromatic ring are active at lower concentrations, which may indicate a better affinity for the site involved in potentiation. Substitution by bromine at the “para” position enhances the apparent affinity of the compound. The most potent compound found in the series was 4-bromo cinnamate (4-BrC) with an EC50 of 21 ± 8 µM (Fig. 5 A and B) when evaluated at pH 7.5. A strong current decay in the presence of the compound, which also potentiates pH 8-elicited currents, occurred during long duration 4-BrC applications, which was reversible during washout at pH 5 (Fig. 5C), indicating a 4-BrC–induced desensitization of sTeLIC. Additionally, a clear indication of cooperativity is seen for most of the potentiating compounds with a Hill number greater than 2. Because X-ray crystallography indicates that 4-BrC fully occupies the five binding sites and that this binding lowers the B-factors of the whole receptor, this large Hill number likely arises from 4-BrC binding to the five binding sites with increasing affinities, defining cooperative behavior as stated in the Monod–Wyman–Changeux theory (45). Subsequently, 4-BrC was used in conjunction with pH activation for many experiments evaluating the channel characteristics of sTeLIC.
Fig. 5.
Modulation of sTeLIC response to pH by 4-BrC. (A) Typical electrophysiological traces in oocytes showing the potentiating effect of increasing concentrations of 4-BrC. Inset shows an enlarged view of 10-μM concentration application which details the sTeLIC response to the protocol used. (B) Corresponding dose–response curve of average ±SD of normalized values with a nonlinear regression fit for 4-BrC at pH 7.5. The EC50 of 21 ± 8 µM and Hill-slope of 2.5 ± 0.5 were found by fitting individual data and averaging the fit values from six oocytes. Values reported as mean ± SD. (C) Current traces obtained from a single oocyte showing the repeated prolonged application (220 s and 60 s) of 30 µM 4-BrC upon pH 8-elicited currents (60 s and 40 s, Left and Right traces, respectively) from pH 5, with a 5 min stimulation at pH 8 only in between (Center). Double lines (//) represent 2- and 12-min respective recording times. The effects observed in the Center and Right traces indicate that the strong 4-BrC–induced current decay observed in the Left trace is reversible, and corresponds to a desensitization.
Structural Characterization of Allosteric Potentiator Binding.
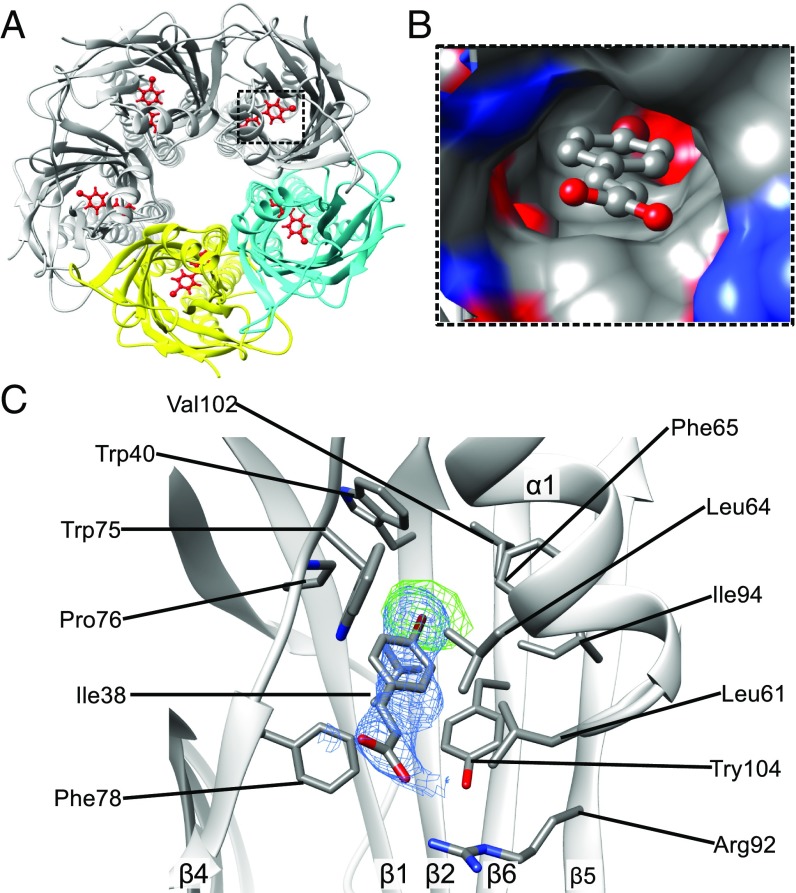
To gain structural information for molecular recognition between sTeLIC and 4-BrC, we solved the cocrystal structural complex of sTeLIC with 4-BrC at 3.0 Å resolution (Fig. 6A). Both the Fourier Fo–Fc map (contoured at 3 σ) and the anomalous map (contoured at 5 σ) using data collected at the absorption edge of bromine allow unambiguous construction of 4-BrC within the density (Fig. 6C). The binding site of 4-BrC is deeply buried in the central vestibular-facing intrasubunit cavity of the β-sandwich, where the hydrophobic head interacts with the hydrophobic core of the β-sandwich via van der Waals interactions (Fig. 6B). Its carboxylate tail forms a salt bridge with R92 from strand β5 and is exposed to the lumen. The overall conformation of this complex is very close to the conformation obtained for sTeLIC in the absence of 4-BrC, with an rmsd of 0.22 Å. However, the distribution of B-factors throughout the structure is strikingly different from the one of the unbound structure.
Fig. 6.
Crystal structure of the complex between sTeLIC and 4-BrC (PDB ID 6FLI). (A) Five symmetrical binding sites of 4-BrC in the sTeLIC ECD vestibule. Two of the subunits are highlighted in yellow and cyan. 4-BrC is colored in red and depicted as sticks. (B) Surface representation of the cavity forming the binding site of 4-BrC with the bound allosteric modulator shown in stick and ball representation. (C) Close-up view of the interaction between 4-BrC and sTeLIC. Residues interacting with 4-BrC are depicted as sticks. The anomalous density map coming from the anomalous signal of a bromine atom is shown in a green mesh contoured at 5 σ and the Fourier difference 2mFo–DFc map around 4-BrC, contoured at 1 σ, is shown as a blue mesh.
The binding site of 4-BrC, with interacting residues indicated in Fig. 6C, largely overlaps with the intrasubunit binding site of flurazepam (FZM) in ELIC, in which FZM also acts as a potentiator at low concentrations (8). Detailed structural comparison of sTeLIC–4-BrC with ELIC-FZM reveals several distinct features. First, 4-BrC inserts more deeply into the center of the β-sandwich, compared with the position of FZM in ELIC (SI Appendix, Fig. S9A). Second, five 4-BrC molecules can be assigned into the pentamer of sTeLIC, with one 4-BrC per monomer (Fig. 6A), whereas just one molecule of FZM could be found in one of the intrasubunit sites for the whole pentamer of ELIC (8). The 4-BrC binding site is also very close to the Br-acetate binding site in GLIC (46) (SI Appendix, Fig. S9B).
It should be mentioned that the difference maps of sTeLIC crystals that were devoid of 4-BrC also revealed a large blob of unexplained electron density in both 2mFo–DFc and mFo–DFc maps at the level of the intrasubunit 4-BrC binding site but closer to ECD lumen. We have tentatively assigned this density to a PEG 200 molecule (SI Appendix, Fig. S9 D and E), the precipitant used for crystallization. This density is localized similarly to the homologous Br-acetate site in GLIC and benzodiazepine FZM in ELIC (SI Appendix, Fig. S9 F and G). This extra density may explain why sTeLIC is maintained in the open state at this pH, as PEG 200, which is present at high concentration (0.9 M) in the crystallization drop, was found to potentiate sTeLIC response, especially in conjunction with DMSO at the concentration used for crystallization (SI Appendix, Fig. S10 and Table S2). It must be emphasized that assignment of the extra density to a PEG 200 molecule is speculative at this stage. Attempts to identify the bound molecule by mass spectrometry were unsuccessful. Strikingly, this density goes away when 4-BrC is bound.
Single-Channel Conductance in the Presence of a Potentiator.
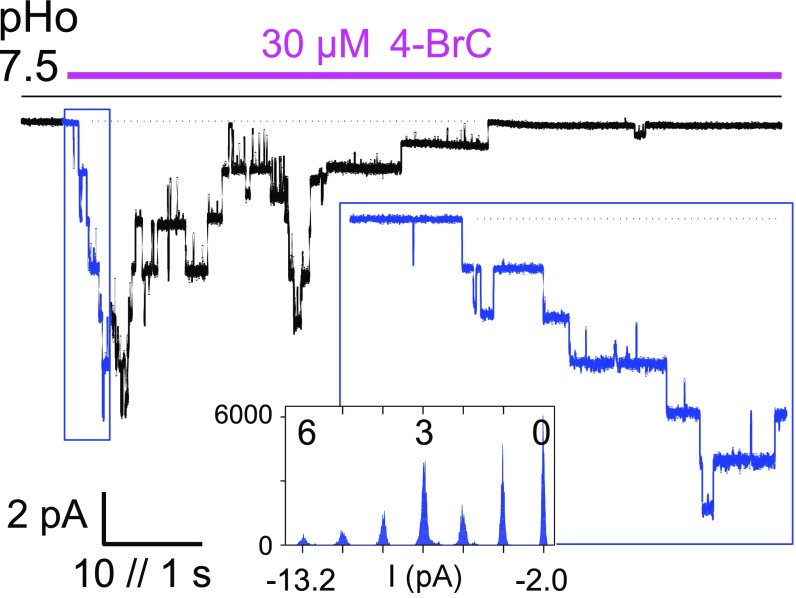
Single-channel currents flowing through individual channels expressed in BHK cells were identified at an extracellular pH of 7.5 using applications of 4-BrC to outside-out patches held at a constant membrane potential. The 4-BrC (30 µM) triggered a burst of sTeLIC activity, with a rising phase followed by a decay in the presence of the compound, synonymous with the desensitization shown in oocyte recordings (Figs. 5C and 7). All bursts displayed large current events such as those emphasized in Fig. 7, Inset. The single-channel current amplitude varied linearly with voltage, and reversed near 0 mV, as expected in the symmetrical NaCl solutions used. This indicated a single-channel conductance of 46 ± 2 pS (n = 4) between −80 and −20 mV. Current bursts from most patches in these conditions also displayed current steps with smaller amplitudes but which are much less frequent than the major 46-pS events, indicating conductance substates, which are not further analyzed here.
Fig. 7.
Identification of sTeLIC single channel current. Current trace from an outside-out patch hold at −40 mV, showing a burst of sTeLIC channels activity triggered by extracellular application of 4-BrC (30 µM) at pH 7.5. Channel activity rose and then decayed in the presence of the compound, as shown from return to all-channels–closed level (dotted line). An expanded trace of the rising phase is shown as Inset, with the corresponding current data points histogram (counts per 0.02-pA interval), showing the current levels corresponding to zero to six channels open simultaneously, as indicated above the histogram.
Inhibition by Divalent Cations but Not by Quaternary Ammonium Cations.
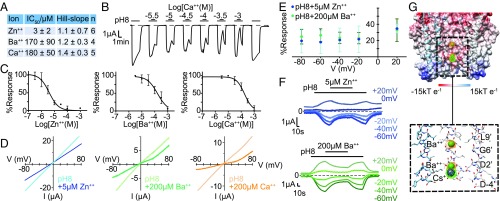
The inhibitory effect of several divalent cations was evaluated with pH 8 activation, and the most potent one was found to be Zn++ with an IC50 of 3 ± 2 µM (Fig. 8A). Calcium and barium ions have similar potencies at 180 ± 50 µM and 170 ± 90 µM, respectively. A current trace showing the effect of increasing concentrations of Ca2+ ions is shown in Fig. 8B and the corresponding dose–response curves for all ions in Fig. 8C. All of the divalent cations tested showed completely reversible inhibition, as can be seen for Ca2+ in Fig. 8B, when long-enough wash times were used. To maintain membrane stability, 1 mM Mg2+ was present in all tests. From the few (n = 4) well-behaving oocytes tested without divalent ions, the addition of Mg2+ was found to inhibit pH 8 currents by 32 ± 9% and 93 ± 8% (1 mM and 30 mM, respectively, mean ± SD), indicating about 100-fold less potency than Ca2+.
Fig. 8.
Inhibition by divalent cations. (A) IC50s (mean ± SD) for the full inhibitory effects of divalent cations Zn++, Ba++, and Ca2+. (B) Typical electrophysiological trace showing the inhibitory effect of Ca2+, at various concentrations. The IC50 is reported in A. Each experiment involved 2.5-min washes at pH 5 with 30-s pH 8 applications encompassing a 1-min application of the indicated Ca2+ concentration at pH 8. (C) Plot of mean ± SD with a nonlinear regression of Zn++, Ba++, and Ca2+ mean inhibition. The values reported in A are averages of individual fit results. (D) Current–voltage relationships for pH 8-elicited responses of sTeLIC with and without the addition of 5 µM Zn++ (cyan), 200 µM Ba++ (green), and 200 µM Ca2+ (orange), respectively. Voltage ramps were achieved from +100 mV to −100 mV over a 10-s interval to allow for divalent ion dissociation. Both I-V curves in the presence and absence of the divalent ion have the baseline pH 5 current, measured before the pH 8 application, subtracted. (E) Voltage dependence of Ba++ and Zn++ inhibition of pH 8 CaCl2-free standard solution elicited currents. Mean ± SD of three replicates represented as the percent current response in the presence of 200 µM Ba++ and 5 µM Zn++, respectively, with respect to the maximal response achieved by pH 8-elicited currents. (F) Representative traces of +20 mV to −60 mV voltage data shown in E. (G) Ba++ binding sites in sTeLIC crystal structure at pH 8. Cartoon representation of sTeLIC viewed parallel to the cell membrane. For clarity, the two monomers in the front are removed. The anomalous electron-density map from Ba++ is superimposed. The Ba++ ions are shown as deep green spheres with the anomalous map shown as an orange mesh and contoured at 4.5 σ. The Cs+ binding site is indicated by a purple sphere, with its anomalous density in green.
We used X-ray crystallography to locate binding sites for Ba++ in the sTeLIC structure due to the anomalous signal of Ba++. Two binding sites were found in the anomalous map using data collected at a longer wavelength from crystals soaked in BaCl2, and both were located in the pore, at the level of D2′ and G6′, respectively (Fig. 8G). This is in contrast with the three different binding sites found for Ba++ in the closed form of ELIC (PDB ID code 2YN6): one on top of the ion channel entrance, one around loop F, and one close to loop Ω in the lumen of the ECD (35). We were unable to collect diffraction data in the presence of Zn++, as the crystals did not stand soaking with this ion.
Because two Ba++ ions are found inside the pore (Fig. 8G), and no other location was identified in the structure, the obvious hypothesis for the mechanism is that inhibition by Ba++ would occur through an open channel block. We therefore tested the influence of voltage on pH 8-elicited currents using Ba++ and Zn++ applications at various constant voltage values (Fig. 8 E and F). We found that the fractions inhibited at from −80 mV to +20 mV were not significantly different (Fig. 8E). Moreover, using a voltage-ramp protocol, we found that the current-to-voltage (I-V) curve in the presence of a divalent cation at nonmaximal concentrations showed no rectification in the negative voltage range (Fig. 8D). These negative results of tests for voltage-dependency do not support an open-channel block mechanism. Moreover, the very slow onset of inhibition, with minutes required even at maximally active concentrations for all divalent ions tested (see traces for Ca2+ in Fig. 8B), is also not in favor of an open-channel block. Finally, the fact that a wash in our standard-solution (which contains 1 mM CaCl2) at pH 7.5 could be used as an alternative to a wash (without Ca2+) at pH 5, as a necessary step, resulting in the recovery of sTeLIC ability to be activated (SI Appendix, SI Materials and Methods), is in favor of an allosteric effect of Ca2+ binding (hypothetically selecting a resting or preactive state). Overall, despite the presence of Ba++ ions in the pore in the structure, our functional data favor an allosteric mechanism of inhibition by divalent cation, rather than an open-channel block mechanism.
Typical pLGIC channel-blockers were also tested on sTeLIC, and neither tetra-methyl ammonium (TMA), tetra-ethyl ammonium (TEA), nor tetra-propyl ammonium (TPA) blocked the channel at up to 50 mM (SI Appendix, Fig. S11). It is interesting to note that similar results were obtained for ELIC with quaternary ammonium compounds (41).
Structural Analysis of Conserved Residues.
As with other prokaryotic members of the pLGIC family, the Pro-loop lacks the disulfide bond conserved in the eukaryotic pLGICs’ Cys-loop. The Pro-loop interacts with the M2–M3 loop through the stacking interactions of F122 and P123 (Y119 and P120 in GLIC, and F119 and P120 in ELIC) and L247 (L246 in GLIC, and L255 in ELIC). The crucial salt bridge between R193 and D125 is the equivalent of R192-D122 in GLIC (R198–D122 in ELIC), whereas the third partner (D32) of this salt bridge in GLIC is replaced by Q25 (T28 in ELIC) in the β1–β2 loop (SI Appendix, Fig. S12E). The stacking interaction of W162 on the hydrophobic part of R193 side chain is also conserved (W160 in GLIC and ELIC) (SI Appendix, Fig. S12D), as well as the primordial role of the Y198 side chain (Y197 in GLIC, Y203 in ELIC). Interestingly, E161 of sTeLIC is also making a salt bridge with R193, opposite from D125. E161 is well conserved in bacterial pLGICs but is absent in GLIC, where its role is fulfilled by D32 from β1–β2 loop.
Strikingly, there is a strong covariation in prokaryotic receptors (10 of 11 cases) of the presence/absence of a negatively charged residue at the positions equivalent to sTeLIC residues 25 and 161 (SI Appendix, Fig. S13). In this respect, GLIC’s covariation results in a different loop F sequence than any other cationic pLGIC for which a structure is resolved, where the conserved short-sequence motif EW in bacteria (extended to GEW in eukaryotes) is changed to TGW. The W of this XEW motif is otherwise well conserved for cationic ion channels, but is absent in the anionic ones, where it is replaced by a conserved P(Q/S)F motif (SI Appendix, Fig. S12E).
At the ECD–TMD interface, the fundamental role of the conserved P205, at the end of the second turn of the M1 π-helix is confirmed in that it induces a characteristic kink that is further stabilized by an interaction of the carbonyl atom of M200 with the side chain of N240 from (+) subunit (SI Appendix, Fig. S14A). In addition, we noted the presence of salt-bridge R249-D246, that connects two adjacent M2–M3 loops, which is not conserved in other pLGICs, as well as R201-D257, somewhat conserved in prokaryotes, that links the pre-M1 region and the top of M3 from (+) subunit (SI Appendix, Figs. S13–S15).
Discussion
sTeLIC Shows Canonical Loop B, Loop C, and Loop F.
The analysis of the known important loops for activation in pLGICs shows that the loop B conformation is “canonical” in sTeLIC, compared with eukaryotic receptors of known structures. With this additional prokaryotic pLGIC structure that conforms to the canonical conformation, the peculiarity of GLIC’s loop B conformations is more evident, marking GLIC’s loop B as an outlier (SI Appendix, Fig. S12B). Loop C in sTeLIC displays no cavity and is in close contact with the complementary subunit, as expected in an active form. It is still unknown what the effect of pH at the level of the agonist site is, although the presence of several histidines around loop C may indicate a possible mechanism for pH-sensing (SI Appendix, Fig. S12A), perhaps through a redistribution of electrostatic energies or dielectric relaxation (47). Loop F maintains the XEW sequence motif of many cationic pLGICs, where the glutamate (E161) has a strong sequence covariation with Q25, the equivalent position of D32 in GLIC, such that the interaction of E161 with the R192-D122 salt bridge is replaced by D32 in GLIC (where XEW is not conserved and replaced by TGW). E161’s role has also been highlighted in ELIC, with the studies of the chlorpromazine binding site (48). In the ELIC structure, loop F contributes to a Ba++ binding site identified in this closed form (35). From the sequence of loop F in sTeLIC, it may be speculated that a homologous binding site involving the side chains of loop F residues E159-E160 may be found in a closed form of sTeLIC, whereas no divalent cation binding site was identified in the present open structure. The assumption that loop F might also play a role in the Ca2+ inhibition of sTeLIC by stabilizing a closed form, as shown in ELIC, is consistent with our observation that sTeLIC inhibition by divalent cations does not show the properties typical of an open-channel block (Fig. 8 D–F). If present at the level of loop F, a divalent cation would also block the alternative lateral cation pathway (see below).
sTeLIC Shares Many Features with ELIC.
The sequence and structural motifs of sTeLIC, such as an amphipathic α1-helix, absent in GLIC, are most similar to ELIC among the pLGICs (SI Appendix, Figs. S3 and S15). We propose that ELIC and sTeLIC are representative members of a subfamily of pLGICs, with specific characteristics of the pore, a possibility already envisaged for ELIC (41).
In addition to the sequence and tertiary structure similarity, sTeLIC belongs to the same gammaproteobacteria family and shares several functional similarities with ELIC. They are both cationic channels, and they are both positively modulated by the same ECD intrasubunit vestibule site (8). Moreover they share additional commonality through their inhibition by divalent ions (35), as well as a lack of inhibition by quaternary ammonium cations, such as TMA, TEA, or TPA (41). However, sTeLIC is activated at alkaline pH, whereas ELIC is activated by amine-containing compounds, such as GABA or cysteamine (49). Here we suggest that sTeLIC is indeed a second member of this subfamily and its high-resolution structure, with a widely open pore, may provide a model for their open-channel conformation.
Comparison with Other Open Conformations.
The widely open pore of sTeLIC matches very well the one of the α1-Gly-R (SI Appendix, Fig. S3L), while it is more open than in the GLIC structure at acidic pH. It has indeed been suggested from electron paramagnetic resonance experiments (50) that this GLIC structure may be only semiopen. This interpretation suggests that GLIC’s open-crystal form may represent an intermediate along the reaction path from closed to open forms, trapped because of crystallization conditions, or that the open form of GLIC and other cationic prokaryotic receptors with the same sequence covariations in loop F may have a different conformation. The extreme open form of sTeLIC’s pore may also result from the binding in both structures of either PEG 200 or 4-BrC to the PAM vestibule site.
Overall, the quaternary ECD structures of sTeLIC and the apparently open conformation of GLIC superimpose well after aligning the TMDs, with very little twist (tangential movement) and little bloom (radial movement) (SI Appendix, Fig. S3 B and E). The same comparison of sTeLIC and the open form of α1-Gly-R gives a similar result (SI Appendix, Fig. S3 C and F). However, superimposing sTeLIC and ELIC in the same way reveals both significant twist and bloom movement in the ECD (SI Appendix, Fig. S3 A and D), while the α1-helix, a secondary structure element present in both of these two structures (but not in GLIC), appears to be shifted by one helical turn along its axis. In conclusion, the quaternary conformation of the ECD of sTeLIC is more similar to the apparently open forms of GLIC and α1-Gly-R than to the closed structures of ELIC, which places at odds the proposal that ELIC’s crystal structure might represent a “decoupled state” with a “relaxed” ECD and a “closed” TMD (40).
Specificities of the Ion Permeation Pathways.
The cation pathway in the pore is well illustrated by crystallographic studies in the presence of Cs+. However, this study also suggests that there might be an additional entry pathway for monovalent cations through a lateral door located at the interface between the ECD and the TMD (Fig. 3B), as observed in GABAA-R (18) as well as 5HT3A-R, based on molecular dynamics studies (51). Changing the rotamers of E160 (loop F) and E28 (loop2) is enough to reveal a lateral tunnel, as shown in SI Appendix, Fig. S16.
In addition, the presence of two constriction rings in the lumen of the ECD (especially at the level of loop Ω) is intriguing, but these constriction rings in the ECD are not uncommon among other members of the family (GluClα, 5HT3A-R). However, we have shown that the residues causing the constriction rings are not essential to sTeLIC function (Fig. 3C), and that the R86A mutation does not significantly alter the receptor conformation (SI Appendix, Fig. S7). They may play a role as an additional filter, or as a valve, forming a temporarily isolated chamber to regulate the flow of ions inside the lumen of the ECD.
To investigate a possible dynamic role of this region, we calculated low-frequency all-atoms normal modes using a recently developed nonlinear implementation in the framework of the Elastic Network Model (52) and looked for specific modes that could potentially open and close this filter or valve. We found several modes that indeed open the constriction ring in the ECD, either in a symmetrical fashion, mode 10, (Movies S1 and S2) or in a nonsymmetrical fashion (e.g., mode 18) (Movies S3 and S4). Interestingly, mode 10 keeps the pore widely open but displays an up-and-down movement of the TMD, mimicking the functioning of a (linear) “peristaltic pump.” Mode 18, on the other hand, involves the coordinated movement of two subunits, in opposition to the three remaining ones, and it actually closes and opens the pore periodically. The normal mode analysis clearly shows that the two constriction rings at the ECD are dynamic but extensive molecular dynamics simulations in a lipid environment would be needed to further assess a possible mechanism. Single-channel recording experiments of the receptors with mutations of K66 and R86, indicating their influence upon the conductance and ion selectivity of the permeation pathway, as well as the open-channel probability, would be required to show the influence of these constriction rings on permeation and gating.
Allosteric Potentiation of sTeLIC.
A single binding site per subunit was present in the cocrystal structure for 4-BrC, the most potent positive modulator identified in this study. Therefore, sTeLIC modulation by 4-BrC, which also occurs at a neutral pH and could therefore alternatively be described as a quasi-agonist effect, occurs almost certainly by binding to this vestibular intrasubunit site in the ECD.
If one directly compares the structures with and without 4-BrC, there is almost no difference (rmsd = 0.22 Å). However, comparison of the B-factors of the two structures, crystallized in the same conditions, shows a striking difference, with the 4-BrC bound form more ordered than the unbound form (SI Appendix, Fig. S17); this is not an effect of the resolution because the form with the lower-resolution dataset has the lowest B-factors. Thus, the binding of a 4-BrC apparently reduces the vibrations around the mean conformation (and the entropy), while the total electrostatic energy is also changed due to the extra carboxylate group, resulting in a different free energy of the PAM-bound state. This could result in a better anchoring of the β-sheets to the top of the ECD, a more stable ECD–TMD interface, and an overall stabilization of the open pore by 4-BrC binding. We note that simple elastic (coarse-grained) models of proteins have been shown to be able to reproduce this effect of modulation of the amplitude of thermal fluctuations around a mean structure (53).
A systematic examination of all available structures of pLGIC in the region of the PAM binding site shows that none of the anionic channels have such a vestibular cavity (GluClα, β3-GABAA-R, α1-Gly-R, α3-Gly-R), due to an extension of loop Ω that blocks its entry. In cationic receptors however, we found, in addition to ELIC and sTeLIC, a clear accessible equivalent cavity in the structure of the eukaryotic 5HT3A-R (SI Appendix, Fig. S18). Strikingly, this last receptor also has the same kind of restriction ring in the ECD through a lysine in loop Ω (SI Appendix, Fig. S6E).
Conclusion
In conclusion, sTeLIC is a bacterial pLGIC that is relatively easy to produce and purify for crystallization and whose functional and structural characterization reveals remarkable similarities with ELIC. It thus provides an apparently open-form model of ELIC that has proved up to now resistant to structural elucidation. Further studies will be needed to indicate whether or not sTeLIC forms, together with ELIC, a new subfamily of bacterial pLGICs (as suggested in ref. 41). Crystallizing and solving the structure of sTeLIC’s resting form would considerably help in assigning a functional state for the currently available closed form of ELIC, which is still uncertain. In addition, the identification of its PAM binding site and the finding that a similar cavity exists in the available structure of 5HT3A-R open the way for the rational design of drugs aimed at this human receptor.
Materials and Methods
sTeLIC was expressed in Escherichia coli with its N-terminal fused to MBP. The fusion protein was extract from E. coli membranes by detergent DDM. The recombinant protein was purified using amylose resin and further purified by Superose TM6 Increase 10/300 GL gel-filtration column. MBP was cleaved by thrombin and removed by a second round of gel filtration. Purified protein was pooled and concentrated to 12 mg/mL. Crystals were grown using hanging-drop method. The phase problem was solved by molecular replacement. Two-electrode voltage-clamp electrophysiology experiments were carried out on sTeLIC and its variants expressed in Xenopus laevis oocytes. Patch-clamp currents were recorded from cells of the BHK cell line driven to express sTeLIC, 1–2 d after DNA-calcium phosphate transfection. Full methods are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Anaïs Menny for sharing expertise on plasmid constructions; the staff of the Crystallography Platform (PF6) in the Institut Pasteur for initial crystal screens; Reinis Reinholds Ruza for help in data collection; Jérôme Loc’h for help during structural refinement and picture preparation; Stacy Gellenoncourt for help in purification and crystallization of the sTeLIC R86A mutant; the staff of synchrotron beamlines in Soleil (Orsay) and European Synchrotron Radiation Facility (Grenoble) for data collection; Pierre Legrand (Soleil synchrotron) for suggestions on refinement in Buster; and Marc Gielen for suggestions during the paper preparation. Á.N. and Z.F. were supported by Agence Nationale de Recherches Grant 13-BSV8-0020 “Pentagate.” H.H. was sponsored by the China Scholarship Council and Institut Pasteur.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.H. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 6FL9, 6FLI, 6FVR, 6FVS, 6FVQ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717700115/-/DCSupplemental.
References
- 1.Corringer P-J, et al. Structure and pharmacology of pentameric receptor channels: From bacteria to brain. Structure. 2012;20:941–956. doi: 10.1016/j.str.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Plested AJR. Structural mechanisms of activation and desensitization in neurotransmitter-gated ion channels. Nat Struct Mol Biol. 2016;23:494–502. doi: 10.1038/nsmb.3214. [DOI] [PubMed] [Google Scholar]
- 3.Jaiteh M, Taly A, Hénin J. Evolution of pentameric ligand-gated ion channels: Pro-loop receptors. PLoS One. 2016;11:e0151934. doi: 10.1371/journal.pone.0151934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steinlein OK. ion channel mutations in neuronal diseases: A genetics perspective. Chem Rev. 2012;112:6334–6352. doi: 10.1021/cr300044d. [DOI] [PubMed] [Google Scholar]
- 5.Morales-Perez CL, Noviello CM, Hibbs RE. X-ray structure of the human α4β2 nicotinic receptor. Nature. 2016;538:411–415. doi: 10.1038/nature19785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howard RJ, et al. Structural basis for alcohol modulation of a pentameric ligand-gated ion channel. Proc Natl Acad Sci USA. 2011;108:12149–12154. doi: 10.1073/pnas.1104480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sauguet L, et al. Structural basis for potentiation by alcohols and anaesthetics in a ligand-gated ion channel. Nat Commun. 2013;4:1697. doi: 10.1038/ncomms2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spurny R, et al. Pentameric ligand-gated ion channel ELIC is activated by GABA and modulated by benzodiazepines. Proc Natl Acad Sci USA. 2012;109:E3028–E3034. doi: 10.1073/pnas.1208208109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fourati Z, et al. Barbiturates bind in the GLIC ion channel pore and cause inhibition by stabilizing a closed state. J Biol Chem. 2017;292:1550–1558. doi: 10.1074/jbc.M116.766964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nury H, Delarue M, Corringer P-J. X-ray structures of general anesthetics bound to their molecular targets. Med Sci. 2011;27:1056–1057. doi: 10.1051/medsci/20112712006. [DOI] [PubMed] [Google Scholar]
- 11.Nury H, et al. X-ray structures of general anaesthetics bound to a pentameric ligand-gated ion channel. Nature. 2011;469:428–431. doi: 10.1038/nature09647. [DOI] [PubMed] [Google Scholar]
- 12.Sauguet L, Shahsavar A, Delarue M. Crystallographic studies of pharmacological sites in pentameric ligand-gated ion channels. Biochim Biophys Acta. 2015;1850:511–523. doi: 10.1016/j.bbagen.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 13.Tasneem A, Iyer LM, Jakobsson E, Aravind L. Identification of the prokaryotic ligand-gated ion channels and their implications for the mechanisms and origins of animal Cys-loop ion channels. Genome Biol. 2005;6:R4. doi: 10.1186/gb-2004-6-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hilf RJC, Dutzler R. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature. 2008;452:375–379. doi: 10.1038/nature06717. [DOI] [PubMed] [Google Scholar]
- 15.Bocquet N, et al. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009;457:111–114. doi: 10.1038/nature07462. [DOI] [PubMed] [Google Scholar]
- 16.Hilf RJC, Dutzler R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature. 2009;457:115–118. doi: 10.1038/nature07461. [DOI] [PubMed] [Google Scholar]
- 17.Hibbs RE, Gouaux E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature. 2011;474:54–60. doi: 10.1038/nature10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller PS, Aricescu AR. Crystal structure of a human GABAA receptor. Nature. 2014;512:270–275. doi: 10.1038/nature13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang X, Chen H, Michelsen K, Schneider S, Shaffer PL. Crystal structure of human glycine receptor-α3 bound to antagonist strychnine. Nature. 2015;526:277–280. doi: 10.1038/nature14972. [DOI] [PubMed] [Google Scholar]
- 20.Du J, Lü W, Wu S, Cheng Y, Gouaux E. Glycine receptor mechanism elucidated by electron cryo-microscopy. Nature. 2015;526:224–229. doi: 10.1038/nature14853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hassaine G, et al. X-ray structure of the mouse serotonin 5-HT3 receptor. Nature. 2014;512:276–281. doi: 10.1038/nature13552. [DOI] [PubMed] [Google Scholar]
- 22.Armstrong CM. Packaging life: The origin of ion-selective channels. Biophys J. 2015;109:173–177. doi: 10.1016/j.bpj.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sauguet L, et al. Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels. EMBO J. 2013;32:728–741. doi: 10.1038/emboj.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sauguet L, et al. Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation. Proc Natl Acad Sci USA. 2014;111:966–971. doi: 10.1073/pnas.1314997111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez-Gutierrez G, Cuello LG, Nair SK, Grosman C. Gating of the proton-gated ion channel from Gloeobacter violaceus at pH 4 as revealed by X-ray crystallography. Proc Natl Acad Sci USA. 2013;110:18716–18721. doi: 10.1073/pnas.1313156110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prevost MS, et al. A locally closed conformation of a bacterial pentameric proton-gated ion channel. Nat Struct Mol Biol. 2012;19:642–649. doi: 10.1038/nsmb.2307. [DOI] [PubMed] [Google Scholar]
- 27.Bertozzi C, Zimmermann I, Engeler S, Hilf RJC, Dutzler R. Signal transduction at the domain interface of prokaryotic pentameric ligand-gated ion channels. PLoS Biol. 2016;14:e1002393. doi: 10.1371/journal.pbio.1002393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Menny A, et al. Identification of a pre-active conformation of a pentameric channel receptor. eLife. 2017;6:e23955. doi: 10.7554/eLife.23955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lev B, et al. String method solution of the gating pathways for a pentameric ligand-gated ion channel. Proc Natl Acad Sci USA. 2017;114:E4158–E4167. doi: 10.1073/pnas.1617567114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basak S, Schmandt N, Gicheru Y, Chakrapani S. Crystal structure and dynamics of a lipid-induced potential desensitized-state of a pentameric ligand-gated channel. eLife. 2017;6:e23886. doi: 10.7554/eLife.23886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laurent B, et al. Sites of anesthetic inhibitory action on a cationic ligand-gated ion channel. Structure. 2016;24:595–605. doi: 10.1016/j.str.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 32.Sauguet L, Fourati Z, Prangé T, Delarue M, Colloc’h N. Structural basis for xenon inhibition in a cationic pentameric ligand-gated ion channel. PLoS One. 2016;11:e0149795. doi: 10.1371/journal.pone.0149795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiara DC, et al. Photoaffinity labeling the propofol binding site in GLIC. Biochemistry. 2014;53:135–142. doi: 10.1021/bi401492k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nemecz Á, et al. Full mutational mapping of titratable residues helps to identify proton-sensors involved in the control of channel gating in the Gloeobacter violaceus pentameric ligand-gated ion channel. PLoS Biol. 2017;15:e2004470. doi: 10.1371/journal.pbio.2004470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zimmermann I, Marabelli A, Bertozzi C, Sivilotti LG, Dutzler R. Inhibition of the prokaryotic pentameric ligand-gated ion channel ELIC by divalent cations. PLoS Biol. 2012;10:e1001429. doi: 10.1371/journal.pbio.1001429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spurny R, et al. Multisite binding of a general anesthetic to the prokaryotic pentameric Erwinia chrysanthemi ligand-gated ion channel (ELIC) J Biol Chem. 2013;288:8355–8364. doi: 10.1074/jbc.M112.424507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kinde MN, et al. Conformational changes underlying desensitization of the pentameric ligand-gated ion channel ELIC. Structure. 2015;23:995–1004. doi: 10.1016/j.str.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonzalez-Gutierrez G, et al. Mutations that stabilize the open state of the Erwinia chrisanthemi ligand-gated ion channel fail to change the conformation of the pore domain in crystals. Proc Natl Acad Sci USA. 2012;109:6331–6336. doi: 10.1073/pnas.1119268109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.daCosta CJB, Dey L, Therien JPD, Baenziger JE. A distinct mechanism for activating uncoupled nicotinic acetylcholine receptors. Nat Chem Biol. 2013;9:701–707. doi: 10.1038/nchembio.1338. [DOI] [PubMed] [Google Scholar]
- 40.Sun J, Comeau JF, Baenziger JE. Probing the structure of the uncoupled nicotinic acetylcholine receptor. Biochim Biophys Acta. 2017;1859:146–154. doi: 10.1016/j.bbamem.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 41.Gonzalez-Gutierrez G, Grosman C. The atypical cation-conduction and gating properties of ELIC underscore the marked functional versatility of the pentameric ligand-gated ion-channel fold. J Gen Physiol. 2015;146:15–36. doi: 10.1085/jgp.201411333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gardebrecht A, et al. Physiological homogeneity among the endosymbionts of Riftia pachyptila and Tevnia jerichonana revealed by proteogenomics. ISME J. 2012;6:766–776. doi: 10.1038/ismej.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Persson I. Hydrated metal ions in aqueous solution: How regular are their structures? Pure Appl Chem. 2010;82:1901–1917. [Google Scholar]
- 44.Althoff T, Hibbs RE, Banerjee S, Gouaux E. X-ray structures of GluCl in apo states reveal a gating mechanism of Cys-loop receptors. Nature. 2014;512:333–337. doi: 10.1038/nature13669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monod J, Wyman J, Changeux J-P. On the nature of allosteric transitions: A plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 46.Fourati Z, Sauguet L, Delarue M. Genuine open form of the pentameric ligand-gated ion channel GLIC. Acta Crystallogr D Biol Crystallogr. 2015;71:454–460. doi: 10.1107/S1399004714026698. [DOI] [PubMed] [Google Scholar]
- 47.Liu J, Nussinov R. Energetic redistribution in allostery to execute protein function. Proc Natl Acad Sci USA. 2017;114:7480–7482. doi: 10.1073/pnas.1709071114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nys M, et al. Allosteric binding site in a Cys-loop receptor ligand-binding domain unveiled in the crystal structure of ELIC in complex with chlorpromazine. Proc Natl Acad Sci USA. 2016;113:E6696–E6703. doi: 10.1073/pnas.1603101113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zimmermann I, Dutzler R. Ligand activation of the prokaryotic pentameric ligand-gated ion channel ELIC. PLoS Biol. 2011;9:e1001101. doi: 10.1371/journal.pbio.1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dellisanti CD, et al. Site-directed spin labeling reveals pentameric ligand-gated ion channel gating motions. PLoS Biol. 2013;11:e1001714. doi: 10.1371/journal.pbio.1001714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Di Maio D, Chandramouli B, Brancato G. Pathways and barriers for ion translocation through the 5-HT3A receptor channel. PLoS One. 2015;10:e0140258. doi: 10.1371/journal.pone.0140258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoffmann A, Grudinin S. NOLB: Nonlinear rigid block normal-mode analysis method. J Chem Theory Comput. 2017;13:2123–2134. doi: 10.1021/acs.jctc.7b00197. [DOI] [PubMed] [Google Scholar]
- 53.McLeish TCB, Rodgers TL, Wilson MR. Allostery without conformation change: Modelling protein dynamics at multiple scales. Phys Biol. 2013;10:056004. doi: 10.1088/1478-3975/10/5/056004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.