Significance
More than 80% of human proteins are N-terminal (Nt)–acetylated during translation. In contrast, actin, the most abundant protein in the cytoplasm of animal cells, is Nt-acetylated posttranslationally and following a unique multistep mechanism that has remained poorly characterized. Here, we describe the discovery of actin’s N-terminal acetyltransferase (NAT), NAA80. We further demonstrate that actin Nt-acetylation plays essential roles in filament assembly, cytoskeleton organization, and cell motility, resulting in a net increase in the ratio of monomeric to filamentous actin and fewer lamellipodia and filopodia. These effects converge to reduce cell hypermotility. This work establishes the role of Nt-acetylation for the most abundant cytoskeletal protein in animals and reveals a NAT acting posttranslationally and on a single dedicated substrate.
Keywords: actin, NAA80, NAT, N-terminal acetylation, cell motility
Abstract
Actin, one of the most abundant proteins in nature, participates in countless cellular functions ranging from organelle trafficking and pathogen motility to cell migration and regulation of gene transcription. Actin’s cellular activities depend on the dynamic transition between its monomeric and filamentous forms, a process exquisitely regulated in cells by a large number of actin-binding and signaling proteins. Additionally, several posttranslational modifications control the cellular functions of actin, including most notably N-terminal (Nt)-acetylation, a prevalent modification throughout the animal kingdom. However, the biological role and mechanism of actin Nt-acetylation are poorly understood, and the identity of actin’s N-terminal acetyltransferase (NAT) has remained a mystery. Here, we reveal that NAA80, a suggested NAT enzyme whose substrate specificity had not been characterized, is Nt-acetylating actin. We further show that actin Nt-acetylation plays crucial roles in cytoskeletal assembly in vitro and in cells. The absence of Nt-acetylation leads to significant differences in the rates of actin filament depolymerization and elongation, including elongation driven by formins, whereas filament nucleation by the Arp2/3 complex is mostly unaffected. NAA80-knockout cells display severely altered cytoskeletal organization, including an increase in the ratio of filamentous to globular actin, increased filopodia and lamellipodia formation, and accelerated cell motility. Together, the results demonstrate NAA80’s role as actin’s NAT and reveal a crucial role for actin Nt-acetylation in the control of cytoskeleton structure and dynamics.
Humans express six highly conserved actin isoforms, which account for ∼5% of the total protein in most cells and ∼20% in muscle cells (1). Actin’s abundance is paralleled by its crucial role in countless cellular functions (2), which are regulated in cells by a large number of actin-binding proteins (ABPs) (3). An essential group of actin regulators is those that control the initiation of actin polymerization, known as “actin nucleators” (4). By overcoming the rate-limiting step in actin polymerization, i.e., nucleation, these proteins control the spatiotemporal polymerization of actin as well as the specific morphology of actin networks (2, 5). Two types of nucleators, actin-related proteins (the Arp2/3 complex) and formins, mediate most nucleation activities in cells. The Arp2/3 complex is responsible for the formation of branched actin networks (6), whereas formins mediate the assembly of unbranched filament structures such as filopodia and stress fibers (7). Other than ABPs, several forms of posttranslational modifications (PTMs) regulate actin’s function, among which N-terminal (Nt)-acetylation is the most widespread. Protein Nt-acetylation, affecting 80–90% of all proteins in humans, is emerging as a key regulator of cell function (8–10) with major roles in the control of protein lifetime (11, 12), protein–protein interaction (13), and protein folding (14). While most eukaryotic proteins are Nt-acetylated cotranslationally by one of several ribosome-associated N-terminal acetyltransferases (NATs), i.e., NatA–NatE (8), animal actins are processed posttranslationally (15). The omnipresence of Nt-acetylation in actin suggests that it can be conceptually viewed as part of its functional sequence and is thus thought to play a major role in cytoskeleton assembly by making actin’s exposed N terminus exceptionally acidic. However, the functional consequences of actin Nt-acetylation remain largely unexplored, and the identity of actin’s NAT has remained a mystery.
Results
NAA80 Nt-Acetylates Actin.
Nat6/Fus-2/NAA80 was previously described as a putative NAT (16), based on sequence analysis that suggested the presence of a NAT-specific (GCN5)-related N-acetyltransferase (GNAT)-fold (Fig. S1A), although no specific substrate had been described. Here we studied the Nt-acetylation activity of NAA80 toward a representative selection of peptides, revealing a marked preference for peptides with acidic N termini (DDDI and EEEI) (Fig. 1A and Fig. S1B). This preference for the DDDI substrate peptide and EEEI as the second best was observed both in NAA80 immunoprecipitated from HeLa cells (Fig. 1A) and in recombinantly expressed and purified NAA80 (Fig. S1B). Among all the proteins expressed in animals, these N termini uniquely correspond to those of cytoplasmic β- and γ-actin, respectively, after the intermediate step of posttranslational cleavage of the N-terminal acetylmethionine (Fig. S1C) (17).
Fig. 1.
NAA80 Nt-acetylates β- and γ-actin. (A) NAA80-V5 was immunoprecipitated from HeLa cells and used in Nt-acetylation assays with [14C]-Ac-CoA and peptides representing a variety of N termini. (B) NAA80-eGFP expressed in HeLa and HAP1 cells shows a cytosolic distribution. (C) Phosphorescence imaging shows a distinct 43-kDa band in NAA80 KO1 cell lysates incubated with [14C]-Ac-CoA and purified recombinant MBP-NAA80, suggesting the incorporation of [14C]-Ac at the N terminus of actin. (D) The isoform and Nt-acetylation specificity of Ac-β-actin and Ac-γ-actin antibodies is confirmed in dot blot assays with peptides representing the unacetylated and acetylated N termini of β- and γ-actin. (E) NAA80 expression is required for actin Nt-acetylation. Immunoblot analysis of HAP1 control and NAA80 KO1 cells transfected with empty V5 plasmid, wild-type NAA80-V5, or the catalytically inactive mutant NAA80mut-V5. The samples were probed with Ac-β-actin, Ac-γ-actin, pan-actin, V5, and GAPDH antibodies. (F) Actin acetylation is eliminated in NAA80-KO cells and restored upon reexpression of active NAA80. HAP1 control cells were transfected with an empty eGFP vector, and NAA80 KO1 cells were additionally transfected with NAA80-eGFP or NAA80mut-eGFP and stained with Ac-β-actin or Ac-γ-actin antibodies and analyzed by IF. The general presence of F-actin in the analyzed cells was visualized by Rhodamine phalloidin staining (Bottom Row). Of note, the cells shown here are from separate samples not subjected to antibody labeling. (Scale bars, 10 µm.)
NAA80-eGFP expressed in human HAP1 cells displayed a diffuse cytosolic distribution (Fig. 1B) and, contrary to most NATs, did not associate with ribosomes (Fig. S1D) (8, 18), suggesting that it targets its substrate(s) posttranslationally. To identify NAA80-specific substrates in cells, we generated two HAP1 NAA80-KO cell lines (KO1 and KO2) using the CRISPR/Cas9 system (Fig. S2A). Phosphoimaging of NAA80 KO1 cell lysates treated with purified NAA80 and isotope-labeled [14C]-acetyl CoA revealed a single band at ∼43 kDa, corresponding to the expected molecular mass of actin (Fig. 1C). Actin’s identity was confirmed using N-terminal proteomics (9, 19) by quantitatively assessing the Nt-acetylation status of 402 unique N termini from control and NAA80 KO1 cell lysates. Only peptides corresponding to the fully processed N termini of β- and γ-actin showed altered Nt-acetylation levels in KO cells (Fig. S2B and Table S1). No other proteins, including Arps (ARPC2, CAZA2, TPM1, and TPM4), displayed any difference between control and KO1 cells (Table S1). This can be explained by the fact that the N termini of these proteins do not match NAA80’s substrate specificity. The lack of matching substrate sequence also applies to other Arps, including all isoforms of the Arp2/3 complex, capping protein, VASP, myosin, tropomyosin, and formin. Furthermore, both actin isoforms were 100% and 0% Nt-acetylated in control and NAA80 KO1 cells, respectively. Isoform- and Nt-acetylation–specific actin antibodies recognized an ∼43-kDa band in control but not NAA80 KO1 cells (Fig. 1 D and E). The Ac-β-actin and Ac-γ-actin antibody signals reappeared in KO cell lysates transfected with NAA80 (Fig. 1E) but not in cells transfected with a catalytically inactive mutant, NAA80mut (W105F, R170Q, G173D, Y205F) (Fig. 1E and Fig. S3), confirming that catalytically active NAA80 is absolutely required for actin Nt-acetylation in cells. Of note, the Ac-β-actin is a weaker antibody in our hands than Ac-γ-actin. Both signals are almost equally strong in rescue experiments (Fig. 1 D and E), suggesting a more effective rescue for β-actin and thus confirming the substrate specificity of NAA80 we observed in our in vitro experiments (Fig. 1A and Fig. S1B). These results were corroborated for both KO cell lines at the single-cell level by immunofluorescence (IF) analysis (Fig. 1F and Fig. S3), showing that only control cells and NAA80-eGFP–transfected NAA80-KO cells were positive for Ac-β-actin and Ac-γ-actin antibody staining.
Actin Nt-Acetylation Controls Cell Motility.
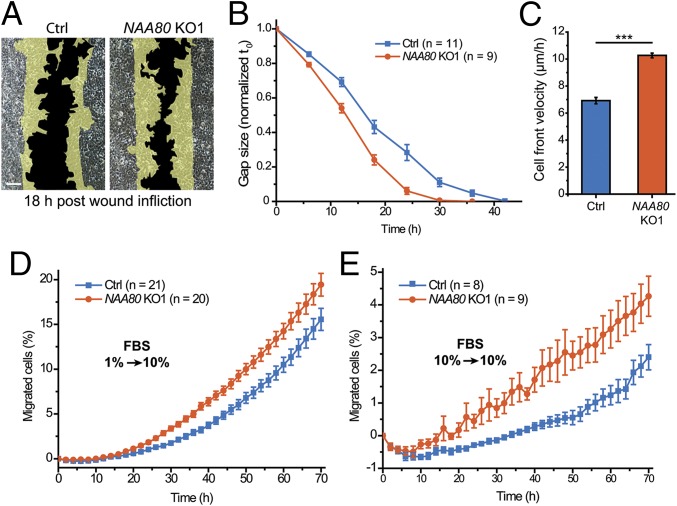
Next, we explored the impact of actin Nt-acetylation in cells. Since the actin cytoskeleton is a major factor in cell motility (20), we tested HAP1 control and NAA80-KO cells in several independent migration assays. We found that the lack of actin Nt-acetylation resulted in increased motility of HAP1 NAA80-KO cells. In wound-healing assays, NAA80 KO1 cells closed the gap ∼12 h faster than control cells (Fig. 2 A and B), with cell-front velocity increasing from 6.4 ± 0.7 to 10 ± 0.6 μm/h (Fig. 2C). We observed similar results for both KO cell lines in an IncuCyte ZOOM system for live-cell imaging, showing an increase of cell-front velocity from 7 to 9 μm/h and 11 μm/h for NAA80 KO1 and KO2 cells, respectively (Fig. S4 and Movie S1). To investigate the migratory properties of single cells, we performed a chemotaxis assay by seeding cells on a transparent porous membrane in medium containing 1% FBS. Cells were attracted to the bottom side by a medium containing 10% FBS, and the number of migrating cells was normalized to the growth rates of each cell line (Fig. S4). NAA80 KO1 cells showed an ∼50% increase in chemotactic motility compared with control cells (Fig. 2D). In addition to the chemotactic assay for directed migration, we also measured random cell migration in parallel wells in which both chambers contained 10% FBS. In this assay, we also observed increased random motility of NAA80 KO1 cells (Fig. 2E), consistent with an increased capacity of these cells to migrate. Similar results were obtained with NAA80 KO2 cells (Fig. S4 D and E). Taken together these data demonstrate that cells deficient in actin Nt-acetylation have increased motility.
Fig. 2.
Actin Nt-acetylation affects cell motility. (A) Representative images of HAP1 control (Ctrl) and NAA80 KO1 cells in the wound-healing assay showing the degree of gap closure after 18 h. (Scale bar, 150 μm.) (B) Quantification of gap size in wound-healing assays relative to the size at t0. (C) Average cell-front velocity calculated from B. (D) Chemotaxis assay showing the percentage of cells that migrated from a 1% FBS chamber to a 10% FBS chamber (see also Fig. S4). (E) Random migration assay showing the percentage of cells migrating from a 10% FBS chamber to a 10% FBS chamber. ***P ≤ 0.001, two-sided Student’s t test. Data are shown as the means ± SEM.
Actin Nt-Acetylation Affects Cytoskeletal Morphology.
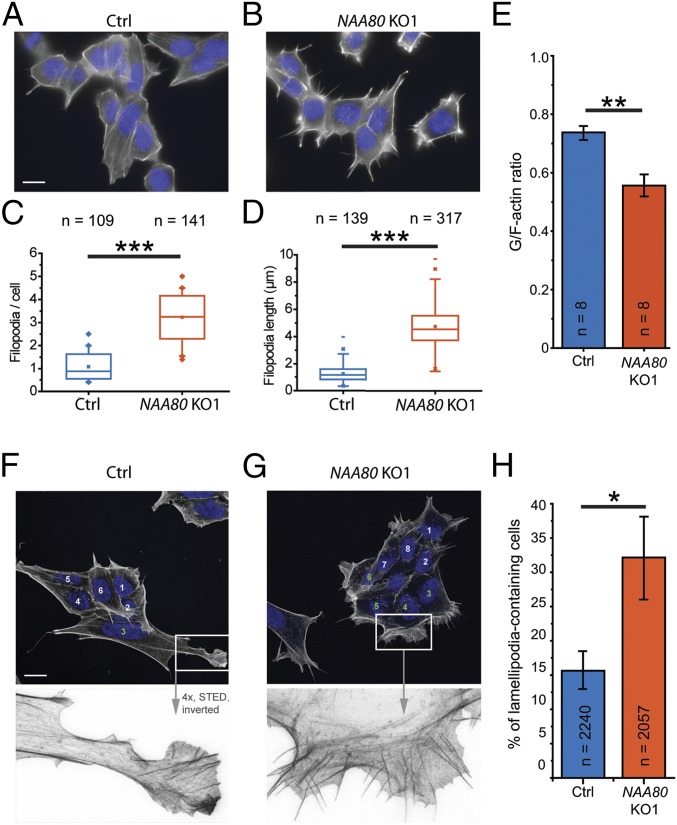
The actin cytoskeleton regulates not only cell motility but also cell shape and morphology (2). Thus, we compared the morphology of the actin cytoskeleton in HAP1 control and NAA80-KO cells by phalloidin staining. Based on the hypermotility phenotype of NAA80-KO cells, we were specifically interested in the formation of cell protrusions, such as filopodia and lamellipodia, that are usually linked to cell motility. As anticipated, NAA80-KO cells displayed an increase in both the number and length of filopodia-like structures (Fig. 3 A–D). These cells showed an approximately fourfold increase in the number of filopodia-like structures, defined as phalloidin-stained protrusions with a length ≥0.5 µm (Fig. 3C). The average filopodia length increased from 1 µm in control cells to 5 µm in NAA80-KO cells (Fig. 3D). Confirming the specificity of this phenotype, the reexpression of NAA80 in NAA80-KO cells reduced the number of filopodia-like structures nearly to control levels, whereas the expression of the catalytically inactive mutant NAA80mut did not (Fig. S5A). As in the other experiments described above, the NAA80 KO2 clone showed similar behavior (Fig. S5B). Consistent with the apparent elevated number of filamentous actin structures in NAA80-KO cells, we observed a significant reduction in the ratio of globular-actin to filamentous-actin (G/F) (21), from 0.73 in HAP1 control cells to 0.55 in NAA80 KO1 cells (Fig. 3E). This effect was reversed by the expression of wild-type NAA80 but not NAA80mut in both NAA80-KO cell lines (Fig. S5C), demonstrating that NAA80-dependent actin Nt-acetylation directly impacts the transition between G- and F-actin.
Fig. 3.
Effects of actin Nt-acetylation on cytoskeletal morphology. (A and B) Representative micrographs illustrating a differential morphological phenotype at the level of filopodia between HAP1 control (Ctrl) (A) and NAA80 KO1 (B) cells where the absence of NAA80 promotes filopodia formation. (C and D) For both cell lines, the number (C) and length (D) of filopodia were determined. (E) G/F-actin ratios from HAP1 control (blue) and NAA80 KO1 (orange) cells. (F and G) Confocal images (Upper) and stimulated emission depletion microscopy (STED) zoom-in frames (Lower) of lamellipodia. The number of cells containing at least one clearly distinguishable lamellipodium supported by filopodia/microspikes was determined. Since HAP1 cells typically grow in clusters, only cells at the borders of clusters (i.e., with a front facing away from the cluster) were considered, as indicated by the numbering. Lamellipodia-positive cells are indicated by green numbers. (H) Quantification of lamellipodia-positive cells. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001, Student’s t test. Data are shown as the means ± SEM.
Since we observed a hypermotility phenotype for NAA80-KO cells (Fig. 3), we quantified the number of cells showing at least one distinguishable lamellipodium supported by microspikes or filopodia-like structures. To efficiently study these cell protrusions in HAP1 cells, which typically grow in clusters, we included only cells at the borders of the clusters in quantifications. These were scored as either positive or negative for the presence of lamellipodia. In agreement with the increased cell-front velocity in the motility assays, we observed that twice as many NAA80-KO cells as control cells formed lamellipodia (32 vs. 16%) (Fig. 3 F–H). These results further emphasize the importance of actin Nt-acetylation in the control of cytoskeletal dynamics.
Nt-Acetylation–Dependent Actin Polymerization and Stability.
To address the mechanism behind the altered cytoskeleton organization phenotypes of NAA80-KO cells, we analyzed the recovery rates of cytoskeletal structures in cells treated with the actin-depolymerizing drug latrunculin A (LatA). Within 60 min of LatA treatment the actin appeared to be fully depolymerized in control and NAA80-KO cells, and washout of the drug resulted in the recovery of actin filament structures, but the time of recovery was significantly delayed for NAA80-KO cells compared with control cells (Fig. S6), consistent with a direct role of actin Nt-acetylation in actin polymerization.
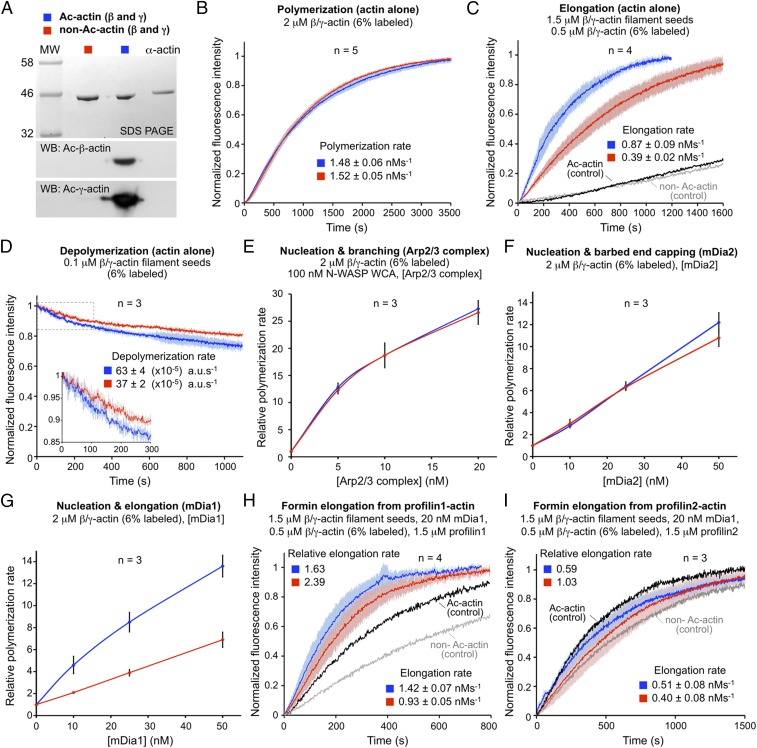
We next explored the in vitro effect of actin Nt-acetylation on the polymerization/depolymerization properties of actin alone or in the presence of some of the most common actin-assembly factors in cells. Cytoplasmic actin (a mixture of β and γ isoforms) was purified from control and NAA80-KO cells, and the presence or absence of Nt-acetylation was verified by Western blotting (Fig. 4A) (22). The time course of actin polymerization measured using the pyrene-actin polymerization assay revealed no significant difference in the polymerization rate of acetylated β/γ-actin (Ac-actin) vs. nonacetylated β/γ-actin (non–Ac-actin) (Fig. 4B). However, the elongation of 1.5-μM β/γ-actin filament seeds (±Nt-acetylation) in the presence of 0.5-μM β/γ-actin monomers (i.e., above the critical concentration for monomer addition at the barbed end but below that of the pointed end) was more than twofold faster for Ac-actin than for non–Ac-actin (Fig. 4C). In contrast, the depolymerization rate of 0.1-μM filament seeds (i.e., below the critical concentration of both the pointed and barbed ends) was approximately twofold faster for Ac-actin than for non–Ac-actin, suggesting that, at least in the absence of other factors, actin filament structures formed by non–Ac-actin might be more stable (Fig. 4D). Because both elongation and depolymerization are approximately twofold faster for Ac-actin than for non–Ac-actin, whereas total polymerization is unchanged, nucleation seems to be mostly unaffected by Nt-acetylation. In cells, nucleation is greatly enhanced by the Arp2/3 complex (3, 5). However, we also found no difference in the relative polymerization rate of Ac-actin and non–Ac-actin induced by Arp2/3 complex (Fig. 4E and Fig. S7 A and B). We next analyzed two formins, mDia1 and mDia2, that, while closely related in sequence, have very different actin-assembly properties. We observed no significant difference in the relative polymerization rates of Ac-actin and non–Ac-actin induced by mDia2 (Fig. 4D and Fig. S7 A and C), a formin that has strong nucleation but poor elongation activity (23, 24). In contrast, for mDia1, which has both strong nucleation and strong elongation activities (23, 24), the polymerization rate was approximately twofold higher for Ac-actin than for non–Ac-actin (Fig. 4G and Fig. S7 A and D). Because Nt-acetylation–dependent differences appear to be emphasized during formin-mediated elongation, we analyzed the role of profilin (isoforms 1 and 2), known to virtually stop pointed-end elongation of actin alone while dramatically accelerating barbed-end elongation by formins (24). As expected, mDia1 accelerated the elongation rate of β/γ-actin filament seeds from profilin1-actin compared with the elongation of seeds from actin alone, but this effect was greater for Ac-actin than for non–Ac-actin (Fig. 4H). Curiously, no significant difference was observed with profilin2-actin (Fig. 4I), suggesting that mDia1 processes profilin1-actin more efficiently than profilin2-actin. We note, however, that these assays detect the incorporation into filaments of the small fraction of pyrene-labeled actin, which binds profilin with weaker affinity than unlabeled actin (24), so the effects of profilin may be far more pronounced than indicated by the bulk assays. Together, these results suggest that the differences in filament assembly between Ac-actin and non–Ac-actin emanate mainly from an Nt-acetylation–dependent increase in the rates of filament depolymerization and elongation, whereas nucleation appears to be mostly unaffected (spontaneous or Arp2/3 complex-driven).
Fig. 4.
Actin Nt-acetylation affects filament elongation and depolymerization. (A) Cytoplasmic actin (a mixture of β and γ isoforms) purified from wild-type and NAA80-KO cells was analyzed by SDS/PAGE and Western blotting using isoform- and Nt-acetylation–specific Ac-β-actin and Ac-γ-actin antibodies. Nt-acetylated α-skeletal actin (15) is shown as reference. (B) The polymerization rate of β/γ-actin alone is unchanged with or without Nt-acetylation. Data are shown as the average curve from five independent experiments (as indicated), with SD error bars in lighter color (Ac-actin, blue; non–Ac-actin, red). (C) The barbed-end elongation rate of β/γ-actin filament seeds is ∼2.2-fold faster for Ac-actin (blue) than for non–Ac-actin (red). The polymerization of Ac-actin (black) and non–Ac-actin (gray) in the absence of seeds is shown as control. (D) The depolymerization rate of β/γ-actin filament seeds is ∼1.7-fold faster for Ac-actin (blue) than for non–Ac-actin (red). (E) The concentration dependence of polymerization rates of actin assembly by Arp2/3 complex (nucleation and branching) shows no difference for Ac-actin (blue) vs. non–Ac-actin (red) (see also Fig. S7B). (F) The concentration dependence of polymerization rates of actin assembly induced by mDia2 (nucleation and barbed-end capping) is unchanged with or without Nt-acetylation (see also Fig. S7C). (G) The concentration dependence of polymerization rates of actin assembly induced by mDia1 (nucleation and barbed-end elongation) is >twofold faster for Ac-actin (blue) than for non–Ac-actin (red) (see also Fig. S7D). (H) The elongation rate of filament seeds by mDia1 from profilin1-actin is ∼35% faster for Ac-actin (blue) than for non–Ac-actin (red). For both actins, the elongation rate of mDia1 from profilin1-actin is faster than the elongation rate of β/γ-actin seeds alone (shown as controls). (I) In contrast, barbed-end elongation by mDia1 from profilin2-actin is unchanged for non–Ac-actin (red) and is reduced ∼40% for Ac-actin (blue) relative to the elongation rate of β/γ-actin seeds. Rates are reported as the mean ± SEM of the number of independent experiments indicated in each panel.
Discussion
The identity of the actin NAT has been a longstanding question since the unique processing of Dictyostelium and mammalian actins was discovered in 1981 and 1983, respectively (15, 25). Based on their N-terminal sequences after processing (26), the other actin isoforms, e.g., skeletal and cardiac muscle α-actin and smooth muscle α- and γ-actin, are also likely to be Nt-acetylated by NAA80 (Fig. S1C). Furthermore, the effects of actin Nt-acetylation likely vary among cell types. For instance, in muscle cells, where actin accounts for ∼20% of the total cellular protein, Nt-acetylation could predictably have rather dramatic effects in muscle contraction and actomyosin force generation (27, 28).
We found that Nt-acetylation is responsible for pronounced and opposite effects on actin filament depolymerization and elongation dynamics, including formin-induced elongation. Thus, while Ac-actin elongates faster, it also depolymerizes faster, suggesting that cytoskeletal structures consisting of non–Ac-actin may last longer. In contrast, actin filament nucleation was mostly unaffected by Nt-acetylation. We note that filament nucleation is a one-time reaction, whereas both elongation and depolymerization are repetitive reactions that thus could be more sensitive to the cumulative effects of Nt-acetylation. The in vitro effects observed here with several actin assembly factors (4, 5, 24) likely converge to produce the global effects on cell migration and monomeric-to-filamentous actin ratios described above (2).
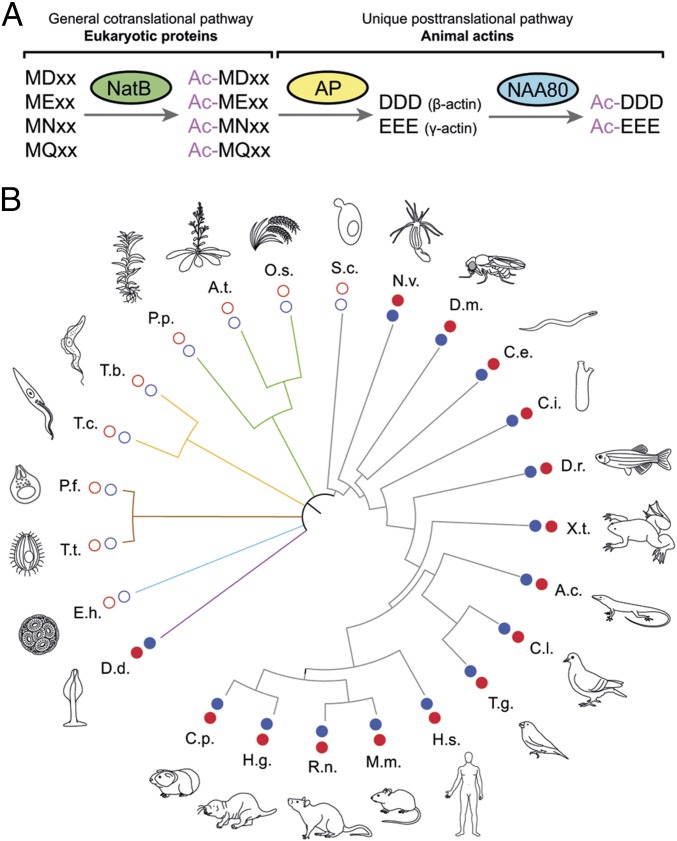
Due to actin’s broad spectrum of cellular activities, its specific N-terminal processing, and its evolution from bacterial precursors such as FtsA and MreB (29–31), we asked whether a correlation existed between actin’s unique N-terminal sequence and the expression of NAA80. Generally, eukaryotic proteins with N-terminal sequences beginning with Met-Asp or Met-Glu are Nt-acetylated cotranslationally by NatB (Fig. 5A) (32). Actin, however, is additionally processed posttranslationally by a unique N-terminal maturation pathway involving a still unidentified aminopeptidase, which, as we show here, is followed by NAA80-mediated Nt-acetylation (Fig. 5A). Interestingly, sequence analysis reveals that the expression of NAA80 is highly correlated with the conservation of this unique actin N-terminal processing mechanism. For instance, an NAA80 ortholog exists in some amoebozoa, including Dictyostelium discoideum (Fig. 5B and Table S2), which share the actin N-terminal processing mechanism present in animals (25, 33). In contrast, NAA80 is absent in fungi and plants, which have either a different actin N terminus or use a different N-terminal processing mechanism (32, 34). Thus, NAA80 appears to have coevolved with the actin sequence in species requiring an acetylated and highly acidic actin N terminus for proper function (29, 35). This offers the possibility to control actin Nt-acetylation in animals by targeting a single gene, NAA80.
Fig. 5.
NAA80 expression is evolutionarily linked to a unique processing mechanism of actin’s N terminus. (A) Processing pathway of animal actins, including an initial step of cotranslational acetylation catalyzed by NatB, which is shared with other eukaryotic proteins beginning with Met-Asp/Met-Glu. (B) Taxonomic tree showing the coevolution of NAA80 and an acidic actin N terminus. For each species, filled red circles indicate the presence of an acidic N terminus in mature actin, and filled blue circles indicate the expression of Naa80. The full names of the organisms are given in Table S2.
The ability to control actin Nt-acetylation enables us to study in more detail the consequences of this modification at the molecular, cellular, and organismal levels in the future. Here we have studied how actin Nt-acetylation affects cell protrusions and migration, but cell migration depends not only on cell protrusions but also on adhesion and contraction. It is possible that adhesion and contraction are directly or indirectly influenced by the presence of actin’s Nt-acetyl group and thus Nt-acetylation may explain the overall impact on cell migration. Further studies will also shed light on the molecular mechanisms for the observed differences described here in actin polymerization by actin alone or driven by formin. Actin Nt-acetylation could directly decrease the affinity of actin toward formin, as was shown for other protein–protein interactions dependent on Nt-acetylation (13). The enhanced actin filament stability and the slower depolymerization rate of purified non–Ac-actin suggest that the actin filament structure itself could be affected. The lack of the Nt-acetyl group and a subsequently altered filament structure could potentially be sterically unfavorable for actin polymerization and depolymerization. Previous studies on yeast actin have shown that the stability of actin filament structures is affected by the negative charge density at the N terminus. Removing the two negatively charged acidic residues in yeast actin (MDSE) caused an increase in the number of actin strands forming filament bundles (36). Comparison of nonmuscle β-actin and γ-actin showed that β-actin has increased polymerization but reduced depolymerization rates. These two isoforms basically differ only in their N termini, with aspartate having a lower pK than glutamate and β-actin thus having a higher negative-charge density (37), supporting the importance of the N terminus for actin functionality. The fact that the removal of the Nt-acetyl group in NAA80-KO cells in this study decreases the negative charge density may explain the phenotypes observed in vitro as well as in vivo.
In this study, the functional and cellular roles of the most conserved actin modification in animals have been revealed. We have also gained a deeper understanding of the cellular roles of Nt-acetylation. In contrast to the established NAT enzymes often acting on the ribosomes and targeting large groups of substrates (8), NAA80 is a specific posttranslational eukaryotic NAT. An accompanying paper defines structural and cellular determinants essential for this specificity towards actin (38). Such a mode of action resembles the enzymes of other major protein modifications such as protein phosphorylation and lysine acetylation, and future studies will reveal whether actin Nt-acetylation also represents a key entry point for cellular regulation.
Materials and Methods
Detailed procedures for cell culturing, microscopy analysis, recombinant protein purification, actin purification, pyrene-based polymerization and depolymerization assays, Nt-acetylation assays, and Western blot analysis are described in SI Materials and Methods and Table S3.
Supplementary Material
Acknowledgments
This work was supported by Research Council of Norway Grants 230865 and 249843 (to T.A.), the Norwegian Cancer Society (T.A.), the Bergen Research Foundation (T.A.), the Norwegian Health Authorities of Western Norway Project 912176, and the NIH Grant R01 GM073791 (to R.D.). F.I. and K.G. acknowledge support from the Flanders Institute for Biotechnology and Odysseus Grant G0F8616N (to F.I.) and Grant 3.G044010.w from the Fund for Scientific Research Flanders (to K.G.). Confocal imaging and IncuCyte scanning were performed at the Molecular Imaging Center and were supported by the Department of Biomedicine and the Faculty of Medicine and Dentistry at the University of Bergen and its partners.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The mass spectrometry proteomics data in Table S1 have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository (dataset ID PXD006856; user name: reviewer89589@ebi.ac.uk; password: LMHVncnM).
See Commentary on page 4314.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718336115/-/DCSupplemental.
References
- 1.Sheterline P, Clayton J, Sparrow J. Actin. Protein Profile. 1995;2:1–103. [PubMed] [Google Scholar]
- 2.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pollard TD. Actin and actin-binding proteins. Cold Spring Harb Perspect Biol. 2016;8:a018226. doi: 10.1101/cshperspect.a018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: Nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci USA. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dominguez R. The WH2 domain and actin nucleation: Necessary but insufficient. Trends Biochem Sci. 2016;41:478–490. doi: 10.1016/j.tibs.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swaney KF, Li R. Function and regulation of the Arp2/3 complex during cell migration in diverse environments. Curr Opin Cell Biol. 2016;42:63–72. doi: 10.1016/j.ceb.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breitsprecher D, Goode BL. Formins at a glance. J Cell Sci. 2013;126:1–7. doi: 10.1242/jcs.107250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aksnes H, Drazic A, Marie M, Arnesen T. First things first: Vital protein marks by N-terminal acetyltransferases. Trends Biochem Sci. 2016;41:746–760. doi: 10.1016/j.tibs.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 9.Arnesen T, et al. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc Natl Acad Sci USA. 2009;106:8157–8162. doi: 10.1073/pnas.0901931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bienvenut WV, et al. Comparative large scale characterization of plant versus mammal proteins reveals similar and idiosyncratic N-α-acetylation features. Mol Cell Proteomics. 2012;11:M111.015131. doi: 10.1074/mcp.M111.015131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang CS, Shemorry A, Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327:973–977. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park SE, et al. Control of mammalian G protein signaling by N-terminal acetylation and the N-end rule pathway. Science. 2015;347:1249–1252. doi: 10.1126/science.aaa3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scott DC, Monda JK, Bennett EJ, Harper JW, Schulman BA. N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science. 2011;334:674–678. doi: 10.1126/science.1209307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holmes WM, Mannakee BK, Gutenkunst RN, Serio TR. Loss of amino-terminal acetylation suppresses a prion phenotype by modulating global protein folding. Nat Commun. 2014;5:4383. doi: 10.1038/ncomms5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rubenstein PA, Martin DJ. NH2-terminal processing of actin in mouse L-cells in vivo. J Biol Chem. 1983;258:3961–3966. [PubMed] [Google Scholar]
- 16.Zegerman P, Bannister AJ, Kouzarides T. The putative tumour suppressor Fus-2 is an N-acetyltransferase. Oncogene. 2000;19:161–163. doi: 10.1038/sj.onc.1203234. [DOI] [PubMed] [Google Scholar]
- 17.Rubenstein PA, Martin DJ. NH2-terminal processing of Drosophila melanogaster actin. Sequential removal of two amino acids. J Biol Chem. 1983;258:11354–11360. [PubMed] [Google Scholar]
- 18.Gautschi M, et al. The yeast N(alpha)-acetyltransferase NatA is quantitatively anchored to the ribosome and interacts with nascent polypeptides. Mol Cell Biol. 2003;23:7403–7414. doi: 10.1128/MCB.23.20.7403-7414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gevaert K, et al. Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat Biotechnol. 2003;21:566–569. doi: 10.1038/nbt810. [DOI] [PubMed] [Google Scholar]
- 20.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 21.Bunnell TM, Burbach BJ, Shimizu Y, Ervasti JM. β-Actin specifically controls cell growth, migration, and the G-actin pool. Mol Biol Cell. 2011;22:4047–4058. doi: 10.1091/mbc.E11-06-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohki T, Ohno C, Oyama K, Mikhailenko SV, Ishiwata S. Purification of cytoplasmic actin by affinity chromatography using the C-terminal half of gelsolin. Biochem Biophys Res Commun. 2009;383:146–150. doi: 10.1016/j.bbrc.2009.03.144. [DOI] [PubMed] [Google Scholar]
- 23.Harris ES, Rouiller I, Hanein D, Higgs HN. Mechanistic differences in actin bundling activity of two mammalian formins, FRL1 and mDia2. J Biol Chem. 2006;281:14383–14392. doi: 10.1074/jbc.M510923200. [DOI] [PubMed] [Google Scholar]
- 24.Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 25.Redman K, Rubenstein PA. NH2-terminal processing of Dictyostelium discoideum actin in vitro. J Biol Chem. 1981;256:13226–13229. [PubMed] [Google Scholar]
- 26.Vandekerckhove J, Weber K. Mammalian cytoplasmic actins are the products of at least two genes and differ in primary structure in at least 25 identified positions from skeletal muscle actins. Proc Natl Acad Sci USA. 1978;75:1106–1110. doi: 10.1073/pnas.75.3.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehman W, Szent-Györgyi AG. Regulation of muscular contraction. Distribution of actin control and myosin control in the animal kingdom. J Gen Physiol. 1975;66:1–30. doi: 10.1085/jgp.66.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rayment I, et al. Structure of the actin-myosin complex and its implications for muscle contraction. Science. 1993;261:58–65. doi: 10.1126/science.8316858. [DOI] [PubMed] [Google Scholar]
- 29.Wickstead B, Gull K. The evolution of the cytoskeleton. J Cell Biol. 2011;194:513–525. doi: 10.1083/jcb.201102065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gunning PW, Ghoshdastider U, Whitaker S, Popp D, Robinson RC. The evolution of compositionally and functionally distinct actin filaments. J Cell Sci. 2015;128:2009–2019. doi: 10.1242/jcs.165563. [DOI] [PubMed] [Google Scholar]
- 31.Pollard TD, Goldman RD. 2017. The Cytoskeleton (Cold Spring Harbor Lab Press, Cold Spring Harbor, NY), pp vii, 391 p.
- 32.Van Damme P, et al. N-terminal acetylome analyses and functional insights of the N-terminal acetyltransferase NatB. Proc Natl Acad Sci USA. 2012;109:12449–12454. doi: 10.1073/pnas.1210303109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vandekerckhove J, Weber K. Vegetative Dictyostelium cells containing 17 actin genes express a single major actin. Nature. 1980;284:475–477. doi: 10.1038/284475a0. [DOI] [PubMed] [Google Scholar]
- 34.Cook RK, Sheff DR, Rubenstein PA. Unusual metabolism of the yeast actin amino terminus. J Biol Chem. 1991;266:16825–16833. [PubMed] [Google Scholar]
- 35.Hightower RC, Meagher RB. The molecular evolution of actin. Genetics. 1986;114:315–332. doi: 10.1093/genetics/114.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cook RK, Blake WT, Rubenstein PA. Removal of the amino-terminal acidic residues of yeast actin. Studies in vitro and in vivo. J Biol Chem. 1992;267:9430–9436. [PubMed] [Google Scholar]
- 37.Bergeron SE, Zhu M, Thiem SM, Friderici KH, Rubenstein PA. Ion-dependent polymerization differences between mammalian beta- and gamma-nonmuscle actin isoforms. J Biol Chem. 2010;285:16087–16095. doi: 10.1074/jbc.M110.110130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goris M, et al. Structural determinants and cellular environment define processed actin as the sole substrate of the N-terminal acetyltransferase NAA80. Proc Natl Acad Sci USA. 2018;115:4405–4410. doi: 10.1073/pnas.1719251115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.