Significance
RNAs, both mRNAs and noncoding RNAs, usually exert their functions in the form of RNA–protein complexes. Although mRNA-binding proteins have been extensively studied, comprehensive identification of coding and noncoding RNA-binding proteins (RBPs) remains challenging. Herein, we developed a click chemistry-assisted RNA interactome capture (CARIC) strategy, which combines metabolic labeling of RNAs with an alkynyl uridine analog and in vivo RNA-protein photocross-linking, followed by click reaction with azide-biotin, affinity enrichment, and proteomic analysis. In HeLa cells, CARIC identified 597 RBPs, including 130 proteins not previously known as RBPs. Since CARIC captures RBPs bound to both mRNAs and noncoding RNAs, the obtained CARIC RBP list provides a valuable resource for studying the posttranscriptional gene regulation network.
Keywords: RNA, RNA–protein interactions, proteomics, bioorthogonal chemistry, noncoding RNA
Abstract
Transcriptome-wide identification of RNA-binding proteins (RBPs) is a prerequisite for understanding the posttranscriptional gene regulation networks. However, proteomic profiling of RBPs has been mostly limited to polyadenylated mRNA-binding proteins, leaving RBPs on nonpoly(A) RNAs, including most noncoding RNAs (ncRNAs) and pre-mRNAs, largely undiscovered. Here we present a click chemistry-assisted RNA interactome capture (CARIC) strategy, which enables unbiased identification of RBPs, independent of the polyadenylation state of RNAs. CARIC combines metabolic labeling of RNAs with an alkynyl uridine analog and in vivo RNA-protein photocross-linking, followed by click reaction with azide-biotin, affinity enrichment, and proteomic analysis. Applying CARIC, we identified 597 RBPs in HeLa cells, including 130 previously unknown RBPs. These newly discovered RBPs can likely bind ncRNAs, thus uncovering potential involvement of ncRNAs in processes previously unknown to be ncRNA-related, such as proteasome function and intermediary metabolism. The CARIC strategy should be broadly applicable across various organisms to complete the census of RBPs.
About 75% of the human genome is transcribed to various kinds of RNAs (1). The protein-coding mRNAs serve as the template for protein synthesis, and hence mediate the flow of genetic information. In addition to mRNAs, which account for only ∼2% of the genome, many classes of noncoding RNAs (ncRNAs) are made in cells. The tRNAs and rRNAs, the two most-studied classes of ncRNAs, participate in protein synthesis by serving as amino acid adaptors and ribosome components, respectively. The past two decades have witnessed the emergence of many previously unannotated ncRNAs, such as microRNAs (miRNAs), Piwi-interacting RNAs, and long ncRNAs (lncRNAs) (2). These ncRNAs carry out a variety of biological functions, including transcription regulation, RNA processing, and genome remodeling. Most of the RNAs, both coding and noncoding, function as ribonucleoprotein particles (RNPs), that is, RNAs in complex with RNA-binding proteins (RBPs) (3). Dysfunction of RBPs has been linked to various human diseases, such as neurodegeneration, muscular disorders, and cancers (4–7).
Large-scale identification of RBPs is a prerequisite for understanding the underlying biological and pathological processes and has recently attracted lots of attentions (3, 8, 9). RBPs can be in vivo photocross-linked to RNAs by 254-nm UV light or to RNAs metabolically labeled with a photoactivatable uridine analog 4-thiouridine (4SU) by 365-nm UV light (4, 10). By combining UV cross-linking with polyadenylated [poly(A)] tail-dependent oligo(dT) enrichment, an mRNA interactome capture approach was developed for proteomic identification of mRNA-binding proteins (mRBPs) in HeLa and HEK293 cells (11, 12). Since UV light is directly applied to living cells, this method allows for covalent cross-linking of native RNP complexes formed by RNAs and their direct binders. Furthermore, the oligo(dT) capture enables detection of low-abundance cross-linked proteins, overcoming the limited efficiency of UV cross-linking. This method has has been applied to profile poly(A) RNA interactome in various mammalian cells (13–18); Saccharomyces cerevisiae (14, 19, 20); Caenorhabditis elegans (20); zebrafish (21); the early embryo of Drosophila melanogaster (22, 23); Arabidopsis thaliana seedlings, leaves, and cultured cells (24–26); and human parasites (27–29). However, the poly(A) tails mostly exist on mature mRNAs, leaving RBPs on nonpoly(A) RNAs, including most ncRNAs and pre-mRNAs, largely undiscovered.
To complete the census of RBPs, methods for transcriptome-wide identification of RBPs, which are independent of the poly(A) tail, are needed. Many RBPs possess well-known RNA-binding domains (RBDs), such as the RNA recognition motif (RRM) and heterogeneous nuclear RNP K-homology (KH) domain (30). Sequence and structure homology have long been used to computationally predict RBPs (31, 32) but cannot uncover the entire RNA interactome because a growing number of RBPs are found to harbor no annotated RBDs (6, 18). Alternatively, in vitro binding of mRNAs to protein microarrays and RBP capture using immobilized mRNAs were employed to identify mRBPs in S. cerevisiae (33, 34). These two methods in principle are not limited to mRBPs but suffer from nonphysiological RNA–protein interactions. Recently, two powerful approaches were reported for large-scale identification of RBPs with no need for oligo(dT) pull-down (35, 36). By demonstrating that the more RBPs a protein interacts with the more likely that protein itself is an RBP, a classification algorithm termed SONAR (support vector machine obtained from neighborhood associated RBPs) was developed to predict RBPs using the existing large-scale protein–protein interaction (PPI) datasets (35). Compared to the previous computational approaches (32), SONAR does not rely on sequence or structure homology, thus allowing for discovery of RBPs with RNA-binding activity through unknown mechanisms. A limitation of SONAR might be that the known PPI networks have not covered all RBPs. The other approach exploits 4SU-dependent UV cross-linking and quantitative MS to identify RBPs with the binding peptide information in the nuclei of mouse embryonic stem cells (36). Termed RBR-ID (proteomic identification of RNA-binding regions), this approach relies on detecting the decreased MS signals of peptides due to their cross-linking to RNAs. Without the need of the oligo(dT) enrichment step, RBR-ID requires many fewer cells and can identify nonpoly(A) RBPs. However, with no enrichment, RBR-ID suffers from high background signals, particularly for RBPs with low RNA binding ratios, and limited detection sensitivity and specificity.
Herein, we report the development of a complementary strategy for transcriptome-wide discovery of both poly(A) and nonpoly(A) RBPs. Termed click chemistry-assisted RNA interactome capture (CARIC), our strategy combines in vivo RNA-protein photocross-linking with metabolic labeling of various RNAs with the alkyne, a bioorthogonal or clickable functional group. Subsequent bioorthogonal reaction (i.e., click chemistry) with a biotin tag enables affinity enrichment and proteomic profiling of RBPs, independent of the polyadenylation state of RNAs. In HeLa cells, CARIC identified 597 RBPs, including 130 proteins that had no prior RNA-binding annotation. The binding targets of these newly discovered RBPs possibly included ncRNAs. Moreover, the newly discovered RBPs included proteasome components, metabolic enzymes, and Mendelian disease-related proteins, thus implicating ncRNAs in the underlying biological processes. The CARIC RBP list provides a rich and valuable resource for analyzing the RNA–RBP interaction networks.
Results
Development of the CARIC Strategy.
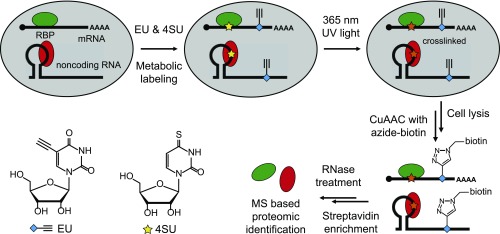
To enrich all RNPs that are photocross-linked, a capture technique independent of the poly(A) tail was required. We exploited an alkyne-containing uridine analog, 5-ethynyluridine (EU), which can be metabolically incorporated into various kinds of RNAs in living cells (37). The alkyne can be chemoselectively reacted with the azide via copper(I)-catalyzed azide-alkyne cycloaddition (CuAAC, also termed click chemistry) (38, 39). CARIC combines EU labeling with photoactivatable-ribonucleoside-enhanced cross-linking (4). The 4SU was metabolized into cellular RNAs together with EU; 365-nm UV light irradiation selectively cross-linked 4SU with bound RBPs. Subsequent click labeling of EU with azide-biotin via CuAAC enabled streptavidin enrichment and MS (Fig. 1 and Fig. S1).
Fig. 1.
Schematic of the workflow of CARIC. EU and 4SU were simultaneously taken up by cells and metabolically incorporated into RNAs. The 365-nm UV light irradiation activated 4SU and covalently cross-linked RNAs with direct binders. The cells were lysed and reacted with azide-biotin to tag EU. After enrichment with streptavidin beads, the eluted RNPs were digested with RNase A and the released RBPs were analyzed by quantitative proteomics using LC-MS/MS.
A series of experiments were performed to optimize the CARIC procedures. UV-visible (UV-Vis) absorption spectroscopy confirmed that 365-nm UV light only activated 4SU but not EU, uridine, cytidine, adenosine, or guanosine (Fig. S2A). Total RNAs were extracted from HeLa cells treated with EU and 4SU together (EU&4SU) and reacted with HPDP-biotin (i.e., N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide, a sulfhydryl-reactive compound) and azide-Cy5 to label 4SU and EU, respectively. The labeled RNAs were captured with streptavidin beads. Flow cytometry analysis of the beads indicated that EU and 4SU were simultaneously incorporated into the same RNA molecules (Fig. S2 B and C). We then treated HeLa cells with EU&4SU at varied concentrations, followed by UV irradiation. In-gel fluorescence scanning of the cell lysates reacted with azide-Cy5 showed the cross-linked RNPs as smeared bands at high molecular weights (>130 kDa) and 1 mM EU and 0.5 mM 4SU, resulting in the highest amount of doubly labeled and cross-linked RNPs (Fig. S2D). Metabolic incorporation of EU&4SU did not cause significant cytotoxicity (Fig. S2E). The photocross-linking was dependent on the energy density of UV light, and 2 J/cm2 was sufficient to produce maximal cross-linking (Fig. S3A). UV light did not cause apparent RNA degradation even at the energy density of 2 J/cm2 (Fig. S3B). Three reported Cu(I) ligands (40–42), BTTAA (2-[4-{(bis[(1-tert-butyl-1H-1,2,3-triazol-4-yl)methyl]amino)methyl}-1H-1,2,3-triazol-1-yl]acetic acid), THPTA (Tris[(1-hydroxypropyl-1H-1,2,3-triazol-4-yl)methyl]amine), and TBTA (Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine), were evaluated for improving the reaction yield of CuAAC on EU&4SU-incorporated cell lysates, and THPTA exhibited a significant improvement on click-labeling efficiency (Fig. S3C). With THPTA, we could lower the concentration of CuSO4 to 0.5 mM, while maintaining enough labeling efficiency (Fig. S3D). Although UV cross-linking did not damage RNAs, Cu(I) could cause fragmentation of RNAs (Fig. S3B), in agreement with previous studies (43). Remarkably, THPTA significantly alleviated this effect (Fig. S3B). Based on these results, experimental conditions including metabolic labeling of HeLa cells with 1 mM EU and 0.5 mM 4SU, UV light irradiation at 2 J/cm2, and THPTA-assisted CuAAC were chosen for CARIC experiments in this work.
Validation of CARIC Capture.
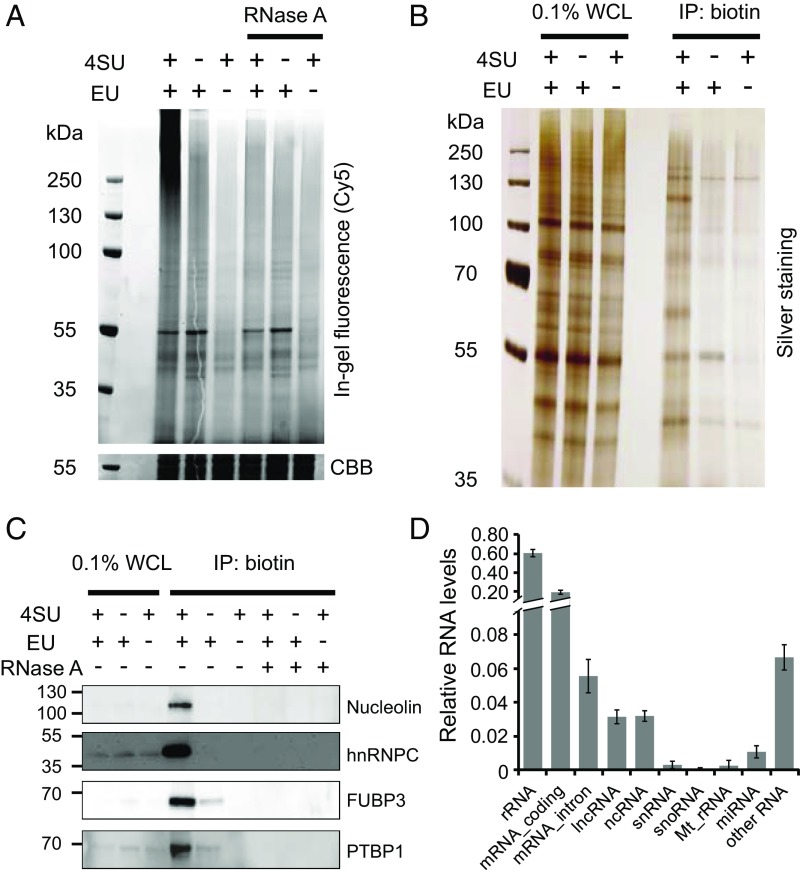
Using the optimized CARIC protocol, HeLa cells were efficiently photocross-linked and the lysates were click-labeled with azide-Cy5. In-gel fluorescence scanning showed that only the doubly labeled RNPs were UV cross-linked (Fig. 2A and Fig. S4A). The bands of cross-linked RNPs (>130 kDa) were completely abolished by RNase A treatment (Fig. 2A) or transcription inhibition with actinomycin D (AD) (Fig. S4 B and C). We next reacted the cross-linked lysates with azide-biotin, followed by enrichment with streptavidin beads. After eluting the beads with a biotin elution buffer, the eluted samples were then digested with RNase A to release RBPs, which were resolved by SDS/PAGE and analyzed by silver staining (Fig. 2B and Fig. S5). Only the doubly labeled samples exhibited a diverse repertoire of RBPs, while omission of 4SU, EU, or UV yielded minimal background signal and several nonspecific bands (Fig. S5A). Specific capture of RBPs was confirmed by RNase A treatment before streptavidin enrichment (Fig. S5B). To further validate the CARIC strategy, the presence of four known RBPs—nucleolin, heterogeneous nuclear ribonucleoproteins C1/C2 (hnRNPC), far upstream element binding protein 3 (FUBP3), and polypyrimidine tract-binding protein 1 (PTBP1)—in the captured samples was detected by immunoblot analysis (Fig. 2C). All of them showed selective enrichment in the EU&4SU-labeled samples and RNase treatment confirmed the RNA dependence of CARIC enrichment.
Fig. 2.
Development and optimization of CARIC. (A) In-gel fluorescence analysis of RNPs. HeLa cells were treated with 1 mM EU and 0.5 mM 4SU, EU, or 4SU alone and irradiated with 365-nm UV light. The lysates were reacted with azide-Cy5 via THPTA-assisted CuAAC. After treatment with or without RNase A, the samples were resolved by SDS/PAGE and visualized by in-gel fluorescence scanning. Coomassie brilliant blue (CBB)-stained gel was used as the loading control. (B) After EU&4SU labeling and UV light irradiation, the cell lysates were reacted with azide-biotin and enriched with streptavidin beads. After elution, the captured RNPs were digested with RNase A to release RBPs, which were resolved by SDS/PAGE and visualized by silver staining. WCL, whole-cell lysate. (C) Western blot analysis of the presence of nucleolin, hnRNPC, FUBP3, and PTBP1 in the CARIC-captured samples. RNase A treatment after click labeling of the lysates with azide-biotin demonstrated selective capture of RNPs doubly labeled with EU and 4SU. (D) Analysis of RNAs isolated by CARIC by next-generation sequencing. The relative RNA level was quantified by normalizing the total mapped reads of each RNA types to the total mapped reads of total RNAs. Error bars represent the SD from three independent biological replicates.
We next analyzed the RNA species contained in the click-labeled and streptavidin bead enriched samples. The elutes were digested with proteinase K and the resulting RNAs were analyzed by next-generation sequencing (Fig. 2D). Besides mRNA, various types of ncRNAs, including lncRNA, small nuclear RNA (snRNA), and miRNA, were mapped. It should be noted that tRNAs, because of their abundant chemical modifications and stable structure, could not be sequenced in our experiments using the standard sequencing method (44). These results demonstrate that CARIC is able to isolate various RNAs, independent of their polyadenylation status, and capture RBPs bound on those RNAs.
Proteomic Identification of CARIC RBPs.
We then analyzed the CARIC-enriched RBPs by quantitative proteomics based on the stable isotope dimethyl labeling strategy (45). To increase the confidence of identification, we performed two sets of experiments, in which the 4SU-omitted and UV-omitted samples were used as negative controls, respectively (Fig. S6). In each set, data from four biological replicates were collected. In two of the replicates, the peptides from the experimental and control samples were labeled with isotopically light and heavy dimethyl labels, respectively. In another two replicates, the “reverse” dimethyl labeling, in which the labeling order was switched (i.e., the experimental samples with heavy label and the control samples with light label), was performed to increase the accuracy and robustness of quantification. The peptides from a total of eight biological replicates were prefractionated by high-pH C18 stop-and-go-extraction tips (StageTips) (46) and analyzed by liquid chromatography tandem MS (LC-MS/MS).
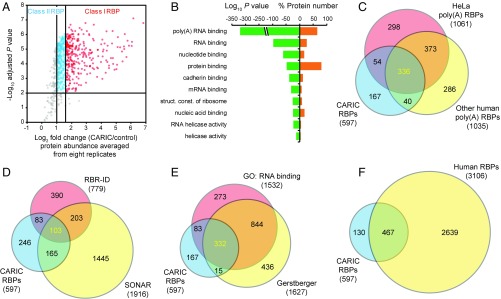
A streamlined MS data analysis procedure was established for RBP identification (Fig. S7). After peptide identification using MaxQuant (47), 1,271 proteins with two or more unique identified peptides were considered as identified proteins (Fig. S8A and Dataset S1). These identified proteins were further filtered by two or more quantified peptides, which resulted in 1,210 proteins with their enrichment ratio quantified as the median of the enrichment ratios of all peptides assigned to that protein. The protein enrichment ratios between biological replicates showed strong correlation, including comparisons between datasets using 4SU-omitted and UV-omitted controls (Fig. S8B). Of the 1,210 quantified proteins, 746 were quantified at least once in both control sets and at least three times from eight replicates. Of these, 691 proteins were enriched in the experimental samples with the log2 enrichment ratio statistically different from 0 (adjusted P < 0.01, empirical Bayes moderated t test), which corresponded to a false discovery rate of 1% (Fig. 3A and Dataset S1). Of the 691 proteins, 597 with a fold change of two or greater were termed CARIC RBPs and subdivided into two classes: class I containing 296 proteins with a fold change greater than three and class II containing 301 proteins with a fold change of three or less (Fig. 3A and Dataset S2).
Fig. 3.
Proteomic analysis of CARIC RBPs. (A) Volcano plot displaying the averaged log2 enrichment ratios from eight replicates (x axis) and their adjusted P values (y axis) for the 746 proteins, which were quantified at least once in both control sets and at least three times from eight replicates. Proteins with adjusted P value <0.01 and a fold change of two or greater were considered as “CARIC RBP.” CARIC RBPs with a fold change greater than three were classified as class I RBPs and the rest were classified as class II RBPs. (B) Molecular function GO terms enriched for the CARIC RBPs. (C) Overlap of the CARIC RBPs with the previously identified human poly(A) RBPs. (D) Overlap of the CARIC RBPs with the RBPs identified by SONAR and RBR-ID. (E) Overlap of the CARIC RBPs with the GO-annotated RBP list and the human RBP list manually curated by Gerstberger et al. (3). (F) Overlap of the CARIC RBPs with the human RBP list. Overlap of CARIC RBPs, subdivided into classes I and II, with known human RBPs is shown in Fig. S10.
Comparison of CARIC RBPs to Known Human RBPs.
Gene Ontology (GO) analysis using DAVID (48, 49) indicated that CARIC RBPs were mostly enriched with the molecular function term “poly(A) RNA binding,” reminiscent of recent extensive efforts on large-scale identification and annotation of poly(A) RBPs (Fig. 3B). Moreover, the “poly(A) RNA binding” term was similarly overrepresented in both class I and II CARIC RBPs (Fig. S9A). Interestingly, several biological process terms were distinctly enriched between the two classes (Fig. S9B). For example, the term “tRNA aminoacylation for protein translation” was highly enriched in class II but not in class I. A total of 12 aminoacyl-tRNA synthetases were identified by CARIC, with 11 belonging to class II (Dataset S3).
To further analyze the CARIC RBPs identified in HeLa cells, we compiled a list of 1,387 human poly(A) RBPs that have so far been identified in HeLa cells (11, 18) as well as three other human cell lines, HEK293 (12), Huh-7 (14), and K562 cells (16), by using the poly(A) tail-dependent capture method (Dataset S4). Of the 597 CARIC RBPs (260 and 170 belonging to classes I and II, respectively) 430 (72%) overlapped with the human poly(A) RBPs, most of which were identified in both HeLa cells and at least one other human cell line (Fig. 3C, Fig. S10A, and Dataset S2). Therefore, CARIC identified 167 RBPs (36 and 131 belonging to classes I and II, respectively) that were not previously identified in human cells by poly(A)-dependent RNA interactome capture (Fig. 3C, Fig. S10A, and Dataset S2).
The CARIC RBPs were then compared with the RBP lists recently generated by the poly(A)-independent methods SONAR and RBR-ID (35, 36). Of the 597 CARIC RBPs (175 and 93 belonging to classes I and II, respectively) 268 (45%) overlapped with the SONAR RBPs (Fig. 3D, Fig. S10B, and Dataset S2). To compare with the RBR-ID RBPs, we converted them to the human orthologs. Of the CARIC RBPs (110 and 76 belonging to classes I and II, respectively) 186 (31%) overlapped with the converted RBR-ID RBPs (Fig. 3D, Fig. S10B, and Dataset S2). The lower overlapping percentage with the RBR-ID RBPs might be because the RBR-ID list was obtained in mouse cells and included only nuclear proteins. We also compared the CARIC RBPs with the GO-annotated RBP list and the human RBP list manually curated by Gerstberger et al. (3). Of the CARIC RBPs (259 and 156 belonging to classes I and II, respectively) 415 (70%) and of the CARIC RBPs (239 and 108 belonging to classes I and II, respectively) 347 (58%) were included in these two databases, respectively (Fig. 3E, Fig. S10C, and Dataset S2).
Moreover, we compiled a human RBP list which combined the human poly(A) RBPs, GO-annotated human RBPs, SONAR RBPs, and the human RBP list by Gerstberger et al. (3) (Dataset S5). By comparing to the human RBP list, CARIC identified 130 (25 and 105 belonging to classes I and II, respectively) candidate RBPs in HeLa cells, which were not previously annotated as RNA binding (Fig. 3F and Fig. S10D). These results demonstrate that CARIC not only confirms a significant portion of known RBPs but also expands the current list of RBPs, thus providing another complementary approach for completing the census of RBPs.
Experimental Validation of RNA-Binding Activity of Several CARIC RBPs.
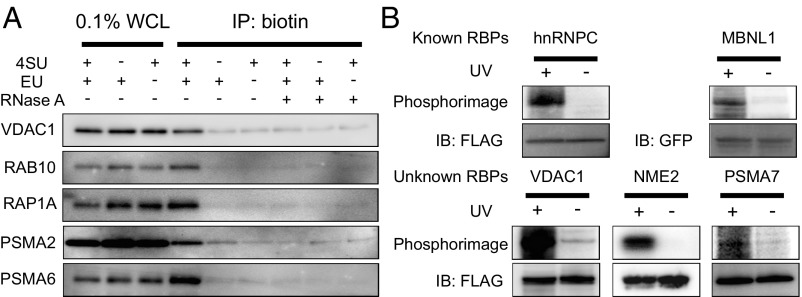
To experimentally validate the RNA-binding activity of the newly identified RBPs in the CARIC RBP list, we examined the RNA dependence of CARIC capture of five selected candidates from the list (one and four from classes I and II, respectively): voltage-dependent anion-selective channel protein 1 (VDAC1), Ras-related protein Rab-10 (RAB10), Ras-related protein Rap-1A (RAP1A), proteasome subunit alpha type-2 (PSMA2), and proteasome subunit alpha thype-6 (PSMA6). Of note, PSMA6 was experimentally found to bind RNAs (50) and therefore is within the list of GO-annotated RBPs. Western blot analysis showed that all five proteins were enriched in the CARIC-isolated RNPs and the enrichment was dependent on RNA labeling with EU&4SU (Fig. 4A). More importantly, RNase treatment depleted the enrichment, confirming that these RBPs were directly bound and cross-linked to RNAs. For an independent validation, we employed conventional cross-linking and immunoprecipitation (CLIP), followed by radiolabeling with T4 polynucleotide kinase. HeLa or HEK293T cells expressing FLAG-tagged or EGFP-tagged RBPs were irradiated with 254-nm UV light. RNPs were immunoprecipitated using anti-FLAG or anti-GFP antibody conjugated magnetic beads, treated with RNase T1 to shorten the length of cross-linked RNAs, and radiolabeled. Phosphorimaging showed that three identified previously unknown RBPs, VDAC1, NME2 and PSMA7, were efficiently cross-linked to RNAs, in a way similar to two known RBPs, hnRNPC and MBNL1, which served as positive controls (Fig. 4B).
Fig. 4.
Validation of RNA-binding activity of several CARIC RBPs. (A) Western blot depicting enrichment of five representative CARIC RBPs. RNase A treatment after click labeling demonstrated the RNA dependence of CARIC capture. (B) Validation of the RNA-binding activity of representative FLAG-tagged CARIC RBPs (VDAC1, NME2, and PSMA7) by CLIP, followed by radiolabeling with T4 polynucleotide kinase and phosphorimaging. Two known RBPs, hnRNPC and MBNL1, were used as positive controls. Anti-FLAG and anti-GFP blots demonstrate equal loading.
pI Values, Binding Domains, and RNA-Binding Specificity of CARIC RBPs.
A characteristic feature of the poly(A) RBPs identified in HeLa cells is a shift of the distribution of pI values toward a more basic pH, compared with all human proteins (11). We therefore analyzed the pI distribution of CARIC RBPs, which were also from HeLa cells (Fig. 5A and Dataset S2). In contrast to the HeLa poly(A) RBPs, the CARIC RBPs were not preferentially represented by basic proteins. Indeed, the combined poly(A) RBPs from various human cells showed a pI distribution similar to the CARIC RBPs (Fig. 5A). The same trend was observed for the human RBP list. These results indicate that the complete RBP interactome, as better represented by the accumulating RBPs, may have a pI distribution similar to that of the whole human proteome.
Fig. 5.
pI values, binding domains, and RNA-binding specificity of CARIC RBPs. (A) Density of pI values of CARIC RBPs, HeLa poly(A) RBPs reported by Castello et al. (11), human poly(A) RBPs, human RBPs, and all human genes. (B) Number of CARIC-annotated RBPs and unknown RBPs containing classical RBDs, nonclassical, or unknown RBDs. (C) Number of CARIC-annotated RBPs and unknown RBPs containing each known RBD. (D) Number of CARIC RBPs with unknown, putative, and validated RNA targets. (E) The RNA targets of CARIC RBPs which were not previously identified as poly(A) RBPs. The RNA binding activity was either experimentally validated or postulated with related evidence in the literature.
A well-studied mode of RNA binding is via the modular RBDs. A limited list of RBDs including 11 classical and 15 nonclassical ones has been experimentally validated (11, 51). About half of the known RBPs in our CARIC RBP list harbor these RBDs (Fig. 5 B and C). In sharp contrast, most (>95%) of the unknown RBPs possess no known RBDs, indicating the existence of distinctive modes of RNA binding. Of note, the recently developed methods for large-scale identification of RBDs and regions should be of use for discovering new RNA binding motifs and binding modes (18, 36).
To shed light on what classes of RNAs the CARIC RBPs bind, we analyzed the ones with reported RNA-binding activities and ones with RNA-related functions. The 430 CARIC RBPs overlapping with the list of human poly(A) RBPs were attributed to bind mRNAs. Of note, although poly(A) tails mostly exist on mRNAs, some ncRNAs, such as rRNAs and lncRNAs, can also be polyadenylated (52, 53). Among the other 167 CARIC RBPs not in the human poly(A) RBP list were six proteins that have been experimentally confirmed as RBPs (Fig. 5D and Dataset S6). For example, exportin-t (XPOT) was found to bind tRNAs with high affinity and mediate tRNA nuclear export (54). WD repeat-containing protein 5 (WDR5) was recently found to bind an lncRNA HOTTIP and regulate long-range gene activation (55). Furthermore, we annotated an additional 58 CARIC RBPs with their putative RNA-binding targets based on their residence within well-characterized RNPs, RNA-related functions, and orthologs in other organisms being confirmed as RBPs (Fig. 5D and Dataset S6). A variety of classes of ncRNAs are targets of CARIC RBPs (Fig. 5E). Of note, in addition to those binding to poly(A) mRNAs, some of the CARIC RBPs also bind pre-mRNAs with no poly(A) tails. These examples support that CARIC can be used for transcriptome-wide identification of coding and noncoding RBPs.
RNA-Binding Activity of Proteasome Proteins and Metabolic Enzymes.
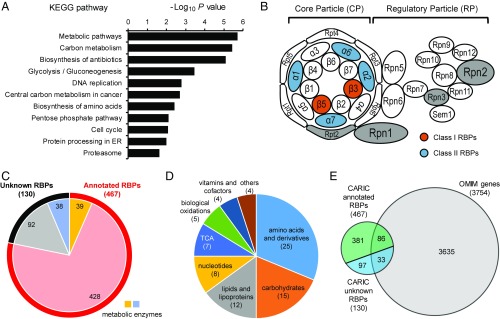
KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis on the previously unknown RBPs identified by CARIC revealed that the term “proteasome” is one of the most enriched pathways (Fig. 6A). A significant portion of the proteasome components were identified by CARIC (Fig. 6B). Among the 14 proteins constituting the 20S proteasome core particle, six were CARIC RBPs (two and four belonging to classes I and II, respectively). In addition, four proteins within the 19S regulatory particle were potential CARIC RBPs (i.e., proteins identified with a fold change between one and two). Importantly, three proteasome proteins, PSMA2, PSMA6, and PSMA7, were experimentally validated to bind RNAs in this work (Fig. 4 A and B).
Fig. 6.
Functional analysis of CARIC RBPs. (A) KEGG pathways enriched in the CARIC unknown RBPs. (B) Various proteasome components were identified as RBPs. In the schematic of the human proteasome, proteins color-coded in orange are CARIC class I RBPs, proteins in blue are CARIC class II RBPs, and proteins in gray are potenital CARIC RBPs (i.e., proteins identified with a fold change between one and two). (C) Number of CARIC-annotated RBPs and unknown RBPs that are metabolic enzymes. (D) Number of CARIC unknown RBPs in specific metabolic pathways. (E) Number of CARIC-annotated RBPs and unknown RBPs listed in OMIM.
Several metabolism-related pathways were also enriched in the KEGG pathway analysis on the unknown RBPs (Fig. 6A). Based on the Reactome pathway database (56), unknown RBPs in the CARIC RBP list harbor 38 metabolic enzymes (Fig. 6C and Dataset S7). These metabolic enzymes are distributed to a variety of metabolism pathways, including nucleotide, amino acid, carbohydrate, and lipid metabolism (Fig. 6D and Dataset S7).
CARIC RBPs in Genetic Diseases.
Many RBPs have been implicated in human Mendelian diseases. Based on the online Mendelian Inheritance in Man (OMIM) database (57), 201 human poly(A) RBPs are disease-related (Fig. S11A). CARIC profiling in HeLa cells identified 76 of those OMIM-listed poly(A) RBPs (Fig. S11A). In total, CARIC identified 119 OMIM-listed proteins (Fig. 6E). More interestingly, 33 of the 130 CARIC-identified previously unknown RBPs are listed in OMIM and associated with various diseases, such as metabolic, neurological, and muscular disorders (Fig. S11B and Dataset S8).
Discussion
Understanding the posttranscriptional gene regulation network requires comprehension of RBPs that dictate the fate of RNAs. Here, we develop CARIC as a high-throughput method for transcriptome-wide identification of RBPs. Among the 597 CARIC RBPs identified in HeLa cells, 78% are previously identified or annotated RBPs, demonstrating the reliability of the CARIC methodology. However, despite the fact that large-scale identification of RBPs has recently been extensively performed using several different methods, CARIC is able to identify 130 unknown RBPs in HeLa cells. Most of these newly identified RBPs do not have known RBDs and probably bind RNAs through alternative mechanisms yet to be investigated. Moreover, these unknown RBPs include the proteasome components, metabolic enzymes, and human Mendelian disease-related proteins, implicating RNAs in the underlying processes. Therefore, CARIC provides a powerful tool for RNA interactome profiling and is complementary to the previously developed strategies.
CARIC shares several nice features with the oligo(dT)-based RNA interactome capture strategy (11, 12) but overcomes its major limitation, being incapable of capturing and identifying RBPs bound on the nonpoly(A) RNAs. In vivo UV cross-linking allows covalent linking of RNAs to their direct binders under physiological conditions and ensures subsequent selective and stringent isolation. The oligo(dT) affinity purification has high affinity and specificity for poly(A) RNPs. In comparison, CARIC exploits metabolic labeling of RNAs with EU and click chemistry to install the biotin tag for isolation. The major advantage of this click chemistry-assisted strategy is the broad coverage of various RNPs, whether or not the RNAs are polyadenylated. In addition to EU, several other clickable nucleosides, such as alkynyl and azido analogs of adenosine (58–61), have been developed to metabolically label RNAs in living cells and may be implemented into CARIC.
Although 254-nm UV light can be used to cross-link natural nucleotides with RBPs, CARIC employs double metabolic labeling with EU and 4SU (which can be activated by 365-nm UV light) to avoid potential contaminants caused by cross-linking free EU and its metabolites (e.g., uridine phosphates) with recognizing proteins. Furthermore, double labeling minimizes photocross-linking on EU so that the click reaction is not blocked. Although 4SU and EU have been widely used for labeling RNAs, we cannot completely rule out the possibility that incorporation of uridine analogs might affect RNA–protein interactions, thus resulting in some false negative and false positive identification.
A potential contaminant in CARIC is the growing polypeptide chains with attached tRNAs that are EU-incorporated. This might contribute to the background signals observed in the 4SU-omitted negative controls (Fig. 2 A and B and Dataset S1). In addition, we observed some nonspecific UV cross-linked bands in the 4SU-omitted samples (Fig. 2A and Fig. S4A). Since these bands could not be removed by RNase A treatment, we suspected that they resulted from nonspecific photoactivation of EU, which exhibited absorbance toward longer wavelength than natural nucleosides (Fig. S2A) during 365-nm UV irradiation. Nevertheless, these background signals were subtracted during MS data analysis.
The RBR-ID method expanded the identification of RBPs to those on nonpoly(A) RNAs by discriminating MS signals between cross-linked and non-cross-linked peptides (36). Although detection of signal loss or decrease is often less optimal, it saves the purification step, thus simplifying the experimental procedures. The trade-off, however, is compromised sensitivity and specificity. Whether RBR-ID can be used to identify RBPs in whole cells remains to be explored, given that more complex proteome samples with many high-abundance proteins, such as actins and tubulins, may cause more false positives. Alternatively, SONAR took a computational approach to address the limitation of oligo(dT) affinity capture (35). One of the strengths of SONAR is obviation of the need of MS-based proteomic identification, which, though it is becoming a routine technique, is still technically demanding. The principle of SONAR relies on accurate and comprehensive information on the PPI networks, which imposes limitations on using SONAR for species whose PPI networks have not been well characterized. Nevertheless, it will be interesting to use the combination of these complementary approaches for obtaining the comprehensive RNA interactome and for comparing different subproteomes. For example, depletion of poly(A) RNPs by oligo(dT) pull-down followed by CARIC may be used to selectively identify nonpoly(A) RBPs.
To experimentally validate the identified RBPs, we used two independent assays on a list of selected RBPs. The first assay confirmed the MS identification by Western blotting. Furthermore, the RNase A treatment before streptavidin enrichment served as a stringent negative control, which confirmed that CARIC capture of RBPs was dependent on RNAs. CLIP was used as an independent assay. In both assays, the signals of unknown RBPs were much weaker compared with the positive controls, several well-known RBPs. One possible explanation is that these proteins might mainly function in other biological processes and moonlight as RBPs under specific regulatory conditions.
The binding specificity of RBPs is dictated by RNA sequences or/and structures. Many mRBPs possess RBDs, such as RRM and KH domain, which recognize specific sequences of single-stranded RNAs (30). It is not uncommon that some RBPs can bind both mRNAs and ncRNAs. For example, some of the snRNA-binding proteins in the spliceosome machinery are in direct contact with mRNAs during the splicing process (62, 63). Accordingly, poly(A) tail-based RNA interactome capture in human cells identified six of the seven Sm proteins, which are common protein components for spliceosome snRNP U1, U2, U4, and U5 (11, 12, 16). It should be noted that poly(A) RBPs identified using this method are not strictly mRBPs, since the oligo(dT) pull-down precipitated a small amount of ncRNAs (11).
An interesting question is whether there are ncRBPs that exclusively bind ncRNAs. The current CARIC protocol does not distinguish poly(A) RBPs from nonpoly(A) RBPs or mRBPs from ncRBPs. One possible solution to this question is to combine CARIC with the oligo(dT) pull-down protocol. Alternatively, the RNA targets of the unknown RBPs identified in this work can be further studied using CLIP followed by next-generation sequencing (4, 64).
The proteasomes were renamed from “prosome” based on the discovery of their proteolytic activity (65). Interestingly, there was early evidence, though not conclusive, indicating that prosomes might associate with RNA species, including mRNAs, tRNAs, and 5S rRNAs (66–69). A recent poly(A) RBP profiling in S. cerevisiae and C. elegans also identified 16 components of the 26S proteasome (20). These results call for reevaluation of the RNA-binding activity of the proteasome complex or its individual protein components and the functional consequences.
The metabolic enzymes with poly(A) RNA-binding activity have also been highlighted in the oligo(dT)-based RNA interactome capture (12), which led to the proposed regulatory interconnections between RNA, enzymes, and metabolites (REM) (70). According to the Reactome pathway database, 4.9% of the human poly(A) RBPs (68 out of 1,387) were annotated as metabolic enzymes. In contrast, metabolic enzymes take up as many as 29.2% of the CARIC unknown RBPs (Fig. 6C). Considering that the newly identified RBPs tend to be nonpoly(A) RNA-specific, we suspect that ncRNAs might also participate in the REM interactions.
In summary, we have demonstrated that CARIC is a poly(A) tail-independent method, which allows for transcriptome-wide identification of RBPs. The HeLa RBP dataset generated in this work, together with previously datasets using other methods, provide invaluable resources for bioinformatics and experimental analysis of RNA–RBP interactions at a system level. Furthermore, the CARIC technique can be readily used in various cell types and organisms to facilitate uncovering the complete RNA–protein interaction network.
Materials and Methods
Details are in SI Materials and Methods, which includes detailed methods for metabolic incorporation of EU and 4SU, in vivo photocross-linking, click chemistry, in-gel fluorescence, Western blot analysis, cell viability assays, RBP isolation by CARIC, RNA sequencing, MS sample preparation, proteomic identification, MS data analysis, validation of CARIC RBPs by CLIP, and functional analysis of CARIC RBPs using online databases.
Supplementary Material
Acknowledgments
We thank Dr. J. Rong and Dr. L. Dong for their early attempt on EU-based RBP capture, X. Zhang and Prof. Y. Huang for help on analyzing RNA sequencing data, Prof. C. Wang for helpful discussions, and Dr. W. Zhou at the mass spectrometry facility of the National Center for Protein Sciences at Peking University for assistance with proteomic analysis. This work is supported by National Natural Science Foundation of China Grants 91753206, 21425204, and 21521003 and National Key Research and Development Project 2016YFA0501500.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718406115/-/DCSupplemental.
References
- 1.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nat Rev Genet. 2014;15:829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castello A, Fischer B, Hentze MW, Preiss T. RNA-binding proteins in Mendelian disease. Trends Genet. 2013;29:318–327. doi: 10.1016/j.tig.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 7.Nussbacher JK, Batra R, Lagier-Tourenne C, Yeo GW. RNA-binding proteins in neurodegeneration: Seq and you shall receive. Trends Neurosci. 2015;38:226–236. doi: 10.1016/j.tins.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jazurek M, Ciesiolka A, Starega-Roslan J, Bilinska K, Krzyzosiak WJ. Identifying proteins that bind to specific RNAs - focus on simple repeat expansion diseases. Nucleic Acids Res. 2016;44:9050–9070. doi: 10.1093/nar/gkw803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hentze MW, Castello A, Schwarzl T, Preiss T. A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol. January 17, 2018 doi: 10.1038/nrm.2017.130. [DOI] [PubMed] [Google Scholar]
- 10.Greenberg JR. Ultraviolet light-induced crosslinking of mRNA to proteins. Nucleic Acids Res. 1979;6:715–732. doi: 10.1093/nar/6.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castello A, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 12.Baltz AG, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell. 2012;46:674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 13.Kwon SC, et al. The RNA-binding protein repertoire of embryonic stem cells. Nat Struct Mol Biol. 2013;20:1122–1130. doi: 10.1038/nsmb.2638. [DOI] [PubMed] [Google Scholar]
- 14.Beckmann BM, et al. The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nat Commun. 2015;6:10127–10135. doi: 10.1038/ncomms10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao Y, et al. The cardiomyocyte RNA-binding proteome: Links to intermediary metabolism and heart disease. Cell Rep. 2016;16:1456–1469. doi: 10.1016/j.celrep.2016.06.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conrad T, et al. Serial interactome capture of the human cell nucleus. Nat Commun. 2016;7:11212–11222. doi: 10.1038/ncomms11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liepelt A, et al. Identification of RNA-binding proteins in macrophages by interactome capture. Mol Cell Proteomics. 2016;15:2699–2714. doi: 10.1074/mcp.M115.056564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castello A, et al. Comprehensive identification of RNA-binding domains in human cells. Mol Cell. 2016;63:696–710. doi: 10.1016/j.molcel.2016.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell SF, Jain S, She M, Parker R. Global analysis of yeast mRNPs. Nat Struct Mol Biol. 2013;20:127–133. doi: 10.1038/nsmb.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matia-González AM, Laing EE, Gerber AP. Conserved mRNA-binding proteomes in eukaryotic organisms. Nat Struct Mol Biol. 2015;22:1027–1033. doi: 10.1038/nsmb.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Despic V, et al. Dynamic RNA-protein interactions underlie the zebrafish maternal-to-zygotic transition. Genome Res. 2017;27:1184–1194. doi: 10.1101/gr.215954.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wessels HH, et al. The mRNA-bound proteome of the early fly embryo. Genome Res. 2016;26:1000–1009. doi: 10.1101/gr.200386.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sysoev VO, et al. Global changes of the RNA-bound proteome during the maternal-to-zygotic transition in Drosophila. Nat Commun. 2016;7:12128. doi: 10.1038/ncomms12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reichel M, et al. In planta determination of the mRNA-binding proteome of Arabidopsis etiolated seedlings. Plant Cell. 2016;28:2435–2452. doi: 10.1105/tpc.16.00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marondedze C, Thomas L, Serrano NL, Lilley KS, Gehring C. The RNA-binding protein repertoire of Arabidopsis thaliana. Sci Rep. 2016;6:29766–29778. doi: 10.1038/srep29766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z, et al. UV crosslinked mRNA-binding proteins captured from leaf mesophyll protoplasts. Plant Methods. 2016;12:42–53. doi: 10.1186/s13007-016-0142-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bunnik EM, et al. The mRNA-bound proteome of the human malaria parasite Plasmodium falciparum. Genome Biol. 2016;17:147–164. doi: 10.1186/s13059-016-1014-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nandan D, et al. Comprehensive identification of mRNA-binding proteins of Leishmania donovani by interactome capture. PLoS One. 2017;12:e0170068. doi: 10.1371/journal.pone.0170068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lueong S, Merce C, Fischer B, Hoheisel JD, Erben ED. Gene expression regulatory networks in Trypanosoma brucei: Insights into the role of the mRNA-binding proteome. Mol Microbiol. 2016;100:457–471. doi: 10.1111/mmi.13328. [DOI] [PubMed] [Google Scholar]
- 30.Ankö ML, Neugebauer KM. RNA-protein interactions in vivo: Global gets specific. Trends Biochem Sci. 2012;37:255–262. doi: 10.1016/j.tibs.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 31.Anantharaman V, Koonin EV, Aravind L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 2002;30:1427–1464. doi: 10.1093/nar/30.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Si J, Cui J, Cheng J, Wu R. Computational prediction of RNA-binding proteins and binding sites. Int J Mol Sci. 2015;16:26303–26317. doi: 10.3390/ijms161125952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scherrer T, Mittal N, Janga SC, Gerber AP. A screen for RNA-binding proteins in yeast indicates dual functions for many enzymes. PLoS One. 2010;5:e15499. doi: 10.1371/journal.pone.0015499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsvetanova NG, Klass DM, Salzman J, Brown PO. Proteome-wide search reveals unexpected RNA-binding proteins in Saccharomyces cerevisiae. PLoS One. 2010;5:e12671. doi: 10.1371/journal.pone.0012671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brannan KW, et al. SONAR discovers RNA-binding proteins from analysis of large-scale protein-protein interactomes. Mol Cell. 2016;64:282–293. doi: 10.1016/j.molcel.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He C, et al. High-resolution mapping of RNA-binding regions in the nuclear proteome of embryonic stem cells. Mol Cell. 2016;64:416–430. doi: 10.1016/j.molcel.2016.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jao CY, Salic A. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc Natl Acad Sci USA. 2008;105:15779–15784. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: Copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed Engl. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 39.Tornøe CW, Christensen C, Meldal M. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(i)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J Org Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 40.Besanceney-Webler C, et al. Increasing the efficacy of bioorthogonal click reactions for bioconjugation: A comparative study. Angew Chem Int Ed Engl. 2011;50:8051–8056. doi: 10.1002/anie.201101817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hong V, Presolski SI, Ma C, Finn MG. Analysis and optimization of copper-catalyzed azide-alkyne cycloaddition for bioconjugation. Angew Chem Int Ed Engl. 2009;48:9879–9883. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan TR, Hilgraf R, Sharpless KB, Fokin VV. Polytriazoles as copper(I)-stabilizing ligands in catalysis. Org Lett. 2004;6:2853–2855. doi: 10.1021/ol0493094. [DOI] [PubMed] [Google Scholar]
- 43.Hermann T, Heumann H. Determination of nucleotide distances in RNA by means of copper phenanthroline-generated hydroxyl radical cleavage pattern. RNA. 1995;1:1009–1017. [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng G, et al. Efficient and quantitative high-throughput tRNA sequencing. Nat Methods. 2015;12:835–837. doi: 10.1038/nmeth.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJR. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc. 2009;4:484–494. doi: 10.1038/nprot.2009.21. [DOI] [PubMed] [Google Scholar]
- 46.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 47.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 48.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 49.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bey F, et al. The prosomal RNA-binding protein p27K is a member of the α-type human prosomal gene family. Mol Gen Genet. 1993;237:193–205. doi: 10.1007/BF00282801. [DOI] [PubMed] [Google Scholar]
- 51.Lunde BM, Moore C, Varani G. RNA-binding proteins: Modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Slomovic S, Laufer D, Geiger D, Schuster G. Polyadenylation of ribosomal RNA in human cells. Nucleic Acids Res. 2006;34:2966–2975. doi: 10.1093/nar/gkl357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 54.Kutay U, et al. Identification of a tRNA-specific nuclear export receptor. Mol Cell. 1998;1:359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- 55.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Joshi-Tope G, et al. Reactome: A knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grammel M, Hang H, Conrad NK. Chemical reporters for monitoring RNA synthesis and poly(A) tail dynamics. ChemBioChem. 2012;13:1112–1115. doi: 10.1002/cbic.201200091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Curanovic D, et al. Global profiling of stimulus-induced polyadenylation in cells using a poly(A) trap. Nat Chem Biol. 2013;9:671–673. doi: 10.1038/nchembio.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng Y, Beal PA. Synthesis and evaluation of an alkyne-modified ATP analog for enzymatic incorporation into RNA. Bioorg Med Chem Lett. 2016;26:1799–1802. doi: 10.1016/j.bmcl.2016.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nainar S, et al. Metabolic incorporation of azide functionality into cellular RNA. ChemBioChem. 2016;17:2149–2152. doi: 10.1002/cbic.201600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Agafonov DE, et al. Molecular architecture of the human U4/U6.U5 tri-snRNP. Science. 2016;351:1416–1420. doi: 10.1126/science.aad2085. [DOI] [PubMed] [Google Scholar]
- 63.Sperling R. 2017. The nuts and bolts of the endogenous spliceosome. Wiley Interdiscip Rev RNA, 8.
- 64.Van Nostrand EL, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP) Nat Methods. 2016;13:508–514. doi: 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arrigo AP, Tanaka K, Goldberg AL, Welch WJ. Identity of the 19S ‘prosome’ particle with the large multifunctional protease complex of mammalian cells (the proteasome) Nature. 1988;331:192–194. doi: 10.1038/331192a0. [DOI] [PubMed] [Google Scholar]
- 66.Castaño JG, Ornberg R, Koster JG, Tobian JA, Zasloff M. Eukaryotic pre-tRNA 5′ processing nuclease: Copurification with a complex cylindrical particle. Cell. 1986;46:377–385. doi: 10.1016/0092-8674(86)90658-6. [DOI] [PubMed] [Google Scholar]
- 67.Nothwang HG, Coux O, Keith G, Silva-Pereira I, Scherrer K. The major RNA in prosomes of HeLa cells and duck erythroblasts is tRNA(Lys,3) Nucleic Acids Res. 1992;20:1959–1965. doi: 10.1093/nar/20.8.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pamnani V, Haas B, Pühler G, Sänger HL, Baumeister W. Proteasome-associated RNAs are non-specific. Eur J Biochem. 1994;225:511–519. doi: 10.1111/j.1432-1033.1994.00511.x. [DOI] [PubMed] [Google Scholar]
- 69.Schmid HP, et al. The prosome: An ubiquitous morphologically distinct RNP particle associated with repressed mRNPs and containing specific ScRNA and a characteristic set of proteins. EMBO J. 1984;3:29–34. doi: 10.1002/j.1460-2075.1984.tb01757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Castello A, Hentze MW, Preiss T. Metabolic enzymes enjoying new partnerships as RNA-binding proteins. Trends Endocrinol Metab. 2015;26:746–757. doi: 10.1016/j.tem.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jiang H, Lei R, Ding SW, Zhu S. Skewer: A fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 2014;15:182–193. doi: 10.1186/1471-2105-15-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cox J, et al. Andromeda: A peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 73.Jain E, et al. Infrastructure for the life sciences: Design and implementation of the UniProt website. BMC Bioinformatics. 2009;10:136. doi: 10.1186/1471-2105-10-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gasteiger E, et al. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The Proteomics Protocols Handbook. Humana; Totowa, NJ: 2005. pp. 571–607. [Google Scholar]
- 75.Finn RD, et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.