Significance
Myocardium thickening at midgestation to late gestation is crucial for the formation of a functionally competent postnatal heart. However, posttranslational mechanisms regulating this process remain unexplored. Here, we uncover a critical role for the ubiquitin-like protein NEDD8 in heart development. By targeting the E1 enzyme that mediates the conjugation of NEDD8 to protein targets, we show that NEDD8 modification is essential for ventricular compaction and heart function. We further identify that the NEDD8 substrate Cullin 7 mediates the degradation of Hippo kinase Mst1, thereby enabling YAP signaling, cardiomyocyte proliferation, and proper heart development. These results reveal a posttranslational regulatory mechanism in ventricular wall maturation and may provide mechanistic insights into the etiology of left ventricular noncompaction cardiomyopathy.
Keywords: NEDD8, Hippo-YAP signaling, Cullin 7, ventricular compaction, cardiomyopathy
Abstract
During development, ventricular chamber maturation is a crucial step in the formation of a functionally competent postnatal heart. Defects in this process can lead to left ventricular noncompaction cardiomyopathy and heart failure. However, molecular mechanisms underlying ventricular chamber development remain incompletely understood. Neddylation is a posttranslational modification that attaches ubiquitin-like protein NEDD8 to protein targets via NEDD8-specific E1-E2-E3 enzymes. Here, we report that neddylation is temporally regulated in the heart and plays a key role in cardiac development. Cardiomyocyte-specific knockout of NAE1, a subunit of the E1 neddylation activating enzyme, significantly decreased neddylated proteins in the heart. Mice lacking NAE1 developed myocardial hypoplasia, ventricular noncompaction, and heart failure at late gestation, which led to perinatal lethality. NAE1 deletion resulted in dysregulation of cell cycle-regulatory genes and blockade of cardiomyocyte proliferation in vivo and in vitro, which was accompanied by the accumulation of the Hippo kinases Mst1 and LATS1/2 and the inactivation of the YAP pathway. Furthermore, reactivation of YAP signaling in NAE1-inactivated cardiomyocytes restored cell proliferation, and YAP-deficient hearts displayed a noncompaction phenotype, supporting an important role of Hippo-YAP signaling in NAE1-depleted hearts. Mechanistically, we found that neddylation regulates Mst1 and LATS2 degradation and that Cullin 7, a NEDD8 substrate, acts as the ubiquitin ligase of Mst1 to enable YAP signaling and cardiomyocyte proliferation. Together, these findings demonstrate a role for neddylation in heart development and, more specifically, in the maturation of ventricular chambers and also identify the NEDD8 substrate Cullin 7 as a regulator of Hippo-YAP signaling.
Cardiac development consists of a series of highly regulated spatiotemporal morphogenic events (1, 2). The maturation of the cardiac chamber during late gestation is a crucial step toward the generation of a functionally competent postnatal heart and involves the coordination of ventricular trabeculation and compaction (3, 4). In the initial stages of heart development, trabeculae (an endocardium-lined myocardial mesh) protrude and expand into the ventricular lumen to increase the myocardial surface area for oxygen and nutrient exchange. From late midgestation to the late fetal and neonatal stage, the compact myocardium grows quickly and fuses with the trabecular myocardium to form a thicker, and eventually mature, compact ventricular wall. Defects in cardiac chamber maturation lead to congenital heart diseases, including left ventricular noncompaction cardiomyopathy (LVNC), the third most commonly diagnosed cardiomyopathy (5, 6). LVNC is characterized by deep and extensive trabeculae in a thin ventricular wall, which can result in dilated cardiomyopathy, heart failure, thromboembolism, arrhythmia, and sudden cardiac death (7). Emerging studies from genetic analysis of LVNC patients and animal models of LVNC have suggested that maturation of the ventricular wall is regulated by a complex signaling network comprised of growth factors, transcription factors, and epigenetic regulators (3, 4), such as NRG1/ERBB2 (8), NOTCH (9, 10), TGFβ (11, 12), Smad7 (13), and HDAC1/2 (14). However, the importance of novel ubiquitin-like protein modifiers in this process has not been demonstrated.
Neural precursor cell expressed, developmentally down-regulated 8 (NEDD8) is a ubiquitin-like protein that shares a high degree of homology with ubiquitin (15). NEDD8 is evolutionarily highly conserved and ubiquitously expressed in all cell types (16). Conjugation of NEDD8 to protein targets, termed neddylation, is a process analogous to ubiquitination and requires NEDD8-specific E1, E2, and E3 enzymes (17, 18). The only identified NEDD8 E1 is a heterodimer of NAE1 (also known as APP-BP1) and UBA3, which together activate NEDD8 in an ATP-dependent manner. With the aid of a NEDD8 E2 (UBC12) enzyme and yet to be fully defined E3 ligases, activated NEDD8 is covalently fused with the lysine residue of substrate proteins via its C-terminal glycine, which can result in the addition of one or more NEDD8 molecules. Neddylation is reversibly regulated; substrate proteins conjugated with NEDD8 can be “deneddylated” by NEDD8-specific proteases, such as the COP9 signalosome (CSN) and NEDP1 (also known as SENP8) (19, 20).
Neddylation controls diverse cellular processes, including transcriptional regulation, cell cycle progression and differentiation, ribosome biogenesis, apoptosis, inflammatory responses, and proteolysis, via altering the activity, stability, subcellular localization, and DNA binding affinity of conjugated proteins (17, 18). Among the growing list of NEDD8 targets, the Cullin proteins (Cullin 1 to 5, 7, and 9) are the best-characterized substrates. Acting as scaffold proteins, Cullins recruit substrate recognition subunits and a RING domain protein to form functional Cullin-RING ubiquitin ligases (CRLs) (21), which together mediate the proteolysis of ∼20% of cellular proteins (22). Neddylation of Cullins induces the assembly of CRLs and thus is essential for their ubiquitin ligase activity (21).
Recent studies have begun to define the physiopathological significance of neddylation in embryonic development (23), synapse formation and maturation (24, 25), adipogenesis (26), tumor development (22), and, more recently, cardiac homeostasis (27–30). Neddylation is dysregulated in the failing human heart and in multiple mouse models of cardiac disease (31). Inactivation of deneddylation in mouse hearts results in the rapid development of heart failure due to increased cardiomyocyte necrosis and deficits in proteasomal and autophagic pathways (27, 29). While these studies highlight the importance of neddylation/deneddylation in adult hearts, the physiological significance of neddylation in developing hearts is completely unknown.
Herein, we show that targeted deletion of NAE1, a regulatory subunit of the NEDD8 E1 enzyme, in mouse cardiomyocytes resulted in loss of neddylation, myocardial hypoplasia, and ventricular noncompaction, leading to heart failure and perinatal lethality. These developmental abnormalities are associated with defective Hippo-YAP signaling, a conserved pathway that is vital to heart development and cardiomyocyte expansion (32, 33). Our study identifies a previously unknown role for neddylation in mediating cardiac chamber maturation and implicates a novel mechanism in the pathogenesis of LVNC.
Materials and Methods
The sources of reagents and detailed methods are described in SI Appendix, Supplemental Materials and Methods. All animal experiments were approved by the Augusta University Institutional Animal Care and Use Committee. A transgenic mouse line bearing an NAE1 “knockout-first” allele (34) (NAE1neoflox) was rederived from targeted frozen mouse embryos (EPD0441_1_C08) created by Eucomm. The mutant mice were first bred to ACTBFLP transgenic mice (35) (005703; The Jackson Laboratory) to remove Frt-flanked neo cassette, and then to αMHCCre mice (36) (011038; The Jackson Laboratory) to generate cardiomyocyte-restricted NAE1 knockout (NAE1CKO) mice. These mice were maintained in the C57BL/6J inbred background for our studies.
Results
Neddylation Is Developmentally Down-Regulated in the Heart.
We first sought to examine the expression of NEDD8 and other ubiquitin-like protein modifiers in the hearts of 2-mo-old mice. We found NEDD8 to be among the most highly expressed genes encoding ubiquitin-like proteins in the heart (SI Appendix, Fig. S1A). Taking advantage of a LacZ reporter mouse line in which LacZ expression is under the control of the endogenous NAE1 promoter, we detected β-galactosidase activity in the whole embryo at embryonic day (E) 14.5 (SI Appendix, Fig. S1B), indicating the ubiquitous expression of NAE1 at this developmental stage. Moreover, we observed a high level of total neddylated proteins in embryonic and perinatal mouse hearts, which was greatly reduced in adult mouse hearts (SI Appendix, Fig. S1C). The temporal reduction of neddylated proteins coincided with the down-regulation of neddylation enzymes NAE1 and UBC12 (SI Appendix, Fig. S1C). These data indicate that neddylation is highly active in developing hearts but down-regulated in adolescent hearts.
Cardiomyocyte-Restricted Knockout of NAE1 (NAE1CKO) Inhibits Neddylation.
Germline deletion of NAE1 or UBC12 in mice caused lethality during early embryonic stage (24). To investigate the role of neddylation in perinatal cardiac development in a cell lineage-specific fashion, we generated a conditional NAE1flox/flox allele by flanking exon 4 with loxP sites (SI Appendix, Fig. S2A). Cardiomyocyte-restricted NAE1 knockout (NAE1CKO, NAE1flox/flox/αMHCCre) mice were then created by crossing NAE1flox/flox mice with αMHC-driven Cre transgenic (αMHCCre) mice, in which Cre recombinase activity can be detected in the heart as early as E9.5 and is present in a majority of ventricular cardiomyocytes at E13.5 (37, 38). PCR and quantitative real-time PCR analyses confirmed the deletion of NAE1 in the NAE1CKO hearts, compared with their littermate control hearts (CTLs) (NAE1flox/flox or NAE1flox/+) (SI Appendix, Fig. S2 B and C). Western blot analyses of whole heart lysates revealed ∼40% and ∼70% reduction of NAE1 proteins in NAE1CKO hearts at E12.5 and postnatal day (P) 1, respectively (SI Appendix, Fig. S2 D and E). The residual NAE1 mRNA and protein in the NAE1CKO hearts are likely from noncardiomyocytes in which NAE1 gene was not deleted and/or NAE1 mRNA and protein that persist in the cardiomyocytes at P1.
We next performed Western blot analyses to determine the impact of loss of NAE1 on neddylation. Cullin family proteins (Cullin 1 to 7) are the best known NEDD8 targets (21). By P1, the NAE1CKO hearts exhibited a significant reduction of neddylated Cullin (Cul) 2 and Cul4, which migrated slower than their native forms due to the conjugation of NEDD8 (SI Appendix, Fig. S2 F and G). Accordingly, the native forms of Cul2 and Cul4 were markedly increased in NAE1CKO hearts. Moreover, total neddylated proteins in the NAE1CKO hearts were reduced to ∼55% of CTL hearts. These data demonstrated that NAE1 is indispensable for neddylation in fetal cardiomyocytes.
NAE1CKO Results in Heart Failure and Perinatal Lethality.
We next analyzed the genotype of the offspring from NAE1flox/flox and NAE1flox/+/αMHCCre intercrosses. NAE1CKO mice were identified at the expected Mendelian ratio at E18.5 and close to the expected ratio at P1 (observed 21% versus expected 25%, nonsignificant difference). No NAE1CKO mice, however, survived to P7 (Fig. 1A). Although NAE1CKO mice were morphologically indistinguishable from their littermate controls at birth, the vast majority of the NAE1CKO mice exhibited several common signs of heart failure, including cyanosis and peripheral edema by 48 to 72 h after birth and died shortly thereafter (Fig. 1B). At P1, the NAE1CKO hearts were readily identified by their larger size compared with the littermate CTL hearts. Consistently, gravimetric analysis confirmed that the NAE1CKO mice exhibited a significant increase in heart to body weight ratio despite their comparable body weights to control mice (Fig. 1C). The enlargement of the NAE1CKO hearts was accompanied by an increase of cardiomyocyte cross-sectional area (Fig. 1D) and the dysregulated expression of several genes associated with hypertrophic remodeling (Nappa, Nappb, and Myh6) (Fig. 1E), indicating that the NAE1-deificent hearts are hypertrophic.
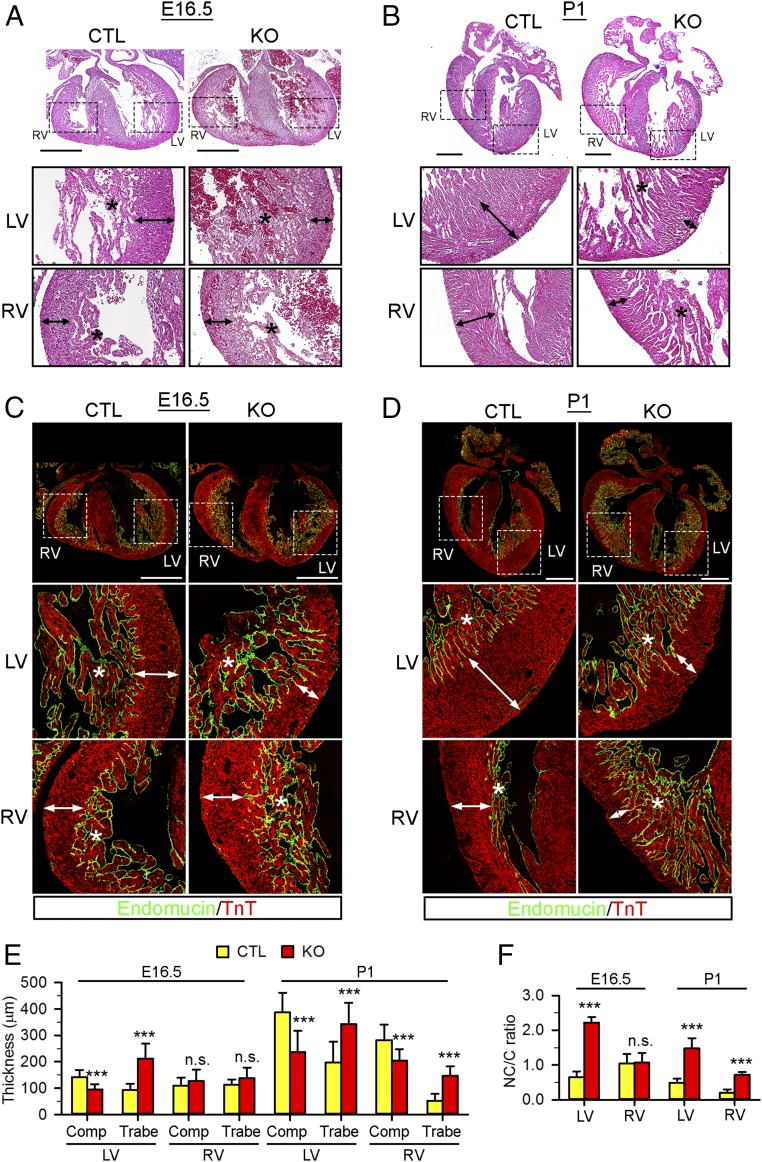
Fig. 1.
NAE1CKO caused heart failure and perinatal lethality of mice. (A) Genotype distribution of offspring from indicated mating pairs. n/%, the number and percentage of the mice with the indicated genotype from the analyzed litters, respectively. The progeny were subjected to χ2 analyses to obtain P value. (B) Gross morphology of CTL and NAE1CKO (KO) mice at P3. NAE1CKO pups displayed cyanosis and peripheral edema. (C) Gross morphology of hearts from P1 CTL and NAE1CKO pups and heart weight (Hw) to body weight (Bw) ratio. (D and E) Wheat-germ agglutinin (WGA, green) staining of myocardial section from P1 mice. The cross-sectional areas of cardiomyocytes were quantified. n = 3 hearts per group (>200 cells per heart). (Scale bars: 20 μm.). (E) qPCR analysis of the indicated hypertrophic genes in the hearts of P1 pups. (F and G) Echocardiography of CTL and NAE1CKO hearts at P1. Representative M-mode images (F) and the quantifications (G) are shown. Double head arrows mark left ventricle (LV) systolic (s) and diastolic (d) internal diameters (LVIDs and LVIDd), respectively. EF, ejection fraction; LVPWs, LV systolic posterior wall thickness. *P < 0.05, ***P < 0.001 vs. CTL.
To assess the function of NAE1CKO hearts, echocardiography was performed on conscious P1 neonates (Fig. 1 F and G and SI Appendix, Table S1). The NAE1CKO mice displayed a striking increase in both left ventricle (LV) systolic and diastolic diameters (1.70 ± 0.53 and 2.04 ± 0.49 in KO versus 0.84 ± 0.16 and 1.40 ± 0.17 in CTL) and a marked decrease in LV systolic wall thickness (0.36 ± 0.07 in KO versus 0.48 ± 0.05 in CTL). Consequently, the ejection fraction of the mutant hearts was reduced to ∼50% of the control hearts (39 ± 14% in KO versus 77 ± 7% in CTL). The heart rate of NAE1CKO mice was also significantly lower than that of control mice (335 ± 45 versus 381 ± 37).
Since parturition is a stressful event associated with abrupt changes in cardiac physiology, we reasoned that cardiac dysfunction in the NAE1CKO embryos renders them susceptible to the development of heart failure. To assess their cardiac function prepartum, we performed echocardiography in utero on a series of E18.5 CTL and NAE1CKO embryos. Our results demonstrated a significant impairment of cardiac function in E18.5 NAE1CKO embryos, which is evidenced by LV wall thinning, LV and right ventricle (RV) dilatation, and significantly decreased ejection fraction (SI Appendix, Fig. S3 and Table S1).
Thus, our data demonstrate that loss of NAE1 in murine cardiomyocytes causes rapid development of dilated cardiomyopathy and heart failure at late gestation, leading to perinatal lethality.
NAE1CKO Hearts Displayed Defects in Ventricular Wall Maturation.
αMHCCre-mediated gene deletion does not typically cause morphogenic defects in developing hearts, likely owing to the relatively late expression of Cre recombinase during development (37). Nevertheless, the rapid development of heart failure in NAE1CKO mice prompted us to determine whether loss of NAE1 disrupts heart morphogenesis. Since NAE1 depletion was discernible starting at E12.5, we temporally assessed the morphometry of the embryonic hearts thereafter. At E14.5, the mutant hearts had intact chamber separation and normal alignment of inflow and outflow tracts. The thickness of trabecular and compact layers was also comparable with that of controls (SI Appendix, Fig. S4 A–C). At E16.5, none of the NAE1CKO embryos (0 of 9) displayed any overt signs of heart failure (cardiac effusion or peripheral edema), and the mutant hearts were grossly indistinguishable from controls (SI Appendix, Fig. S4 D–F). However, both H&E staining and endomucin immunostaining, which outlines endocardial cell-lined trabeculae (39), showed that the NAE1CKO hearts exhibited thinner compact myocardium and a substantially thicker trabecular layer in the LV, but not in the RV (Fig. 2 A, C, E, and F). At P1, the control hearts exhibited a thick and compacted epicardial myocardium layer with few subendocardial trabeculae, indicating maturation of the ventricular wall. In contrast, the enlarged NAE1CKO hearts displayed a remarkably thinner compact layer in both ventricles associated with prominent trabeculae that deeply penetrated into the compact layer of both ventricles, consequently leading to a marked increase of LV noncompaction-to-compaction ratio (Fig. 2 B and D–F). The pronounced myocardial hypoplasia and hypertrabeculation, observed in all examined P1 NAECKO hearts (12 of 12), are consistent with a defect in ventricular maturation attributed to LVNC (7). These findings indicate that NAE1 deletion causes ventricular hypoplasia and noncompaction at late gestation, which precedes and eventually leads to peripartum heart failure.
Fig. 2.
NAE1CKO mice displayed ventricular noncompaction. (A and B) Hematoxylin/eosin staining in coronal sections of CTL and NAE1CKO hearts at E16.5 (A) and P1 (B). The Bottom show magnified views of boxed areas in the Top. Double head arrows and asterisks mark the thickness of the compact layer and trabeculae, respectively. (Scale bars: 200 μm.) (C and D) E16.5 (C) and P1 (D) heart sections stained with endomucin (green) and cardiac troponin T (TnT, red) antibodies to delineate the trabecular (covered by endomucin-positive endocardium) and compact layer. (Scale bars: 500 μm.) (E) Quantification of the thickness of the compact (Comp) and trabecular (Trabe) layer in CTL and NAE1CKO hearts. (F) Trabecular layer/compact layer (NC/C) ratio in CTL and NAE1CKO hearts at E16.5 and P1. For morphological analysis, n > 4 hearts per group at each time point were analyzed. Bar graphs are presented as mean + SD. n.s., not significant, ***P < 0.005 versus CTL in unpaired two-tailed t test.
Neddylation Is Required for Cardiomyocyte Proliferation.
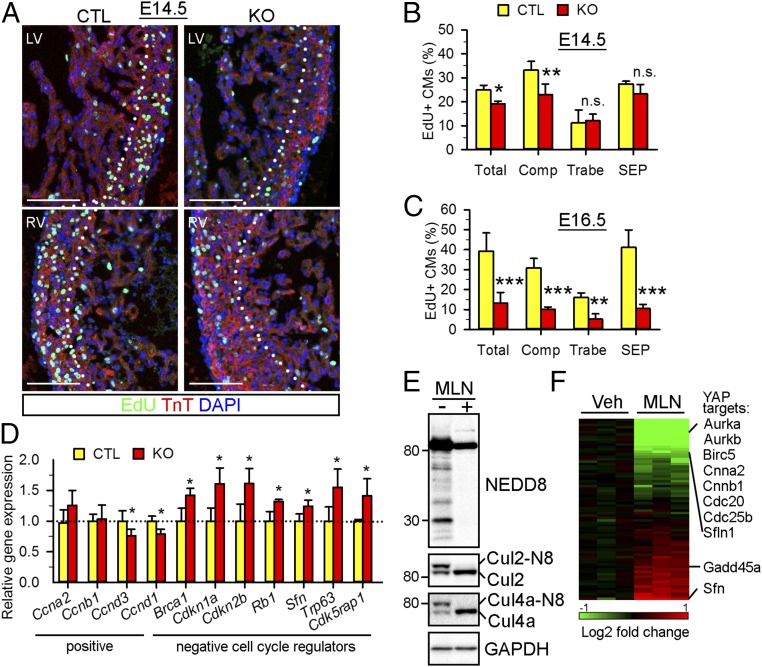
Active cardiomyocyte proliferation is essential for embryonic cardiac development (3, 4). Perturbation of cardiomyocyte proliferation has been shown to disrupt cardiac chamber maturation and lead to LVNC in mouse models (11, 40, 41). We thus investigated whether neddylation regulates cardiomyocyte proliferation during development. An EdU incorporation assay demonstrated that, at E14.5, the NAE1CKO hearts had significantly fewer EdU+ cardiomyocytes compared with CTL hearts (19.1 ± 1.1% versus 24.9 ± 1.9% in CTL) (Fig. 3 A and B). Interestingly, cardiomyocyte proliferation was mainly diminished in the compact layer of the NAE1CKO hearts (23.0 ± 4 0.4% versus 33.2 ± 3.7% in CTL) as opposed to the trabecular layer (12.0 ± 2.8% versus 11.1 ± 5.5% in CTL) or the septum (23.2.0 ± 4.0% versus 27.4 ± 1.1% in CTL). At E16.5, the proliferation capacity was further impaired (∼70% reduction at E16.5 versus ∼20% reduction at E14.5), and an EdU incorporation assay and pH3 immunostaining showed fewer proliferating cardiomyocytes, not only in the compact layer but also in the trabecular layer and the septum of the NAE1CKO hearts (Fig. 3C and SI Appendix, Fig. S5 A–C). Suppression of cardiomyocyte proliferation persisted at P1 in the NAE1CKO hearts (SI Appendix, Fig. S5 D and E). In contrast, a TUNEL assay demonstrated a comparable percentage of apoptotic cardiomyocytes in CTL and NAE1CKO hearts at P1 (0.29 ± 0.25% in CTL versus 0.14 ± 0.16% in NAE1CKO) (SI Appendix, Fig. S5 F and G), suggesting a minimal effect of NAE1 deficiency on embryonic cardiomyocyte survival.
Fig. 3.
Neddylation is indispensable for cardiomyocyte proliferation. (A) EdU (green)-immunostained heart sections from E14.5 embryos. Nuclei and cardiomyocytes were counterstained with DAPI (blue) and TnT (red) antibodies. (Scale bars: 100 μm.) (B and C) Quantification of EdU-positive cardiomyocytes (CMs) in E14.5 and E16.5 heart sections. n > 4 hearts per genotype at each time point were analyzed. (D) PCR array analysis of cell cycle gene expression in P1 hearts (n = 3 per genotype). Genes dysregulated in the NAE1CKO hearts are shown. (E) Western blot of neddylated proteins in vehicle (Veh)-treated and MLN4924 (MLN, 1 μM, a specific NAE inhibitor)-treated neonatal cardiomyocytes. (F) Heat map of PCR array data for differentially expressed cell cycle genes in MLN-treated neonatal cardiomyocytes. YAP downstream targets are listed. n.s., not significant; *P < 0.05, **P < 0.01, ***P < 0.001 versus CTL in unpaired two-tailed t test.
To investigate how neddylation regulates cardiomyocyte proliferation, we performed qPCR array analysis of cell cycle gene expression in P1 CTL and NAE1CKO hearts. Compared with CTL hearts, NAE1CKO hearts exhibited down-regulated expression of two cell cycle activators, Cnnd1 and Cnnd3, and significant up-regulation of numerous cell cycle inhibitors: Brca1, Cdkn1a, Cdkn2b, Rb1, Sfn, Trp63, and Cdk5rap1 (Fig. 3D). Since neonatal hearts contain many noncardiomyocytes that retain NAE1 expression and undergo active proliferation, we reasoned that the qPCR array results from whole heart lysates may underestimate the effect of neddylation in cardiomyocyte cell cycle regulation. We thus inhibited neddylation in cultured neonatal rat ventricular cardiomyocytes (NRVCs) with MLN4924 (MLN), a potent and specific NAE1 inhibitor that abolishes NAE1-mediated NEDD8 activation and thereby neddylation (22). Indeed, MLN robustly reduced neddylated proteins in NRVCs (Fig. 3E). Moreover, qPCR array analysis identified a broader range of dysregulated cell cycle genes, with a greater extent of dysregulation in MLN-treated cardiomyocytes (Fig. 3F and SI Appendix, Fig. S6). These data suggest that neddylation modulates the expression of a wide array of cell cycle regulators in a cell-autonomous manner.
Neddylation Regulates Hippo-YAP Signaling.
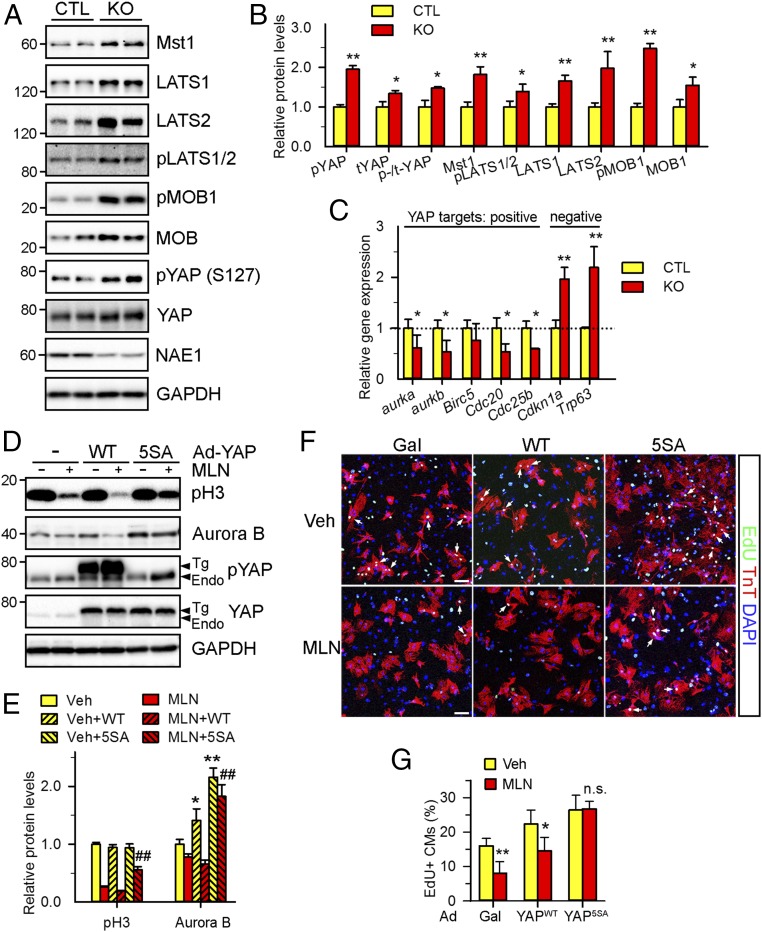
We observed that a number of the dysregulated genes in MLN-treated cardiomyocytes, such as Aukb, Cnna2, Cnnb1, Sfn, etc., are the direct targets of Yes-Associated Protein (YAP) (Fig. 3F and SI Appendix, Fig. S6). The Hippo-YAP pathway is known to control heart morphogenesis by regulating cardiomyocyte proliferation (42). In this pathway, sequential activation of Hippo kinases Mst1/2 and LATS1/2 mediates phosphorylation of YAP and inhibits its nuclear translocation and transcriptional activity. Western blot analysis of myocardial lysates from NAE1CKO hearts demonstrated an increase in Hippo kinases (Mst1, LATS1, and LATS2) and their associating partner MOB1 compared with CTL (Fig. 4 A and B). We also observed increased phosphorylation of LAST1/2 and MOB1 and elevated phosphorylation of YAP at serine 127 in NAE1CKO hearts, suggesting the activation of Hippo signaling. Phosphorylation of YAP at serine 127 causes its cytoplasmic sequestration and inhibits its transcriptional activity (43). Indeed, immunostaining analysis of E16.5 heart sections revealed that YAP nuclear localization was largely abrogated in NAE1CKO hearts (SI Appendix, Fig. S7). qPCR analysis of direct YAP targets using gene-specific primers showed that several YAP-activated genes (Auroka, Aurokb, Cdc20, and Cdc25b) were down-regulated in P1 NAE1CKO hearts while YAP-repressed genes (Cdkn1a and Trp63) were up-regulated (Fig. 4C), indicating the impairment of YAP signaling in NAE1CKO hearts.
Fig. 4.
Neddylation controls Hippo-YAP signaling. (A and B) Western blot of the key components of the Hippo-YAP pathway in P1 hearts (A) and the quantification (B). (C) qPCR analysis of the expression of cell cycle genes that are either positively or negatively regulated by YAP in P1 hearts. (D–G) NRVCs were infected with adenoviruses (Ad) expressing WT (YAPWT) or LATS2-insenstive mutant (YAP5SA) form of YAP for 24 h, followed by MLN (1 μM) treatment for additional 24 h. (D and E) Western blots (D) of the indicated proteins and the quantification (E). (F and G) Representative images (F) and the quantification (G) of EdU (green)-labeled cardiomyocytes (arrows). Cardiomyocytes and nuclei labeled by TnT (red) and DAPI (blue), respectively. (Scale bars: 100 μm.) n = 4 fields per group (∼500 cardiomyocytes per field). n.s., not significant; *P < 0.05, **P < 0.01 versus CTL or Veh, ##P < 0.01 versus MLN in unpaired two-tailed t test.
Consistent with the in vivo data, the NAE1 inhibitor MLN increased total and phosphorylated Mst1, Mst2, LATS1, LATS2, and MOB1 proteins in NRVCs and consequently induced YAP phosphorylation (SI Appendix, Fig. S8 A and B). Moreover, MLN reduced the expression of Aurora B and Cyclin B1 (known to be induced by YAP) (44) and increased the expression of cell cycle inhibitor Gpr132 (known to be repressed by YAP) (45) (SI Appendix, Fig. S8C). qPCR analysis using gene-specific primers confirmed dysregulated expression of YAP targets in MLN-treated cardiomyocytes: down-regulation of Aurob, Birc5, Cnnb1, Cdc20, Cyr61, and Ctgf that are activated by YAP and up-regulation of Gdd45a and Sfn that are repressed by YAP (SI Appendix, Fig. S8D). In line with the diminished YAP signaling, MLN treatment led to ∼80% reduction in NRVC proliferation, as demonstrated by an EdU incorporation assay (SI Appendix, Fig. S8 E and F). Meanwhile, genetic inhibition of neddylation through codepletion of the neddylation enzymes NAE1 and UBC12 also increased Mst1 and LATS2 proteins, promoted phosphorylation of LATS1/2 and YAP, and suppressed cardiomyocyte proliferation in NRVCs (SI Appendix, Fig. S8 G–J). Together, these in vivo and in vitro data demonstrate that inhibition of neddylation activates Hippo kinases and represses YAP signaling.
Reactivation of YAP Signaling Attenuates MLN-Inhibited Cardiomyocyte Proliferation.
To determine whether the activation of Hippo signaling contributes to reduced cardiomyocyte proliferation consequent to NAE1 inhibition, we performed two sets of rescue experiments. First, we infected NRVCs with adenovirus expressing a WT YAP (YAPWT) or a Hippo kinase-resistant YAP mutant in which LATS-phosphorylation sites are mutated (constitutively active, YAP5SA) (46). Interestingly, overexpression of YAPWT was unable to overcome the MLN-induced repression of cardiomyocyte proliferation (as indicated by expression of the mitotic marker pH3 and the cell cycle regulator Aurora B), likely due to extensive phosphorylation and inactivation of the overexpressed YAPWT protein (Fig. 4 D and E). An EdU incorporation assay confirmed that MLN markedly inhibited the proliferation of β-Gal- and YAPWT-infected cardiomyocytes. In contrast, overexpression of YAP5SA at equivalent levels partially restored the expression of pH3 and Aurora B and blocked the inhibition of NRVC proliferation by MLN (Fig. 4 F and G). Secondly, we silenced LATS1 and LATS2 in NRVCs via specific siRNAs. Silencing of LATS2, but not LATS1, abolished MLN-induced YAP phosphorylation and inhibition of proliferation (SI Appendix, Fig. S9). These lines of evidence collectively support a direct role of Hippo kinases in inhibiting YAP signaling and cell proliferation in NAE1-inactived cardiomyocytes.
YAP Signaling Has a Crucial Role in Ventricular Chamber Maturation.
Given the critical role of Hippo-YAP signaling in embryonic heart development, we asked whether defective YAP signaling suffices to perturb cardiac chamber maturation. Cardiac-specific deletion of YAP by Nkx2-5Cre or Tnnt2Cre caused embryonic lethality at early gestation or midgestation due to cardiac failure (44, 47) while αMHCCre-mediated YAP deletion did not affect cardiomyocyte proliferation or cardiac development (48). Therefore, neither mouse model is suitable to study the role of YAP in regulating ventricular compaction. Since SM22αCre is specifically expressed in fetal cardiomyocytes and smooth muscle cells starting at E9.5, and a small percentage of SM22αCre-driven YAP knockout (YAPCKO) mice survived to and even beyond late gestation (45), we assessed the compaction of YAPCKO hearts from mice surviving to E18.5 and P1, when myocardial compaction is prominent. Consistent with previous reports (45), YAPCKO hearts showed severe ventricular hypoplasia, as evidenced by ventricular wall thinning and decreased cardiomyocyte proliferation (SI Appendix, Fig. S10 A and B). Moreover, the embryonic and perinatal YAPCKO hearts retained prominent trabeculae in both ventricles compared with the control hearts (SI Appendix, Fig. S10 C–F), indicating a defect in trabeculae regression and compaction. Notably, cardiomyocyte proliferation arrest was discernable by as early as E11.5 in YAPCKO hearts, which appears to cause a more severe noncompaction phenotype at P1 compared with NAE1CKO hearts (45). Our observations are consistent with a recent report, in which loss of YAP in compact myocardium was found to cause extensive noncompaction in postnatal hearts (49). Together, our data, along with that of others (49), support the contention that defective YAP signaling contributes to impaired ventricular maturation in NAE1CKO hearts.
NEDD8 Substrate Cullin 7 Controls the Ubiquitination and Degradation of Mst1.
We next investigated how neddylation regulates Hippo signaling. Neddylation of Cullins (Cul1 to -5, -7, and -9) is a key event that triggers the assembly of functional Cullin-RING ubiquitin ligases (CRLs), which typically consist of one of the Cullin proteins, a RING finger protein, and a substrate-recognizing protein, and degrade many cellular proteins (Fig. 5A) (21, 22). In light of the increase in Mst1 and LATS2 proteins in NAE1-deficient/inactivated cardiomyocytes, we hypothesized that neddylation is required for their ubiquitination and turnover. A cycloheximide-based pulse–chase assay demonstrated that MLN significantly increased the stability of Mst1 and LATS2 in NRVCs (Fig. 5B and SI Appendix, Fig. S11). An immunoprecipitation assay showed that MLN robustly decreased the ubiquitinated forms of Mst1 (Fig. 5C), indicating that neddylation controls the ubiquitination of Mst1.
Fig. 5.
Cullin 7 (Cul7) ubiquitin ligase mediates the ubiquitination and degradation of Mst1. (A) A schematic diagram illustrating the regulation of Cullin-RING ubiquitin (Ub) ligases (CRLs) by neddylation (see main text). (B) NRVCs were treated with vehicle (Veh) or MLN (1 μM) for 12 h and subsequently with cycloheximide (CHX, 50 μM) for the indicated times. Cell lysates were collected for Western blot. Representative Western blot images (Top) and the quantification (Bottom) are shown. *P < 0.05 versus Veh at the indicated time point. (C) NRVCs were treated with Veh or MLN (1 μM) for 12 h in the presence of proteasome inhibitor bortezomib (100 nM). Cell lysates were used for immunoprecipitation (IP), followed by Western blot. Representative images are shown. H.C., IgG heavy chain. (D) Western blots showing Mst1 expression levels in NRVCs transfected siRNAs against either luciferase (siLuci) or the indicated Cullins. The relative abundance of Mst1 is denoted under the blot. (E) HEK293 cells were transfected with hemagglutinin (HA)-tagged Mst1 and myc-Cul7 for 24 h. Cell lysates were used for immunoprecipitation, followed by Western blot. Representative images are shown. (F) A CHX-based pulse–chase assay revealed the increased half-life of Mst1 in NRVCs transfected with siRNA against Cul7. Representative Western blots (Top) and the quantification (Bottom) are shown. (G) Immunoprecipitation assay followed by Western blot demonstrates a decrease in the ubiquitinated form of Mst1 in NRVCs transfected with siCul7. (H) Western blots of Hippo pathway proteins from NRVCs transfected with the indicated siRNAs. Some of the cells were treated with MLN for 24 h before harvest. #, a nonspecific band. (I and J) Representative confocal images (I) of EdU (green, arrow)-labeled cardiomyocytes and the quantification (J). (Scale bars: 100 μm.) n = 6 fields per group (∼500 cardiomyocytes per field). *P < 0.05, **P < 0.01 versus Veh or siLuci.
To determine which CRL mediates the ubiquitination of Mst1, we silenced individual Cullins in NRVCs. Our results showed that silencing Cullin 7 (Cul7), a ubiquitin ligase previously reported to mediate the degradation of insulin receptor substrate 1 (IRS1) (50), led to accumulation of Mst1 in cultured NRVCs while silencing Cullins 2, 3, and 5 did not (Fig. 5D). Reciprocal immunoprecipitation assay in HEK293 cells further revealed that Cul7 interacts with Mst1, and this interaction was diminished in the presence of MLN (Fig. 5E), suggesting that inhibition of neddylation disrupts Cul7 and Mst1 interactions. A pulse–chase assay demonstrated that silencing of Cul7 slowed down the degradation of Mst1 (Fig. 5F) and decreased the ubiquitinated forms of Mst1 (Fig. 5G), indicating that Cul7 mediates the ubiquitination and degradation of Mst1.
In line with the accumulation of Mst1, Cul7 depletion induced the phosphorylation of its downstream targets, including LATS1/2, MOB1, and YAP (Fig. 5H), indicating the activation of Hippo signaling. Moreover, down-regulation of Cul7 robustly inhibited cardiomyocyte proliferation (Fig. 5 I and J), presumably due to the defects in YAP signaling.
These data collectively suggest that NEDD8 substrate Cul7 acts as a ubiquitin ligase to promote Mst1 degradation, thereby activating YAP signaling and cardiomyocyte proliferation.
Discussion
This study provides evidence that neddylation is essential for proper cardiac development through inhibition of the Hippo pathway and activation of YAP signaling. Our results support a working model (SI Appendix, Fig. S12) wherein neddylation is required to support the activity of Cullin-RING ubiquitin ligases (CRLs), which degrade Hippo kinases in a spatiotemporal manner, leading to dephosphorylation and nuclear retention of YAP. This, in turn, enables cardiomyocyte proliferation, thus permitting ventricular compaction and normal cardiac development.
Besides ubiquitin, the ubiquitin superfamily contains a number of ubiquitin-like proteins (UBLs) including NEDD8, SUMO, ISG15, ufm1, etc. (15). Eukaryotic cells utilize ubiquitin/UBL-mediated posttranslational modifications to tightly control protein function and thus regulate a wide array of cellular processes (15). Recent studies have unveiled the importance of some of these protein modifiers in the heart (51, 52). Ubiquitin-mediated proteolysis of misfolded proteins ensures protein quality control in cardiomyocytes and protects against desmin-related cardiomyopathy and myocardial ischemia-reperfusion injury (53, 54). A tightly controlled level of SUMOylation is essential for heart formation, possibly through regulation of the activity of several cardiogenic transcription factors during embryonic development (55–57). In this study, we showed that neddylation activity is temporally regulated during cardiac development (SI Appendix, Fig. S1) and that inhibition of neddylation in embryonic hearts results in the rapid development of heart failure and neonatal lethality (Fig. 1). These findings not only establish an indispensable role of NEDD8 in developing hearts and add neddylation into the complex regulatory network that governs cardiogenesis, but also have implications for heart function in adults. We have previously reported that the blockade of deneddylation in the heart causes heart failure and premature death (27, 29). Therefore, a balance between neddylation and deneddylation is crucial for the maintenance of normal cardiac function. Notably, MLN4924 (Pevonedistat) is under clinical investigation for the treatment of certain cancers (58), and our findings highlight a need to monitor its potential cardiotoxicity.
Cardiac chamber maturation at late gestation stage involves the expansion and compaction of myocardium. This final step of chamber development is critical for ventricular wall thickening, proper patterning of the myocardium, and formation of the conduction system (3, 4). Perturbation of this process is known to cause LVNC, a commonly diagnosed form of cardiomyopathy (5–7). Although the molecular mechanisms that orchestrate ventricular maturation are not well understood, animal models of LVNC have indicated that alterations in growth signals, such as Notch (10), TGFβ (11), and NRG1/ERBB2 (59), may be contributing factors. As evidenced in mice with deficient 14-3-3σ and Cdc42 and exaggerated TGFβ signaling (11, 40, 41), the pathogenesis of LVNC may associate with the arrest of embryonic cardiomyocyte proliferation. In line with these reports, our study identifies a previously unrecognized role of neddylation in cardiac chamber maturation, at least in part through sustaining cardiomyocyte proliferation. The NAE1-deficient hearts replicate multiple aspects of human LVNC, including a lack of compaction, thin ventricular walls, deep intertrabecular recesses, and increased trabeculae (Fig. 2). The defects in cardiomyocyte proliferation precede and intensify with the progression of noncompaction (Fig. 3), which eventually results in heart failure in NAE1CKO mice. Interestingly, the impact of NAE1 deficiency on cardiomyocyte proliferation was restricted to the compact layer of the myocardium at E14.5 (Figs. 2A and 3B). One possible reason for this is that NAE1 depletion at this time point fails to robustly impact the region with a lower proliferation rate (trabeculae) compared with those with higher proliferation rate (compact layer) (37). Since trabeculae begin to regress and fuse with the compact myocardium after E14.5 (3, 4), inhibition of neddylation by NAE1CKO does not influence trabeculation but rather suppresses the expansion of the compact layer and trabecular compaction, leading to the noncompaction phenotype. Additional studies using Cre transgenes (Tnnt2Cre and Nkx2-5Cre) that are expressed at earlier time points (60, 61) will help clarify whether neddylation controls trabeculation.
The Hippo-YAP pathway is crucial for cardiac morphogenesis (32, 33). During development, various growth factors (IGFs, BMPs, Wnts, etc.) sequentially suppress the Hippo kinases Mst1/2 and LATS1/2, leading to the diminished phosphorylation of YAP. Dephosphorylated YAP translocates into the nucleus and binds to TEAD transcription factors to drive the expression of genes involved in cardiomyocyte differentiation and proliferation. Our study identifies Hippo-YAP signaling as a primary mechanism through which neddylation influences cardiac development. We present multiple lines of evidence in vitro and in vivo demonstrating that neddylation suppresses the Hippo pathway and activates YAP signaling in cardiomyocytes. First, suppression of neddylation resulted in increased levels of both phosphorylated (active) and total Hippo kinases (Mst1/2, LATS1/2) and their associating factors (MOB1) (Fig. 4A and SI Appendix, Fig. S8). Second, inhibition of neddylation increased the phosphorylation (serine 127, inactive form) and consequent cytoplasmic retention of YAP (Fig. 4A and SI Appendix, Figs. S7 and S8). Third, loss of neddylation regulated the expression of cell cycle genes downstream of YAP (Fig. 4C and SI Appendix, Fig. S8C). Fourth, rescue of YAP signaling by silencing LATS2 or overexpression of a Hippo kinase-insensitive YAP mutant (YAP5SA) overcame the blockade of cardiomyocyte proliferation induced by the inhibition of neddylation (Fig. 4 D–G and SI Appendix, Fig. S9). Moreover, we found that, in addition to cardiac hypoplasia (44, 45, 47), YAP-deficient hearts also exhibited ventricular noncompaction (SI Appendix, Fig. S10), which is consistent with the changes observed in NAE1CKO hearts and indicates a function of YAP signaling in ventricular compaction. It is conceivable that YAP signaling could modulate ventricular wall maturation through interplay with related signaling pathways, such as Wnt, Notch, and/or TGFβ pathways (62–64). Whether inhibiting the Hippo pathway is sufficient to rescue the chamber maturation defects in NAE1CKO mice remains to be determined. Additionally, given the well-documented role of Hippo-YAP in regulating neoplasia and tissue development (65), our findings may provide novel insight into how neddylation controls tumor growth and spine formation (22, 24).
Accumulating evidence suggests that the Hippo-YAP pathway is tightly regulated by various types of posttranslational modifications, including phosphorylation, ubiquitination, sumoylation, acetylation, etc. (66). Our study reveals a mechanistic link between neddylation and Hippo signaling. Although it remains unclear whether Hippo pathway components are direct NEDD8 targets, we have demonstrated that neddylation regulates the stability of Hippo kinases by modulating the activity of CRLs. This concept is supported by the accumulation of Hippo pathway components (Mst1, LATS1, LATS2, and MOB1) (Fig. 4) and by the increased half-life of Mst1 and LATS2 (Fig. 5 and SI Appendix, Fig. S11) when neddylation is inhibited. Since LATS1 and -2 are substrates of Cul4A ubiquitin ligase (67), it is very likely that neddylation controls the degradation of LATS1/2 in a Cul4A-dependent manner in the heart. Moreover, we provide evidence demonstrating that Cul7 is a regulator of Hippo-YAP signaling by acting as the ubiquitin ligase of Mst1 (Fig. 5). Cul7 is a relatively cryptic NEDD8 substrate that mediates the ubiquitination of certain cellular proteins, such as IRS1 (50). Cul7 appears to be oncogenic for several types of cancer (68, 69). Mice deficient in Cul7 displayed growth retardation and perinatal lethality (70). Mutations in Cul7 are associated with autosomal recessive 3-M and Yakut short stature syndromes that are characterized by pre- and postnatal growth retardation (71). These studies underscore the importance of Cul7 in growth control and are consistent with a role for Cul7 in regulating Hippo-YAP signaling. Thus, our findings may provide new insights into the underlying pathogenic mechanisms of these diseases.
Supplementary Material
Acknowledgments
This study is supported by US National Institutes of Health Grants R01HL124248 (to H.S.) and F31HL139079 (to R.L.) and American Heart Association Grants 17POST33410592 (to J. Zou) and 16SDG30940002 (to J.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719309115/-/DCSupplemental.
References
- 1.Moorman AF, Christoffels VM. Cardiac chamber formation: Development, genes, and evolution. Physiol Rev. 2003;83:1223–1267. doi: 10.1152/physrev.00006.2003. [DOI] [PubMed] [Google Scholar]
- 2.Xin M, Olson EN, Bassel-Duby R. Mending broken hearts: Cardiac development as a basis for adult heart regeneration and repair. Nat Rev Mol Cell Biol. 2013;14:529–541. doi: 10.1038/nrm3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Samsa LA, Yang B, Liu J. Embryonic cardiac chamber maturation: Trabeculation, conduction, and cardiomyocyte proliferation. Am J Med Genet C Semin Med Genet. 2013;163C:157–168. doi: 10.1002/ajmg.c.31366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang W, Chen H, Qu X, Chang CP, Shou W. Molecular mechanism of ventricular trabeculation/compaction and the pathogenesis of the left ventricular noncompaction cardiomyopathy (LVNC) Am J Med Genet C Semin Med Genet. 2013;163C:144–156. doi: 10.1002/ajmg.c.31369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daubeney PE, et al. National Australian Childhood Cardiomyopathy Study Clinical features and outcomes of childhood dilated cardiomyopathy: Results from a national population-based study. Circulation. 2006;114:2671–2678. doi: 10.1161/CIRCULATIONAHA.106.635128. [DOI] [PubMed] [Google Scholar]
- 6.Nugent AW, et al. National Australian Childhood Cardiomyopathy Study Clinical features and outcomes of childhood hypertrophic cardiomyopathy: Results from a national population-based study. Circulation. 2005;112:1332–1338. doi: 10.1161/CIRCULATIONAHA.104.530303. [DOI] [PubMed] [Google Scholar]
- 7.Finsterer J, Stöllberger C, Towbin JA. Left ventricular noncompaction cardiomyopathy: Cardiac, neuromuscular, and genetic factors. Nat Rev Cardiol. 2017;14:224–237. doi: 10.1038/nrcardio.2016.207. [DOI] [PubMed] [Google Scholar]
- 8.Liu J, et al. A dual role for ErbB2 signaling in cardiac trabeculation. Development. 2010;137:3867–3875. doi: 10.1242/dev.053736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grego-Bessa J, et al. Notch signaling is essential for ventricular chamber development. Dev Cell. 2007;12:415–429. doi: 10.1016/j.devcel.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luxán G, et al. Mutations in the NOTCH pathway regulator MIB1 cause left ventricular noncompaction cardiomyopathy. Nat Med. 2013;19:193–201. doi: 10.1038/nm.3046. [DOI] [PubMed] [Google Scholar]
- 11.Kodo K, et al. iPSC-derived cardiomyocytes reveal abnormal TGF-β signalling in left ventricular non-compaction cardiomyopathy. Nat Cell Biol. 2016;18:1031–1042. doi: 10.1038/ncb3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartram U, et al. Double-outlet right ventricle and overriding tricuspid valve reflect disturbances of looping, myocardialization, endocardial cushion differentiation, and apoptosis in TGF-beta(2)-knockout mice. Circulation. 2001;103:2745–2752. doi: 10.1161/01.cir.103.22.2745. [DOI] [PubMed] [Google Scholar]
- 13.Chen Q, et al. Smad7 is required for the development and function of the heart. J Biol Chem. 2009;284:292–300. doi: 10.1074/jbc.M807233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montgomery RL, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamitani T, Kito K, Nguyen HP, Yeh ET. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem. 1997;272:28557–28562. doi: 10.1074/jbc.272.45.28557. [DOI] [PubMed] [Google Scholar]
- 17.Enchev RI, Schulman BA, Peter M. Protein neddylation: Beyond cullin-RING ligases. Nat Rev Mol Cell Biol. 2015;16:30–44. doi: 10.1038/nrm3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kandala S, Kim IM, Su H. Neddylation and deneddylation in cardiac biology. Am J Cardiovasc Dis. 2014;4:140–158. [PMC free article] [PubMed] [Google Scholar]
- 19.Gan-Erdene T, et al. Identification and characterization of DEN1, a deneddylase of the ULP family. J Biol Chem. 2003;278:28892–28900. doi: 10.1074/jbc.M302890200. [DOI] [PubMed] [Google Scholar]
- 20.Menon S, et al. COP9 signalosome subunit 8 is essential for peripheral T cell homeostasis and antigen receptor-induced entry into the cell cycle from quiescence. Nat Immunol. 2007;8:1236–1245. doi: 10.1038/ni1514. [DOI] [PubMed] [Google Scholar]
- 21.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 22.Soucy TA, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 23.Tateishi K, Omata M, Tanaka K, Chiba T. The NEDD8 system is essential for cell cycle progression and morphogenetic pathway in mice. J Cell Biol. 2001;155:571–579. doi: 10.1083/jcb.200104035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogl AM, et al. Neddylation inhibition impairs spine development, destabilizes synapses and deteriorates cognition. Nat Neurosci. 2015;18:239–251. doi: 10.1038/nn.3912. [DOI] [PubMed] [Google Scholar]
- 25.Li L, et al. Enzymatic activity of the scaffold protein rapsyn for synapse formation. Neuron. 2016;92:1007–1019. doi: 10.1016/j.neuron.2016.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park HS, et al. PPARγ neddylation essential for adipogenesis is a potential target for treating obesity. Cell Death Differ. 2016;23:1296–1311. doi: 10.1038/cdd.2016.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su H, et al. Perturbation of cullin deneddylation via conditional Csn8 ablation impairs the ubiquitin-proteasome system and causes cardiomyocyte necrosis and dilated cardiomyopathy in mice. Circ Res. 2011;108:40–50. doi: 10.1161/CIRCRESAHA.110.230607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su H, Li F, Ranek MJ, Wei N, Wang X. COP9 signalosome regulates autophagosome maturation. Circulation. 2011;124:2117–2128. doi: 10.1161/CIRCULATIONAHA.111.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su H, et al. The COP9 signalosome is required for autophagy, proteasome-mediated proteolysis, and cardiomyocyte survival in adult mice. Circ Heart Fail. 2013;6:1049–1057. doi: 10.1161/CIRCHEARTFAILURE.113.000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su H, et al. COP9 signalosome controls the degradation of cytosolic misfolded proteins and protects against cardiac proteotoxicity. Circ Res. 2015;117:956–966. doi: 10.1161/CIRCRESAHA.115.306783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, et al. Nedd8 ultimate buster 1 long (nub1l) protein suppresses atypical neddylation and promotes the proteasomal degradation of misfolded proteins. J Biol Chem. 2015;290:23850–23862. doi: 10.1074/jbc.M115.664375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Q, Li L, Zhao B, Guan KL. The hippo pathway in heart development, regeneration, and diseases. Circ Res. 2015;116:1431–1447. doi: 10.1161/CIRCRESAHA.116.303311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wackerhage H, Del Re DP, Judson RN, Sudol M, Sadoshima J. The Hippo signal transduction network in skeletal and cardiac muscle. Sci Signal. 2014;7:re4. doi: 10.1126/scisignal.2005096. [DOI] [PubMed] [Google Scholar]
- 34.Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodríguez CI, et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- 36.Agah R, et al. Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J Clin Invest. 1997;100:169–179. doi: 10.1172/JCI119509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye B, et al. APC controls asymmetric Wnt/β-catenin signaling and cardiomyocyte proliferation gradient in the heart. J Mol Cell Cardiol. 2015;89:287–296. doi: 10.1016/j.yjmcc.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McFadden DG, et al. The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development. 2005;132:189–201. doi: 10.1242/dev.01562. [DOI] [PubMed] [Google Scholar]
- 39.D’Amato G, et al. Sequential Notch activation regulates ventricular chamber development. Nat Cell Biol. 2016;18:7–20. doi: 10.1038/ncb3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kosaka Y, et al. 14-3-3ε plays a role in cardiac ventricular compaction by regulating the cardiomyocyte cell cycle. Mol Cell Biol. 2012;32:5089–5102. doi: 10.1128/MCB.00829-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, et al. Essential role of Cdc42 in cardiomyocyte proliferation and cell-cell adhesion during heart development. Dev Biol. 2017;421:271–283. doi: 10.1016/j.ydbio.2016.12.012. [DOI] [PubMed] [Google Scholar]
- 42.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 43.Basu S, Totty NF, Irwin MS, Sudol M, Downward J. Akt phosphorylates the yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol Cell. 2003;11:11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- 44.von Gise A, et al. YAP1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. Proc Natl Acad Sci USA. 2012;109:2394–2399. doi: 10.1073/pnas.1116136109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, et al. Deletion of yes-associated protein (YAP) specifically in cardiac and vascular smooth muscle cells reveals a crucial role for YAP in mouse cardiovascular development. Circ Res. 2014;114:957–965. doi: 10.1161/CIRCRESAHA.114.303411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao B, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xin M, et al. Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci Signal. 2011;4:ra70. doi: 10.1126/scisignal.2002278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Del Re DP, et al. Yes-associated protein isoform 1 (Yap1) promotes cardiomyocyte survival and growth to protect against myocardial ischemic injury. J Biol Chem. 2013;288:3977–3988. doi: 10.1074/jbc.M112.436311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tian X, et al. Identification of a hybrid myocardial zone in the mammalian heart after birth. Nat Commun. 2017;8:87. doi: 10.1038/s41467-017-00118-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu X, et al. The CUL7 E3 ubiquitin ligase targets insulin receptor substrate 1 for ubiquitin-dependent degradation. Mol Cell. 2008;30:403–414. doi: 10.1016/j.molcel.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mendler L, Braun T, Müller S. The ubiquitin-like sumo system and heart function: From development to disease. Circ Res. 2016;118:132–144. doi: 10.1161/CIRCRESAHA.115.307730. [DOI] [PubMed] [Google Scholar]
- 52.Wang X, Pattison JS, Su H. Posttranslational modification and quality control. Circ Res. 2013;112:367–381. doi: 10.1161/CIRCRESAHA.112.268706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li J, et al. Enhancement of proteasomal function protects against cardiac proteinopathy and ischemia/reperfusion injury in mice. J Clin Invest. 2011;121:3689–3700. doi: 10.1172/JCI45709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhuiyan MS, et al. Enhanced autophagy ameliorates cardiac proteinopathy. J Clin Invest. 2013;123:5284–5297. doi: 10.1172/JCI70877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang J, et al. Myocardin sumoylation transactivates cardiogenic genes in pluripotent 10T1/2 fibroblasts. Mol Cell Biol. 2007;27:622–632. doi: 10.1128/MCB.01160-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim EY, et al. Expression of sumoylation deficient Nkx2.5 mutant in Nkx2.5 haploinsufficient mice leads to congenital heart defects. PLoS One. 2011;6:e20803. doi: 10.1371/journal.pone.0020803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kang X, et al. SUMO-specific protease 2 is essential for suppression of polycomb group protein-mediated gene silencing during embryonic development. Mol Cell. 2010;38:191–201. doi: 10.1016/j.molcel.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shah JJ, et al. Phase I study of the novel investigational nedd8-activating enzyme inhibitor pevonedistat (mln4924) in patients with relapsed/refractory multiple myeloma or lymphoma. Clin Cancer Res. 2016;22:34–43. doi: 10.1158/1078-0432.CCR-15-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fuller SJ, Sivarajah K, Sugden PH. ErbB receptors, their ligands, and the consequences of their activation and inhibition in the myocardium. J Mol Cell Cardiol. 2008;44:831–854. doi: 10.1016/j.yjmcc.2008.02.278. [DOI] [PubMed] [Google Scholar]
- 60.Jiao K, et al. An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes Dev. 2003;17:2362–2367. doi: 10.1101/gad.1124803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stanley EG, et al. Efficient Cre-mediated deletion in cardiac progenitor cells conferred by a 3'UTR-ires-Cre allele of the homeobox gene Nkx2-5. Int J Dev Biol. 2002;46:431–439. [PubMed] [Google Scholar]
- 62.Saito A, Nagase T. Hippo and TGF-β interplay in the lung field. Am J Physiol Lung Cell Mol Physiol. 2015;309:L756–L767. doi: 10.1152/ajplung.00238.2015. [DOI] [PubMed] [Google Scholar]
- 63.Wu N, et al. The Hippo signaling functions through the Notch signaling to regulate intrahepatic bile duct development in mammals. Lab Invest. 2017;97:843–853. doi: 10.1038/labinvest.2017.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heallen T, et al. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332:458–461. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu FX, Zhao B, Guan KL. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.He M, et al. New insights into posttranslational modifications of Hippo pathway in carcinogenesis and therapeutics. Cell Div. 2016;11:4. doi: 10.1186/s13008-016-0013-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li W, et al. Merlin/NF2 loss-driven tumorigenesis linked to CRL4(DCAF1)-mediated inhibition of the hippo pathway kinases Lats1 and 2 in the nucleus. Cancer Cell. 2014;26:48–60. doi: 10.1016/j.ccr.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paradis V, et al. Cullin7: A new gene involved in liver carcinogenesis related to metabolic syndrome. Gut. 2013;62:911–919. doi: 10.1136/gutjnl-2012-302091. [DOI] [PubMed] [Google Scholar]
- 69.Men X, Wang L, Yu W, Ju Y. Cullin7 is required for lung cancer cell proliferation and is overexpressed in lung cancer. Oncol Res. 2015;22:123–128. doi: 10.3727/096504014X14198596979742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Arai T, et al. Targeted disruption of p185/Cul7 gene results in abnormal vascular morphogenesis. Proc Natl Acad Sci USA. 2003;100:9855–9860. doi: 10.1073/pnas.1733908100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huber C, et al. Identification of mutations in CUL7 in 3-M syndrome. Nat Genet. 2005;37:1119–1124. doi: 10.1038/ng1628. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.