Significance
Auxin signaling components participate in ethylene-mediated inhibition of root elongation. However, the interplay between TIR1/AFB2-auxin-Aux/indole acetic acid (IAA) signaling and ethylene response remains to be elucidated in detail. In this study, we report an E3 ubiquitin ligase soil-surface rooting 1 (SOR1), which targets a noncanonical Aux/IAA protein OsIAA26 for 26S proteasome-mediated degradation. The E3 ligase activity of SOR1 can be repressed by the canonical Aux/IAA protein OsIAA9, which is the target of OsTIR1/AFB2. Our study identifies a potential regulator that modulates auxin-mediated ethylene response at the auxin signaling level.
Keywords: rice, root elongation, ethylene response, Aux/IAA protein, ubiquitination
Abstract
Plant hormones ethylene and auxin synergistically regulate plant root growth and development. Ubiquitin-mediated proteolysis of Aux/IAA transcriptional repressors by the E3 ubiquitin ligase SCFTIR1/AFB triggers a transcription-based auxin signaling. Here we show that rice (Oryza sativa L.) soil-surface rooting 1 (SOR1), which is a RING finger E3 ubiquitin ligase identified from analysis of a rice ethylene-insensitive mutant mhz2/sor1-2, controls root-specific ethylene responses by modulating Aux/IAA protein stability. SOR1 physically interacts with OsIAA26 and OsIAA9, which are atypical and canonical Aux/IAA proteins, respectively. SOR1 targets OsIAA26 for ubiquitin/26S proteasome-mediated degradation, whereas OsIAA9 protects the OsIAA26 protein from degradation by inhibiting the E3 activity of SOR1. Auxin promotes SOR1-dependent degradation of OsIAA26 by facilitating SCFOsTIR1/AFB2-mediated and SOR1-assisted destabilization of OsIAA9 protein. Our study provides a candidate mechanism by which the SOR1-OsIAA26 module acts downstream of the OsTIR1/AFB2-auxin-OsIAA9 signaling to modulate ethylene inhibition of root growth in rice seedlings.
Both the plant hormones ethylene and auxin regulate root growth. Recent studies in Arabidopsis have portrayed an ethylene–auxin interaction in regulating root growth, in which the ethylene elevates auxin accumulation and then triggers TIR1/AFB2-mediated signal transduction (1–4). The molecular details that ethylene promotes auxin accumulation have been studied comprehensively, involving ethylene-induced WEI2/ASA-, WEI7/ASB1-, and TAA1-dependent auxin biosynthesis (5, 6) as well as PIN2- and AUX1-facilitated auxin transport (7, 8) in roots. However, the mechanism underlying auxin signaling-mediated ethylene regulation of root growth still requires elucidation.
Rice is a monocotyledonous and semiaquatic plant, and ethylene plays essential roles in the adaptive growth of rice in water-saturated environments (9–11). Previously, we have screened for mutants with altered ethylene sensitivity to identify modulators in the regulation of ethylene responses. A set of mutants with an abnormal growth of the roots in the presence of ethylene was identified and named as mao hu zi (mhz, a Chinese name with an English meaning of cat whiskers) (12). Through map-based cloning, several corresponding genes have been identified, including MHZ7/OsEIN2 (12), MHZ6/OsEIL1 (13), MHZ5 (14), and MHZ4/OsABA4 (15). MHZ7/OsEIN2 and MHZ6/OsEIL1 encode proteins homologous to the EIN2 and EIN3 of the Arabidopsis ethylene signaling pathway, suggesting that the major components of ethylene signaling are conserved. Moreover, ethylene induces expressions of the carotenoid isomerase gene MHZ5 and the ABA biosynthesis gene MHZ4/OsABA4 to drive the metabolic flux into the ABA biosynthesis for regulation of rice root growth (14, 15). These analyses revealed that other plant hormones may modulate ethylene-regulated root growth in rice.
In this study, we report identification of MHZ2/SOR1 from analysis of a rice root-specific ethylene-insensitive mutant mhz2/sor1-2. MHZ2/SOR1 encodes an E3 ubiquitin ligase and affects root growth by modulating Aux/indole acetic acid (IAA) protein stability. SOR1 interacts with and ubiquitinates OsIAA26, an atypical Aux/IAA protein, for degradation. OsIAA9, a canonical Aux/IAA protein, connects the SOR1-OsIAA26 signal module to the OsTIR1/AFB2-mediated auxin signaling pathway by interacting with SOR1.
Results
Identification of Rice mhz2/sor1 Mutants.
A set of rice ethylene-response mutants (mhz) have been identified in our previous study and the mhz2 was analyzed in this study. The mhz2 seedlings showed root-specific defects in ethylene-inhibited root growth at a wide range of hormone concentrations (Fig. 1 A and B). The mhz2 seedlings also display defects in gravity response (Fig. 1A and SI Appendix, Fig. S1A) and soil-surface rooting phenotypes in the field (SI Appendix, Fig. S1B).
Fig. 1.
Characterization of mhz2/sor1 mutant and SOR1 protein. (A) Response of wild-type (WT) and sor1-2 etiolated seedlings to 10 ppm ethylene (ET). (Scale bar, 10 mm.) (B) Quantification of the ethylene response of WT and sor1-2 roots. The root lengths relative to the controls (0 ppm) are shown. Bars indicates SD (n ≥ 30). (C) Diagrams of SOR1 and its truncated version SOR1T used for E3 ligase activity studies. Conserved amino acids of RING domain and their positions in full-length protein are shown. (D) GST-SOR1T protein (∼45 kDa) displays E3 ubiquitin ligase activity in vitro. GST itself was used as a negative control. Coomassie Brilliant Blue (CBB) staining served as a loading control. (E) C127S mutation disrupts the E3 ligase activity of GST-SOR1T. (F) Phenotypic analysis of sor1-2 transformed with SOR1-GFP with or without mutations in the RING domain. SOR1SY harbors C127S and H148Y double mutations. (Scale bar, 10 mm.) (G) Quantification of the root ethylene response in WT, mutant, and rescued lines shown in F. The data show mean ± SD (n ≥ 30).
We cloned the MHZ2 gene using a map-based approach (SI Appendix, Fig. S2). As MHZ2 (Os04g01160) is the same as the rice SOR1 (16), we renamed the MHZ2 as SOR1. Our mutant mhz2 was also renamed as sor1-2 (SI Appendix, Fig. S2B) following the original mutant sor1-1 (16). Three additional mutant alleles were also identified (SI Appendix, Fig. S2B). SOR1 encodes a plant-specific RING-type protein (17), containing a nuclear localization signal (NLS), a RING domain, and a von Willebrand factor type A (VWA) domain (Fig. 1C and SI Appendix, Fig. S2C). SOR1 protein is conserved in other plant species (16) (SI Appendix, Fig. S2D) and is similar to Arabidopsis WAV3, which is an E3 ubiquitin ligase and regulates root gravitropism by affecting auxin response in Arabidopsis (18). To further confirm that the SOR1 mutation was responsible for the ethylene-insensitive phenotype, the full-length genomic DNA fragment of SOR1 was transformed into the sor1-2 mutant (SI Appendix, Fig. S2E), and the transgene-positive seedlings restored the ethylene-response phenotype and gravity response phenotype (SI Appendix, Fig. S2F). These results indicate that the Os04g01160 locus corresponds to the SOR1 gene.
We further examined the genetic relationship between SOR1 and MHZ7/OsEIN2, the central component of ethylene signaling in rice (12). The SOR1 mutation suppressed the short root phenotype of the OsEIN2-overexpressing seedlings under both normal and ethylene-treated conditions (SI Appendix, Fig. S3 A and B), suggesting that the SOR1 gene acts downstream of OsEIN2 to regulate root growth.
SOR1 Functions as an E3 Ubiquitin Ligase in Modulating Ethylene-Inhibited Root Growth.
Because proteins with a RING domain usually function as E3 ubiquitin ligases (18–20), we tested whether SOR1 also possesses E3 ligase activity. We expressed a truncated SOR1 version (amino acids 52–241) containing the RING domain, which is tagged with GST and referred to as GST-SOR1T (Fig. 1C), since expression of the recombinant full-length SOR1 is not successful. In the presence of E1, E2, and ubiquitin, the GST-SOR1T had the ability of self-ubiquitination (Fig. 1D). We further introduced mutations at the Cys-127 (C127S) and/or His-148 (H148Y) positions in the RING domain. The C127S mutation in the GST-SOR1T version completely disrupted the E3 ligase activity; however, the H148Y mutation does not affect the activity (Fig. 1E). These results indicate that the conserved residue Cys-127 is required for SOR1 E3 ligase activity.
To address whether E3 ligase activity is required for the SOR1-mediated root responses to ethylene and gravity, the 35S promoter-driven SOR1-coding sequence or its mutant (SOR1SY) harboring the C127S and H148Y double mutations was fused with green fluorescent protein (GFP) and transformed into the sor1-2 mutant. SOR1-GFP rescued the root sensitivity to both ethylene and gravity; however, SOR1SY-GFP did not rescue the ethylene-inhibited root growth at 1 ppm but partially rescued at 10 ppm ethylene, and SOR1SY-GFP largely restored the gravity response (Fig. 1 F and G and SI Appendix, Fig. S3 C and D). These results indicate that the SOR1 E3 ligase activity is substantially required for the ethylene inhibition of root growth especially at lower ethylene concentration.
The Ethylene-Insensitivity of sor1 Is Related to Alteration of Auxin Response.
In Arabidopsis, mutations of several proteins required for auxin functions (4–8, 21) have been identified to cause defects of ethylene response phenotypes. The defects of the sor1-2 roots in both ethylene and gravity responses suggest that SOR1 may mediate ethylene-inhibited root growth through auxin functions. We then tested whether auxin is required for ethylene response in rice roots and found that application of l-kynurenine (l-Kyn), an inhibitor of auxin biosynthesis (22), repressed the ethylene-induced inhibition of root growth (SI Appendix, Fig. S4 A and B). Furthermore, the effect of l-Kyn on ethylene-regulated root growth could be overcome by supplementation with the auxin analog naphthalene acetic acid (NAA, 20 nM) (SI Appendix, Fig. S4 C and D). These results indicate that auxin is required for ethylene inhibition of root growth.
To further examine whether TIR1-mediated nuclear auxin signaling is required for root response to ethylene, the MiR393a-overexpressed transgenic plants (23, 24), with the reduction of the auxin receptor gene OsTIR1/AFB2 expression were analyzed (SI Appendix, Fig. S5A). We found that these transgenic seedling roots displayed insensitivity to ethylene at a concentration of 1 ppm (SI Appendix, Fig. S5 B and C). Furthermore, the application of inhibitors (4-CI-PEO-IAA and auxinole) (25) blocking the interaction of the receptor and the Aux/IAA protein also reduced the ethylene inhibition of the rice roots (SI Appendix, Fig. S6). These results indicate that the auxin pathway is required for ethylene-inhibited root growth in rice, similar to previous findings in Arabidopsis.
We then explored whether the SOR1 mutation would change the auxin accumulation and/or sensitivity and found that sor1-2 roots had similar auxin accumulation and similar transporter gene expression in comparison with the wild-type (WT) roots (SI Appendix, Fig. S7 A and B). Moreover, supplementation of exogenous NAA, which is able to override the effect of l-Kyn in rice roots (SI Appendix, Fig. S4 C and D) and restore the root ethylene sensitivity in mutants defective in auxin production and transport in Arabidopsis (6, 26), did not restore the root ethylene response and gravity response in the sor1-2 mutant (SI Appendix, Fig. S7 C and D). The sor1-2 roots showed resistance to lower concentrations of NAA in comparison with the wild-type roots (Fig. 2A), suggesting that SOR1 mutation may alter auxin response in rice roots.
Fig. 2.
SOR1 mutation alters auxin response in rice root. (A) Quantification of the auxin response of WT and sor1-2 roots. The rates of root growth inhibition relative to the controls (0 µM) are shown. Bars indicate SD (n ≥ 30). (B) Ethylene inductions of OsIAA26, OsIAA9, and OsIAA20 fully require SOR1 function. Relative expression of Aux/IAA genes in WT and sor1-2 root tips was evaluated in response to 100 ppm ethylene. (C) Auxin inductions of these genes partially require SOR1 function. NAA (10 µM) was used for treatment. (D) Ethylene inductions of OsIAA26, OsIAA9, and OsIAA20 require auxin function. The auxin biosynthesis inhibitor l-Kyn (10 µM) was used for the treatment. The data show mean ± SE. Four independent biological repeats were performed and analyzed. All of the mRNA levels were normalized to OsACTIN2. (E–G) Venn diagram shows overlaps between genes induced by NAA or by ethylene (ET) treatment and the genes related by SOR1 or OsEIN2 in rice root tips. The genes regulated more than twofold were used for analysis. ET EIGs, ET ethylene-induced genes; NAA NIGs, NAA-induced genes; SOR1 Reg EIGs, SOR1-regulated ethylene-induced genes; SOR1 Reg NIGs, SOR1-regulated NAA-induced genes; and OsEIN2 Reg EIGs, OsEIN2-regulated ethylene-induced genes.
From our microarray and transcriptome data of the root tips treated with ethylene (National Center for Biotechnology Information: GSE51152 and SRP041468), three Aux/IAA genes, OsIAA26, OsIAA9, and OsIAA20, relevant to the auxin response were identified as ethylene-responsive genes, and the ethylene-induced expression of these genes fully depended on SOR1 functions (Fig. 2B). However, only around half of auxin-induced expression of these three genes depended on the SOR1 function (Fig. 2C). These results imply that SOR1 is fully required for the ethylene response but only partially required for the auxin response at the molecular level in roots. Moreover, inhibition of auxin synthesis by l-Kyn application or repression of auxin signaling by knockdown of auxin receptor genes OsTIR1/AFB2 also blocked ethylene-induced expression of OsIAA26, OsIAA9, and OsIAA20 (Fig. 2D and SI Appendix, Fig. S5D). Collectively, these results suggest that SOR1 may modulate root responses to ethylene through affecting a portion of the auxin response.
Transcriptome analysis (BIG Data Center: CRA000688) was further performed to dissect SOR1-modulated gene expression in root tips. We found that SOR1 controlled ∼77% of the OsEIN2-mediated ethylene-induced genes. It also mediated ∼70% of auxin-induced genes, among which ∼56% were ethylene-induced genes (Fig. 2 E–G). These analyses imply that SOR1 affects ethylene response genes, among which a subset belongs to auxin response genes.
SOR1 Interacts with and Ubiquitinates OsIAA26.
Since mutation of SOR1 leads to deficiency of auxin-dependent ethylene response in roots (Fig. 2 A–D), and the reduced root responses of the MiR393aOE (SI Appendix, Fig. S5B) and sor1-2/SOR1SY-GFP (Fig. 1F) seedlings to 1 ppm ethylene were very similar, we hypothesized that SOR1 may be involved in the OsTIR1/AFB2-mediated auxin signaling pathway. There are 31 members in the rice Aux/IAA family, and most of these have four conserved domains, i.e., domains I–IV. However, a few of these do not have conserved domain II (27, 28). Among SOR1-dependent ethylene-induced Aux/IAA genes, the OsIAA9 and OsIAA20 encode Aux/IAA proteins with conserved domains, whereas the OsIAA26 encodes an atypical Aux/IAA protein with substitutions in domain II (Fig. 3A). OsIAA26 has canonical domain I contributing to transcriptional regulation and shows transcriptional repression activity (SI Appendix, Fig. S8). In the presence or absence of IAA, SOR1 could interact with both OsIAA26 and OsIAA9, but not OsIAA20, in a yeast two-hybrid assay. In contrast, in the presence of IAA, the auxin receptor OsTIR1 interacted with OsIAA20, and the auxin receptor OsAFB2 interacted with both OsIAA9 and OsIAA20 (Fig. 3A).
Fig. 3.
SOR1 interacts with and ubiquitinates OsIAA26 protein. (A) SOR1 interacts with OsIAA26 and OsIAA9 in a yeast two-hybrid assay. Yeast transformants were grown on the DDO media [synthetic defined (SD) –Trp −Leu] and on the DQO/X/A media (SD −Trp −Leu −His −Ade plus X-α-gal and aureobasidin A) with or without 100 µM IAA. Greenish blue indicates positive interactions. OsIAA9 and OsIAA20 are canonical Aux/IAA proteins with a conserved domain II. OsIAA26 is a noncanonical Aux/IAA protein. Interactions of the three Aux/IAA proteins with auxin receptors OsTIR1 and OsAFB2 were also tested. (B) The truncated SOR1 version containing the RING domain (GST-SOR1T) ubiquitinates OsIAA26, but not OsIAA9. Asterisks indicate ubiquitinated MBP-OsIAA26 proteins. (C) Ubiquitination of OsIAA26 depends on the intact RING finger of SOR1. (D) SOR1 associates with OsIAA26 in a pull-down assay. The assay was performed using MBP-OsIAA26 recombinant protein and extracts were prepared from tobacco leaves expressing Myc-SOR1 protein. (E) SOR1C127S, but not SOR1, can be detected to interact with OsIAA26 in the BiFC assay. Yellow fluorescence indicates positive interactions. (Scale bar, 50 µm.)
We further investigated whether SOR1 can ubiquitinate OsIAA26 and OsIAA9. We performed in vitro ubiquitination experiments using the GST-SOR1T and the OsIAA26 and OsIAA9 proteins tagged with maltose-binding protein (MBP). While MBP-OsIAA9 was not ubiquitinated, MBP-OsIAA26 can be ubiquitinated by SOR1T, and the C127S mutation in the RING domain of SOR1T disrupted OsIAA26 ubiquitination (Fig. 3 B and C).
The association of SOR1 with OsIAA26 is further supported by a pull-down assay. Full-length SOR1 proteins expressed in tobacco leaves can be pulled down by incubation with MBP-OsIAA26 proteins (Fig. 3D). Furthermore, we employed bimolecular fluorescence complementation (BiFC) assays in tobacco leaf cells to check the interaction between SOR1 and OsIAA26. The yellow fluorescence that represents a positive interaction can be detected in the combination of OsIAA26 and SOR1C127S, a version without E3 ligase activity (Fig. 3E). However, the combination between normal SOR1 and OsIAA26 did not generate a positive interaction signal, probably due to rapid degradation of the ubiquitinated proteins.
SOR1 Targets OsIAA26 for Degradation.
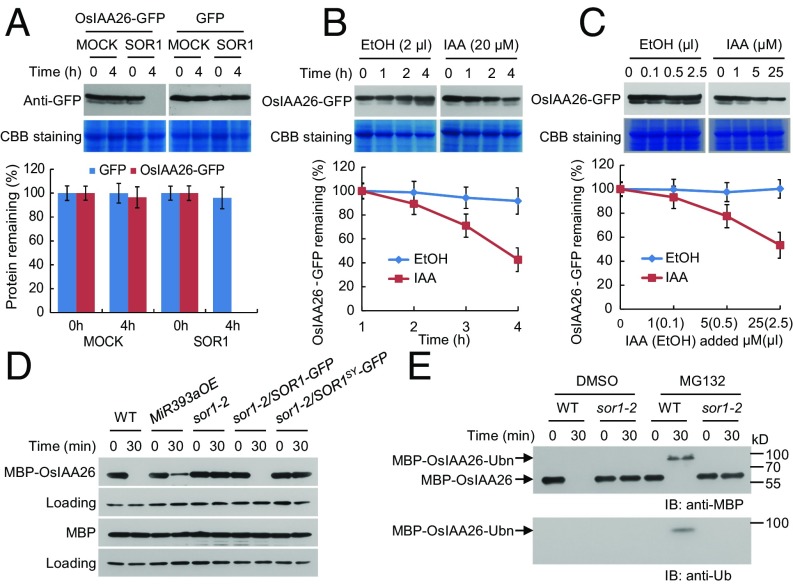
To reveal whether OsIAA26 can be targeted directly for degradation by SOR1, the OsIAA26-GFP and SOR1 were coexpressed in tobacco leaves to perform in vitro degradation assays. Results showed that SOR1 promoted degradation of the OsIAA26-GFP (Fig. 4A). Moreover, the atypical Aux/IAA protein OsIAA26 can be destabilized by auxin with different incubation times or with different concentrations (Fig. 4 B and C), supporting that Aux/IAA proteins with substitutions in domain II could also be degraded slowly by auxin treatment (29). These results indicate that SOR1 facilitates OsIAA26 degradation and auxin may play a positive role in this process.
Fig. 4.
OsIAA26 is a substrate of SOR1. (A) OsIAA26-GFP protein levels in the absence or presence of SOR1. Crude plant protein extracts from tobacco leaves coexpressing SOR1 and OsIAA26-GFP or GFP were incubated at 30 °C for degradation assays. Coomassie Brilliant Blue (CBB) staining served as a loading control. Three independent biological repeats were performed and analyzed, and the relative protein levels were measured using ImageJ software. (B) OsIAA26-GFP protein levels in response to IAA at different time points. Ethanol solvent (EtOH) was used as a control. (C) OsIAA26-GFP protein levels in response to different concentrations of IAA. Crude plant protein extracts from tobacco leaves expressing OsIAA26-GFP were incubated with IAA for 4 h. (D) Stability of MBP-OsIAA26 in protein extracts from various indicated rice seedling roots. E. coli-expressed MBP-OsIAA26 proteins were incubated with different plant protein extracts prepared from indicated materials. MBP itself was used as a control. (E) Analysis of MBP-OsIAA26 ubiquitination in protein extracts prepared from WT and sor1-2 with or without MG132 (100 µM) treatment. DMSO was used as a control. MBP-OsIAA26-Ubn indicates ubiquitinated MBP-OsIAA26.
We further employed a cell-free degradation assay system (30–32) to explore whether SOR1 regulates OsIAA26 degradation in rice. In these assays, Escherichia coli-expressed MBP-OsIAA26 proteins were incubated with protein extracts from various rice materials for different times and the remaining MBP-OsIAA26 levels were examined. We found that the loss of SOR1 in the sor1-2 mutant or the disruption of the SOR1 E3 ligase activity in the sor1-2/SOR1SY-GFP transgenic plants prevented the degradation of the MBP-OsIAA26 protein (Fig. 4D). The reduction of auxin receptors in the MiR393OE transgenic plants also slightly prevented MBP-OsIAA26 degradation (Fig. 4D). We further used the MG132 to inhibit the 26S proteasome-mediated protein degradation and checked the MBP-OsIAA26 ubiquitination status. We found that, in the presence of MG132, the MBP-OsIAA26 is polyubiquitinated in the WT protein extracts but not ubiquitinated in the sor1-2 extracts (Fig. 4E). These results indicate that SOR1 is responsible for the MBP-OsIAA26 ubiquitination and facilitates its degradation by the 26S proteasome pathway, and auxin receptors OsTIR1/AFB2 also play some roles in the OsIAA26 degradation process.
OsIAA9 Interacts with SOR1 and Inhibits Its E3 Ligase Activity.
From the above experiments, we found that SOR1 could interact with both OsIAA26 and OsIAA9 in the yeast two-hybrid assays (Fig. 3A); however, the SOR1 could not ubiquitinate OsIAA9 (Fig. 3B). We further studied the interaction of SOR1 and OsIAA9 by BiFC assay, and found that they could also associate (Fig. 5A). However, unlike the case for SOR1 and OsIAA26, the interaction signal between SOR1 and OsIAA9 was not altered when SOR1 had the E3 ubiquitin ligase activity. Using the pull-down assays, we further found that OsIAA9, different from OsIAA26, interacted with truncated SOR1 version SOR1T (Fig. 5B and SI Appendix, Fig. S9), and this interaction required an intact RING finger (SI Appendix, Fig. S9). These results indicated that OsIAA9 associates with SOR1 at the RING domain, possibly affecting the E3 ubiquitin ligase activity of SOR1.
Fig. 5.
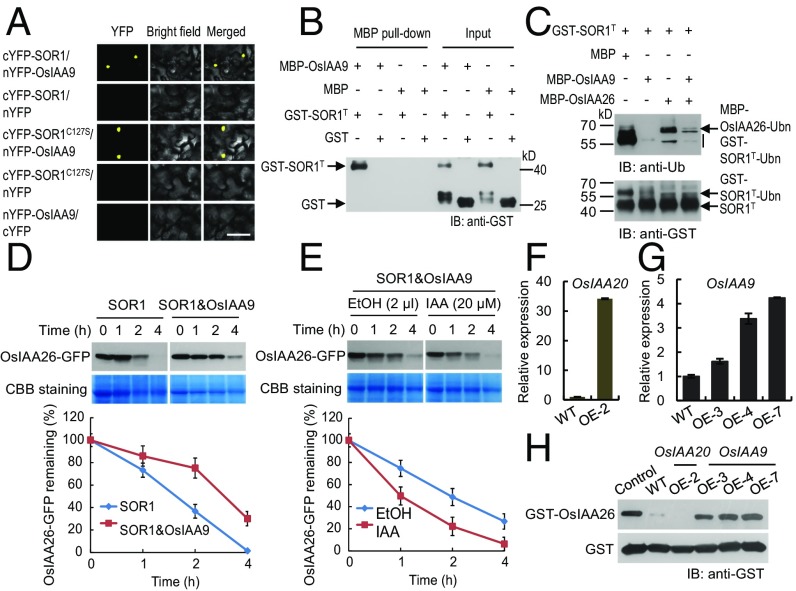
OsIAA9 protects OsIAA26 from SOR1-mediated degradation. (A) SOR1 interacts with OsIAA9 in a BiFC assay. Yellow fluorescence indicates positive interactions. (Scale bar, 50 µm.) (B) The truncated SOR1 version containing the RING domain (GST-SOR1T) interacts with MBP-OsIAA9 in a pull-down assay. (C) OsIAA9 inhibits the E3 ligase activity of GST-SOR1T in vitro. MBP was used as a control. (D) OsIAA9 reduces SOR1-mediated degradation of OsIAA26. Assays were performed using crude plant protein extracts from tobacco leaves coexpressing SOR1 and OsIAA26-GFP, with or without OsIAA9. Coomassie Brilliant Blue (CBB) staining served as a loading control. Three independent biological repeats were performed and analyzed, and the relative protein levels were measured using ImageJ software. (E) Degradation of OsIAA26 in the presence of both SOR1 and OsIAA9 in response to auxin. Assays were performed using protein extracts from tobacco leaves coexpressing SOR1, OsIAA26-GFP, and OsIAA9. Ethanol solvent (EtOH) was used as a control. (F–G) Real-time qPCR analysis for expression of OsIAA20 and OsIAA9 in their corresponding overexpressed transgenic lines. The mRNA levels were normalized to OsACTIN2. (H) Stability of GST-OsIAA26 in protein extracts from indicated transgenic rice seedling roots. E. coli-expressed GST-OsIAA26 proteins were incubated with plant protein extracts prepared from indicated materials. GST itself was used as a control.
To test whether OsIAA9 would affect SOR1 E3 ligase activity, we performed in vitro ubiquitination assays, and found that the OsIAA9 can inhibit the E3 ubiquitin ligase activity of SOR1 and thus prevent SOR1-mediated OsIAA26 ubiquitination (Fig. 5C). We further tested the OsIAA9 effect on SOR1-mediated OsIAA26 degradation in in vitro degradation assays using tobacco leaves transiently coexpressing SOR1, OsIAA26, and OsIAA9, and found that OsIAA9 reduced the SOR1-mediated degradation of OsIAA26 (Fig. 5D). When the auxin IAA was included in the assay, the OsIAA9-dependent stabilization of OsIAA26-GFP was affected (Fig. 5E). Moreover, we found that OsIAA26 protein was more stable in OsIAA9-overexpressed rice than in WT or OsIAA20-overexpressed rice (Fig. 5 F–H). These results indicate that the OsIAA9 can protect OsIAA26 from SOR1-mediated degradation by inhibiting the E3 ligase activity of SOR1.
OsIAA9 Degradation Is Mediated by OsTIR1/AFB2 and Facilitated by SOR1.
Because OsIAA9 is a canonical Aux/IAA protein (Fig. 3A), and OsIAA9 regulates OsIAA26 stability (Fig. 5 D, E, and H), we propose that auxin may regulate the SOR1-dependent degradation of OsIAA26 through possible recruitment of the OsIAA9 protein from SOR1 to OsTIR1/AFB2 for ubiquitination and degradation. Pull-down assays were performed and the results showed that the interaction between OsIAA9 and auxin receptor OsAFB2 is enhanced in the presence of NAA (Fig. 6A).
Fig. 6.
Regulation of OsIAA9 stability and a working model for SOR1. (A) OsIAA9 interacts with OsAFB2 in the presence of auxin. Crude protein extracts prepared from tobacco leaves expressing OsAFB2 were used to do MBP pull-down assays. (B) Stability of MBP-OsIAA9 in protein extracts from various rice materials. E. coli-expressed MBP-OsIAA9 proteins were incubated with different plant protein extracts prepared from indicated materials. MBP itself was used as a control. (C) A working model for SOR1 functions in auxin-mediated ethylene response. In the absence of ethylene and auxin (Left), OsIAA9 inhibits the E3 ligase activity of SOR1 and thus stabilizes OsIAA26, facilitating normal root elongation. In the presence of ethylene and auxin (Right), ethylene-triggered IAA accumulation enhanced OsAFB2-mediated degradation of OsIAA9, with the help of SOR1. OsIAA9 removal released the E3 ligase activity of SOR1 and then promoted SOR1-mediated degradation of OsIAA26. The normal root elongation is thus repressed.
We also examined the stability of OsIAA9 when incubated with protein extracts from various rice materials. The MBP-OsIAA9 can be degraded in the WT protein extracts. However, when the auxin receptor gene OsTIR1/AFB2 expression was reduced in the MiR393aOE extracts, the MBP-OsIAA9 protein showed increased stability (Fig. 6B). Similarly, the MBP-OsIAA9 in the sor1-2 extracts also showed increased longevity (Fig. 6B). In the sor1-2/SOR1-GFP plant extracts, the MBP-OsIAA9 degradation was recovered. It is interesting to note that when the SOR1 E3 ligase activity was disrupted in the sor1-2/SOR1SY-GFP, the MBP-OsIAA9 degradation remained (Fig. 6B). Together, these results indicate that degradation of OsIAA9 protein is mediated by auxin receptor OsTIR1/AFB2 in rice, and OsIAA9 degradation is also facilitated by the presence of SOR1; however, the E3 ligase activity of SOR1 is not required for OsIAA9 degradation.
Alteration of OsIAA9 and OsIAA26 Expression Affects Ethylene-Regulated Root Growth.
Because OsIAA26 is ubiquitinated by SOR1 for degradation and OsIAA9 affects this process (Figs. 3–5), we further tested whether OsIAA26 and OsIAA9 have any roles in ethylene inhibition of root growth. The OsIAA26- and OsIAA9-overexpressed transgenic plants were generated and three higher expressers for each transgene were analyzed (SI Appendix, Fig. S10 A and B). Both the OsIAA26- and OsIAA9-overexpressed seedlings showed reduced ethylene sensitivity in roots than the wild-type seedlings did (SI Appendix, Fig. S10 C and D). Furthermore, mutations of OsIAA9 and OsIAA26 were generated by genome editing using the CRISPR/Cas9 system to further study their roles in root ethylene response (SI Appendix, Fig. S11A). Both Osiaa9 and Osiaa26 mutants showed similar ethylene response to WT (SI Appendix, Fig. S11 B and C). However, mutation of the OsIAA26 protein caused root wave growth in the presence of 10 ppm ethylene (SI Appendix, Fig. S11D). These results suggest that both OsIAA26 and OsIAA9 may function in the reduction of ethylene effects on root growth, and other Aux/IAA proteins may also be involved in ethylene-regulated root growth.
Discussion
E3 ubiquitin ligase-mediated protein degradation plays a central role in ethylene and auxin signaling. The stability of ethylene signaling protein EIN2 is modulated by the two F-box proteins ETP1/2 (33), while the EIN3 is regulated by another two F-box proteins EBF1/2 (34–36). The auxin receptor is a small family of related F-box proteins (37), which triggers nuclear auxin signaling by targeting transcriptional repressor Aux/IAA proteins for degradation (38). Here we find that SOR1, a RING finger E3 ubiquitin ligase (Fig. 1 C–E), ubiquitinates a noncanonical Aux/IAA protein OsIAA26 for degradation (Figs. 3 and 4), and the OsTIR1/AFB2-auxin-OsIAA9 signaling affects this process (Figs. 5 and 6 A and B). Our study identifies a potentially unique component of auxin signaling to regulate ethylene-inhibited root growth.
Noncanonical Aux/IAA proteins can regulate auxin-related phenotypes in Arabidopsis (39); however, their protein stability cannot be directly modulated by SCFTIR1 E3 ubiquitin ligase (28, 40). In the present study, we discovered that SOR1 ubiquitinates a noncanonical Aux/IAA protein OsIAA26, for degradation (Figs. 3 and 4). In the in vitro assay, the OsIAA26 appeared to be monoubiquitinated by the truncated SOR1 version SOR1T (Fig. 3 B and C). However, in the rice protein extracts, the OsIAA26 can be polyubiquitinated by full-length SOR1 as judged from the protein size (Fig. 4E). This difference may be due to the different assay systems used or different SOR1 versions involved. Interaction assays showed that OsIAA26 interacts with full-length SOR1 but not with the truncated SOR1T (Fig. 3D and SI Appendix, Fig. S9); however, the SOR1T still ubiquitinates OsIAA26. This fact may suggest that the extra sequences outside the RING finger likely facilitate the ubiquitination process. Other possibilities could not be excluded.
Moreover, auxin also regulates the OsIAA26 protein degradation likely by OsTIR1/AFB2 receptors through SOR1, because in the MiR393aOE plants with reduced OsTIR1/AFB2 expression, the OsIAA26 can be partially stabilized (Fig. 4D). This fact may be explained by the stabilization of OsIAA9 in MiR393aOE plants, which leads to SOR1 inactivation and OsIAA26 stabilization (Figs. 5 C, D, and H and 6 A and B). It is worth noting that among six noncanonical Aux/IAA proteins in rice, we only studied the OsIAA26, whose gene expression is induced by ethylene and the induction fully depends on SOR1 (Fig. 2B). Other noncanonical and/or canonical Aux/IAA proteins should be examined to test whether they are also the substrates of SOR1. It is also interesting to study whether SOR1 modulates other components of ethylene/auxin signaling pathways.
Aux/IAA family proteins usually function as transcriptional repressors (41, 42) and bind to ARF transcription factors to repress their transcriptional activity (43). Previous study has shown that HaIAA27 (44), one of the Aux/IAA proteins in sunflower, can interact with HaHSFA9, a non–ARF-type transcription factor, and represses its transcriptional activity. In this study, we show that OsIAA9, a canonical Aux/IAA protein (Figs. 3A and 6 A and B), can function as a repressor for inhibition of the E3 ligase activity of SOR1 (Fig. 5C). Considering that SOR1 can interact with both OsIAA26 and OsIAA9 but only ubiquitinate the former, the OsIAA9 may play some inhibitory roles in the SOR1 E3 ubiquitin ligase activity toward its substrate OsIAA26. This conclusion is further supported by the fact that OsIAA9 protects OsIAA26 from degradation (Fig. 5 D and H). This finding widens our understanding of the specific roles of each Aux/IAA protein. The mechanism involving the interplay of the three proteins is still not clear and requires further investigation.
It should be noted that although SOR1 cannot ubiquitinate OsIAA9 (Fig. 3B), it assists OsIAA9 degradation, and this assistance does not require E3 ubiquitin ligase activity (Fig. 6B). Considering that OsIAA9 can also interact with OsAFB2 in the presence of auxin and be targeted by these receptors for degradation (Figs. 3A and 6 A and B), SOR1 may act as a facilitator for OsIAA9 degradation in this process. The VWA domain of SOR1 may function to recognize and promote ubiquitinated OsIAA9 for 26S proteasome-mediated degradation. This speculation may be supported by a previous study. Verma et al. (45) reported that, during delivery of ubiquitinated substrates to the 26S proteasome, deubiquitination and degradation of substrates delivered by Rad23 require a facilitator activity within the VWA domain of Rpn10. Further studies should clarify the possible functions of the SOR1 VWA domain in the process of Aux/IAA degradation.
While our study finds that ethylene affects root growth through SOR1-mediated auxin signaling, ethylene may also inhibit the second rate-limiting step at the YUCCA gene of auxin biosynthesis to confer auxin levels suboptimal for root cell elongation as in the cases of Brachypodium (46) and rice (47), providing an alternative mechanism of ethylene–auxin interaction.
From our current studies, we propose a model in which SOR1 acts as an E3 ligase in regulating the rice root response to ethylene by modulating Aux/IAA protein stability (Fig. 6C). In the absence of ethylene, the canonical Aux/IAA protein OsIAA9 associates with SOR1 and blocks the SOR1-mediated ubiquitin-related degradation of OsIAA26. In the presence of ethylene, ethylene-triggered auxin accumulation facilitates the interaction of the auxin receptors OsTIR1/AFB2 with OsIAA9, leading to OsIAA9 degradation. A decrease of the OsIAA9 protein releases the E3 ligase activity of SOR1, thereby targeting OsIAA26 for degradation via the ubiquitin/26S proteasome pathway.
Materials and Methods
Details of the experimental programs for ethylene response assay, quantitative real-time PCR analysis, recombinant protein purification, yeast two-hybrid assay, pull-down assay, BiFC assay, in vitro ubiquitination assay, and cell-free degradation assay are provided in SI Appendix, SI Materials and Methods. The primers used in this study are listed in SI Appendix, Tables S1 and S2.
Supplementary Material
Acknowledgments
We thank M. Y. Zhu (Zhejiang University) and M. Y. Zhang [South China Botanical Garden, Chinese Academy of Sciences (CAS)] for providing the seeds of MiR393OE; K. Hayashi (Okayama University of Science) for providing the chemicals PEO-IAA, 4-CI-PEO-IAA, and auxinole; and J. F. Chu (Center for Plant Hormone Determination, Institute of Genetics and Developmental Biology, CAS) for IAA measurement. This work is supported by the National Natural Science Foundation of China (Grant 31530004), the 973 Project (Grant 2015CB755702), and the State Key Laboratory of Plant Genomics.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE51152 and SRP041468) and BIG Data Center: CRA000688.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719387115/-/DCSupplemental.
References
- 1.Swarup R, et al. Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell. 2007;19:2186–2196. doi: 10.1105/tpc.107.052100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stepanova AN, Yun J, Likhacheva AV, Alonso JM. Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell. 2007;19:2169–2185. doi: 10.1105/tpc.107.052068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Růzicka K, et al. Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell. 2007;19:2197–2212. doi: 10.1105/tpc.107.052126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alonso JM, et al. Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc Natl Acad Sci USA. 2003;100:2992–2997. doi: 10.1073/pnas.0438070100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stepanova AN, Hoyt JM, Hamilton AA, Alonso JM. A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell. 2005;17:2230–2242. doi: 10.1105/tpc.105.033365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stepanova AN, et al. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell. 2008;133:177–191. doi: 10.1016/j.cell.2008.01.047. [DOI] [PubMed] [Google Scholar]
- 7.Luschnig C, Gaxiola RA, Grisafi P, Fink GR. EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 1998;12:2175–2187. doi: 10.1101/gad.12.14.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marchant A, et al. AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J. 1999;18:2066–2073. doi: 10.1093/emboj/18.8.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu K, et al. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature. 2006;442:705–708. doi: 10.1038/nature04920. [DOI] [PubMed] [Google Scholar]
- 10.Hattori Y, et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature. 2009;460:1026–1030. doi: 10.1038/nature08258. [DOI] [PubMed] [Google Scholar]
- 11.Ma B, Chen SY, Zhang JS. Ethylene signaling in rice. Chin Sci Bull. 2010;55:2204–2210. [Google Scholar]
- 12.Ma B, et al. Identification of rice ethylene-response mutants and characterization of MHZ7/OsEIN2 in distinct ethylene response and yield trait regulation. Mol Plant. 2013;6:1830–1848. doi: 10.1093/mp/sst087. [DOI] [PubMed] [Google Scholar]
- 13.Yang C, et al. MAOHUZI6/ETHYLENE INSENSITIVE3-LIKE1 and ETHYLENE INSENSITIVE3-LIKE2 regulate ethylene response of roots and coleoptiles and negatively affect salt tolerance in rice. Plant Physiol. 2015;169:148–165. doi: 10.1104/pp.15.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin CC, et al. Ethylene responses in rice roots and coleoptiles are differentially regulated by a carotenoid isomerase-mediated abscisic acid pathway. Plant Cell. 2015;27:1061–1081. doi: 10.1105/tpc.15.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma B, et al. Ethylene-induced inhibition of root growth requires abscisic acid function in rice (Oryza sativa L.) seedlings. PLoS Genet. 2014;10:e1004701. doi: 10.1371/journal.pgen.1004701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanzawa E, et al. Isolation of a novel mutant gene for soil-surface rooting in rice (Oryza sativa L.) Rice (N Y) 2013;6:30. doi: 10.1186/1939-8433-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stone SL, et al. Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 2005;137:13–30. doi: 10.1104/pp.104.052423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakai T, et al. The wavy growth 3 E3 ligase family controls the gravitropic response in Arabidopsis roots. Plant J. 2012;70:303–314. doi: 10.1111/j.1365-313X.2011.04870.x. [DOI] [PubMed] [Google Scholar]
- 19.Lorick KL, et al. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie Q, et al. SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature. 2002;419:167–170. doi: 10.1038/nature00998. [DOI] [PubMed] [Google Scholar]
- 21.Roman G, Lubarsky B, Kieber JJ, Rothenberg M, Ecker JR. Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: Five novel mutant loci integrated into a stress response pathway. Genetics. 1995;139:1393–1409. doi: 10.1093/genetics/139.3.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He W, et al. A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell. 2011;23:3944–3960. doi: 10.1105/tpc.111.089029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bian H, et al. Distinctive expression patterns and roles of the miRNA393/TIR1 homolog module in regulating flag leaf inclination and primary and crown root growth in rice (Oryza sativa) New Phytol. 2012;196:149–161. doi: 10.1111/j.1469-8137.2012.04248.x. [DOI] [PubMed] [Google Scholar]
- 24.Xia K, et al. OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS One. 2012;7:e30039. doi: 10.1371/journal.pone.0030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayashi K, et al. Rational design of an auxin antagonist of the SCF(TIR1) auxin receptor complex. ACS Chem Biol. 2012;7:590–598. doi: 10.1021/cb200404c. [DOI] [PubMed] [Google Scholar]
- 26.Rahman A, Amakawa T, Goto N, Tsurumi S. Auxin is a positive regulator for ethylene-mediated response in the growth of Arabidopsis roots. Plant Cell Physiol. 2001;42:301–307. doi: 10.1093/pcp/pce035. [DOI] [PubMed] [Google Scholar]
- 27.Jain M, et al. Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa) Funct Integr Genomics. 2006;6:47–59. doi: 10.1007/s10142-005-0005-0. [DOI] [PubMed] [Google Scholar]
- 28.Calderón Villalobos LI, et al. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol. 2012;8:477–485. doi: 10.1038/nchembio.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dreher KA, Brown J, Saw RE, Callis J. The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell. 2006;18:699–714. doi: 10.1105/tpc.105.039172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- 31.Kim HJ, et al. Control of plant germline proliferation by SCF(FBL17) degradation of cell cycle inhibitors. Nature. 2008;455:1134–1137. doi: 10.1038/nature07289. [DOI] [PubMed] [Google Scholar]
- 32.Wang F, et al. Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell. 2009;21:2378–2390. doi: 10.1105/tpc.108.065433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiao H, Chang KN, Yazaki J, Ecker JR. Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev. 2009;23:512–521. doi: 10.1101/gad.1765709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo H, Ecker JR. Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell. 2003;115:667–677. doi: 10.1016/s0092-8674(03)00969-3. [DOI] [PubMed] [Google Scholar]
- 35.Potuschak T, et al. EIN3-dependent regulation of plant ethylene hormone signaling by two arabidopsis F box proteins: EBF1 and EBF2. Cell. 2003;115:679–689. doi: 10.1016/s0092-8674(03)00968-1. [DOI] [PubMed] [Google Scholar]
- 36.Gagne JM, et al. Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc Natl Acad Sci USA. 2004;101:6803–6808. doi: 10.1073/pnas.0401698101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dharmasiri N, et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell. 2005;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 38.Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature. 2001;414:271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- 39.Sato A, Yamamoto KT. Overexpression of the non-canonical Aux/IAA genes causes auxin-related aberrant phenotypes in Arabidopsis. Physiol Plant. 2008;133:397–405. doi: 10.1111/j.1399-3054.2008.01055.x. [DOI] [PubMed] [Google Scholar]
- 40.Shimizu-Mitao Y, Kakimoto T. Auxin sensitivities of all Arabidopsis Aux/IAAs for degradation in the presence of every TIR1/AFB. Plant Cell Physiol. 2014;55:1450–1459. doi: 10.1093/pcp/pcu077. [DOI] [PubMed] [Google Scholar]
- 41.Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ. AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell. 2001;13:2809–2822. doi: 10.1105/tpc.010289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tiwari SB, Hagen G, Guilfoyle TJ. Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell. 2004;16:533–543. doi: 10.1105/tpc.017384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salehin M, Bagchi R, Estelle M. SCFTIR1/AFB-based auxin perception: Mechanism and role in plant growth and development. Plant Cell. 2015;27:9–19. doi: 10.1105/tpc.114.133744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carranco R, Espinosa JM, Prieto-Dapena P, Almoguera C, Jordano J. Repression by an auxin/indole acetic acid protein connects auxin signaling with heat shock factor-mediated seed longevity. Proc Natl Acad Sci USA. 2010;107:21908–21913. doi: 10.1073/pnas.1014856107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 46.Pacheco-Villalobos D, Sankar M, Ljung K, Hardtke CS. Disturbed local auxin homeostasis enhances cellular anisotropy and reveals alternative wiring of auxin-ethylene crosstalk in Brachypodium distachyon seminal roots. PLoS Genet. 2013;9:e1003564. doi: 10.1371/journal.pgen.1003564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yin CX, et al. Endogenous auxin is required but supraoptimal for rapid growth of rice (Oryza sativa L.) seminal roots, and auxin inhibition of rice seminal root growth is not caused by ethylene. J Plant Growth Regul. 2011;30:20–29. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.