Significance
Comparing gene regulatory programs throughout developmental time and across species allows us to reveal their constraints and flexibilities. Here we study the organism Acrobeloides nanus, a clade IV nematode, by sequencing its genome, identifying its developmental transcriptome, and studying the patterns of embryonic conservation and divergence through a comparison with Caenorhabditis elegans. The gene regulatory programs of these two species show many differences early in development, but significantly converge at the middevelopmental transition. Moreover, the genes most conserved in their expression during development arose at the dawn of the superphylum Ecdysozoa. Our work shows that variation is not evenly distributed but, rather, that developmental and evolutionary constraints act to shape gene regulatory programs.
Keywords: evolution, development, gene expression, Nematoda, developmental constraints
Abstract
The evolution of development has been studied through the lens of gene regulation by examining either closely related species or extremely distant animals of different phyla. In nematodes, detailed cell- and stage-specific expression analyses are focused on the model Caenorhabditis elegans, in part leading to the view that the developmental expression of gene cascades in this species is archetypic for the phylum. Here, we compared two species of an intermediate evolutionary distance: the nematodes C. elegans (clade V) and Acrobeloides nanus (clade IV). To examine A. nanus molecularly, we sequenced its genome and identified the expression profiles of all genes throughout embryogenesis. In comparison with C. elegans, A. nanus exhibits a much slower embryonic development and has a capacity for regulative compensation of missing early cells. We detected conserved stages between these species at the transcriptome level, as well as a prominent middevelopmental transition, at which point the two species converge in terms of their gene expression. Interestingly, we found that genes originating at the dawn of the Ecdysozoa supergroup show the least expression divergence between these two species. This led us to detect a correlation between the time of expression of a gene and its phylogenetic age: evolutionarily ancient and young genes are enriched for expression in early and late embryogenesis, respectively, whereas Ecdysozoa-specific genes are enriched for expression during the middevelopmental transition. Our results characterize the developmental constraints operating on each individual embryo in terms of developmental stages and genetic evolutionary history.
An insight regarding the embryo that continues to provide understanding is the notion that evolutionary constraints have shaped development (1, 2). Indeed, the field of evolutionary developmental biology posits that these two concepts are intertwined and mutually illuminating (3). The comparative approach of analyzing distant species has shed light on many processes, including the evolution and development of the bilaterian body plan by HOX genes (4, 5). Although it might be naively expected that comparing two closely related species would result in only a few genomic and transcriptomic changes, the last two decades have provided plenty of evidence that the genome and its phenotypes are extremely plastic (6, 7). These changes are manifest, but they are not random, and we require an understanding of how constraints act on possible genomic changes.
Transcriptomics methods, beginning with DNA microarrays, later followed by RNA-Seq (8, 9), have been transformative for biological research, as they afford a comprehensive view of gene expression. Whereas previous methods examined individual genes, with the simultaneous knowledge of the expression of all the genes in a given sample, a highly resolved state of the system emerged, enabling the study of cellular, developmental, and evolutionary biology. Using transcriptomics, sharp changes in gene expression were detected throughout embryogenesis, suggesting the existence of developmental milestones (10). These were observed by gene expression changes that are not gradual but, rather, punctuate the embryo. Moreover, it was shown that different stages show different levels of expression conservation, suggesting different levels of expression constraints. The different stages also showed different compositions of genes in terms of their ages (11), which supported the notion that the stages of embryogenesis have unique evolutionary histories.
One particular stage during embryogenesis stood out in comparative transcriptomics studies. Studying a collection of Caenorhabditis species, the ventral enclosure stage was found to correspond to a period of intense changes in gene expression (10). Studies in arthropods and chordates revealed a similar middevelopmental stage. Interestingly, the stage in each of these phyla corresponded to the phylotypic stage: a period in which the species appear the most similar, morphologically. This middevelopmental transition between an early gastrulation module and a late morphogenesis module was observed in seven additional phyla in a recent study (12). Also, when studying this middevelopmental transition using mutation accumulation lines, it was observed that genes expressed during this stage are less likely to be different within a population of C. elegans species (2), suggesting that the middevelopmental transition is under severe developmental constraints.
The rate of development varies drastically in nematodes, even between those that are closely related (13–18). Although C. elegans has a generation time of 3–7 days, other nematode species can take anywhere from days to a year (19, 20). The clade IV species Acrobeloides nanus has a rate of embryogenesis that is four times longer than that of C. elegans (at 20 °C) and differs substantially from C. elegans in many aspects of life cycle, mode of living, and phenotype. Although it was initially assumed that C. elegans development is archetypic for nematodes, it has now been shown that early development in A. nanus is far more regulative (21) and that, for example, gastrulation in the enoplean species Tobrilus stefanskii is much more similar to nonnematode Bilateria (13). It has also become apparent that the molecular toolkit of development varies across the phylum, and even between closely related taxa (22, 23).
In particular, A. nanus blastomeres remain multipotent until at least the five-cell stage, able to reassign their cell fates to compensate for the death of a neighboring blastomere (21). A. nanus also differs from C. elegans in its ability to tolerate a wider range of environmental stresses: it develops optimally at 25 °C, whereas C. elegans, typically cultured at 20 °C, is negatively affected by such a high temperature (17). Moreover, A. nanus has an increased tolerance to desiccation and toxins (24, 25). Finally, A. nanus is one of many obligate parthenogens in the nematode phylum, and as such, its development is, unlike that of C. elegans, initiated without sperm input (26).
Here we compare the embryogenesis of A. nanus and C. elegans at the gene expression level. We describe the genome and transcriptome of A. nanus and show how they allow for the study of transcriptional differences of cells and developmental stages in this species. We compare at the single-cell level the two-cell stage and find a tremendous amount of variation. Comparing the temporal developmental transcriptomes of these two species, we find that there are similar sharp changes at developmental milestones. In particular, we find that the middevelopmental transition is the stage at which gene expression differences between the pair of species begin to significantly decrease. In general, the genes that are more conserved are those that arose at the origin of the Nematode phylum and the superphylum Ecdysozoa. Further examining this observation, we found a relationship between the timing of expression of a gene and its phylogenetic origin. Genes that originated along with the superphylum Ecdysozoa are expressed during the middevelopmental transition, which can explain their increased conservation over evolutionary time. Our analysis illustrates how species with key phylogenetic distances may be leveraged to address evolutionary developmental biology, using molecular tools.
Results
Genome Analysis of Acrobeloides nanus.
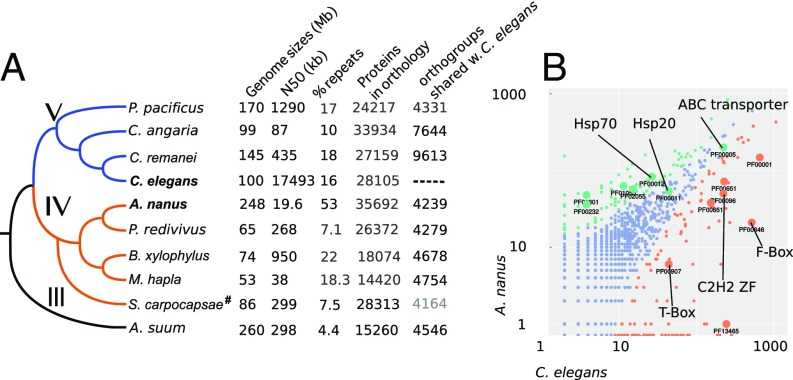
To study the evolution of embryogenesis, we sought to compare C. elegans and the clade IV nematode Acrobeloides nanus at the molecular and developmental level (Fig. 1A). We assembled the A. nanus genome on the basis of Illumina sequencing of DNA and RNA (SI Experimental Procedures). Our genome assembly encompassed 248 Mbp comprising 30,759 contigs with an N50 of 19,614 bp. As Fig. 1A shows, A. nanus has a fairly large genome relative to the other species. To account for this difference, we investigated repetitive DNA and estimated that it constitutes ∼50% of the genome, with 43% of these repeats being unclassified (Table S1). A driver for this might be parthenogenetic reproduction in A. nanus, as parthenogenetic species are thought not to be able to efficiently remove repeats from the population (27). Recent studies, however, did not find an inflation of transposable elements in several parthenogenetic arthropod species (28), nor in another parthenogenetic nematode (29). Thus, we propose that the accumulation of repeats in A. nanus is random, as observed in other species with small effective population sizes (30, 31).
Fig. 1.
The genome of the nematode A. nanus in comparison with that of other nematodes. (A) Phylogenetic tree of the indicated species. Roman numerals indicate clades according to ref. 20. Genome sizes, N50 of the assembly, repeats (23, 50), protein count, and number of orthologs with A. nanus are indicated in the table (see SI Experimental Procedures, #except for S. carpocapsae data, where 1–1 orthologs from ref. 43 are given). (B) Scatter plot of gene family sizes between A. nanus and C. elegans. Differentially enriched families are indicated by color. Larger circles indicate specific families: PF00001, Rhodopsin-like receptors; PF00001, ABC transporters; PF00011, Hsp20/alpha crystallin family; PF00012, Hsp70 protein; PF00096, zinc finger; C2H2 type; PF000232, glycosyl hydrolase family 1, transcription factors; PF00651, overrepresented Pfam domains between A. nanus and C. elegans.
Running the BUSCO3 pipeline (32) on our A. nanus assembly revealed that it is 89% complete and 95% partial complete for the Eukaryote gene set. We obtained 35,692 gene predictions using Augustus (33), trained on the RNA-Seq data (SI Experimental Procedures). We annotated 20,281 of the A. nanus proteins with Pfam domains, using InterProScan and in a bispecies comparison with C. elegans screened for gene family inflations (Fig. 1B). Finally, employing OrthoFinder (34), we identified A. nanus orthologs across eight species selected on the basis of their phylogenetic position, with 4,240 groups of orthologs containing A. nanus and C. elegans proteins.
The A. nanus genome shows dramatic variation at the level of gene families relative to C. elegans (Fig. 1B). Pfam analysis shows more Brachyury-like (T-box) genes in C. elegans (22 genes) relative to A. nanus (six genes). The C. elegans genome is also overrepresented in other transcription factor families; namely, Zinc fingers of the C2H2 and C4 type, F-Box domains, and BTB/POZ domains. In contrast, A. nanus has more glycosyl hydrolase family genes, Hsp70, and Hsp20, as well as ABC transporters (P < 0.05, Fisher’s exact test, false discovery rate-corrected). Interestingly, consistent with the expansion of the Hsp gene family, A. nanus develops into normal adults in large numbers when kept at 30 °C; a temperature at which C. elegans quickly becomes sterile (35).
Studying A. nanus Blastomeres Using Single-Cell RNA-Seq.
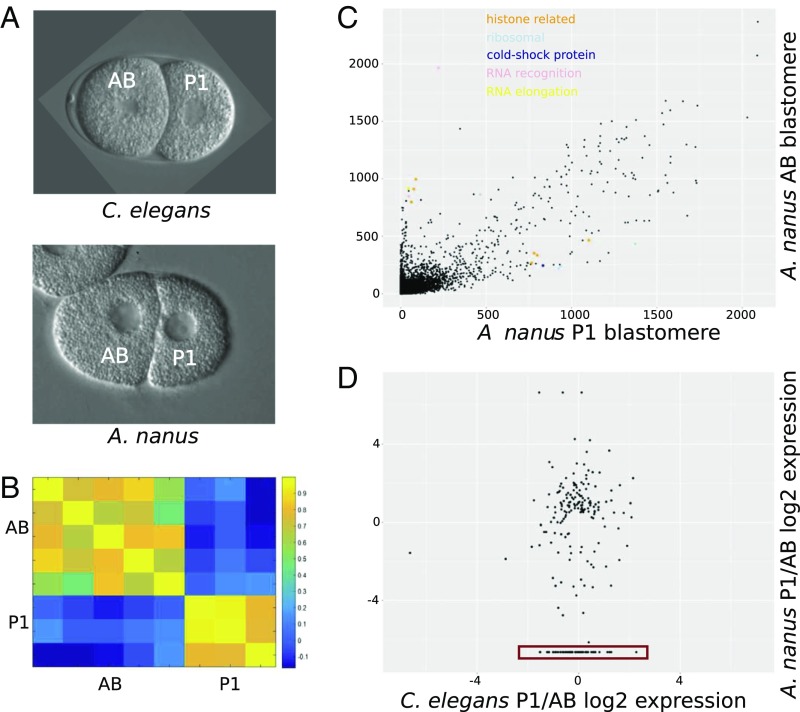
We sought to use the genome assembly to study the early stages of embryogenesis. We collected individual AB and P1 blastomeres (Fig. 2A) and sequenced their transcriptomes using single-cell RNA-Seq (SI Experimental Procedures). The identity of the blastomeres could be clearly distinguished morphologically, as well as from their transcriptomes (Fig. 2B). To study the transcriptomes at the gene level, we identified the differentially segregated genes between the AB and P1 blastomeres. We found that transcripts of heat shock genes are found in greater numbers in AB, whereas ribosomal genes are higher in P1 (Fig. 2C). Interestingly, this was not observed in C. elegans (36).
Fig. 2.
Single-cell A. nanus blastomere analysis. (A) The two-cell stage in A. nanus and C. elegans, indicating also the AB and P1 blastomeres. Embryos are 50 µm in length. (B) Heat map showing correlation coefficients among the A. nanus transcriptomes of five AB blastomeres and three P1 blastomeres. (C) Comparison of the A. nanus gene expression levels between the AB and P1 blastomeres. Expression levels are computed as transcripts per million (tpm; SI Experimental Procedures). Genes of the indicated functional groups are highlighted. (D) Ratios of expression between AB and P1 in C. elegans and A. nanus, respectively. The red box indicates genes with high P1 expression only in A. nanus.
We next compared the overall pattern of gene expression at the two-cell stage between C. elegans and A. nanus. For this, we compared with previously published C. elegans single-cell RNA-Seq data (36) and found genes with conserved and divergent AB- P1 segregations (Fig. 2D). P-granule–associated genes are expressed in the same direction (36). skn-1 transcripts are evenly expressed between AB and P1 in C. elegans; however, our previous analysis using in situ staining of skn-1 mRNA (22) showed a higher expression of this gene in the AB cell in the A. nanus two-cell stage embryo. Our single-cell transcriptomics data are in accordance with this previous finding, supporting the validity of the approach.
We found a small number of genes to be highly expressed in either the A. nanus AB or P1 blastomere that had no expression in the C. elegans two-cell stage. Screening these genes for enriched functional groups according to their gene ontology terms, we found terms relating to reproduction, body morphogenesis, molting, regulation of growth, and transcription initiation (P < 0.001, hypergeometric distribution). This last functional description is particularly of interest because the slow and regulative development of A. nanus might not rely on many maternally deposited transcripts and proteins, similar to C. elegans, but, rather, on primarily zygotic expression. This is in accordance with the prediction that the fast development seen in C. elegans requires the deposition of a higher amount of maternal factors in general (37). Because comparison of the two-cell stage showed differences between the clade IV species and the model organism from clade V, we wanted to quantify the divergence in embryonic development between A. nanus and C. elegans on a global level.
Developmental Dynamics in A. nanus Reveal Distinct Stages.
To identify the gene expression of all genes throughout embryogenesis, we assayed expression in individual embryos throughout A. nanus development. In contrast to the two-cell stage analysis, in this analysis, we focused exclusively on temporal resolution for the entire developmental process (Fig. 3A). Morphologically, A. nanus stages differ from those of C. elegans; however, at the 102-cell stage, the two species appear to have converged in their cell locations (38).
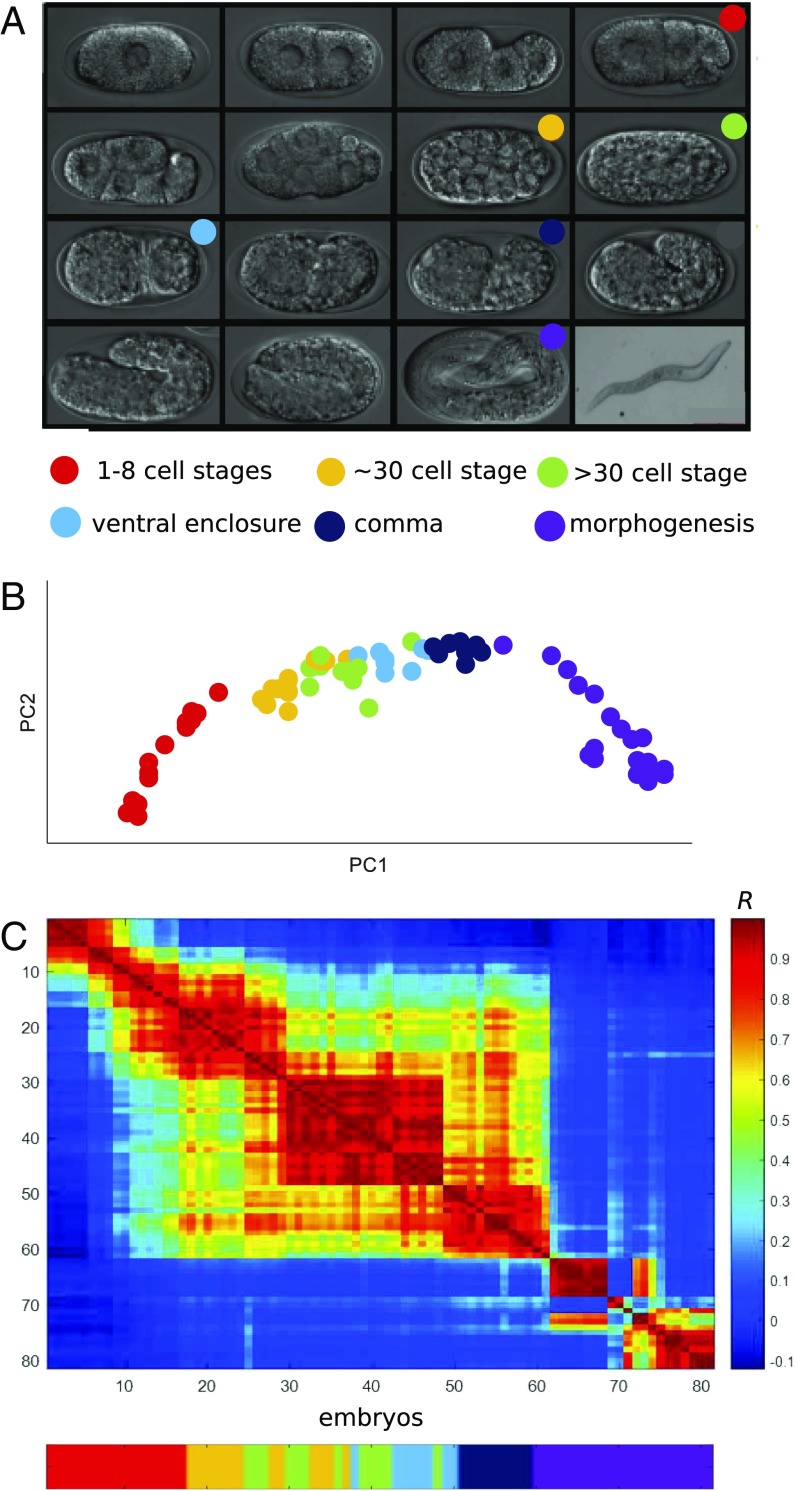
Fig. 3.
A gene expression developmental time-course for A. nanus embryogenesis. (A) Micrographs of A. nanus embryos at the indicated stages. (B) RNA-Seq of 81 randomly collected A. nanus embryos. The embryos were sorted according to BLIND. (C) A correlation matrix of the BLIND-sorted A. nanus transcriptomes. Note the sharp transitions after the one- to eight-cell stages and then again at morphogenesis.
We produced a gene expression time-course dataset according to our previously described BLIND method, in which embryos are randomly collected and sorted by their transcriptomes (39). We collected 81 A. nanus embryos and processed each individually, using CEL-Seq (36), to obtain an expression matrix (Fig. 3B). For each embryo, we also noted the apparent morphological stage of development: one- to eight-cell stages, ∼30-cell stage, >30-cell stage, ventral enclosure, comma, or morphogenesis. Examining the transcriptomes using principal components analysis, we found that the overall ordering of the embryonic transcriptomes corresponded to the morphological stages (Fig. 3B). This principal components analysis on 1,314 dynamically expressed genes (SI Experimental Procedures) accounted for 49.8% (PC1) and 13.6% (PC2) of the gene expression variation. PC1 clearly captures developmental time, and PC2 distinguishes between the stages of the middevelopmental transition and the ends of embryogenesis. Thus, from randomly collected worm embryos, we obtained a time-course of expression throughout embryogenesis.
Studying the correlation among the transcriptomes, we found sharp developmental transitions (Fig. 3C). To annotate the stage of each transition, we compared these results with our morphological annotations and found that each transition corresponded to a shift between developmental stages (Fig. 3D). The first transition occurs after the likely degradation of the maternal transcriptome at the end of the 8-cell stage, and the next transition between early gastrulation (∼30-cell stage) and midgastrulation (>30-cell stage). Another transition occurs at the end of the ventral enclosure stage. Finally, the comma stage was found to express a major transcriptomic transition after ventral enclosure and before morphogenesis. Thus, despite differences in the timing of embryonic development, we find a conservation in the pattern of gene expression transitions in A. nanus and C. elegans (10).
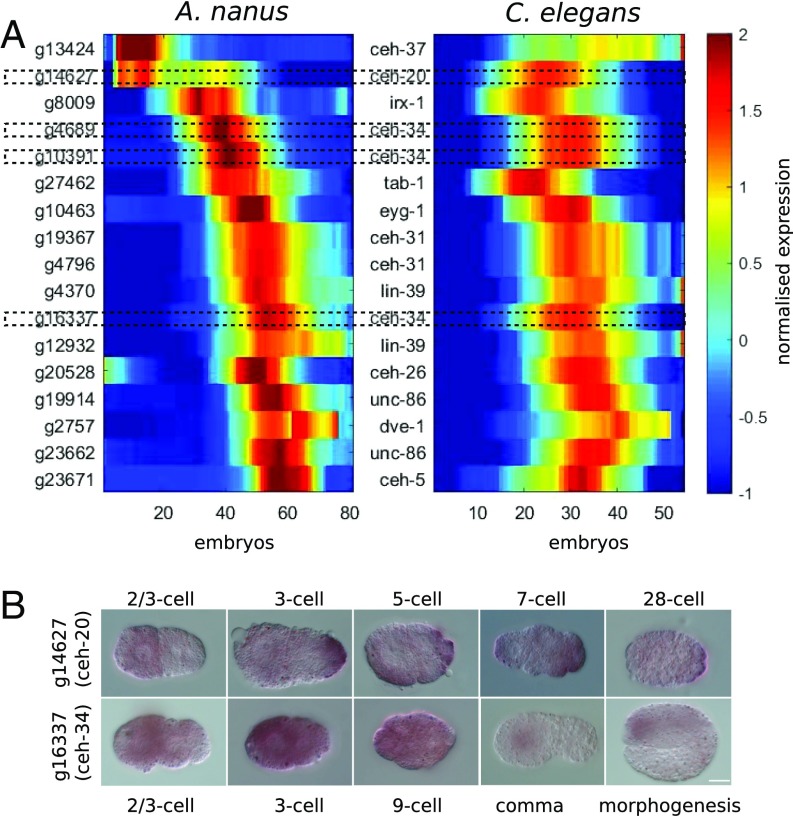
To validate the RNA-Seq data, we further examined the expression of homeodomain genes, known to play important developmental roles, between A. nanus and C. elegans (Fig. 4A). We found that although many genes are expressed at similar developmental stages between the two species, there were also some interesting divergences. One example is the ceh-20 gene, which encodes one of the three C. elegans homeodomain proteins (CEH-20, CEH-40, and CEH-60) homologous to Drosophila Extradenticle (Exd/Pbx). In C. elegans, this gene is expressed during the ventral enclosure stage (40), whereas in A. nanus, the ortholog is expressed earlier, during the one- to eight-cell stage. To validate this difference, we performed an in situ hybridization against the ceh-20 ortholog in A. nanus (Fig. 4B). The in situ confirmed the early A. nanus expression. Moreover, an additional in situ of the ceh-34 gene, which is homologous to human SIX2, revealed expression consistent with our RNA-Seq analysis (Fig. 4B). This analysis further supports the quality of the gene expression time-course.
Fig. 4.
Expression of homeodomain genes between A. nanus and C. elegans. (A) Comparison of temporal expression of selected orthologous genes in A. nanus and C. elegans. Specific homeodomain genes that were further analyzed by in situ (B) are emphasized with dotted outlines. (B) In situ hybridizations for ceh-20 and ceh-34 orthologs in A. nanus.
Comparison of the A. nanus and C. elegans Developmental Transcriptomes.
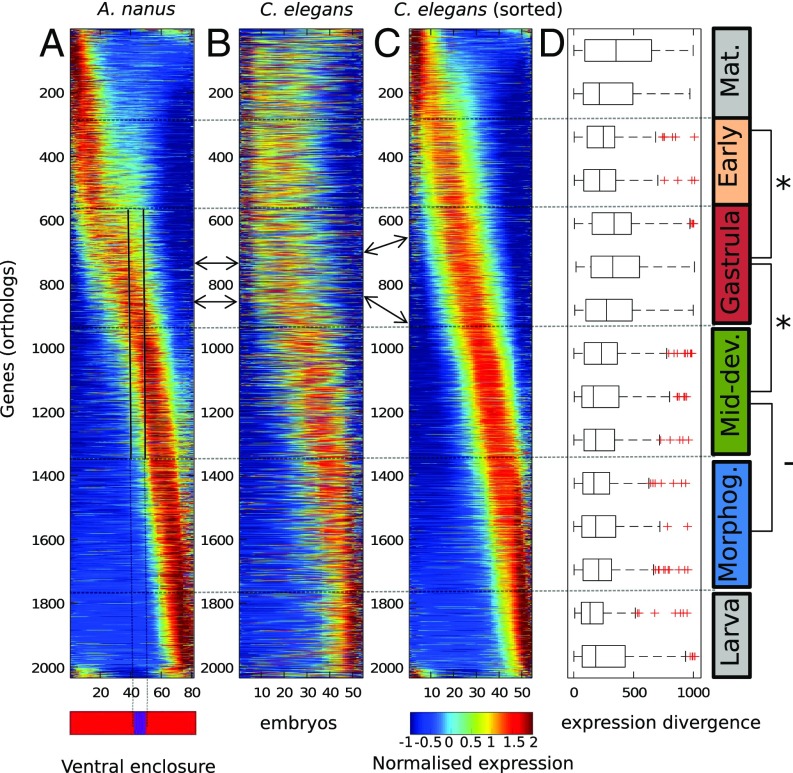
Seeking to compare the developmental transcriptomes of A. nanus and C. elegans in their entireties, we applied our previous approach in which dynamically expressed genes are first sorted according to their temporal expression (Fig. 5A) (2). Examining expression profiles of orthologous C. elegans genes, sorted according to expression of the corresponding A. nanus orthologs (Fig. 5A), we found an immediately apparent correspondence (Fig. 5B), suggesting general conservation of gene expression programs.
Fig. 5.
Expression divergence between the developmental transcriptomes of C. elegans and A. nanus. (A) Developmental transcriptome of A. nanus. Genes are sorted by the Zavit method (2). (B) Developmental transcriptome of the C. elegans orthologs of A. nanus, sorted as in A. nanus. Arrows indicate orthologs. (C) Developmental transcriptome of the C. elegans orthologs sorted independent of A. nanus. Arrows indicate corresponding genes, sorted in C according to C. elegans time. (D) Box plots indicating the expression divergences between genes in A and C for stages along development. Developmental stages are indicated on the right (Mat., maternal; early; gastrula; Mid-dev., middevelopmental transition; Morphog., morphogenesis; and larva). Note the increased relative conservation of genes expressed early and at middevelopmental transition.
We asked whether gene expression at particular developmental stages is more evolvable than at other stages. To address this, we also sorted the C. elegans genes according to their temporal expression (Fig. 5C). For each pair of orthologs, we computed the difference between the relative order in which each gene appears in its respective time-course, which we refer to as the expression divergence index. We then examined whether at different stages of development, genes show different overall expression patterns between species. Proceeding from the earliest to the latest expression, we examined the distributions of expression divergence scores for A. nanus genes within the nonoverlapping windows shown in Fig. 5D.
As the distributions show, expression divergence is not uniform for genes expressed at different times. Genes expressed at the earliest stage may be considered maternal transcripts, and these appear to be highly divergent (Fig. 5D). The earliest zygotically expressed genes appear to be significantly more conserved in their expression (Fig. 5D, early) than the gastrula expressed genes, whereas genes expressed during the middevelopmental transition show significantly less divergence than those expressed at the gastrula stage (P < 10−8, Wilcoxon test). Interestingly, this level of conservation continues throughout morphogenesis and does not increase, as would be expected from the hourglass model. This suggests a more complicated, funnel-like pattern of developmental constraints than previously recognized, although the reduction in divergence during the middevelopmental transition does mark a period of increased conservation, as expected.
Phylostratigraphic Analysis of Expression Divergence.
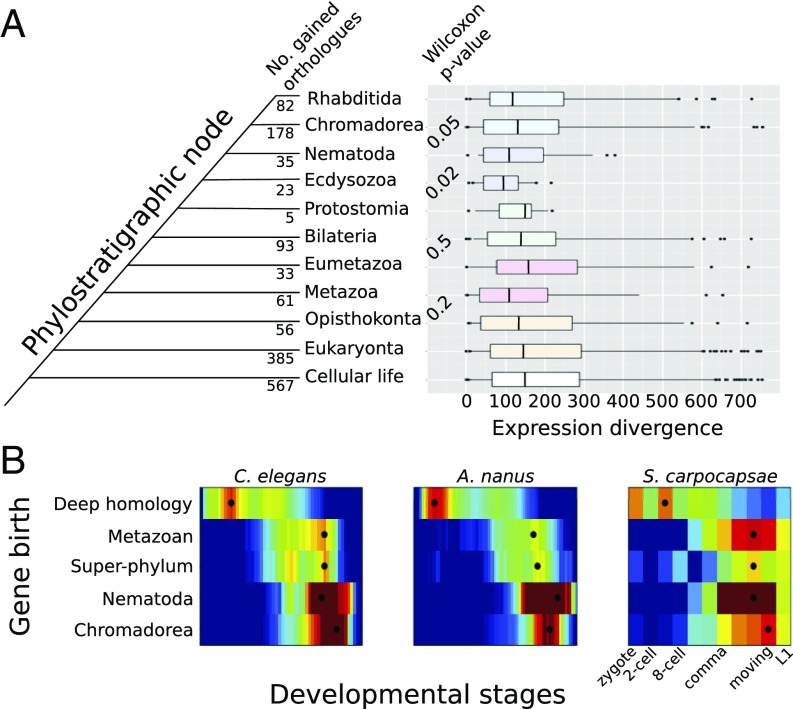
Previous studies across animals separated by hundreds of millions of years of independent evolution have revealed that temporal expression of genes during animal development is correlated to the evolutionary age of genes (41, 42). We sought to investigate whether a similar pattern is observable between the closer-related clade IV and clade V nematode species examined here. For each pair of orthologs, we inferred the phylostratigraphic age (11), ranging from cellular life (common to all studied organisms) to Rhabditida, and restricted to this class of roundworms. To study whether genes differed in their evolvability throughout development, we studied the distributions of expression divergence for each class of gene ages (Fig. 6).
Fig. 6.
Ecdysozoan- and Nematode-specific genes are more conserved in their expression between C. elegans and A. nanus. (A) Genes were grouped according to their phylostratigraphic age (Left, see SI Experimental Procedures). Expression divergence index (ED) of C. elegans and A. nanus orthologs in comparison with their phylostratigraphic age. Phylostratigraphic age was calculated by blasting against a previously reported database (47) using the Phylostratigraphy software (https://github.com/AlexGa/Phylostratigraphy.git). A statistical test of difference in ED distributions for phylostratigraphic nodes revealed significance of divergence for comparisons in Nematoda, but not for genes that evolved before the phylum. The ED appears to follow an hourglass shape through evolutionary time, with evolutionary very old and young genes showing less constrained ED than those acquired on intermediate nodes in Nematoda. (B) Average expression profiles of genes of a common phylostratigraphic age for the three indicated species. Black dots indicate the stage for each category at which average expression is at its maximum.
We observed a restriction of expression divergence for genes originating at superphylum, phylum, and class levels within Nematoda. The sample sizes did not allow for direct statistical comparisons of phylostratigraphic nodes. However, a Wilcoxon ranks-sum test confirmed that the distributions were significantly different between neighboring phylostrata for genes that evolved at the base of the superphylum Ecdysozoa and the phylum Nematoda (Fig. 6). Thus, in addition to genes expressed at or after the middevelopmental transition, genes originating at the dawn of the Nematode phylum are also more conserved in their expression across species than expected.
We hypothesized that the reason that genes of distinct phylostratigraphic ages are conserved in their gene expression between species at different levels follows from their expression at distinct periods during embryogenesis. In other words, if genes of different ages are expressed at different developmental stages, then their expression would evolve at different rates following our results shown in Fig. 5. Interestingly, we found that deeply conserved genes are expressed early in both C. elegans and A. nanus. Meanwhile, genes specific to the Chromadorea class or more specific taxa (SI Experimental Procedures) are expressed later in development, during differentiation (Fig. 6B). However, genes that originated in the early metazoan lineage and with the ecdysozoan superphylum are expressed during the middevelopmental transition. We further tested this result by examining the expression of genes of different ages in the recently published developmental transcriptome of the parasitic clade IV species Steinernema carpocapsae (43). Again, we found the same pattern (Fig. 6B), suggesting that a relationship between phylogenetic age and developmental expression may be general to the Nematode phylum.
Discussion
In this work, we compared the developmental transcriptomes of two distantly related nematodes. C. elegans is a clade V nematode of the Rhabditoidea superfamily, whereas A. nanus belongs to the Cephaloboidea superfamily within clade IV. The lineages of both species most likely diverged not more than 200 million years ago (44). Although the embryogenesis of A. nanus has been analyzed in classical cell biological studies, here we report for the first time its genome, transcriptome, and developmental gene regulation. Compared with C. elegans, we found important differences at the two-cell stage, in terms of transcription factor expression during the course of development and the overall pattern of development. We also compared the divergence in gene expression in terms of the phylostratigraphy and found that genes specific to Nematodes and the Ecdysozoa superphylum are more conserved. In this section, we discuss our results in light of the methodologies for evaluating developmental transcriptomics, the middevelopmental transition, developmental constraints, and phylostratigraphy.
As in other species examined by transcriptomics, we identified a clear middevelopmental transition in A. nanus, depicted as a sharp transition in the heat map of correlations between transcriptomes. We also observed that at this stage in development, the transcriptomes of C. elegans and A. nanus begin to converge. Interestingly, the transcriptomes do not diverge in an hourglass shape after the middevelopmental transition, as was initially suggested for vertebrates (45), and later for a variety of invertebrates (46) and plants (47). This is similar to a previous observation of two frog species (48) that converged at the tailbud stage (the phylotypic stage of chordates) and then did not diverge again. This may be a result of the large number of cell types present at this stage. These results also somewhat mirror what was seen when examining mutation accumulation strains of C. elegans (2), as well as the results of a recent study examining the developmental transcriptomes of the parasitic clade IV species S. carpocapsae (43).
Our phylostratigraphical analysis shows that genes that emerged during the origin of the superphylum Ecdysozoa and the Nematoda are more conserved in their developmental expression. We found that this may follow from a relationship between the age of a gene and its expression during development. Although Domazet-Lošo and Tautz found that a middevelopmental stage has an overall older transcriptome when computed by the transcriptomic age index (42), we found that genes of older origin tend to be expressed early in development. We attribute this difference to us having studied separately groups of genes of distinct ages, rather than combining ages to specify an age of the transcriptome. In our analysis, genes of the superphylum and phylum age category are enriched for expression during the middevelopmental transition. This suggests that genetic pathways originating at the dawn of the Ecdysozoa superphylum are more conserved in their expression program during embryogenesis because they have been integrated into the more conserved middevelopmental transition stage.
Importantly, our finding that genes of intermediate evolutionary age show a restriction in their developmental expression divergence is in line with the inference that these genes are definitive of superphyla and phyla within the sphere of animal diversity. Moreover, it has been argued that taxon specific (“orphan”) genes contribute most to the differentiation of developmental between taxa (49). Thus, our evidence that evolutionarily young genes are more variable in their developmental expression and expressed at later stages, indeed suggests that these genes drive the differentiation of developmental gene expression programs. Our detailed study of the developmental gene expression and genome of A. nanus will allow for detailed comparative studies into these patterns, and enable deeper insights into the evolvability and constraint of molecular pathways in animal development.
Experimental Procedures
We used Illumina technology to sequence the A. nanus genome and transcriptome, and followed the CEL-Seq protocol to establish a developmental time course and single blastomere transcriptomes. Details of the procedure and the short-read cleaning and assembly pipelines can be found in the SI Experimental Procedures. We annotated the genome with Augustus, inferred orthology with OrthoFinder, and analyzed expression data using Matlab, R, and Python as described in the SI Experimental Procedures.
Supplementary Material
Acknowledgments
We gratefully acknowledge the support and inspiration of Einhard Schierenberg throughout this project. We acknowledge the Technion Genome Center for assistance with the sequencing. We thank Maayan Baron for help in the processing of the data. We thank Ali Mortazavi and Marissa Macchietto for kindly providing supplementary data on orthologous genes between C. elegans and S. carpocapsae. P.H.S. was funded by the VolkswagenStiftung in their initiative for evolutionary biology and was supported by a grant from the European Research Council (ERC-2012-AdG 322790 to Max Telford).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All raw data are deposited in the NCBI Sequence Read Archive (Bioproject PRJNA354072). The genome, transcriptome, and annotations are available at genomehubs.org.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720817115/-/DCSupplemental.
References
- 1.Gould SJ, Lewontin RC. The spandrels of San Marco and the Panglossian paradigm: A critique of the adaptationist programme. Proc R Soc Lond B Biol Sci. 1979;205:581–598. doi: 10.1098/rspb.1979.0086. [DOI] [PubMed] [Google Scholar]
- 2.Zalts H, Yanai I. Developmental constraints shape the evolution of the nematode mid-developmental transition. Nat Ecol Evol. 2017;1:113. doi: 10.1038/s41559-017-0113. [DOI] [PubMed] [Google Scholar]
- 3.Arthur W. A Theory of the Evolution of Development. John Wiley & Sons Incorporated; Hoboken, NJ: 1988. [Google Scholar]
- 4.Valentine JW, Jablonski D, Erwin DH. Fossils, molecules and embryos: New perspectives on the Cambrian explosion. Development. 1999;126:851–859. doi: 10.1242/dev.126.5.851. [DOI] [PubMed] [Google Scholar]
- 5.Angelini DR, Kaufman TC. Comparative developmental genetics and the evolution of arthropod body plans. Annu Rev Genet. 2005;39:95–119. doi: 10.1146/annurev.genet.39.073003.112310. [DOI] [PubMed] [Google Scholar]
- 6.Lynch M. The Origins of Genome Architecture. Sinauer Associates Incorporated; Sunderland, MA: 2007. [Google Scholar]
- 7.Yanai I, Lercher M. The Society of Genes. Harvard Univ Press; Cambridge, MA: 2016. [Google Scholar]
- 8.Slonim DK, Yanai I. Getting started in gene expression microarray analysis. PLoS Comput Biol. 2009;5:e1000543. doi: 10.1371/journal.pcbi.1000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 10.Levin M, Hashimshony T, Wagner F, Yanai I. Developmental milestones punctuate gene expression in the Caenorhabditis embryo. Dev Cell. 2012;22:1101–1108. doi: 10.1016/j.devcel.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Domazet-Lošo T, Brajković J, Tautz D. A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet. 2007;23:533–539. doi: 10.1016/j.tig.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 12.Levin M, et al. The mid-developmental transition and the evolution of animal body plans. Nature. 2016;531:637–641. doi: 10.1038/nature16994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schulze J, Schierenberg E. Evolution of embryonic development in nematodes. Evodevo. 2011;2:18. doi: 10.1186/2041-9139-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldstein B, Frisse LM, Thomas WK. Embryonic axis specification in nematodes: Evolution of the first step in development. Curr Biol. 1998;8:157–160. doi: 10.1016/s0960-9822(98)70062-4. [DOI] [PubMed] [Google Scholar]
- 15.Laugsch M, Schierenberg E. Differences in maternal supply and early development of closely related nematode species. Int J Dev Biol. 2004;48:655–662. doi: 10.1387/ijdb.031758ml. [DOI] [PubMed] [Google Scholar]
- 16.Skiba F, Schierenberg E. Cell lineages, developmental timing, and spatial pattern formation in embryos of free-living soil nematodes. Dev Biol. 1992;151:597–610. doi: 10.1016/0012-1606(92)90197-o. [DOI] [PubMed] [Google Scholar]
- 17.Schierenberg E. Three sons of fortune: Early embryogenesis, evolution and ecology of nematodes. BioEssays. 2001;23:841–847. doi: 10.1002/bies.1119. [DOI] [PubMed] [Google Scholar]
- 18.Houthoofd W, et al. Different roads to form the same gut in nematodes. Evol Dev. 2006;8:362–369. doi: 10.1111/j.1525-142X.2006.00108.x. [DOI] [PubMed] [Google Scholar]
- 19.Houthoofd W, et al. Embryonic cell lineage of the marine nematode Pellioditis marina. Dev Biol. 2003;258:57–69. doi: 10.1016/s0012-1606(03)00101-5. [DOI] [PubMed] [Google Scholar]
- 20.Blaxter ML, et al. A molecular evolutionary framework for the phylum Nematoda. Nature. 1998;392:71–75. doi: 10.1038/32160. [DOI] [PubMed] [Google Scholar]
- 21.Wiegner O, Schierenberg E. Regulative development in a nematode embryo: A hierarchy of cell fate transformations. Dev Biol. 1999;215:1–12. doi: 10.1006/dbio.1999.9423. [DOI] [PubMed] [Google Scholar]
- 22.Schiffer PH, et al. Developmental variations among Panagrolaimid nematodes indicate developmental system drift within a small taxonomic unit. Dev Genes Evol. 2014;224:183–188. doi: 10.1007/s00427-014-0471-2. [DOI] [PubMed] [Google Scholar]
- 23.Schiffer PH, et al. The genome of Romanomermis culicivorax: Revealing fundamental changes in the core developmental genetic toolkit in Nematoda. BMC Genomics. 2013;14:923. doi: 10.1186/1471-2164-14-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doroszuk A, Wojewodzic M, Kammenga J. Rapid adaptive divergence of life-history traits in response to abiotic stress within a natural population of a parthenogenetic nematode. Proc Biol Sci. 2006;273:2611–2618. doi: 10.1098/rspb.2006.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bird AF, Ryder MH. Feeding of the nematode Acrobeloides nanus on bacteria. J Nematol. 1993;25:493–499. [PMC free article] [PubMed] [Google Scholar]
- 26.Heger P, Kroiher M, Ndifon N, Schierenberg E. Conservation of MAP kinase activity and MSP genes in parthenogenetic nematodes. BMC Dev Biol. 2010;10:51. doi: 10.1186/1471-213X-10-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arkhipova I, Meselson M. Deleterious transposable elements and the extinction of asexuals. BioEssays. 2005;27:76–85. doi: 10.1002/bies.20159. [DOI] [PubMed] [Google Scholar]
- 28.Bast J, et al. No accumulation of transposable elements in asexual arthropods. Mol Biol Evol. 2016;33:697–706. doi: 10.1093/molbev/msv261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schiffer PH, et al. Signatures of the evolution of parthenogenesis and cryptobiosis in the genomes of panagrolaimid nematodes. bioRXiv. July 3, 2017 doi: 10.1101/159152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 31.Szitenberg A, et al. Genetic drift, not life history or RNAi, determine long term evolution of transposable elements. Genome Biol Evol. 2016;8:2964–2978. doi: 10.1093/gbe/evw208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 33.Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19:ii215–ii225. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- 34.Emms DM, Kelly S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wittenburg N, Baumeister R. Thermal avoidance in Caenorhabditis elegans: An approach to the study of nociception. Proc Natl Acad Sci USA. 1999;96:10477–10482. doi: 10.1073/pnas.96.18.10477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: Single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 37.Schierenberg E. Embryological variation during nematode development. WormBook. 2006:1–13. doi: 10.1895/wormbook.1.55.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schulze J. 2008. Vergleichende Untersuchungen zur Embryonalentwicklung basaler und abgeleiteter Nematoden. Ein Beitrag zum Verständnis der Evolution von Entwicklungsprozessen. PhD thesis (University of Cologne, Cologne, Germany), 153 p.
- 39.Anavy L, et al. BLIND ordering of large-scale transcriptomic developmental timecourses. Development. 2014;141:1161–1166. doi: 10.1242/dev.105288. [DOI] [PubMed] [Google Scholar]
- 40.Hashimshony T, Feder M, Levin M, Hall BK, Yanai I. Spatiotemporal transcriptomics reveals the evolutionary history of the endoderm germ layer. Nature. 2015;519:219–222. doi: 10.1038/nature13996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moyers BA, Zhang J. Phylostratigraphic bias creates spurious patterns of genome evolution. Mol Biol Evol. 2015;32:258–267. doi: 10.1093/molbev/msu286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Domazet-Lošo T, Tautz D. A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature. 2010;468:815–818. doi: 10.1038/nature09632. [DOI] [PubMed] [Google Scholar]
- 43.Macchietto M, et al. Comparative transcriptomics of Steinernema and Caenorhabditis single embryos reveals orthologous gene expression convergence during late embryogenesis. Genome Biol Evol. 2017;9:2681–2696. doi: 10.1093/gbe/evx195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rota-Stabelli O, Daley AC, Pisani D. Molecular timetrees reveal a Cambrian colonization of land and a new scenario for ecdysozoan evolution. Curr Biol. 2013;23:392–398. doi: 10.1016/j.cub.2013.01.026. [DOI] [PubMed] [Google Scholar]
- 45.Duboule D. Temporal colinearity and the phylotypic progression: A basis for the stability of a vertebrate Bauplan and the evolution of morphologies through heterochrony. Dev Suppl. 1994:135–142. [PubMed] [Google Scholar]
- 46.Kalinka AT, Tomancak P. The evolution of early animal embryos: Conservation or divergence? Trends Ecol Evol. 2012;27:385–393. doi: 10.1016/j.tree.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 47.Drost HG, Gabel A, Grosse I, Quint M. Evidence for active maintenance of phylotranscriptomic hourglass patterns in animal and plant embryogenesis. Mol Biol Evol. 2015;32:1221–1231. doi: 10.1093/molbev/msv012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yanai I, Peshkin L, Jorgensen P, Kirschner MW. Mapping gene expression in two Xenopus species: Evolutionary constraints and developmental flexibility. Dev Cell. 2011;20:483–496. doi: 10.1016/j.devcel.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tautz D, Domazet-Lošo T. The evolutionary origin of orphan genes. Nat Rev Genet. 2011;12:692–702. doi: 10.1038/nrg3053. [DOI] [PubMed] [Google Scholar]
- 50.Kikuchi T, et al. Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus. PLoS Pathog. 2011;7:e1002219. doi: 10.1371/journal.ppat.1002219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.