Significance
Wnt/β-catenin signaling is crucial for adult homeostasis and stem cell maintenance, and its dysregulation is strongly associated to cancer. Upon Wnt binding, Wnt receptors assemble into large complexes called signalosomes that provide a platform for interactions with downstream effector proteins. The assembly and regulation of these signalosomes remains largely elusive. Here, we use internally tagged Wnt ligands as a tool to isolate and analyze the composition and regulation of endogenous Wnt receptor complexes. We identify a positive regulator of Wnt signaling that facilitates signalosome formation by promoting intramembrane receptor interactions. Our results reveal that the assembly of multiprotein Wnt signalosomes proceeds along well-ordered steps and involves regulated intramembrane interactions.
Keywords: Wnt signaling, signalosome, Frizzled, multimerization, protein–protein interactions
Abstract
Wnt/β-catenin signaling controls development and adult tissue homeostasis by regulating cell proliferation and cell fate decisions. Wnt binding to its receptors Frizzled (FZD) and low-density lipoprotein-related 6 (LRP6) at the cell surface initiates a signaling cascade that leads to the transcription of Wnt target genes. Upon Wnt binding, the receptors assemble into large complexes called signalosomes that provide a platform for interactions with downstream effector proteins. The molecular basis of signalosome formation and regulation remains elusive, largely due to the lack of tools to analyze its endogenous components. Here, we use internally tagged Wnt3a proteins to isolate and characterize activated, endogenous Wnt receptor complexes by mass spectrometry-based proteomics. We identify the single-span membrane protein TMEM59 as an interactor of FZD and LRP6 and a positive regulator of Wnt signaling. Mechanistically, TMEM59 promotes the formation of multimeric Wnt–FZD assemblies via intramembrane interactions. Subsequently, these Wnt–FZD–TMEM59 clusters merge with LRP6 to form mature Wnt signalosomes. We conclude that the assembly of multiprotein Wnt signalosomes proceeds along well-ordered steps that involve regulated intramembrane interactions.
Wnt/β-catenin signaling controls stem cell maintenance, cell proliferation, and cell fate decisions during development, tissue homeostasis, and regeneration. In healthy tissues, signaling strength is tightly controlled by extensive feedback mechanisms. Inappropriate pathway activation due to mutations leads to excessive tissue growth and is strongly linked to cancer formation. Central to the signaling events in the Wnt/β-catenin cascade is the proteolytic regulation of the transcriptional coactivator β-catenin (1–3). In the absence of Wnt, a cytosolic destruction complex, composed of the scaffolds Axin and adenomatous polyposis coli and kinases GSK3β and CK1, phosphorylates β-catenin to earmark it for rapid proteasomal degradation (4–8). Binding of Wnt to its receptors Frizzled (FZD) and low-density lipoprotein-related 6 (LRP6) initiates a series of molecular rearrangements that converge on the inhibition of β-catenin proteolysis (9). As a result, β-catenin accumulates in the cytosol and migrates to the nucleus to activate target gene expression (1, 2).
Wnt stimulation induces the receptors to assemble into large and highly dynamic protein assemblies termed signalosomes (10–13). Such signalosomes markedly increase local concentrations of signaling proteins, which enhances avidity for low-affinity interactors and improves the temporal and spatial regulation of signaling amplitude. In current models, the formation of higher-order complexes is initiated via binding of Wnt, which bridges the extracellular domains of the seven-span FZD and single-span LRP6 coreceptors (14). In a next step, the cytosolic effector protein Dishevelled (Dvl) is recruited via interaction of its DEP domain with a discontinuous cytosolic binding surface in FZD (11, 15). The resulting increase in local Dvl concentration triggers the formation of swapped-DEP dimers that enable self-polymerization of Dvl via its weakly interacting DIX domains (11). In turn, Dvl DIX multimers license the recruitment of Axin through heterotypic interactions with the Axin DIX domain (16). The locally increased concentrations of Axin and its associated kinases facilitate the phosphorylation of PPPSPxS/T motifs in the LRP6 cytosolic tail (17). These phosphorylated motifs provide additional high-affinity docking sites for Axin and mediate subsequent inhibition of GSK3 kinase activity (18–20). Collectively, these events lead to the stabilization of β-catenin.
Over recent years, the Wnt receptor complex has emerged as a key node for signaling regulation. In stem cells, the membrane-bound E3 ligases RNF43 and ZNRF3 down-regulate signaling via ubiquitination, internalization, and subsequent lysosomal degradation of FZD (21, 22). This negative regulation is counterbalanced by R-spondin and its receptor Lgr5 that capture RNF43/ZNRF3 away from the Wnt receptors to permit augmented levels of Wnt signaling required for stem cell maintenance (21, 23–25). Additionally, the deubiquitinating enzymes UBPY and USP6 may promote the recycling of endocytosed FZD back to the surface and restore Wnt signaling (26, 27). Another important negative regulator is the Wnt antagonist Dickkopf that uses the Kremen receptor to rapidly capture and remove phosphorylated LRP6 from the cell surface via induced endocytosis (28, 29). Notably, endocytosis might also exert positive effects on Wnt signaling (30, 31). Recent models suggest that the increased concentration of receptors in either clathrin-coated pits or lipid rafts may promote local protein–protein interactions required for signalosome formation (32).
Despite tremendous progress, we are still lacking a comprehensive understanding of signalosome formation, composition, and regulation. Progress to address this issue has been hampered by the lack of molecular tools to isolate and analyze endogenous Wnt-bound components. Here, we used an active, internally tagged Wnt3a variant that allowed for the detection and isolation of Wnt-bound receptors. Utilizing this molecular tool, we investigated the Wnt receptor interactome by mass spectrometry-based proteomics and identified the class I transmembrane (TM) protein TMEM59 as a regulatory Wnt signaling component. We show that TMEM59 facilitates FZD oligomerization via intramembrane interactions. Incoming Wnts first bind these preassembled FZD clusters, after which LRP6 is recruited to form large, mature signalosomes to initiate full-blown Wnt signaling. Thus, we identify TMEM59 as a modifier of Wnt signaling that promotes the formation of multimerized FZD building blocks that assemble into larger signalosomes to potentiate Wnt signal transduction.
Results
Internally Flag-Tagged Wnt3a Binds to FZD5 and LRP6 and Activates Wnt Signaling.
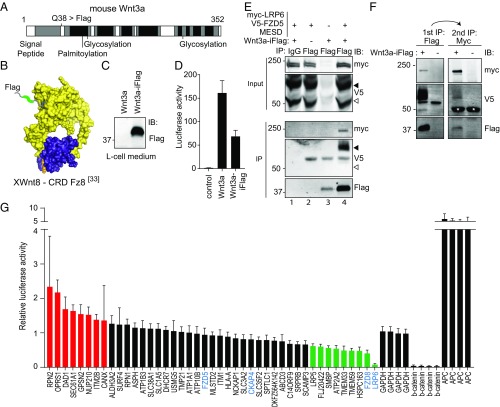
To identify regulators of Wnt signalosome formation, we set out to examine the composition of the endogenous Wnt receptor interactome by using tagged Wnt3a proteins that remain signaling competent. We inserted a Flag-tag in a flexible region of murine Wnt3a that is nonconserved and mapped opposite to the FZD binding domain (Fig. 1 A and B and ref. 33). Previously, we have shown that mice carrying an HA tag knock-in at the same position in Wnt3 are fertile and viable, indicating full retention of functional Wnt3 protein activity (34). Like wild-type Wnt3a (35), the Wnt3a–iFlag protein was efficiently secreted from transfected L cells, allowing us to produce conditioned media that activate Wnt/β-catenin signaling in HEK293T cells (Fig. 1 C and D). To verify that Wnt3a–iFlag interacts with its cognate receptors, we incubated HEK293T cells overexpressing V5–FZD5 and Myc–LRP6 with Wnt3a–iFlag-conditioned medium (Wnt3a–iFlag–CM) for 3 h before lysis. We chose to stimulate cells for 3 h to allow for robust formation of signalosomes before commencing with signalosome isolation (10). Subsequent anti-Flag pulldown experiments clearly revealed coprecipitation of both FZD5 and LRP6 with Wnt3a–iFlag (Fig. 1E, lane 4). As expected, Wnt3a–iFlag selectively interacted with the mature, fully glycosylated receptor forms present at the cell surface, but not with the immature forms that were derived from the endoplasmic reticulum (ER) (22) (Fig. 1E, lane 4). Furthermore, sequential immunoprecipitation of Wnt3a–iFlag and Myc–LRP6 from lysates of Wnt3a–iFlag-stimulated cells confirmed the formation of a Wnt3a/FZD5/LRP6 complex (Fig. 1F). Importantly, Wnt3a–iFlag also bound to nontransfected cells, indicating its feasible use for the detection of endogenous Wnt receptors (Fig. 1E, lane 3). Together, our results show that Wnt3a–iFlag is secreted from L cells, forms a complex with FZD5 and LRP6, activates Wnt signaling, and can be used to isolate endogenous Wnt receptors.
Fig. 1.
Isolation and analysis of components of the endogenous Wnt receptor complexes using internally tagged Wnt3a. (A) Schematic overview of mouse Wnt3a amino acid conservation (black, highly conserved; gray, intermediate; white, nonconserved). The internal site for Flag insertion (Q38) is indicated. (B) Structure of the XWnt8 (yellow)–XFZD8CRD (purple) complex (33). The inserted Flag-tag (green) flanks the crystal structure in the indicated flexible region of the protein. The palmitoleic acid (PAM) group is indicated in orange. (C) Western blot showing secreted Wnt3a–iFlag in the culture medium of stably transfected L cells. (D) Wnt3a–iFlag-conditioned medium induces Wnt luciferase-reporter activity in HEK293T cells. Graph shows average luciferase reporter activities ±SD. (E) FZD5 and LRP6 receptors are coimmunoprecipitated with Wnt3a–iFlag. HEK293T cells were either left untreated or transfected with V5–FZD5, myc-LRP6, and MESD and stimulated with Wnt3a–iFlag for 3 h. Immature (open arrowhead) and fully glycosylated mature (filled arrowhead) FZD forms are indicated. (F) Sequential immunoprecipitation of Wnt3a–iFlag and myc–LRP6. HEK293T cells were transfected with V5–FZD5, myc–LRP6, and MESD and stimulated with Wnt3a–iFlag for 3 h. Consecutive immunoprecipitations show that Wnt3a, FZD5, and LRP6 are residing in one complex. Asterisks indicate nonspecific signal. (G) Wnt luciferase-reporter activity in HEK293T cells transfected with the indicated smartpool siRNAs. Known Wnt pathway components in the screen are marked in blue. Graph shows average luciferase activities normalized to Wnt3a-treated and control siRNA-treated cells (±SD). Putative novel Wnt pathway modifiers are colored in red (negative modifiers) and green (positive modifiers).
Examination of the Endogenous Wnt3a Interactome.
To examine constituents of endogenous Wnt receptor complexes, we stimulated HEK293T cells with Wnt3a–iFlag for 3 h and investigated the interactomes of immunoprecipitated, activated receptors. For comparison, we included HEK293T cells overexpressing FZD5 and LRP6, to enrich for Wnt receptor complexes and associated factors, while cells stimulated with untagged Wnt3a served as a negative control (Fig. S1). Wnt3a–iFlag-bound proteins were analyzed by using mass spectrometry-based proteomic analysis, yielding a total of 1,883 identified proteins. Of these hits, we detected 217 proteins that were identified with at least two unique peptides and displayed a twofold or greater enrichment in the overexpression sample compared with the control. Among these 217 hits, several core Wnt receptor complex components and previously reported Wnt pathway modifiers were present, including LRP5, FZD5, FZD8, WLS, Wnt5A, CKAP4, ATP6V1A, Flot2, EDD1, and RACK1 (Dataset S1, highlighted in green; refs. 36–41). Hence, the Wnt3a–iFlag protein provides a robust tool to isolate and investigate endogenous Wnt-associated protein complexes and identify potential Wnt pathway modifiers.
TMEM59 Interacts with the Wnt Receptors and Potentiates Wnt/β-Catenin Signaling.
To investigate the role of TM interactors, we focused on a subset of 43 proteins with predicted TM domains for further investigation. We used siRNA pools (composed of four independent siRNAs) to deplete the corresponding gene products in HEK293T cells and investigate consequences for Wnt3a-induced cellular responses using a TOP-flash luciferase reporter assay (42). The results uncovered 17 TM proteins as potential Wnt pathway modifiers, based on a threshold of 35% signal enhancement or repression compared with samples subjected to control siRNAs (Fig. 1G). To confirm the regulatory effect of these 17 candidates, we used the individual nonoverlapping siRNAs and assessed their effects on Wnt signaling. For eight membrane proteins, three or more individual siRNAs reproduced the results derived from the siRNA pool (Tables S1–S3).
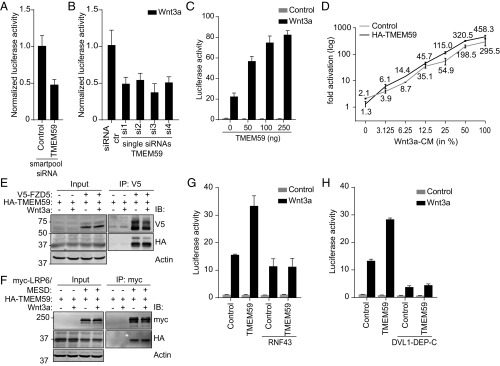
The type I single-span TM protein TMEM59 that associated with both endogenous and overexpressed Wnt receptor complexes (Dataset S1, highlighted in orange), provided one of the most promising hits. Depletion of TMEM59 by multiple nonoverlapping siRNAs substantially reduced Wnt3a-induced signaling in HEK293T cells (Fig. 2 A and B and Fig. S2A), while its overexpression potentiated responses to Wnt3a in a dose-dependent manner (Fig. 2C). Notably, both the potency and efficacy of Wnt signaling responses was increased by TMEM59 (Fig. 2D). Thus, we identify TMEM59 as a positive regulator of Wnt/β-catenin signaling.
Fig. 2.
TMEM59 is a positive regulator of Wnt signal transduction. (A and B) Wnt luciferase-reporter activity in Wnt3a-stimulated HEK293T cells using a smartpool of four siRNAs (A) or four individual nonoverlapping TMEM59 siRNAs (B) targeted against TMEM59. siRNA against GAPDH was used as a control. Graph shows average luciferase reporter activities ±SD. (C) Wnt luciferase-reporter activity in HEK293T cells transfected with increasing doses of TMEM59 expression plasmid. Graph shows average luciferase reporter activities ±SD. (D) Wnt luciferase-reporter activity in HEK293T cells expressing empty vector (control) or HA–TMEM59 stimulated with increasing concentrations of Wnt3a-conditioned medium (Wnt3a-CM). Graph shows relative luciferase activities normalized to the unstimulated control ±SD. (E and F) Coimmunoprecipitation of HA–TMEM59 and V5–FZD5 (E) or myc–LRP6 (F). Cells were transfected with the indicated constructs and stimulated with Wnt3a-conditioned (+) or control (−) medium for 3 h as indicated. (G and H) Wnt luciferase-reporter activity in HEK293T cells coexpressing TMEM59 and RNF43 (G) or the dominant negative DVL1-derived fragment DEP-C (DVL1–DEP-C) (H). Graphs show average luciferase reporter activities ±SD.
The association of TMEM59 with Wnt3a–iFlag-bound complexes suggests that the protein interacts with Wnt receptors. Indeed, TMEM59 robustly interacted with both FZD5 and FZD4 in coimmunoprecipitation experiments and remained unaffected by Wnt3a stimulation (Fig. 2E and Fig. S2B). In addition, TMEM59 coprecipitated with the coreceptor LRP6 (Fig. 2F). Two unrelated TM proteins, EGFR and LAIR1, did not bind TMEM59 (Fig. S2C). We conclude that TMEM59 associates with the canonical Wnt receptor complex.
The earliest known Wnt-induced signaling step involves the binding of FZD to the cytosolic effector protein DVL (43–45). To assess whether TMEM59 promotes Wnt signaling in a FZD-dependent manner, we overexpressed the E3 ligase RNF43, which was previously shown to efficiently remove FZD5 from the cell surface (22). In parallel, we overexpressed a dominant negative fragment of DVL1 (DVL1–DEP-C) that blocks Wnt-mediated signal relay via the cytosolic domains of FZD (15). TMEM59 was unable to enhance signaling in the presence of either of these proteins (Fig. 2 G and H), indicating that TMEM59 depends on FZD–DVL signaling for its Wnt signaling potentiating role.
TMEM59-Induced Alterations in FZD5 Glycosylation Do Not Affect Wnt Signal Relay.
Given the described inhibitory role of TMEM59 on the glycosylation of amyloid precursor protein (APP) (46), we wondered if TMEM59-mediated alterations in the glycosylation pattern of FZD5 may influence Wnt signaling outcome. Indeed, pulse-chase experiments showed that TMEM59 coexpression inhibited the modification of FZD5 with complex glycans (Fig. S3A). FZD5 turnover rates, however, remained unaffected (Fig. S3B). Using enzymes that selectively removed either high-mannose (EndoH) or all N-glycans (PNGaseF) from substrate proteins, we confirmed that the majority of FZD5 in TMEM59 coexpressing cells is modified with high mannose (EndoH-sensitive fraction) and failed to acquire complex glycosylation (EndoH-resistant fraction) (Fig. S3 C and D). Thus, TMEM59 prevents Golgi-dependent complex glycosylation of FZD5.
Next, we assessed the consequences of TMEM59-mediated alterations in FZD5 glycosylation for Wnt signaling. FZD5 trafficking to the plasma membrane (PM) proceeded at similar rates in control and TMEM59-coexpressing cells, as determined by comparing the cell surface-localized fraction to the total pool of intracellular FZD5 (Fig. S4A). In addition, binding of Wnt3a–iFlag to cell surface-expressed FZD5 was unaffected by TMEM59 expression (Fig. S4B). Moreover, FZD5 variants that lacked N-glycosylation sites (N47Q/N151Q) were equally active as wild-type FZD5 in luciferase reporter assays and were still responsive to TMEM59-induced Wnt pathway potentiation (Fig. S4 C and D). Of note, such N47Q/N151Q FZD5 variants were no longer sensitive to the N-glycosylation inhibitor Swainsonine (SWA), indicating removal of all N-glycosylation sites (Fig. S4E). Finally, SWA or the O-glycosylation inhibitor benzyl 2-acetamido-2-deoxy-α-d-galactopyranoside (BADGP) did not cause substantial alterations in Wnt responses, and TMEM59 was still able to enhance Wnt signaling in these cells (Fig. S4 F and G). We conclude that inhibition of complex N- or O-glycosylation does not affect FZD turnover rates, Wnt binding, and subsequent signal relay and therefore does not explain the potentiating effect of TMEM59 on Wnt signaling.
TMEM59 Is Cointernalized with the Canonical Wnt Receptors FZD5 and LRP6.
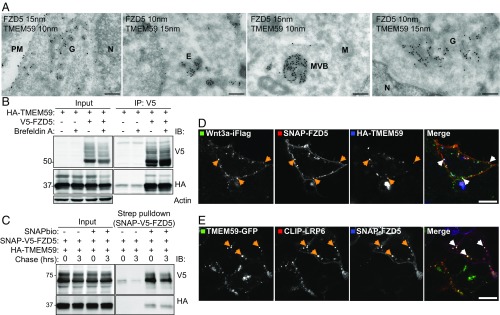
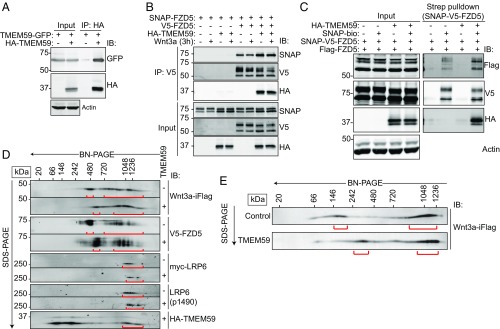
Next, we examined the subcellular compartments in which TMEM59 engages Wnt receptors using cryo-immunoelectron microscopy (immuno-EM) and confocal microscopy. TMEM59 showed a wide distribution among various membrane compartments including ER and Golgi, as well as endosomes and lysosomes (Fig. S5A and ref. 47). In HEK293T cells coexpressing TMEM59 and FZD5, we observed substantial colocalization of both proteins in both biosynthetic and endocytic compartments (Fig. 3A). These findings suggest that TMEM59 already forms a complex with FZD5 in early biosynthetic stages. To investigate this issue, we treated cells with Brefeldin A to block ER exit of both proteins (Fig. S5 B and C and ref. 48). Under these conditions, TMEM59 and FZD5 readily coprecipitated (Fig. 3B). To investigate whether TMEM59 associates with FZD5 at the PM, we used SNAP–V5-tagged FZD5 (SNAP–V5–FZD5) that allowed for selective cell surface labeling of FZD5 by using a membrane-impermeable biotinylated SNAP probe (22). Streptavidin pull-down experiments of HEK293T cell expressing both SNAP–V5–FZD5 and HA–TMEM59 showed that TMEM59 resides in a complex with cell surface localized SNAP–V5–FZD5 (Fig. 3C). Thus, TMEM59 forms a complex with FZD during or immediately after ER biosynthesis and remains associated with FZD after its arrival at the PM.
Fig. 3.
TMEM59 associates with FZD5 after synthesis and remains in complex during endocytosis. (A) EM images of cells coexpressing HA–TMEM59 and V5–FZD5, labeled with 15- and 10-nm gold particles, respectively. Endosomes (E), nucleus (N), PM, Golgi (G), mitochondria (M), and multivesicular bodies (MVB) are indicated. (Scale bars, 200 nm.) (B) Coimmunoprecipitation of HA–TMEM59 and V5–FZD5 in the presence of Brefeldin A (BFA). Cells were transfected with the indicated constructs and treated with 1 μg/mL M BFA for 4 h. (C) Streptavidin pull-down of HEK293T cells expressing HA–TMEM59 and SNAP–V5–FZD. Cells were transfected with the indicated constructs. Cell surface SNAP–V5–FZD5 was labeled with SNAP–biotin and chased for 0 or 3 h. (D) Confocal microscopy of HEK293T cells expressing HA–TMEM59 and SNAP–FZD5. Cell-surface SNAP–FZD5 was labeled with surface SNAP549, and cells were chased for 3 h in the presence of Wnt3a–iFlag. Arrows indicate colocalization of TMEM59 and FZD5 in endosomes. (Scale bar, 10 μm.) (E) Confocal microscopy of HEK293T cells expressing TMEM59–GFP, CLIP–LRP6, and SNAP–FZD5. Cells were labeled with surface CLIP547 and surface SNAP647, and chased for 3 h in the presence of Wnt3a. Arrows indicate colocalization of both proteins in endosomes. (Scale bar, 10 μm.)
To investigate whether TMEM59 cointernalizes with active Wnt-bound receptors, we used SNAP-tagged FZD5 and CLIP-tagged LRP6 receptors that allowed for selective fluorescent labeling at the cell surface and subsequent monitoring of internalization and subcellular trafficking (22). First, cells expressing both SNAP–FZD5 and HA–TMEM59 were labeled with a membrane-impermeable fluorescent SNAP probe and incubated with Wnt3a–iFlag for 3 h. During this period, Wnt3a–iFlag-bound FZD5 receptors cointernalized with TMEM59 and accrued in peripheral endocytic compartments (Fig. 3D). In addition, cell surface-labeled CLIP–LRP6 accumulated together with both FZD5 and TMEM59 in similar endocytic structures after a chase of 3 h (Fig. 3E). Together, these findings argue that TMEM59 encounters and associates with FZD5 at an early stage after biosynthesis, in the ER. After arrival at the cell surface, a pool of TMEM59 remains associated with FZD and cointernalizes with the active, Wnt3a-bound receptor complex.
The TMEM59 TM Domain Mediates Wnt Signaling Potentiation.
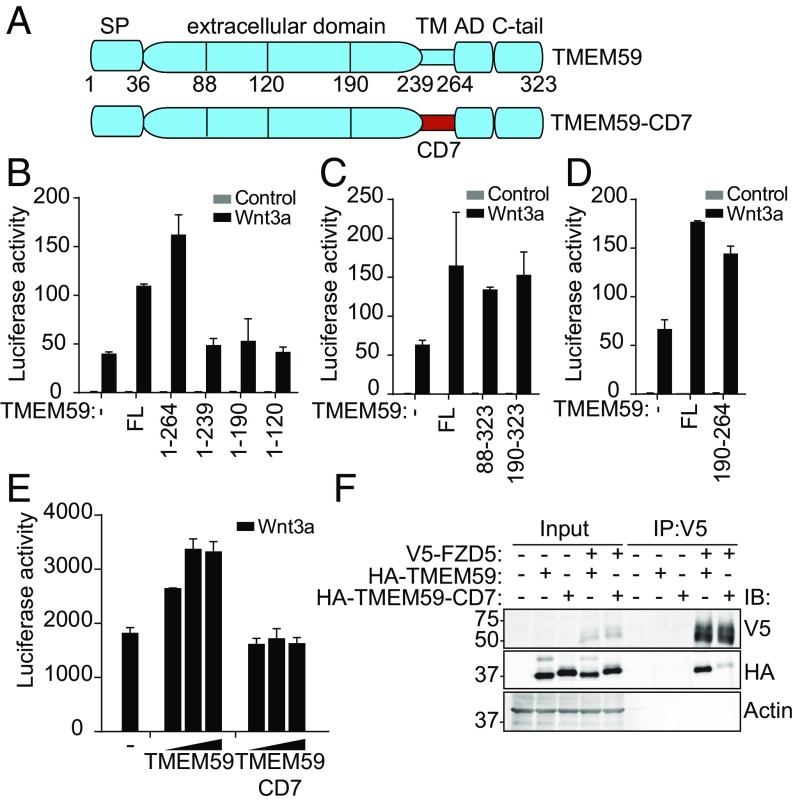
To identify the region of TMEM59 that is responsible for Wnt pathway potentiation, we generated multiple truncated TMEM59 variants and determined their ability to enhance Wnt signaling (Fig. 4A). C-terminal truncations showed that the entire intracellular domain was dispensable for signal amplification. By contrast, progressive truncations that also removed the TM domain fully abrogated the enhancing effect of TMEM59 on Wnt signaling (Fig. 4B). Subsequently, truncations starting from the N terminus revealed that the largest part of the extracellular region of TMEM59 could be removed as well without any consequence for its Wnt potentiating effect (Fig. 4C). By truncating both the N and C termini, we mapped the Wnt-potentiating activity of TMEM59 to an 8-kDa fragment (amino acids 190–264), which includes the TM domain and a short 49-aa extracellular sequence (Fig. 4D).
Fig. 4.
Identification of a TMEM59 minimal fragment that potentiates Wnt signaling. (A) Schematic representation of TMEM59 and TMEM59–CD7. AD, autophagy domain and the C terminus; CD7, CD7 TM domain; SP, signal peptide, extracellular domain; TM, TM domain. Amino acids at truncation positions are indicated. (B–D) Wnt luciferase-reporter activity in HEK293T cells expressing the indicated TMEM59 truncations. Average luciferase reporter activities ±SD are shown. (E) Wnt luciferase-reporter activity in HEK293T cells expressing increasing amounts of wild-type TMEM59 or the TMEM59–CD7 variant, in which the TM domain is replaced with that of CD7. Graph shows averages of luciferase activities ±SD. (F) Coimmunoprecipitation of HA–TMEM59 or HA–TMEM59–CD7 with V5–FZD5.
The above experiments suggest a potential role of the TM domain of TMEM59 in regulating Wnt receptor activity. To investigate the contribution of the TM domain in more detail, we designed a chimeric TMEM59 protein that harbors the TM domain of CD7, an unrelated protein (Fig. 4A; TMEM59–CD7). Subcellular distribution, colocalization with FZD5, and presence at the PM of TMEM59–CD7 was indistinguishable from wild-type TMEM59, both in the presence and absence of Wnt3a (Fig. S6). Strikingly, however, the TMEM59–CD7 mutant was no longer able to potentiate Wnt signaling, suggesting an important functional role for the TM domain (Fig. 4E). In strong agreement, the interaction of TMEM59–CD7 with FZD5 was largely abolished (Fig. 4F). Thus, these results revealed that intramembrane interactions with FZD5 underlie TMEM59-mediated Wnt signaling potentiation.
TMEM59 Facilitates the Assembly of Wnt Receptors in Higher-Order Complexes.
In current models, oligomerization of Wnt receptors is central to signalosome formation and largely mediated by self-association of interaction effector proteins such as Dvl (10, 11). As TMEM59 associates with FZD5, but does not alter its overall surface expression or levels of Wnt binding (Fig. S4 A and B), we reasoned that TMEM59 might facilitate the assembly of Wnt receptors into oligomeric structures. Coimmunoprecipitation experiments indeed revealed strong self-associating properties of TMEM59 (Fig. 5A). Moreover, the interaction of SNAP- and V5-tagged FZD5 was enhanced in the presence of TMEM59, in a Wnt-independent manner (Fig. 5B). In addition, increased self-association of FZD5 via TMEM59 occurred at the PM (Fig. 5C).
Fig. 5.
TMEM59 promotes the assembly of multimerized FZD receptor complexes. (A) Coimmunoprecipitation of GFP–TMEM59 with HA–TMEM59. (B) Coimmunoprecipitation of V5–FZD5 with SNAP–FZD5 in the absence or presence of HA–TMEM59. (C) Analysis of TMEM59-mediated promotion of an FZD–FZD interaction at the cell surface of HEK293T cells. A SNAP–biotin-mediated pull-down was performed to isolate SNAP–V5–FZD5, and levels of associated Flag–FZD5 were analyzed in the presence or absence of HA–TMEM59. Labeling with SNAP–biotin was done for 4 h. (D) BN/SDS/PAGE analysis of Wnt3a–iFlag-bound receptor complexes in the presence or absence of HA–TMEM59. Cells transfected with V5–FZD5, myc–LRP6, and either empty vector or HA–TMEM59 were stimulated for 4 h with Wnt3a–iFlag medium, after which Flag-immunoprecipitations were performed. BN/SDS/PAGE analysis of the eluted complexes is shown. Red markings show approximate size distribution. (E) BN/SDS/PAGE analysis of Wnt3a–iFlag-bound endogenous receptor complexes in the presence and absence of HA–TMEM59. HEK293T cells were transfected with either empty vector or HA–TMEM59 and stimulated for 4 h with Wnt3a–iFlag medium, after which Flag-immunoprecipitations were performed. BN/SDS/PAGE analysis of the eluted complexes are shown. Red markings show approximate size distribution.
To obtain information on the size and composition of Wnt-bound receptor complexes, we stimulated FZD5- and LRP6-overexpressing HEK293T cells with Wnt3a–iFlag for 3 h and, after lysis, isolated Wnt-associated receptor complexes using Flag-immunoprecipitation. Following elution with Flag peptides, we used 2D blue-native PAGE and SDS/PAGE (BN/SDS/PAGE) to analyze the size range of the isolated Wnt-bound complexes (49). Thus, in the first dimension, Coomassie-charged native protein complexes were PAGE-separated based on complex size, while in the second dimension, denaturing conditions allowed for analysis of individual subunits by using conventional SDS/PAGE. In cells that were not transfected with TMEM59, Wnt3a–iFlag-bound FZD5 receptors were enriched in complexes ranging from 400 to 500 kDa as well as 700 kDa to 1.5 MDa (Fig. 5D). Upon TMEM59 co-overexpression, both complexes shifted to a higher-molecular-mass range (Fig. 5D, red markings). The large overlap in size distribution of Wnt3a- and FZD5-based complexes indicated that the largest fraction of precipitated Wnt was bound by FZD5 (Fig. 5D). Full-blown Wnt signaling requires activation of both FZD and LRP6 coreceptors (10, 50, 51). Notably, LRP6 was only detected in complexes with a high molecular mass range (>1 MDa). Together, these findings indicate that after initial formation of Wnt–FZD complexes, mature Wnt–FZD–LRP6 signalosomes are formed and accumulate in the highest molecular mass fraction (Fig. 5D, red markings). Indeed, these high-molecular-mass complexes also contained phosphorylated S1490 LRP6 (pLRP6), a hallmark of Wnt pathway activation (Fig. 5D and Fig. S7A) (10, 32, 52, 53). In TMEM59-coexpressing cells, LRP6-containing complexes displayed a slight but consistent shift in molecular mass as well. Although we used Wnt3a–iFlag to isolate the receptor complex, and TMEM59 itself does not detectably bind Wnt3a (Fig. S4B), we noted that a low-molecular-mass fraction of TMEM59 did not comigrate with FZD or LRP6 (Fig. 5D and Fig. S7B). A likely explanation is that TMEM59 partially dissociates from receptor complexes during detergent-based lysis and complex isolation. As an alternative method to analyze the consequences of TMEM59 expression for Wnt receptor signalosome formation, we used sucrose gradients, as described (10). Whole-cell lysates of Wnt3a-treated cells that overexpress FZD5 and LRP6 were separated on a linear sucrose gradient of 10–40%. In agreement with the above findings, a fraction of the total pool of cellular FZD and LRP6 complexes shifted toward higher molecular mass in TMEM59-expressing cells (Fig. S7 C–E, lanes 6–8).
Next, we aimed to assess if TMEM59 also modulates the size distribution of endogenous, native Wnt receptor complexes by BN/SDS/PAGE (Fig. 5E). In the absence of TMEM59 overexpression, Wnt-bound complexes were enriched in complexes of ∼150–200 kDa as well as in complexes of >800 kDa (Fig. 5 E, Upper) in a pattern resembling that of FZD5- and LRP6-overexpressing cells. Again, the relative distribution of Wnt3a-bound complexes clearly shifted toward a higher molecular mass when TMEM59 was overexpressed (Fig. 5 E, Lower and Fig. S7B). Thus, TMEM59 promotes the formation of large higher-order molecular-mass Wnt receptor complexes. Together, these results show that TMEM59 promotes the assembly of active signalosomes by facilitating the formation of higher-order Wnt/FZD complexes that subsequently can interact with LRP6.
Discussion
The molecular mechanisms underlying the assembly of Wnt receptors into higher-order signalosomes and the regulation of this process at the endogenous level remain important, but poorly understood, issues. Here, we used a modified Wnt ligand to isolate endogenous, activated Wnt receptor complexes and identify Wnt receptor-associated components using a mass spectrometry-based proteomics approach. We identified multiple known core components of the Wnt receptor complex, confirming robustness of the method. Our results reveal a strong preference of Wnt3a to bind FZD5 and FZD8, while no binding was found for FZD1, FZD2, and FZD3 that are known to be expressed in HEK293T cells as well (22, 54). In addition, we identified several proteins that were previously implicated in the regulation of Wnt signaling amplitude, including RACK1, CKAP4, and the E3 ligase EDD1 (38, 40, 41).
We set out to improve our understanding of the regulation of Wnt receptor complex assembly and activity by focusing on TM proteins associated with the Wnt receptor complex. We identified the single-span TM protein TMEM59 as an interactor of the FZD receptor and a positive regulator of FZD–Dvl mediated Wnt signal relay. In search of the underlying mechanism, we first investigated the role of known molecular activities of the TMEM59 protein. Initially, TMEM59 was identified as a ubiquitous modulator of complex glycosylation of APP, inhibiting its shedding via Golgi retention (46). Although we show that TMEM59 expression also diminishes glycan modification of FZD, we were unable to link these glycosylation alterations to the Wnt-potentiating activity of TMEM59. In addition, TMEM59 was reported to induce unconventional autophagy in response to bacterial infection and to promote apoptotic cell death in neuronal cells (47, 55, 56). Both these functions require the TMEM59 intracellular domain that is dispensable for its role in Wnt signaling. Thus, we conclude that the mechanism underlying TMEM59-mediated potentiation of Wnt signaling relies on a previously undescribed molecular activity of the protein. We noted that TMEM59 forms a complex with FZD at early stages during biogenesis. Both proteins were found to be associated upon their arrival at the PM as well as during endocytosis and their trafficking to late endosomal compartments. Moreover, we mapped both the FZD interacting and Wnt enhancing activities of TMEM59 to its TM domain, indicating that TMEM59 promotes Wnt responses via intramembrane interactions with FZD. In agreement with these findings, the TM domain of TMEM59 appears strongly conserved across evolution compared with other type-I TM proteins (47), signifying an additional functional role besides anchoring the protein to the membrane.
To uncover the consequences of TMEM59 binding for Wnt signaling initiation, we used BN/SDS/PAGE to portray the size and composition of Wnt-activated receptor complexes. Introduction of TMEM59 in cells readily favored the formation of enlarged Wnt–FZD assemblies. This is illustrated by the prominent shift of Wnt-bound receptor complexes to a higher molecular mass upon overexpression of TMEM59, both at the endogenous level as well as in cells overexpressing FZD5 and LRP6. In receptor overexpression experiments, we identified a substantial fraction of Wnt–FZD5 complexes of 500–600 kDa that did not yet interact with LRP6. This suggests that the Wnt–FZD5 complex formation precedes the recruitment of LRP6, which is only present in Wnt–FZD5 complexes in a considerably higher-molecular-mass size range, ∼1–1.5 MD. The well-demarcated presence of phosphorylated LRP6 in this region suggests that these high-molecular-mass complexes represent mature Wnt signalosomes (10). BN/SDS/PAGE analysis upon Wnt ligand-mediated isolation of Wnt receptor complexes thus provides a read-out for the assembly of endogenous signalosomes.
How does TMEM59 mediate enhanced Wnt receptor clustering at the molecular level? First, we show that TMEM59 is able to self-associate. Second, TMEM59 robustly interacts with FZD5 and promotes the interaction between different FZDs, both in the absence and presence of Wnt. Similar to other G-protein-coupled receptors (GPCRs) (57), FZDs are able to form dimers or multimers (58, 59). Moreover, forced dimerization of xFZD7 via extracellular protein interfaces induced Wnt/β-catenin signaling in Xenopus (60), further highlighting the importance of FZD multimerization for canonical signaling. Multiple ways of FZD dimerization have been described. Two recent studies showed that the lipid moiety of Wnt can bridge two cysteine-rich domain (CRD) monomers via binding to a discontinuous lipid-binding groove of two adjacent CRDs, thereby facilitating FZD dimerization (61, 62). Besides ligand-dependent dimerization, several FZDs dimerized in a ligand-independent manner at early stages during biosynthesis in the ER (59). For this mode of dimerization, both the CRD and cytosolic tail are dispensable, consistent with a role of TM domain-mediated interactions. Indeed, recently the TM domains IV and V were implicated in FZD6 dimerization (63), similar to what was shown for the Smoothened receptor (64, 65). Notably, homodimerization and heterodimerization via TM domains were described for several other GPCRs (57, 66–68). The molecular basis of ligand-independent FZD dimerization and its dependency on auxiliary proteins in the membrane remains unknown.
Here, we show that the single-span protein TMEM59 induces ligand-independent FZD5 multimerization via heterotypic intramembrane interactions immediately following synthesis of both proteins in the ER. A similar mode of action was described for the family of tetraspanins that consists of a special class of membrane proteins that facilitate clustering of their associated proteins via intramembrane domains. A striking example is TSPAN12, a TM protein that potentiates Norrin-mediated, but not Wnt-mediated, signaling of FZD4 via the induction of intramembrane receptor multimerization (69). FZD oligomerization via TM interactions therefore appears to be a means to boost signaling in different systems, while retaining ligand dependency and specificity.
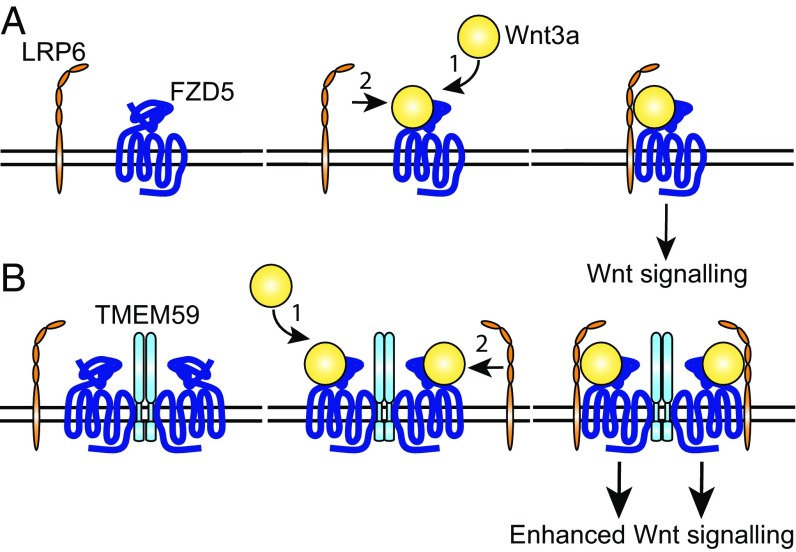
Emerging evidence suggests that Wnt-induced signalosome formation depends on multiple protein interactions that bridge both the extracellular and intracellular interfaces of the receptor complex (70, 71). At the extracellular side, Wnt proteins facilitate dimerization of the receptors, while the recruitment of multimerizing adaptor proteins to intracellular receptor interfaces further facilitate the formation of higher-order assemblies. Our results here reveal that intramembrane interactions provide additional anchor points for Wnt receptor clustering and signaling amplification. In our summarizing model, TMEM59 drives the preassembly of multimerized FZD5 complexes in the ER, after which the complex traffics to the cell surface (Fig. 6). Incoming Wnts first interact with these preassembled FZD5 complexes at the PM to form low-molecular-mass Wnt–FZD5–TMEM59 complexes. These complexes are poised for engagement with LRP6 and continue to form high-molecular-mass mature Wnt–FZD5–TMEM59–LRP6 signalosomes that now acquire full signaling activity, recruit downstream effectors, and drive LRP6 phosphorylation. Thus, we propose that signalosome formation proceeds along well-ordered steps in which initially formed Wnt–FZD building blocks assemble into larger Wnt–FZD5–LRP6 signaling units. By applying a range of Wnt concentrations, we show that TMEM59 increases both the potency and efficacy of cellular Wnt responses. At the same time, levels of Wnt binding to its receptors remain unaffected by TMEM59 expression. Together, these data suggest that signaling efficiency of Wnt-bound receptors is increased by TMEM59.
Fig. 6.
Schematic model of Wnt signaling in the absence (A) and presence (B) of TMEM59. TMEM59 facilitates the preassembly of FZD oligomers. Incoming Wnt ligands first bind to FZD–TMEM59 clusters, after which these clusters recruit LRP6 to form mature signalosomes that drive full-blown Wnt stimulation.
In what biological context might TMEM59-mediated regulation of Wnt signaling play a role? To address this issue, the following points will need to be considered. First, the expression level of TMEM59 varies among tissues, which might affect its role in Wnt signaling regulation in different tissue contexts (47). Second, the molecular activity of TMEM59 might display redundancy with the highly homologous gene TMEM59-like (TMEM59L) (56). Third, in contrast to core Wnt pathway components, a sensitized genetic background might be required for gene deletion approaches to reveal the biological role of TMEM59, as shown for other recently identified Wnt signaling modifiers (72–74). Nonetheless, our findings predict that enhanced expression of TMEM59 in cells potentiates responses to Wnt. Interestingly, amplification of TMEM59 was recently reported in a significant fraction of neuroendocrine prostate cancers (75), for which alterations in Wnt receptor signaling have emerged as a key pathway for cancer progression (76). Whether TMEM59 overexpression sensitizes these cancer cells to Wnts produced by surrounding stromal cells to promote cell growth and survival deserves further investigation.
In summary, we demonstrate the application of internally tagged Wnt ligands as a tool to isolate and analyze the composition and regulation of endogenous Wnt receptor complexes. By means of this tool, we identified a positive regulator of Wnt signaling that facilitates signalosome formation by promoting intramembrane interactions. Our findings thus reveal a regulatory step in the assembly of higher-order Wnt receptor complexes and highlight the underlying protein interactions as key targets for pathway regulation.
Materials and Methods
DNA Constructs and Reagents.
Murine V5–FZD5 (77), murine SNAP–FZD5 (22), human RNF43–2xFlag_2xHA (22), murine Flag–DVL–DEP-C (15), and human TMEM59–GFP were described (47). Human myc-LRP6 was a gift from C. Niehrs, Institute of Molecular Biology, Mainz, Germany; human EGFR-GFP was a gift from P. van Bergen en Henegouwen, University of Utrecht, Utrecht, The Netherlands; human LAIR1a-GFP was a gift from L. Meyaard, University Medical Center Utrecht, Utrecht, The Netherlands; the LRP6 chaperone MESD was a gift from B. C. Holdener, Stony Brook University, Stony Brook, NY; and human TMEM59 and HA–TMEM59 were a gift of S. Lichtenthaler, Ludwig-Maximilians University Munich, Munich (46). HA–TMEM59–CD7 was generated by replacing the TMEM59 TM domain by that of human CD7 via standard PCR methods. Murine SNAP–V5–FZD5 was generated by placing the SNAP domain derived from an nSNAP construct (Bioke) between the mouse MHC class I H2-Kb signal sequence and a V5-tag of murine V5–FZD5 into pcDNA4T/O (Invitrogen). Flag–FZD5 was generated by cloning a Flag-tag between the H2-Kb signal sequence and mature murine FZD5. FZD5–GFP was generated by cloning a GFP-tag after the C terminus of murine FZD5. CLIP–LRP6 (human) was generated by placing CLIP (Bioke) between the H2-Kb signal sequence and mature human LRP6. C-terminally truncated HA–TMEM59 constructs were generated by the introduction of a stop codons by using site-directed mutagenesis. N-terminally truncated HA–TMEM59 constructs were generated by PCR amplification of the indicated constructs using N-terminal primers containing an HA tag and subcloning of the amplified regions after the H2-Kb signal peptide in pcDNA4T/O. Wnt3a–iFlag was generated by overlap-extension PCR and subcloning into pcDNA4T/O. At later stages of this study, we obtained a functional N-terminal Flag-tagged Wnt3a as well, which was used for some replicates. This variant was generated by inserting a Flag-epitope tag between the native signal sequence and the first mature codon of murine Wnt3a. This variant displayed comparable activity as wild-type Wnt3a protein and faithfully copied results obtained with Wnt3a–iFlag. LRP6 was always coexpressed with its chaperone MESD.
The following antibodies were used: mouse and rabbit anti-Flag (Sigma), rat anti-HA (3F10; Roche), mouse anti-V5 (Life Technologies), rabbit anti-V5 (Sigma), mouse anti-myc (9E10; Sigma and produced from hybridoma), rabbit anti-LRP6 (Cell Signaling Technologies), rabbit antiphosphorylated LRP6 pS1490 (Cell Signaling Technologies), rabbit anti-SNAP (Bioke), mouse anti-GFP (Sigma-Aldrich), and mouse anti-actin (MP Biomedicals).
Cell Culture and Transfection.
Human embryonic kidney 293T (HEK293T) cells (ATCC CRL-3216) were cultured in RPMI GlutaMAX (Invitrogen) supplemented with 10% FBS (GE Healthcare) and 100 units/mL penicillin and 100 μg/mL streptomycin (P/S; Invitrogen). L cells (ATCC CRL-2648) were cultured in DMEM containing 1 g/L glucose (Life Technologies), supplemented with 10% FCS and P/S. All cells were maintained at 37 °C in 5% CO2. L cells stably expressing and secreting Wnt3a or Wnt3a–iFlag were cultured in the presence of 125 μg/mL Zeocine (Life Technologies) to obtain Wnt3a-conditioned medium (35). For Luciferase reporter assays, microscopy, and BN/PAGE, transfections were performed by using the FuGENE6 transfection reagent according to the manufacturer’s instructions (Promega). For immunoprecipitations, cells were transfected by using polyethylenimine. siRNA transfections were performed with Dharmafect1 (Dharmacon) transfection reagent, according to the manufacturer’s instructions. TMEM59 siRNAs and GAPDH control siRNAs were from Dharmacon and used at a concentration of 10 or 20 nM.
Immunofluorescence and Confocal Microscopy.
HEK293T or HeLa cells were grown on glass coverslips coated with laminin (Sigma) in 24-well plates. After overnight transfection, cells were fixed in ice-cold methanol or in 4% paraformaldehyde (PFA) in 50 mM sodium phosphate (pH 7.4) buffer. For SNAP and CLIP cell surface staining, live cells were labeled with 1 μM SNAP–Alexa549, SNAP–Alexa647, or CLIP–Alexa547 (Bioke) for 15 min at room temperature (RT). Subsequently, cells were chased for the indicated times at 37 °C and fixed in 4% PFA. After being quenched in 50 mM NH4Cl and incubated in blocking buffer containing 2% BSA and 0.1% saponin in PBS for 30 min, cells were incubated with primary and secondary antibodies for 1 h at RT in blocking buffer. Cells were mounted in Prolong Gold with DAPI (Life technologies), and images were acquired with Zeiss LSM510 or LSM700 confocal microscopes. Images were analyzed and processed with ImageJ.
EM.
Cells were fixed according to the Tokuyasu method by adding 4% freshly prepared formaldehyde (wt/vol) (Polysciences) and 0.4% glutaraldehyde in 0.1 M phosphate buffer (NaH2PO4 and Na2HPO4, pH 7.4) to an equal volume of culture medium for 5 min, followed by postfixation in 2% formaldehyde and 0.2% glutaraldehyde for 2 h at RT or overnight at 4 °C in 0.1 M phosphate buffer (pH 7.4). Processing of cells for ultrathin cryosectioning and immunolabeling according to the protein A-gold method was done as described (78). Fixed cells were incubated with PBS containing 0.15% glycine, gently scraped, and embedded in 12% gelatin in PBS at 37 °C. The cell pellet was solidified on ice and cut into small blocks. For cryoprotection, blocks were infiltrated overnight with 2.3 M sucrose at 4 °C and then mounted on pins and frozen in liquid nitrogen. Ultrathin cryosections (70 nm) were prepared on a Leica ultracut UCT ultra cryomicrotome and picked up with a freshly prepared 1:1 mixture of 2.3 M sucrose and 2% methylcellulose. Sections were then immunogold-labeled and examined by using a JEOL TEM 1010 electron microscope at 80 kV.
Luciferase Reporter Assays.
Luciferase TOPflash and FOPflash reporter assays were performed as described (77). Plasmid DNA was transfected in a single well of a 24-well plate as indicated with a total of 250 ng of transfected DNA. Transfection efficiency was controlled and normalized by the addition of a constant amount of TK-Renilla reporter plasmid. Control or Wnt3a-conditioned medium was added 16 h before cells were lysed in passive lysis buffer (Promega). Luciferase activities were measured with the dual luciferase reporter assay (Promega) according to the manufacturer’s instructions in a Centro XS 960 microplate luminometer (Berthold). Graphs show average luciferase activities normalized to Renilla values of duplicates with the SDs.
Immunoprecipitation, SNAP Biotin Pull-Down, and Western Blotting.
Cells were grown to 80% confluency. After washing cells with PBS, cells were scraped and lysed in cell lysis buffer containing 100 mM NaCL, 50 mM Tris (pH 7.5), 0.25% Triton X-100, 10% glycerol, 50 mM NaF, 10 mM Na3VO4, 10 μM leupeptin, 10 μM aprotinin, and 1 mM PMSF. Lysates were cleared by centrifugation at 16,000 × g for 15 min at 4 °C. Lysates were taken up in SDS sample buffer. For immunoprecipitation, lysates were incubated with the indicated antibodies at 4 °C for 2 h or overnight, followed by incubation with protein G beads (Millipore) or protein A beads (Repligen) for 1 h. For Flag-immunoprecipitations, precoupled Flag-M2 beads (Sigma) were added to lysates and incubated overnight at 4 °C. After washing, beads were eluted with sample buffer and heated for 5 min at 100 °C or incubated at 37 °C for 30 min in the case of FZD5 analysis. For SNAP–biotin pulldowns, cells were incubated with 1 μM SNAP–biotin for 15 min at room temperature. After washing with RPMI, cells were chased for the indicated time periods. Lysates were prepared as described above and incubated with streptavidin–agarose (Thermo Fisher Scientific) for 3 h or overnight. After SDS/PAGE, proteins were transferred via Western blotting onto Immobilon-FL PVDF membranes (Milipore). After blocking with Odyssey blocking buffer (LI-COR), proteins were labeled with the indicated primary antibodies that were detected with goat anti-mouse/rabbit Alexa 680 (Invitrogen), donkey anti-rat Alexa 680 (Invitrogen), or goat anti-mouse/rabbit IRDye 800 (Rockland) by using an Odyssey Infrared Imaging System (LI-COR).
siRNA Screen for Wnt Pathway Regulators.
A total of 1 pmol of On-target plus siRNAs for the selected targets (Dharmacon) was spotted in 96-well plates. A total of 10 µL of RMPI containing 0.2 µL of Dharmafect1 transfection reagent was added to the wells and incubated for 30 min. A total of 80 µL of a 200,000 cells per mL of RPMI (Life Technologies) and 10% FCS (Sigma) cell suspension were added to the wells. After 48 h, the cells were transfected, stimulated with Wnt3a, and analyzed according to the Luciferase reporter protocol described above. All knockdowns were performed in triplicate on distinct plates, each containing two control siRNAs (GAPDH). The effects on Wnt pathway activity were determined by dividing the sample measurements by the two controls from the same assay plate and calculating the average and SD of all sample/control ratios. A level of 35% up or down-regulation, compared with the controls, was set as the threshold for pathway regulation. Hit selection was performed using the unrounded values.
BN/SDS/PAGE.
Cells were grown to 80% confluency in 10-cm dishes, scraped in PBS, and pelleted. Cells were resuspended and lysed in 600 µL of BN-PAGE buffer (pH 7.0), containing 20 mM Bis-Tris, 20 mM NaCl, 500 mM ε-aminocaproic acid, 2 mM EDTA, 10% glycerol, 0,1% Triton X-100, 10 μM leupeptin, 10 μM aprotinin, and 1 mM PMSF. Lysates were centrifuged at 16,000 × g for 30 min at 4 °C. Supernatants were collected and subjected twice to buffer exchange using fresh BN-PAGE buffer and 30 K buffer exchange columns (Millipore). Samples were concentrated to a final volume of 120, and 20 μL was loaded on a 4–12% Bis-Tris NativePAGE gel (Life Technologies). NativeMark unstained protein standards (Life Technologies) were used to determine complex size. Gel runs were performed overnight at 22 V by using NativePAGE anode and cathode buffers (Life Technologies). BN-PAGE gel slices were excised and incubated for 45 min at 37 °C in 2D sample buffer containing 12.5 mM Tris (pH 6.8), 4% SDS, 20% glycerol, 0.02% bromophenol blue, and 2.5% β-mercaptoethanol. The NativeMark gel slice was fixed and destained in 40% methanol and 10% acetic acid. Sample gel slices were placed on 2D-preparative SDS/PAGE gels, and electrophoresis was performed at 10 mA. Western blotting was performed according to standard procedures.
For analysis of the Wnt receptor complex, cells were grown to 80% confluency in 625-cm2 cell culture dishes. Four dishes per sample were used for the analysis of the complex of overexpressed receptors. Seven dishes per sample were used for the analysis of the endogenous Wnt receptor complexes. Cells were stimulated for 4 h with Wnt3a–iFlag-conditioned medium, washed with 25 mL of PBS, scraped in 6 mL of PBS per plate, and collected. Cell pellets were thoroughly resuspended and lysed for 1 h in cell lysis buffer containing 100 mM NaCL, 50 mM Tris (pH 7.5), 0.25% Triton X-100, 10% glycerol, 50 mM NaF, 10 mM Na3VO4, 10 μM leupeptin, 10 μM aprotinin, and 1 mM PMSF. Cell lysates were cleared by centrifugation for 20 min at 20,000 × g at 4 °C. Cleared lysates were incubated with 75 µL of precoupled anti-Flag M2 beads (Sigma) overnight at 4 °C. After collection of the beads, the lysates were incubated with new anti-Flag beads for 3 h. All beads were collected, transferred to Eppendorf tubes, and washed four times with cell lysis buffer and three times with PBS. Native, Wnt3a-associated complexes were eluted from the washed beads by incubation with 450 µL of elution buffer containing 10 mM Tris (pH 7.4), 0.2% Triton X-100, and 166 µg/mL Flag peptides (Sigma) for 90 min at 37 °C. After collection of the eluates, fresh elution buffer was added to the beads, and elution was performed another time. Eluates were pooled, and the buffer was exchanged for BN-PAGE buffer by using 10 K centrifugal filter units (Millipore). BN/SDS/PAGE analysis was performed as described above.
Supplementary Material
Acknowledgments
We thank Luca Ferrari for technical assistance with the sucrose gradients and René Scriwanek for processing the EM pictures. This work is part of the Oncode Institute which is partly financed by the Dutch Cancer Society. This work was supported by European Research Council Starting Grant 242958 (to M.M.M.), European Union Grant FP7 Marie Curie ITN 608180 “WntsApp” (to M.M.M.), and the Netherlands Organization for Scientific Research NWO VICI Grant 91815604 and ECHO Grant 711.013.012 (to M.M.M.). T.Y.L. and A.J.R.H. are supported by large-scale proteomics facility Proteins@Work Project 184.032.201 embedded in the Netherlands Proteomics Centre and supported by the Netherlands Organization for Scientific Research. They were additionally supported through European Union Horizon 2020 Program FET-OPEN Project MSmed, Project 686547. F.X.P. was supported by Grants SAF2014-53320R and SAF2017-88390R from the Spanish Government.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721321115/-/DCSupplemental.
References
- 1.Behrens J, et al. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 2.Molenaar M, et al. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 3.van de Wetering M, et al. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- 4.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. Beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behrens J, et al. Functional interaction of an axin homolog, conductin, with beta-catenin, APC, and GSK3beta. Science. 1998;280:596–599. doi: 10.1126/science.280.5363.596. [DOI] [PubMed] [Google Scholar]
- 6.Ikeda S, et al. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3beta and beta-catenin and promotes GSK-3beta-dependent phosphorylation of beta-catenin. EMBO J. 1998;17:1371–1384. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kishida S, et al. Axin, a negative regulator of the wnt signaling pathway, directly interacts with adenomatous polyposis coli and regulates the stabilization of beta-catenin. J Biol Chem. 1998;273:10823–10826. doi: 10.1074/jbc.273.18.10823. [DOI] [PubMed] [Google Scholar]
- 8.Sakanaka C, Weiss JB, Williams LT. Bridging of beta-catenin and glycogen synthase kinase-3beta by axin and inhibition of beta-catenin-mediated transcription. Proc Natl Acad Sci USA. 1998;95:3020–3023. doi: 10.1073/pnas.95.6.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Kappel EC, Maurice MM. Molecular regulation and pharmacological targeting of the β-catenin destruction complex. Br J Pharmacol. 2017;174:4575–4588. doi: 10.1111/bph.13922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bilic J, et al. Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science. 2007;316:1619–1622. doi: 10.1126/science.1137065. [DOI] [PubMed] [Google Scholar]
- 11.Gammons MV, Renko M, Johnson CM, Rutherford TJ, Bienz M. Wnt signalosome assembly by DEP domain swapping of dishevelled. Mol Cell. 2016;64:92–104. doi: 10.1016/j.molcel.2016.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwarz-Romond T, et al. The DIX domain of dishevelled confers Wnt signaling by dynamic polymerization. Nat Struct Mol Biol. 2007;14:484–492. doi: 10.1038/nsmb1247. [DOI] [PubMed] [Google Scholar]
- 13.Schwarz-Romond T, Metcalfe C, Bienz M. Dynamic recruitment of axin by dishevelled protein assemblies. J Cell Sci. 2007;120:2402–2412. doi: 10.1242/jcs.002956. [DOI] [PubMed] [Google Scholar]
- 14.Schweizer L, Varmus H. Wnt/Wingless signaling through beta-catenin requires the function of both LRP/Arrow and frizzled classes of receptors. BMC Cell Biol. 2003;4:4. doi: 10.1186/1471-2121-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tauriello DV, et al. Wnt/β-catenin signaling requires interaction of the dishevelled DEP domain and C terminus with a discontinuous motif in frizzled. Proc Natl Acad Sci USA. 2012;109:E812–E820. doi: 10.1073/pnas.1114802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bienz M. Signalosome assembly by domains undergoing dynamic head-to-tail polymerization. Trends Biochem Sci. 2014;39:487–495. doi: 10.1016/j.tibs.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Tamai K, et al. A mechanism for Wnt coreceptor activation. Mol Cell. 2004;13:149–156. doi: 10.1016/s1097-2765(03)00484-2. [DOI] [PubMed] [Google Scholar]
- 18.Cselenyi CS, et al. LRP6 transduces a canonical Wnt signal independently of axin degradation by inhibiting GSK3's phosphorylation of beta-catenin. Proc Natl Acad Sci USA. 2008;105:8032–8037. doi: 10.1073/pnas.0803025105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piao S, et al. Direct inhibition of GSK3beta by the phosphorylated cytoplasmic domain of LRP6 in Wnt/beta-catenin signaling. PLoS One. 2008;3:e4046. doi: 10.1371/journal.pone.0004046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu G, Huang H, Garcia Abreu J, He X. Inhibition of GSK3 phosphorylation of beta-catenin via phosphorylated PPPSPXS motifs of Wnt coreceptor LRP6. PLoS One. 2009;4:e4926. doi: 10.1371/journal.pone.0004926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hao HX, et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature. 2012;485:195–200. doi: 10.1038/nature11019. [DOI] [PubMed] [Google Scholar]
- 22.Koo BK, et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature. 2012;488:665–669. doi: 10.1038/nature11308. [DOI] [PubMed] [Google Scholar]
- 23.Chen PH, Chen X, Lin Z, Fang D, He X. The structural basis of R-spondin recognition by LGR5 and RNF43. Genes Dev. 2013;27:1345–1350. doi: 10.1101/gad.219915.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng WC, et al. Structures of Wnt-antagonist ZNRF3 and its complex with R-spondin 1 and implications for signaling. PLoS One. 2013;8:e83110. doi: 10.1371/journal.pone.0083110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zebisch M, Jones EY. Crystal structure of R-spondin 2 in complex with the ectodomains of its receptors LGR5 and ZNRF3. J Struct Biol. 2015;191:149–155. doi: 10.1016/j.jsb.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madan B, et al. USP6 oncogene promotes Wnt signaling by deubiquitylating frizzleds. Proc Natl Acad Sci USA. 2016;113:E2945–E2954. doi: 10.1073/pnas.1605691113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mukai A, et al. Balanced ubiquitylation and deubiquitylation of frizzled regulate cellular responsiveness to Wg/Wnt. EMBO J. 2010;29:2114–2125. doi: 10.1038/emboj.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakane H, Yamamoto H, Kikuchi A. LRP6 is internalized by Dkk1 to suppress its phosphorylation in the lipid raft and is recycled for reuse. J Cell Sci. 2010;123:360–368. doi: 10.1242/jcs.058008. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto H, Sakane H, Yamamoto H, Michiue T, Kikuchi A. Wnt3a and Dkk1 regulate distinct internalization pathways of LRP6 to tune the activation of beta-catenin signaling. Dev Cell. 2008;15:37–48. doi: 10.1016/j.devcel.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 30.Blitzer JT, Nusse R. A critical role for endocytosis in Wnt signaling. BMC Cell Biol. 2006;7:28. doi: 10.1186/1471-2121-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seto ES, Bellen HJ. Internalization is required for proper wingless signaling in Drosophila melanogaster. J Cell Biol. 2006;173:95–106. doi: 10.1083/jcb.200510123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim I, et al. Clathrin and AP2 are required for PtdIns(4,5)P2-mediated formation of LRP6 signalosomes. J Cell Biol. 2013;200:419–428. doi: 10.1083/jcb.201206096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janda CY, Waghray D, Levin AM, Thomas C, Garcia KC. Structural basis of wnt recognition by frizzled. Science. 2012;337:59–64. doi: 10.1126/science.1222879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farin HF, et al. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature. 2016;530:340–343. doi: 10.1038/nature16937. [DOI] [PubMed] [Google Scholar]
- 35.Willert K, et al. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature. 2003;423:448–452. doi: 10.1038/nature01611. [DOI] [PubMed] [Google Scholar]
- 36.Bhavanasi D, Speer KF, Klein PS. CKAP4 is identified as a receptor for Dickkopf in cancer cells. J Clin Invest. 2016;126:2419–2421. doi: 10.1172/JCI88620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cruciat CM, et al. Requirement of prorenin receptor and vacuolar H+-ATPase-mediated acidification for Wnt signaling. Science. 2010;327:459–463. doi: 10.1126/science.1179802. [DOI] [PubMed] [Google Scholar]
- 38.Hay-Koren A, Caspi M, Zilberberg A, Rosin-Arbesfeld R. The EDD E3 ubiquitin ligase ubiquitinates and up-regulates beta-catenin. Mol Biol Cell. 2011;22:399–411. doi: 10.1091/mbc.E10-05-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katanaev VL, et al. Reggie-1/flotillin-2 promotes secretion of the long-range signaling forms of wingless and hedgehog in Drosophila. EMBO J. 2008;27:509–521. doi: 10.1038/sj.emboj.7601981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kimura H, et al. CKAP4 is a Dickkopf1 receptor and is involved in tumor progression. J Clin Invest. 2016;126:2689–2705. doi: 10.1172/JCI84658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li S, et al. Rack1 is required for Vangl2 membrane localization and planar cell polarity signaling while attenuating canonical Wnt activity. Proc Natl Acad Sci USA. 2011;108:2264–2269. doi: 10.1073/pnas.1013170108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Korinek V, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 43.Rothbächer U, et al. Dishevelled phosphorylation, subcellular localization and multimerization regulate its role in early embryogenesis. EMBO J. 2000;19:1010–1022. doi: 10.1093/emboj/19.5.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Umbhauer M, et al. The C-terminal cytoplasmic Lys-thr-X-X-X-Trp motif in frizzled receptors mediates Wnt/beta-catenin signaling. EMBO J. 2000;19:4944–4954. doi: 10.1093/emboj/19.18.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang-Snyder J, Miller JR, Brown JD, Lai CJ, Moon RT. A frizzled homolog functions in a vertebrate Wnt signaling pathway. Curr Biol. 1996;6:1302–1306. doi: 10.1016/s0960-9822(02)70716-1. [DOI] [PubMed] [Google Scholar]
- 46.Ullrich S, et al. The novel membrane protein TMEM59 modulates complex glycosylation, cell surface expression, and secretion of the amyloid precursor protein. J Biol Chem. 2010;285:20664–20674. doi: 10.1074/jbc.M109.055608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boada-Romero E, et al. TMEM59 defines a novel ATG16L1-binding motif that promotes local activation of LC3. EMBO J. 2013;32:566–582. doi: 10.1038/emboj.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fujiwara T, Oda K, Yokota S, Takatsuki A, Ikehara Y. Brefeldin A causes disassembly of the Golgi complex and accumulation of secretory proteins in the endoplasmic reticulum. J Biol Chem. 1988;263:18545–18552. [PubMed] [Google Scholar]
- 49.Gerlach JP, Emmink BL, Nojima H, Kranenburg O, Maurice MM. Wnt signaling induces accumulation of phosphorylated β-catenin in two distinct cytosolic complexes. Open Biol. 2014;4:140120. doi: 10.1098/rsob.140120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cong F, Schweizer L, Varmus H. Wnt signals across the plasma membrane to activate the beta-catenin pathway by forming oligomers containing its receptors, frizzled and LRP. Development. 2004;131:5103–5115. doi: 10.1242/dev.01318. [DOI] [PubMed] [Google Scholar]
- 51.Mao J, et al. Low-density lipoprotein receptor-related protein-5 binds to Axin and regulates the canonical Wnt signaling pathway. Mol Cell. 2001;7:801–809. doi: 10.1016/s1097-2765(01)00224-6. [DOI] [PubMed] [Google Scholar]
- 52.Gammons MV, Rutherford TJ, Steinhart Z, Angers S, Bienz M. Essential role of the dishevelled DEP domain in a Wnt-dependent human-cell-based complementation assay. J Cell Sci. 2016;129:3892–3902. doi: 10.1242/jcs.195685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zeng X, et al. A dual-kinase mechanism for Wnt co-receptor phosphorylation and activation. Nature. 2005;438:873–877. doi: 10.1038/nature04185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Atwood BK, Lopez J, Wager-Miller J, Mackie K, Straiker A. Expression of G protein-coupled receptors and related proteins in HEK293, AtT20, BV2, and N18 cell lines as revealed by microarray analysis. BMC Genomics. 2011;12:14. doi: 10.1186/1471-2164-12-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie Y, et al. Overexpression of DCF1 inhibits glioma through destruction of mitochondria and activation of apoptosis pathway. Sci Rep. 2014;4:3702. doi: 10.1038/srep03702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng Q, et al. The neuron-specific protein TMEM59L mediates oxidative stress-induced cell death. Mol Neurobiol. 2017;54:4189–4200. doi: 10.1007/s12035-016-9997-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Milligan G. G protein-coupled receptor dimerization: Function and ligand pharmacology. Mol Pharmacol. 2004;66:1–7. doi: 10.1124/mol.104.000497.. [DOI] [PubMed] [Google Scholar]
- 58.Dann CE, et al. Insights into Wnt binding and signaling from the structures of two Frizzled cysteine-rich domains. Nature. 2001;412:86–90. doi: 10.1038/35083601. [DOI] [PubMed] [Google Scholar]
- 59.Kaykas A, et al. Mutant frizzled 4 associated with vitreoretinopathy traps wild-type frizzled in the endoplasmic reticulum by oligomerization. Nat Cell Biol. 2004;6:52–58. doi: 10.1038/ncb1081. [DOI] [PubMed] [Google Scholar]
- 60.Carron C, et al. Frizzled receptor dimerization is sufficient to activate the Wnt/beta-catenin pathway. J Cell Sci. 2003;116:2541–2550. doi: 10.1242/jcs.00451. [DOI] [PubMed] [Google Scholar]
- 61.DeBruine ZJ, et al. Wnt5a promotes frizzled-4 signalosome assembly by stabilizing cysteine-rich domain dimerization. Genes Dev. 2017;31:916–926. doi: 10.1101/gad.298331.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nile AH, Mukund S, Stanger K, Wang W, Hannoush RN. Unsaturated fatty acyl recognition by Frizzled receptors mediates dimerization upon Wnt ligand binding. Proc Natl Acad Sci USA. 2017;114:4147–4152. doi: 10.1073/pnas.1618293114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Petersen J, et al. Agonist-induced dimer dissociation as a macromolecular step in G protein-coupled receptor signaling. Nat Commun. 2017;8:226. doi: 10.1038/s41467-017-00253-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang C, et al. Structure of the human smoothened receptor bound to an antitumour agent. Nature. 2013;497:338–343. doi: 10.1038/nature12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao Y, Tong C, Jiang J. Hedgehog regulates smoothened activity by inducing a conformational switch. Nature. 2007;450:252–258. doi: 10.1038/nature06225. [DOI] [PubMed] [Google Scholar]
- 66.Harikumar KG, Pinon DI, Miller LJ. Transmembrane segment IV contributes a functionally important interface for oligomerization of the Class II G protein-coupled secretin receptor. J Biol Chem. 2007;282:30363–30372. doi: 10.1074/jbc.M702325200. [DOI] [PubMed] [Google Scholar]
- 67.Hebert TE, et al. A peptide derived from a beta2-adrenergic receptor transmembrane domain inhibits both receptor dimerization and activation. J Biol Chem. 1996;271:16384–16392. doi: 10.1074/jbc.271.27.16384. [DOI] [PubMed] [Google Scholar]
- 68.Ng GY, et al. Dopamine D2 receptor dimers and receptor-blocking peptides. Biochem Biophys Res Commun. 1996;227:200–204. doi: 10.1006/bbrc.1996.1489. [DOI] [PubMed] [Google Scholar]
- 69.Junge HJ, et al. TSPAN12 regulates retinal vascular development by promoting norrin- but not Wnt-induced FZD4/beta-catenin signaling. Cell. 2009;139:299–311. doi: 10.1016/j.cell.2009.07.048. [DOI] [PubMed] [Google Scholar]
- 70.DeBruine ZJ, Xu HE, Melcher K. Assembly and architecture of the Wnt/β-catenin signalosome at the membrane. Br J Pharmacol. 2017;174:4564–4574. doi: 10.1111/bph.14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gammons M, Bienz M. Multiprotein complexes governing Wnt signal transduction. Curr Opin Cell Biol. 2017;51:42–49. doi: 10.1016/j.ceb.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 72.Azzolin L, et al. YAP/TAZ incorporation in the β-catenin destruction complex orchestrates the Wnt response. Cell. 2014;158:157–170. doi: 10.1016/j.cell.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 73.Huang S, et al. DDB2 is a novel regulator of Wnt signaling in colon cancer. Cancer Res. 2017;77:6562–6575. doi: 10.1158/0008-5472.CAN-17-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ladang A, et al. Elp3 drives Wnt-dependent tumor initiation and regeneration in the intestine. J Exp Med. 2015;212:2057–2075. doi: 10.1084/jem.20142288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Beltran H. Update on the biology and management of neuroendocrine prostate cancer. Clin Adv Hematol Oncol. 2016;14:513–515. [PubMed] [Google Scholar]
- 76.Murillo-Garzón V, Kypta R. WNT signaling in prostate cancer. Nat Rev Urol. 2017;14:683–696. doi: 10.1038/nrurol.2017.144. [DOI] [PubMed] [Google Scholar]
- 77.Tauriello DV, et al. Loss of the tumor suppressor CYLD enhances Wnt/beta-catenin signaling through K63-linked ubiquitination of Dvl. Mol Cell. 2010;37:607–619. doi: 10.1016/j.molcel.2010.01.035. [DOI] [PubMed] [Google Scholar]
- 78.Slot JW, Geuze HJ. Cryosectioning and immunolabeling. Nat Protoc. 2007;2:2480–2491. doi: 10.1038/nprot.2007.365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.