Significance
The lobules are the functional units of the liver. They consist of 15–25 layers of hepatocytes with specialized metabolic functions and gene expression patterns relative to their position along the lobule, a phenomenon referred to as metabolic zonation. The Wnt/β-catenin pathway regulates hepatocyte function but how the zonation is controlled to meet the metabolic demands of the liver is unclear. Glucagon regulates hepatic function. We now demonstrate that glucagon contributes to liver zonation by interacting and opposing the actions of the Wnt/β-catenin pathway.
Keywords: glucagon, glucagon receptor, liver zonation, Wnt
Abstract
Liver zonation characterizes the separation of metabolic pathways along the lobules and is required for optimal function. Wnt/β-catenin signaling controls metabolic zonation by activating genes in the perivenous hepatocytes, while suppressing genes in the periportal counterparts. We now demonstrate that glucagon opposes the actions of Wnt/β-catenin signaling on gene expression and metabolic zonation pattern. The effects were more pronounced in the periportal hepatocytes where 28% of all genes were activated by glucagon and inhibited by Wnt/β-catenin. The glucagon and Wnt/β-catenin receptors and their signaling pathways are uniformly distributed in periportal and perivenous hepatocytes and the expression is not regulated by the opposing signal. Collectively, our results show that glucagon controls gene expression and metabolic zonation in the liver through a counterplay with the Wnt/β-catenin signaling pathway.
The liver performs diverse functions essential for energy homeostasis, lipid and protein synthesis, biotransformation of xenobiotics and endogenous byproducts. The proper function of the liver depends on its structure, which consists of small units called lobules. Each lobule is composed of concentric layers of hepatocytes expanding from the central vein toward the periportal vein. Hepatocytes perform different metabolic functions along the periportal–central axis, a phenomenon referred to as metabolic zonation (1, 2). The hepatocytes closest to the branches of the portal vein are located in the periportal (PP) zone, whereas hepatocytes surrounding the central vein are in the perivenous (PV) zone (1, 2). Blood flows from the periportal to the central vein and consists of a mixture of nutrient-rich but poorly oxygenated blood from the portal vein and highly oxygenated blood from the hepatic artery (1). Due to the metabolic functions and signaling properties of the hepatocytes along the periportal–central axis, gradients of oxygen, nutrients, and hormones are created as the blood flows through the sinusoids. For these reasons, oxygen demanding pathways colocalize in the PP zone, which has the highest partial oxygen pressure. The functional division of hepatocytes also prevents futile cycling when fulfilling anabolic and catabolic requirements of opposing pathways and avoids competition for common substrates between pathways. Furthermore, efficient xenobiotic metabolism and biotransformation of endogenous byproducts of metabolism is accomplished by spatial separation of complementary pathways (1–4).
Zonation of liver functions is supported by pronounced heterogeneity in gene expression between hepatocytes along the sinusoid. RNA sequencing (RNA-seq) of single mouse hepatocytes revealed that half of the detected transcripts are nonrandomly expressed (3,496 of 7,277 genes) (5). This suggests that liver zonation is a highly regulated process. Diffusible Wnt morphogens are secreted by the endothelial cells surrounding the central vein and regulate liver zonation by activation of the Wnt/β-catenin signaling pathway (6–9). Gene expression profiles of livers with hyperactivated [adenomatous polyposis coli (Apc) knockout mice] or inactivated (β-catenin–deficient mice) Wnt signaling revealed that 25% (884 of 3,496 genes) of all zonated genes are regulated by Wnt/β-catenin (5–7). Another 9% (298 genes) are regulated by hypoxia, Ras-dependent signaling pathways and pituitary hormones (5). Thus, the regulation of expression of two-thirds of the zonated liver genes remains to be established.
Glucagon is secreted from the α-cells of the endocrine pancreas and promotes hepatic glucose output to maintain normal blood glucose levels during fasting (10). In this study, we generated glucagon deficient (Gcg−/−) mice and demonstrate that glucagon is an important regulator of metabolic zonation in the liver. Unexpectedly, we found that glucagon modulates expression of a number of genes that are also regulated by the Wnt/β-catenin signaling pathway. Consistent with the drop in the blood glucagon concentration in the sinusoid along the periportal–central axis, we show that glucagon primarily affects gene expression and metabolic zonation in the periportal hepatocytes.
Results
Characterization of Gcg−/− Mice.
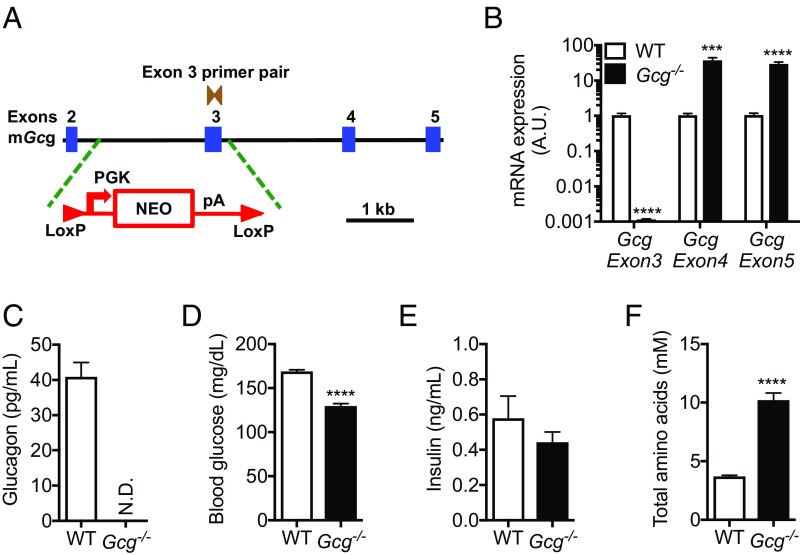
Gcg−/− mice were generated using Velocigene technology (11). The targeting vector contained a specific deletion of the glucagon coding exon 3 leaving the glucagon-like peptide 1 (GLP-1) and 2 (GLP-2) exons (exons 4 and 5) to be transcribed and translated in-frame (Fig. 1A). To verify specific absence of glucagon coding exon 3 in pancreas from Gcg−/− mice, real-time quantitative RT-PCR (qRT-PCR) analysis was performed with three primer pairs, each recognizing a specific exon. Gcg exon 3 mRNA was not detected in Gcg−/− mice (Fig. 1B). Lack of glucagon was confirmed in plasma of Gcg−/− mice and by pancreas α-glucagon immunostaining (Fig. 1C and Fig. S1A). Disruption of glucagon signaling induces compensatory increase in proglucagon (Pgcg) gene transcription (12–14). This is also the case in Gcg−/− mice, since exon 4 and 5 mRNA was increased >30-fold, resulting in high plasma levels of active GLP-1 (Fig. 1B and Fig. S1B). Gcg−/− mice showed an increased number of GLP-1+ islet cells compared with control mice. GLP-1 was used as a surrogate for glucagon to detect α-cells (Fig. S1A). Lack of glucagon and elevated plasma GLP-1 lowered fed and fasted blood glucose and fed insulin levels in the Gcg−/− mice (Fig. 1 D and E and Fig. S1C). Similar to previous studies of disrupted glucagon signaling (15–18), we found that Gcg−/− mice have elevated circulating amino acid levels (Fig. 1F and Fig. S1 D and E).
Fig. 1.
Targeting construct and phenotyping of Gcg−/− mice. (A) Cartoon of the targeting construct. Gcg-specific exon 3 was replaced by a Lox P-flanked transcriptional blocking cassette consisting of a neomycin resistance-encoding sequence with a poly(A) signal (pA). (B) Taqman qPCR analysis demonstrating Gcg-coding exon 3 deletion, but not deletion of exons 4 and 5 in Gcg−/− mice pancreas. (C) No glucagon detected in Gcg−/− mice plasma. (D) Decreased blood glucose in Gcg−/− mice. (E) Slightly decreased insulin in Gcg−/− mice plasma. (F) Increased plasma amino acids in Gcg−/− mice. Data are mean ± SEM. ***P < 0.001, ****P < 0.0001. N.D., not detected.
Gcg−/− Mice Have Perturbed Liver Gene Expression and Zonation Pattern.
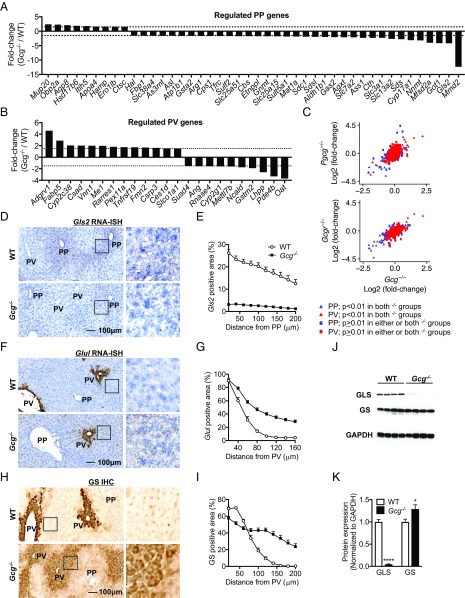
Using published data (3, 5), we identified 286 PP genes and 485 PV genes. Fig. S2 shows the contribution and overlap of the PP and PV genes from each study and their average scaled expression in the zones. Dataset S1 lists names of all PP and PV genes. We found 297 genes with perturbed expression in Gcg−/− mice (Dataset S2). Of these genes, 67 (23%) had PP or PV zonated expression. Two-thirds of the genes (44) were detected in PP hepatocytes. The majority of the genes (35) had reduced expression (Fig. 2A). Of the PV regulated genes, slightly more than half of the genes had increased expression in Gcg−/− mice (Fig. 2B). It is unlikely that the compensatory increases in GLP-1 and GLP-2 expression and secretion in the Gcg−/− mice affects liver zonation. This is based on the observations that the receptors for these ligands were expressed at very low levels in mouse livers [Glp1r, ≤0.06 reads per kilobase per million (RPKM) and Glp2r, ≤0.01 RPKM] and were not detected in single cell sequencing of hepatocytes (5). Furthermore, we observed a strong correlation of PP and PV zone gene expression regulation between Gcg−/− and Pgcg−/− (Glp-1, Glp-2, and Gcg deficiency) mice as well as between Gcg−/− and Gcgr−/− mice (Fig. 2C).
Fig. 2.
Glucagon contributes to metabolic zonation. (A) Regulated PP genes in Gcg−/− mice liver. (B) Regulated PV genes in Gcg−/− mice liver. (C) The correlation of PP and PV gene expression between Gcg−/− and Pgcg−/− or Gcgr−/− mice. (D) Gls2 RNA-ISH demonstrating down-regulation of Gls2 expression in Gcg−/− mice liver. (E) Quantification of Gls2 RNA-ISH positive area in liver. (F) Glul RNA-ISH, demonstrating extended Glul expression in Gcg−/− mice liver. (G) Quantification of Glul RNA-ISH positive area in liver. (H) GS immunohistochemistry (IHC) demonstrating extended GS expression in Gcg−/− mice liver. (I) Quantification of GS IHC positive area in liver. (J) Western blot demonstrating decreased expression of GLS and increased expression of GS in Gcg−/− mice liver. (K) Quantification of Western blot of GLS and GS in liver. All images are the same magnification. (Scale bar: 100 μm.) The insertions in D, F, and H were amplified with 4× magnification. Data are mean ± SEM *P < 0.05, ****P < 0.0001. RNA-seq comparison in A and B was between Gcg−/− and WT mouse liver samples.
To investigate the effects of reduced expression of PP genes on liver zonation, we used RNA in situ hybridization (RNA-ISH) to show RNA localization of glutaminase 2 (RNA, Gls2; protein, GLS) in Gcg−/− and WT mice. GLS catalyzes the hydrolysis of glutamine to stoichiometric amounts of glutamate and ammonia. It is a landmark gene for the PP zone. The Gls2 RNA-ISH signal in WT mice was localized to the six to eight layers of hepatocytes surrounding the periportal vein and no signal was detected in the hepatocytes surrounding the central vein. The Gls2 RNA-ISH signal was barely detectable in the PP area of Gcg−/− mice (Fig. 2 D and E). Quantification of the Gls2 RNA-ISH signal in increments of 20 μm (approximately the diameter of a hepatocyte; Fig. S3) from the periportal vein revealed a reduction in Gls2 positive area from 26% to 13% within 200 μm in WT mice, whereas only 1–3% positive area was detected in Gcg−/− mice at any distance from the periportal vein (Fig. 2 D and E). These data are consistent with reduced Gls2 mRNA expression in Gcg−/− mice (Fig. 2A) and GLS protein determined by Western blotting (Fig. 2 J and K). Similar Gls2 zonation pattern and gene expression changes were detected in female Gcg−/− mice (Fig. S4 A and B).
Glutamine synthetase (RNA, Glul; protein, GS) is a key landmark gene for the PV zone and involved in ammonia detoxification (7). The Glul RNA-ISH signal was detected in the one to three cell layers surrounding the central vein in WT mice (Fig. 2F). Interestingly, in the Gcg−/− mice the Glul RNA-ISH signal was extended by five to six cell layers toward the PP zone (Fig. 2F). Quantification of the Glul RNA-ISH signal from the central vein revealed a sharp reduction in positive area from 90% to 14% in 80 μm in WT mice, whereas 28% positive area was maintained until 160 μm from the central vein in Gcg−/− mice (Fig. 2 F and G). GS immunohistochemistry (IHC) showed similar but more clear expansion of the PV zone in Gcg−/− mice (Fig. 2 H and I). This was associated with slightly higher GS protein level in Gcg−/− mice (Fig. 2 J and K). Similar GS protein zonation pattern changes were detected in female Gcg−/− mice (Fig. S4C). Despite the interference with genes involved in amino acid metabolism and ammonia handling, we did not detect changes in plasma ammonia and urea levels in the Gcg−/− mice (Fig. S1 F and G). This is consistent with spare capacity of the urea cycle, which normally runs at 20–50% of capacity and most of the enzymes operate at or below their half-maximal capacity (19). Collectively, these data show that lack of glucagon reduces expression of PP genes, resulting in contraction of the PP zonation pattern. On the contrary, glucagon deficiency expanded the PV zone as illustrated by the detection of Glul mRNA and GS protein in additional cell layers surrounding the central vein.
Glucagon Infusion Restores Liver Zonation in Gcg−/− Mice.
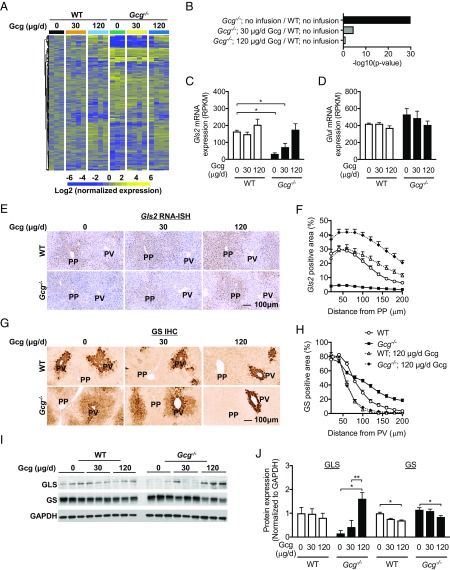
To test whether glucagon application restores liver zonation in Gcg−/− mice, dosages of 30 and 120 μg/d glucagon were infused to Gcg−/− and WT mice for 7 d, resulting in a dose-dependent increase in blood glucagon and glucose levels (Fig. S5). Fig. 3A shows a dose-dependent reversal of expression of the 297 affected genes in Gcg−/− mice. Gene pathway analysis revealed a normalization of PP gene expression in Gcg−/− mice (Fig. 3B). Glucagon infusion normalized Gls2 expression and zonation pattern in Gcg−/− mice (Fig. 3 C, E, and F). Western blotting confirmed normalization of GLS protein in Gcg−/− mice (Fig. 3 I and J). Contrary to the effects in the PP zone, glucagon infusion decreased GS protein by 30% in both WT and Gcg−/− mice (Fig. 3 I and J), which was secondary to retraction of GS expression to the two to three cell layers closest to the central vein (Fig. 3 G and H). Glul RNA levels in the 120 μg/d dose trended lower without a significant difference (Fig. 3D). Collectively, these data show that glucagon restores Gcg−/− mouse liver PP gene expression and zonation pattern. Glucagon also reduced expression of the GS protein except in cells in the immediate vicinity to the central vein.
Fig. 3.
Gcg infusion restored Gcg−/− mice liver zonation pattern. (A) Heat map of 297 affected genes in Gcg−/− mice demonstrated dose-dependent effects of Gcg infusion. (B) Gene pathway analysis demonstrated dosage-dependent rescue effects of Gcg infusion on regulated PP gene signature. (C) Gls2 mRNA liver expression (RPKM) demonstrated Gcg induced dosage dependent up-regulation of Gls2 expression in Gcg−/− mice. (D) Glul mRNA liver expression with Gcg infusion (RPKM). (E) Representative Gls2 RNA-ISH images in liver demonstrated Gcg infusion restored Gls2 liver zonation pattern in Gcg−/− mice. (F) Quantification of RNA-ISH Gls2-positive area in liver with Gcg infusion. (G) Representative GS IHC images in liver demonstrated Gcg infusion restored GS liver zonation pattern in Gcg−/− mice and high dosage pushed GS toward the central vein further in WT mice. (H) Quantification of GS IHC-positive area in liver with Gcg infusion. (I) GLS and GS protein expression with Gcg infusion demonstrated Gcg inducing GLS expression but inhibiting GS expression. (J) Quantification of protein expression of GLS and GS in liver with Gcg infusion. All images are the same magnification. (Scale bar: 100 μm.) Data are mean ± SEM. *P < 0.05, **P < 0.01, ****P < 0.0001. In C and D, WT glucagon 0 μg/d group was compared with the other five groups. Significant genes were defined as fold change > 1.5 in either up or down direction and with P values <0.01, and indicated by *.
Glucagon Receptors Are Expressed Throughout the Liver Lobules.
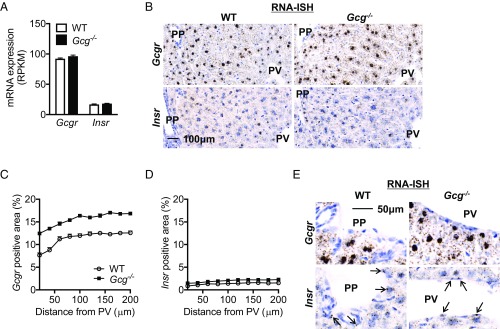
Next, we wanted to test whether the effects of glucagon on metabolic zonation in the liver is secondary to zonated expression of the glucagon receptor (Gcgr) and its downstream signaling components and whether the expression of the glucagon signaling pathway is compromised in Gcg−/− mice. Fig. 4A shows unperturbed hepatic expression of Gcgr mRNA in Gcg−/− mice. RNA-ISH detected Gcgr mRNA at comparable levels throughout the lobules (Fig. 4 B and C). The Gcgr RNA-ISH signal was slightly higher in Gcg−/− mice (Fig. 4C). Next, we extracted and plotted published expression data for Gcgr and 21 downstream signaling genes (5). The results confirm that expression of Gcgr and its signaling components are evenly distributed throughout the liver lobules (Fig. S6 A and B).
Fig. 4.
Expression of Gcgr and Insr in liver. (A) Gcgr and Insr mRNA liver expression (RPKM) did not show changes in Gcg−/− mouse. (B) Representative Gcgr and Insr RNA-ISH images in liver did not reveal zonation pattern or changes in Gcg−/− mouse. Images are the same magnification. (Scale bar: 100 μm.) (C) Quantification of Gcgr RNA-ISH positive area in liver. (D) Quantification of Insr RNA-ISH positive area in liver. (E) RNA-ISH images showed Insr but not Gcgr expressed in endothelial cells at liver PP and PV zone. Arrows indicate positive endothelial cells. Images are the same magnification. (Scale bar: 50 μm.)
Insulin is the counterregulatory hormone of glucagon in the liver. Insulin receptor (Insr) RNA-ISH revealed even expression throughout the liver lobules and unchanged expression in Gcg−/− mice (Fig. 4 A, B, and D). Uniform distribution of Insr and its downstream 35 signaling genes was confirmed by reanalysis of published RNA expression data (5) (Fig. S6 A and C). Interestingly, Insr, but not Gcgr, is also expressed in the endothelial cells lining the periportal and central veins (Fig. 4E).
Glucagon and Wnt/β-Catenin Regulated Genes Overlap in PP and PV Hepatocytes.
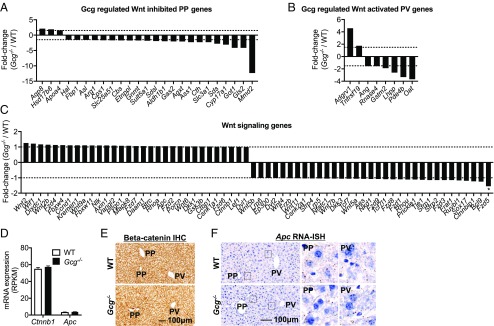
Our data show that glucagon contributes to liver zonation, although the glucagon receptor and its signaling genes do not show zonated expression. This raises the possibility that glucagon may interact with the Wnt/β-catenin signaling pathway to regulate gene expression and the metabolic zonation pattern. It has previously been reported that Wnt/β-catenin–inhibited genes (88 genes) are primarily expressed in PP hepatocytes, whereas Wnt/β-catenin–activated genes (87 genes) are found in their PV counterparts (5). Supporting the hypothesis that the glucagon and Wnt/β-catenin signaling pathways interact, we found that 28% of the 88 PP expressed and Wnt/β-catenin–inhibited genes were regulated in the Gcg−/− mice (Fig. 5A). Furthermore, expression of 9% of the PV and Wnt/β-catenin–activated genes were affected in the Gcg−/− mice (Fig. 5B). These data suggest that many liver genes are regulated by both glucagon and Wnt/β-catenin, particularly in the PP hepatocytes.
Fig. 5.
Gcg regulated Wnt target genes but not Wnt signaling genes. (A) Fold changes of glucagon regulated Wnt inhibited PP genes. (B) Fold changes of glucagon regulated Wnt activated PV genes. (C) Fold changes of all Wnt signaling genes expressed in Gcg−/− mouse liver. *P < 0.01 indicated the only regulated gene. (D) Ctnnb1 and Apc2 mRNA liver expression (RPKM). (E) Representative β-catenin IHC images in liver. (F) Representative Apc2 RNA-ISH images in liver. Middle and Right images are sixfold magnification from the circled area in the Left images. All images are the same magnification. (Scale bar: 100 μm.) The RNA-seq analysis is the same as in Fig. 2 A and B. Significant genes were defined as fold change > 1.5 in either up or down direction and with P value <0.01, and indicated by *.
Wnt/β-Catenin Signaling Genes Are Not Zonated or Regulated by Glucagon.
Among 41 Wnt/β-catenin signaling genes in the mouse liver, only Fzd5 (−1.55) was slightly down-regulated in Gcg−/− mice (Fig. 5C). Reanalysis of published data (5) revealed that 40 of these genes are express evenly throughout the lobules, whereas Ctnnbip1 shows a trend toward higher expression in the PP zone (Fig. S6D).
Further analysis revealed that the distribution of the β-catenin (RNA, Ctnnb1; protein, β-catenin) protein was even throughout the liver lobules and the mRNA expression was similar in WT and Gcg−/− mouse livers (Fig. 5 C–E). RNA-ISH of Apc (RNA, Apc; protein, APC), a negative regulator of Wnt/β-catenin signaling was also evenly detected throughout the liver lobules and the expression was similar in WT and Gcg−/− mice (Fig. 5 C, D, and F). In summary, the expression of Wnt/β-catenin signaling genes was not altered in the Gcg−/− mouse liver. This suggests that glucagon and Wnt/β-catenin likely regulate gene expression by cross-talk at the transcriptional level.
Wnt Signaling Inhibition Contracts the PV Zone in Gcg−/− Mice.
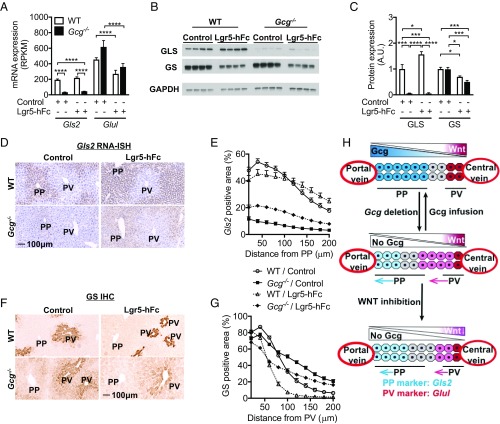
Leucine-rich repeat-containing G protein-coupled receptor 5 (Lgr5) binds to Rspondin (RSPO) to promote Wnt/β-catenin signaling (20–22). Therefore, to inhibit Wnt/β-catenin signaling, we delivered cDNA of the extracellular domain of Lgr5 fused with hFc by hydrodynamic DNA delivery (HDD) to livers of Gcg−/− and WT mice. Lgr5–hFc slightly increased Gls2 mRNA and GLS protein levels in Gcg−/− and WT mice (Fig. 6 A–E). Consistent with the primary action of Wnt/β-catenin in the PV zone, Lgr5–hFc reduced Glul mRNA and GS protein in Gcg−/− and WT mice (Fig. 6 A–C). Furthermore, the GS zonation pattern in Gcg−/− mice was retracted toward the central vein and was limited to one to two cell layers around the central vein in WT mice (Fig. 6 F and G). The finding that the GS zone was retracted but not missing is consistent with Lgr5–hFc binding to RSPO being a partial inhibitor of Wnt/β-catenin signaling (23, 24).
Fig. 6.
Wnt inhibition down-regulated GS expression and up-regulated GLS expression. (A) Gls2 and Glul mRNA liver expression with Lgr5-hFc HDD (RPKM). (B) GLS and GS protein liver expression with Lgr5-hFc HDD demonstrated Wnt inhibition down-regulated GS expression but up-regulated GLS expression. (C) Quantification of protein expression of GLS and GS in liver with Lgr5-hFc HDD. (D) Representative Gls2 RNA-ISH images in liver with Lgr5-hFc HDD. (E) Quantification of RNA-ISH Gls2 positive area in liver with Lgr5-hFc HDD. (F) Representative GS IHC images in liver with Lgr5-hFc HDD. (G) Quantification of GS IHC positive area in liver with Lgr5-hFc HDD. (H) Model for Gcg and Wnt cross-talk to regulate liver zonation. All images are the same magnification. (Scale bar: 100 μm.) Data are mean ± SEM. *P < 0.05, ***P < 0.001, ****P < 0.0001.
Discussion
Zonation of liver metabolic functions is supported by pronounced heterogeneity in gene expression between hepatocytes along the liver lobules. A recent single hepatocyte mRNA sequencing study revealed that half of the detected transcripts in mouse liver have zonated expression patterns, suggesting that liver zonation is a highly regulated process (5). Recent studies have highlighted that the Wnt/β-catenin signaling pathway is a key determinant of liver zonation (6–9). In fact, 25% of all zonated genes are regulated by Wnt/β-catenin (5). The regulation of only 9% of the other zonated genes is understood, leading to the question of which other hormones and mechanisms control liver zonation. Our data reveal an interaction between the glucagon and Wnt/β-catenin signaling pathways. Specifically, we demonstrate that these pathways counteract to regulate liver metabolic functions. Wnt/β-catenin signaling is more dominant in the PV zone regulating Wnt/β-catenin–activated gene expression, while glucagon action is more important in the PP zone.
Although the target gene expression of glucagon and Wnt/β-catenin showed restricted zonation patterns, the genes involved in their signaling were evenly distributed throughout the liver lobules. Previous studies have indicated that Apc IHC-positive staining was localized to the PP zone to inhibit Wnt/β-catenin signaling and that its counterplay maintains the homeostasis of liver zonation (6). Our RNA-ISH results show that Apc mRNA is evenly distributed throughout the liver lobules. Reanalysis of published single hepatocyte RNA sequencing data confirms that Apc mRNA is evenly distributed along the lobules (5).
Despite the established role of Wnt/β-catenin as the gatekeeper of liver metabolic zonation, the instructive mechanism controlling its spatiotemporal regulation is not known. With the even (nonzoned) distribution of the glucagon and Wnt/β-catenin signaling genes, the restricted expression patterns of their target genes could be explained by a combination of concentration gradients of the ligands between the PP and PV zones and by interactions between the signaling pathways at the transcriptional level. Diffusible Wnt morphogens are secreted by the endothelial cells surrounding the central vein and act upon nearby hepatocytes (25, 26). The efficiency of Wnt/β-catenin targeting depends on the local Wnt and Wnt regulator concentrations. The same accounts for glucagon but in the PP zone. Glucagon is released from the pancreatic α-cells and is distributed to the liver lobules with nutrient-rich blood from the portal vein where it flows through the PP and PV zones before reaching the central vein. Importantly, 24% of glucagon is removed from the blood during the passage of the PP and PV zones (27). The concentration gradients of glucagon and Wnt ligands may only partially explain the zoned regulation of their target genes. The counterplay between them, as demonstrated in our study, also contributes to the regulation. Similar opposing transcriptional interactions have been described for Wnt/β-catenin and hepatocyte transcription factor 4α (28). Thus, liver zonation may be influenced by the rate of glucagon secretion from the α-cells, a highly regulated process which involves neurohormonal factors and nutrients (10). Our understanding of the regulation of Wnt release from endothelial cells is quite limited. It has been hypothesized that injury-related chemokines may regulate release of Wnt ligands (29, 30).
In conclusion, our data show that glucagon is required for the maintenance of liver metabolic zonation and that it counteracts the Wnt/β-catenin signaling pathway to regulate the expression of glucagon and Wnt/β-catenin–activated or –inhibited genes.
Materials and Methods
Generation of Mouse Lines.
Mice were generated using BAC-based homologous recombination technology (11). In brief, we modified a BAC containing the Gcg coding sequence by replacing the 1.86-kb region, consisting of introns flanking both ends of exon 3, with the neomycin cassette. After germline transmission was established, F1 Gcg+/− mice were bred together to generate F2 Gcg+/+ and Gcg−/− mice. Also, F1 Gcg+/− mice were backcrossed to C57BL6/J to generate N2 breeding heterozygote pairs that were used to generate N2F2 Gcg+/+ and Gcg−/− mice. The studies reported here were conducted on F2 or N2F2 littermates (8–12 wk of age) that were housed under 12 h of light per day in a temperature-controlled environment (22 ± 1 °C, 60–70% humidity). Mice deficient in the proglucagon gene (Pgcg−/−) (87.5% C57BL/6NTac; 12.5% 129S6/SvEvTac) were generated by a 4.9-kb deletion of proglucagon gene exons 1–4 by homologous recombination using Regeneron’s VelociGene technology (11). Mice deficient in the glucagon receptor (Gcgr−/−) (87.5% C57BL/6NTac; 12.5% 129S6/SvEvTac) were generated as described (16). All procedures were conducted in compliance with protocols approved by the Regeneron Institutional Animal Care and Use Committee. Animals had free access to standard chow (LabDiet 5001).
Hydrodynamic DNA Delivery of Lgr5–hFc.
Extracellular domain of Lgr5 was fused with hFc and cloned into expression vector pRG977 construct. The construct was verified by DNA sequencing and secretion of the corresponding gene product was confirmed by ELISA analysis of culture media from transiently transfected HEK293 cells. To establish a baseline for serum chemistry parameters, serum samples were collected 7 d before HDD construct administration. Mice (n = 4–5 per group) were administered a single IV injection of Lgr5–hFc or control construct. One week after the injection, mice were killed and liver samples were collected for histology and Western blotting. LGR5–hFc expression was confirmed by plasma ELISA.
Glucagon Pump Infusion.
Glucagon microosmotic pump implantation surgery was performed as described (31). Glucagon (Sigma) was dissolved in a cetrimide solution (Sigma) at a ratio of 6 mol cetrimide to 1 mol glucagon to maintain long-term solubility (31). Glucagon solutions were made to concentrations necessary to deliver 30 or 120 μg glucagon per day for 7-d microosmotic pumps (Alzt). Control littermates were implanted with a pump containing the same concentration of cetrimide solution alone. Briefly, animals were anesthetized and a microosmotic pump (model 1002) was implanted s.c. in the back of the mice. Blood glucose was measured using a hand-held glucometer. One week later, blood was collected by cardiac puncture and liver was collected and processed for histology and RNA preparation.
RNA-Seq Preparation and Analysis.
Total RNA was purified from all samples using MagMAX-96 for Microarrays Total RNA Isolation Kit (Ambion by Life Technologies) according to manufacturer’s specifications. Genomic DNA was removed using MagMAXTurboDNase Buffer and TURBO DNase from the MagMAX Kit listed above (Ambion by Life Technologies). mRNA was purified from total RNA using Dynabeads mRNA Purification Kit (Invitrogen). Strand-specific RNA-seq libraries were prepared using KAPA mRNA-Seq Library Preparation Kit (Kapa Biosystems). Twelve-cycle PCR was performed to amplify libraries. Sequencing was performed on Illumina HiSeq2000 by a multiplexed single-read run with 33 cycles. Raw sequence data (BCL files) were converted to FASTQ format via Illumina Casava 1.8.2. Reads were decoded based on their barcodes and read quality was evaluated with FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were mapped to the mouse transcriptome (NCBI GRCm38) using ArrayStudio software (OmicSoft) allowing two mismatches. Reads mapped to the exons of a gene were summed at the gene level. Differentially expressed genes were identified by the DESeq2 package and significantly perturbed genes were defined with fold changes no less than 1.5 in either up or down direction and with P values of at least 0.01.
Quantitative Real-Time PCR Analysis.
qRT-PCR analysis was performed using TaqMan qRT-PCR chemistry and detection system (Applied Biosystems) with the primer pairs and labeled probes for Gcg gene exons 3–5. Relative mRNA levels were calculated by the ΔΔCt method, using a housekeeping gene (cyclophilin A) for normalization and the mean value of WT mice as the reference value.
Liver Zonation Genes.
PP- and PV-enriched genes were obtained from refs. 3 and 5. A total of 285 PP and 485 PV genes were identified. Gls-2 was manually added to the PP gene list. Wnt-activated (635 genes) or inhibited genes (249 genes) were extracted from liver gene signatures from Apc−/− and β-cat−/− mice (28). Wnt-signaling genes were obtained from SABiosciences. Glucagon-signaling pathway genes were obtained according to the pathway diagram from https://rgd.mcw.edu/rgdweb/pathway/pathwayRecord.html?acc_id=PW:0000676. Insulin-signaling pathway genes were obtained according to the pathway diagram from https://www.intechopen.com/books/type-2-diabetes/mitochondrial-metabolism-and-insulin-action.
Annotation of Gcg−/−, Pgcg−/−, and Gcgr−/− Mouse Signature.
Gcg−/− mouse signature is annotated by the four compiled gene sets (PP, PV, Wnt-inhibited, and Wnt-activated) and Biological Process (BP) GO terms with P value <10−5. None of the glucagon and insulin signaling genes were regulated in the Gcg−/− mouse. Pgcg−/− and Gcgr−/− mouse signature were generated using the same approch.
Tissue Morphology.
Liver samples were collected, fixed in 4% paraformaldehyde for 48 h, equilibrated in 20% sucrose for 24 h, and then in 30% sucrose for 24 h at 4 °C before cryosectioning. The 10-μm sections were freshly prepared. IHC and RNA-ISH were done on separate sections. For RNA-ISH, sections were permeabilized and hybridized with mRNA probes to Gls2, Glul, Gcgr, Insr, or Apc (ACD Bio). Following probe hybridization and amplification, mRNA was detected using RNAscope 2.5 HD Assay, brown kit (n = 3–4 mice from each genotype, n = 4–6 random pictures from each slide). Slides were scanned using a Zeiss Axio Scan Z1 slide scanner and the images were analyzed using Halo software (Indica Labs). For IHC, sections from each animal were stained with either α-GS or α-β-catenin antibodies (BD).
Blood Chemistry.
Blood glucose was determined using ACCU-CHEK Compact Plus (Roche Diagnostics). Plasma glucagon (Mercodia), active-GLP-1 (Meso Scale Discovery), and insulin (Mercodia) levels were determined using ELISA. Plasma total amino acid levels were measured using l-Amino Acid Quantification Kit (Sigma-Aldrich), which detects l-amino acids with exception of l-glycine. Concentrations of individual amino acids were measured by Metabolon with the use of gas chromatography–mass spectrometry. Ammonia and urea were assayed in a Siemens ADVIA Chemistry XPTB Clinical System.
Western Blotting.
Liver samples were lysed with ice-cold RIPA buffer (50 mM Tris, 150 mM NaCl, 1 mM EDTA, 50 mM NaF, 10 mM β-glycerophosphate, 5 mM sodium pyrophosphate dibasic and 1% Nonidet P-40) in the presence of protease and phosphatase inhibitor mixtures (Thermo Fisher Scientific), 1 mM DTT and 2 mM Na3VO4. Total sample lysates were mixed with 6× SDS loading buffer (Alfa-Aesar) and boiled for 5 min. Protein samples (10–100 μg) were loaded and separated on 4–20% gradient SDS/PAGE gels (Bio-Rad) and transferred to polyvinylidene difluoride membranes. The membranes were blocked for 1 h with 5% BSA in 1× TBS supplemented with 0.1% Tween 20 (Bio-Rad) and incubated with the following antibodies: GS (610518, BD), GLS2 (150474, Abcamab), and GAPDH (14C10, Cell Signaling). Bound antibodies were detected using horseradish peroxidase-conjugated anti-rabbit or anti-mouse secondary antibodies (1:10,000; Jackson ImmunoResearch) and enhanced chemiluminescence reagent (Thermo Fisher Scientific). Band intensities were quantified in ImageJ software.
Data Analysis.
All data are mean ± SEM. Statistical analyses were performed utilizing GraphPad software Prism 6.0. All parameters were analyzed by Student’s t test, one-way ANOVA, or two-way ANOVA; a threshold of P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Erqian Na for processing pancreas IHC and Kevin Barringer for generating the Lgr5–hFc HDD construct.
Footnotes
Conflict of interest statement: All authors are employees and shareholders of Regeneron Pharmaceuticals, Inc.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. 110674).
See Commentary on page 4308.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721403115/-/DCSupplemental.
References
- 1.Jungermann K, Katz N. Functional specialization of different hepatocyte populations. Physiol Rev. 1989;69:708–764. doi: 10.1152/physrev.1989.69.3.708. [DOI] [PubMed] [Google Scholar]
- 2.Jungermann K, Kietzmann T. Zonation of parenchymal and nonparenchymal metabolism in liver. Annu Rev Nutr. 1996;16:179–203. doi: 10.1146/annurev.nu.16.070196.001143. [DOI] [PubMed] [Google Scholar]
- 3.Braeuning A, et al. Differential gene expression in periportal and perivenous mouse hepatocytes. FEBS J. 2006;273:5051–5061. doi: 10.1111/j.1742-4658.2006.05503.x. [DOI] [PubMed] [Google Scholar]
- 4.Gebhardt R, Matz-Soja M. Liver zonation: Novel aspects of its regulation and its impact on homeostasis. World J Gastroenterol. 2014;20:8491–8504. doi: 10.3748/wjg.v20.i26.8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halpern KB, et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature. 2017;542:352–356. doi: 10.1038/nature21065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benhamouche S, et al. Apc tumor suppressor gene is the “zonation-keeper” of mouse liver. Dev Cell. 2006;10:759–770. doi: 10.1016/j.devcel.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 7.Sekine S, Lan BY, Bedolli M, Feng S, Hebrok M. Liver-specific loss of beta-catenin blocks glutamine synthesis pathway activity and cytochrome p450 expression in mice. Hepatology. 2006;43:817–825. doi: 10.1002/hep.21131. [DOI] [PubMed] [Google Scholar]
- 8.Burke ZD, et al. Liver zonation occurs through a beta-catenin-dependent, c-Myc-independent mechanism. Gastroenterology. 2009;136:2316–2324.e1-3. doi: 10.1053/j.gastro.2009.02.063. [DOI] [PubMed] [Google Scholar]
- 9.Gebhardt R, Hovhannisyan A. Organ patterning in the adult stage: The role of Wnt/beta-catenin signaling in liver zonation and beyond. Dev Dyn. 2010;239:45–55. doi: 10.1002/dvdy.22041. [DOI] [PubMed] [Google Scholar]
- 10.Gromada J, Franklin I, Wollheim CB. Alpha-cells of the endocrine pancreas: 35 years of research but the enigma remains. Endocr Rev. 2007;28:84–116. doi: 10.1210/er.2006-0007. [DOI] [PubMed] [Google Scholar]
- 11.Valenzuela DM, et al. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat Biotechnol. 2003;21:652–659. doi: 10.1038/nbt822. [DOI] [PubMed] [Google Scholar]
- 12.Gelling RW, et al. Lower blood glucose, hyperglucagonemia, and pancreatic alpha cell hyperplasia in glucagon receptor knockout mice. Proc Natl Acad Sci USA. 2003;100:1438–1443. doi: 10.1073/pnas.0237106100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parker JC, Andrews KM, Allen MR, Stock JL, McNeish JD. Glycemic control in mice with targeted disruption of the glucagon receptor gene. Biochem Biophys Res Commun. 2002;290:839–843. doi: 10.1006/bbrc.2001.6265. [DOI] [PubMed] [Google Scholar]
- 14.Conarello SL, et al. Glucagon receptor knockout mice are resistant to diet-induced obesity and streptozotocin-mediated beta cell loss and hyperglycaemia. Diabetologia. 2007;50:142–150. doi: 10.1007/s00125-006-0481-3. [DOI] [PubMed] [Google Scholar]
- 15.Solloway MJ, et al. Glucagon couples hepatic amino acid catabolism to mTOR-dependent regulation of α-cell mass. Cell Rep. 2015;12:495–510. doi: 10.1016/j.celrep.2015.06.034. [DOI] [PubMed] [Google Scholar]
- 16.Okamoto H, et al. Glucagon receptor blockade with a human antibody normalizes blood glucose in diabetic mice and monkeys. Endocrinology. 2015;156:2781–2794. doi: 10.1210/en.2015-1011. [DOI] [PubMed] [Google Scholar]
- 17.Dean ED, et al. Interrupted glucagon signaling reveals hepatic α cell axis and role for L-glutamine in α cell proliferation. Cell Metab. 2017;25:1362–1373.e5. doi: 10.1016/j.cmet.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J, et al. Amino acid transporter Slc38a5 controls glucagon receptor inhibition-induced pancreatic α cell hyperplasia in mice. Cell Metab. 2017;25:1348–1361.e8. doi: 10.1016/j.cmet.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meijer AJ, Lamers WH, Chamuleau RA. Nitrogen metabolism and ornithine cycle function. Physiol Rev. 1990;70:701–748. doi: 10.1152/physrev.1990.70.3.701. [DOI] [PubMed] [Google Scholar]
- 20.Glinka A, et al. LGR4 and LGR5 are R-spondin receptors mediating Wnt/β-catenin and Wnt/PCP signalling. EMBO Rep. 2011;12:1055–1061. doi: 10.1038/embor.2011.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carmon KS, Gong X, Lin Q, Thomas A, Liu Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc Natl Acad Sci USA. 2011;108:11452–11457. doi: 10.1073/pnas.1106083108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Lau W, et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- 23.Rocha AS, et al. The angiocrine factor Rspondin3 is a key determinant of liver zonation. Cell Rep. 2015;13:1757–1764. doi: 10.1016/j.celrep.2015.10.049. [DOI] [PubMed] [Google Scholar]
- 24.Planas-Paz L, et al. The RSPO-LGR4/5-ZNRF3/RNF43 module controls liver zonation and size. Nat Cell Biol. 2016;18:467–479. doi: 10.1038/ncb3337. [DOI] [PubMed] [Google Scholar]
- 25.Nejak-Bowen K, Monga SP. Wnt/beta-catenin signaling in hepatic organogenesis. Organogenesis. 2008;4:92–99. doi: 10.4161/org.4.2.5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J, et al. β-catenin signaling in murine liver zonation and regeneration: A Wnt-Wnt situation! Hepatology. 2014;60:964–976. doi: 10.1002/hep.27082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dobbins RL, et al. Compartmental modeling of glucagon kinetics in the conscious dog. Metabolism. 1995;44:452–459. doi: 10.1016/0026-0495(95)90051-9. [DOI] [PubMed] [Google Scholar]
- 28.Gougelet A, et al. T-cell factor 4 and β-catenin chromatin occupancies pattern zonal liver metabolism in mice. Hepatology. 2014;59:2344–2357. doi: 10.1002/hep.26924. [DOI] [PubMed] [Google Scholar]
- 29.Monga SP. Role and regulation of β-catenin signaling during physiological liver growth. Gene Expr. 2014;16:51–62. doi: 10.3727/105221614X13919976902138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lengfeld JE, et al. Endothelial Wnt/β-catenin signaling reduces immune cell infiltration in multiple sclerosis. Proc Natl Acad Sci USA. 2017;114:E1168–E1177. doi: 10.1073/pnas.1609905114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Webb GC, Akbar MS, Zhao C, Swift HH, Steiner DF. Glucagon replacement via micro-osmotic pump corrects hypoglycemia and alpha-cell hyperplasia in prohormone convertase 2 knockout mice. Diabetes. 2002;51:398–405. doi: 10.2337/diabetes.51.2.398. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.