Significance
We demonstrate yeast’s capacity for biosynthesis of complex plant natural products by reconstructing a de novo noscapine biosynthetic pathway and optimizing noscapine production toward scalable manufacturing. Engineered strain contains 25 heterologous plant, bacteria, and mammalian genes and 6 mutant or overexpressed yeast genes. The noscapine biosynthetic pathway incorporates seven endomembrane-localized plant enzymes, highlighting yeast’s ability to functionally express and properly localize large numbers of heterologous enzymes into the endoplasm reticulum. Noscapine titers were improved by 18,000-fold (to low mg/L levels) via a combination of enzyme engineering, pathway and strain engineering, and fermentation optimization. We demonstrated that microbial fermentation can be used to produce halogenated alkaloid derivatives, which can ultimately serve as potential drug leads, through feeding amino acid derivatives to strains.
Keywords: benzylisoquinoline alkaloids, metabolic engineering, plant alkaloids, Saccharomyces cerevisiae
Abstract
Microbial biosynthesis of plant natural products from simple building blocks is a promising approach toward scalable production and modification of high-value compounds. The pathway for biosynthesis of noscapine, a potential anticancer compound, from canadine was recently elucidated as a 10-gene cluster from opium poppy. Here we demonstrate the de novo production of noscapine in Saccharomyces cerevisiae, through the reconstruction of a biosynthetic pathway comprising over 30 enzymes from plants, bacteria, mammals, and yeast itself, including 7 plant endoplasmic reticulum (ER)-localized enzymes. Optimization directed to tuning expression of pathway enzymes, host endogenous metabolic pathways, and fermentation conditions led to an over 18,000-fold improvement from initial noscapine titers to ∼2.2 mg/L. By feeding modified tyrosine derivatives to the optimized noscapine-producing strain we further demonstrated microbial production of halogenated benzylisoquinoline alkaloids. This work highlights the potential for microbial biosynthetic platforms to support the synthesis of valuable and novel alkaloid compounds, which can advance alkaloid-based drug discovery and development.
Noscapine is a natural phthalideisoquinoline alkaloid drug that is extracted from opium poppy (Papaver somniferum). Although present in opium, noscapine is a nonnarcotic drug that has been used worldwide for over 50 years as a safe, nonnarcotic antitussive. Noscapine has also been demonstrated to exhibit anticancer activity (1, 2); however, the dose required for treating cancer is much higher than when it is used as a cough suppressant (1,000–2,250 mg/d for cancer treatment vs. 45–200 mg/d for cough treatment). Due to its long-established safety record, noscapine is a compelling candidate anticancer drug and has been used off-label to treat cancers in some countries (3). In addition, a number of noscapine derivatives have been shown to exhibit higher therapeutic potential toward certain cancer cell lines resulting from enhanced in vitro and in vivo activities or water solubility (4, 5). Noscapine and its analogs (collectively referred to as noscapinoids) serve as an emerging class of microtubule-modulating anticancer agents that exhibit fewer side effects than traditional chemotherapy drugs (6).
All noscapinoids are currently derived from noscapine isolated from the opium poppy. Tens of tons of noscapine are extracted from thousands of tons of raw narcotic poppy straw annually to meet medical and scientific demand (7). Industrial poppy farming is susceptible to environmental factors such as pests, disease, and climate, resulting in instability and variability in this geographically concentrated supply chain (8). Relying on a narcotic supply chain (i.e., opium poppy) to produce a nonnarcotic drug introduces additional unnecessary hurdles and costs to procurement, which are amplified due to the small size of the noscapinoid market relative to the opioid market. Despite these challenges, poppy farming remains the sole source of noscapinoids, in part because chemical synthesis of these complex molecules is not commercially competitive. A number of chemical syntheses of noscapine and derivatives have been reported, but none are currently feasible for large-scale production (5, 9).
A microbial-based manufacturing process for noscapinoids, which are part of a larger class of benzylisoquinoline alkaloids (BIAs), has the potential to address many of the challenges associated with the current poppy-based supply chain. Industrial cultivation of microorganisms, such as the baker’s yeast Saccharomyces cerevisiae, occurs over days, whereas poppies are annuals. Also, because microbes are grown in closed fermentation vessels, the production process is not susceptible to external environmental factors and could provide greater consistency in product composition and impurity profiles across batches. Finally, by engineering a microorganism that does not make the broader profile of narcotic alkaloids present naturally in the opium poppy, one can remove the unnecessary hurdles associated with a tightly controlled supply chain.
In recent years, a number of pioneering studies have demonstrated the reconstruction of complex plant-based pathways in yeast, including the complete biosynthesis of strictosidine (10) and opioids (11). A 10-gene cluster encoding enzymes responsible for the biosynthesis of noscapine from (S)-scoulerine in opium poppy was recently identified (12, 13). We reconstructed the noscapine gene cluster in yeast to achieve microbial biosynthesis of noscapine from (S)-canadine and norlaudanosoline (14). Separately, we engineered the first part of the biosynthetic pathway from glucose to (S)-reticuline (15) and (S)-reticuline to (S)-canadine (16) into yeast (Fig. 1A). These earlier studies suggested that yeast might be capable of synthesizing noscapinoids via fermentation of simple carbon and nitrogen sources. However, functionally expressing the more than 25 heterologous enzymes required for complete biosynthesis of these complex molecules is nontrivial because of the decreases in titer observed with each additional enzymatic step. In addition, microbial expression of multiple endomembrane-localized plant enzymes, such as cytochrome P450s, has remained challenging (17), and full reconstruction of the noscapine biosynthetic pathway requires expression of seven membrane-localized enzymes, including five plant cytochrome P450s.
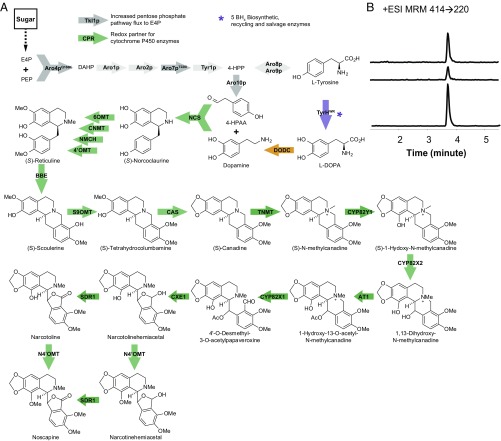
Fig. 1.
De novo noscapine biosynthesis in S. cerevisiae. (A) The proposed biosynthetic pathway of noscapine in yeast. Block arrows indicate enzyme-catalyzed steps. Light gray arrows, unmodified yeast enzymes; dark gray arrows, overexpressed and modified yeast enzymes; purple arrows, mammalian (Rattus norvegicus) enzymes; orange arrows, bacterial (Pseudomonas putida) enzymes; green arrows, plant (Papaver somniferum, Coptis japonica) enzymes. Aro10p, phenylpyruvate decarboxylase; Aro1p, pentafunctional arom enzyme; Aro2p, bifunctional chorismate synthase and flavin reductase; Aro4pQ166K, DAHP synthase; Aro7pT226I, chorismate mutase; Aro8p, aromatic aminotransferase I; Aro9p, aromatic aminotransferase II; AT1, 1,13-dihydroxy-N-methylcandine 13-O-acetyltransferase; BBE, berberine bridge enzyme; BH4, 5,6,7,8-tetrahydrobiopterin; CAS, canadine synthase; CNMT, coclaurine N-methyltransferase; CPR, cytochrome P450 reductase; CXE1, 3-O-acetylpapaveroxine carboxylesterase; CYP82X1, 1-hydroxy-13-O-acetyl-N-methylcanadine 8-hydroxylase; CYP82X2, 1-hydroxy-N-methylcanadine 13-hydroxylase; CYP82Y1, 1-hydroxy-N-methylcanadine synthase; DAHP, 3-deoxy-d-arabino-2-heptulosonic acid 7-phosphate; DODC, l-dopa decarboxylase; E4P, erythrose 4-phosphate; MT2/MT3, narcotoline-4′-O-methyltransferase. NCS, (S)-norcoclaurine synthase; NMCH, N-methylcoclaurine hydroxylase; PEP, phosphoenolpyruvate; S9OMT, scoulerine 9-O-methyltransferase; SDR1, short-chain dehydrogenase/reductase; Tkl1p, transketolase; TNMT, tetrahydroprotoberberine cis-N-methyltransferase; Tyr1p, prephenate dehydrogenase; TyrHWR, feedback inhibition-resistant tyrosine hydroxylase (R37E, R38E, W166Y); 4-HPAA, 4-hydroxyphenylacetaldehyde; 4-HPP, 4-hydroxyphenylpyruvate; 4′OMT, 3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase; and 6OMT, norcoclaurine 6-O-methyltransferase. (B) EIC MRM using noscapine’s highest characteristic precursor ion/product ion transition (414 → 220) of (i) 0.5 μg/L noscapine standard, (ii) CSY1149 media, and (iii) CSY1150 media. CSY1150 was cultured in SD with 2% dextrose in the presence of 10 mM ascorbic acid at 25 °C for 72 h. Growth medium was analyzed by LC–MS/MS MRM. Traces are representative of three biological replicates.
We demonstrate the de novo production of the anticancer alkaloid noscapine via a single engineered yeast strain. The initial yeast strain was engineered to express 29 enzymes from plant, bacteria, mammals, and yeast and produced ∼120–230 ng/L noscapine. Through engineering rate-limiting pathway enzymes, optimizing enzyme expression levels, introducing modifications to the endogenous yeast metabolism to enhance NADPH supply, and optimizing fermentation conditions, we improved noscapine titers by over 18,000-fold to ∼2.2 mg/L. We also demonstrated the biosynthesis of halogenated benzylisoquinoline alkaloid molecules by feeding tyrosine derivatives to the optimized noscapine-producing strain. Further optimization of the pathway and process will enable scale up to a commercially relevant, cost-effective fermentation process, offering a supply chain for noscapine and related molecules that does not rely on opium poppy farming. In addition, the yeast-based platform also supports the synthesis of novel benzylisoquinoline alkaloids and can be used to advance drug discovery, particularly for new chemotherapeutics.
Results
Construction of a de Novo Noscapine Biosynthetic Pathway in Yeast.
We started the construction of a de novo biosynthetic pathway to noscapine with two previously described platform strains that produce (S)-reticuline (11, 15), a key biosynthetic intermediate to many benzylisoquinoline alkaloids including the noscapinoids. The reticuline-producing strains encode the expression of 17 enzymes responsible for the biosynthesis of reticuline across five genetic modules (Fig. 1B and Fig. S1). The five modules were integrated into select chromosomal loci in a wild-type haploid strain CEN.PK2-1D. Module I is designed to increase accumulation of l-tyrosine and 4-hydroxyphenylacetaldehyde (4-HPAA) and encodes the overexpression of four yeast proteins—mutants of 3-deoxy-d-arabino-2-heptulosonic acid 7-phosphate (DAHP) synthase and chorismate mutase (Aro4pQ166K and Aro7pT226I) that are less susceptible to feedback inhibition by l-tyrosine, and transketolase (TKL1p) and phenylpyruvate decarboxylase (Aro10p). Module II is designed to synthesize and recycle tetrahydrobiopterin (BH4), the cofactor for tyrosine hydroxylase (TyrH), and encodes expression of four proteins from Rattus norvegicus—sepiapterin reductase (SepR), 6-pyruvoyl tetrahydrobiopterin synthase (PTPS), quinonoid dihydropteridine reductase (QDHPR), and pterin carbinolamine dehydratase (PCD). Module III is designed to synthesize norcoclaurine and encodes expression of four proteins—a mutant (R37E, R38E, and W166Y) of tyrosine hydroxylase (TyrHWR) that is more resistant to inhibition by l-tyrosine and catecholamines and the BH4 salvage enzyme dihydrofolate reductase (DHFR) from R. norvegicus, DOPA decarboxylase (DoDC) from Pseudomonas putida, and norcoclaurine synthase (NCS) from Coptis japonica. Module IV is designed to synthesize reticuline and encodes expression of five plant proteins—norcoclaurine 6-O-methyltransferase (6OMT), coclaurine-N-methyltransferase (CNMT), 4′-O-methyltransferase (4′OMT), cytochrome P450 reductase (CPR) from P. somniferum, and N-methylcoclaurine hydroxylase (NMCH) from Eschscholzia californica. Finally, module V encodes the overexpression of three identified bottleneck proteins—TyrHWR, 4′OMT, and NCS—and was integrated either to knockout the native ZWF1 gene (zwf1Δ; CSY1061) or into a separate locus (CSY1060).
Initial noscapine-producing strains were constructed by introducing 12 additional plant enzymes into the reticuline-producing platform strains (CSY1060 and CSY1061) encoded across three additional genetic modules. Module VI encodes the expression of canadine synthase (CAS) from Coptis japonica and berberine bridge enzyme (BBE), scoulerine 9-O-methyltransferase (S9OMT), and an N-terminal engineered variant of 1-hydroxy-N-methylcanadine synthase (CYP82Y1A) from P. somniferum. Module VII encodes the expression of tetrahydroprotoberberine cis-N-methyltransferase (TNMT), 1-hydroxy-13-O-acetyl-N-methylcanadine 8-hydroxylase (CYP82X1), 3-O-acetylpapaveroxine carboxylesterase (CXE1), and narcotoline-4′-O-methyltransferase (I) (MT2) from P. somniferum. Finally, module VIII encodes the expression of 1-hydroxy-N-methylcanadine 13-hydroxylase (CYP82X2), 1,13-dihydroxy-N-methylcandine 13-O-acetyltransferase (AT1), short-chain dehydrogenase/reductase (SDR1), and narcotoline-4′-O-methyltransferase (II) (MT3) from P. somniferum. The introduction of these three genetic modules into chromosomal loci in CSY1060 and CSY1061 resulted in two base noscapine-producing strains (CSY1149: zwf1Δ and CSY1150: ZWF1).
We assayed noscapine production by growing yeast strains in synthetic defined medium (SD) supplemented with 10 mM ascorbic acid for 72 h at 25 °C. Because the intracellular concentrations of most metabolites along the noscapine biosynthetic pathway are relatively low (14), we used the extracellular titer to indicate the production of noscapine and pathway intermediates from the engineered yeast strains. The growth media was analyzed for BIA molecules by liquid chromatography coupled with tandem mass spectrometry (LC–MS/MS). The synthesis of noscapine was confirmed through comparison with the chemical reference standard (Fig. 1B). The base noscapine-producing strains, incorporating modules I–VIII, produced 120.0 ng/L (CSY1149; zwf1Δ) and 227.1 ng/L (CSY1150; ZWF1) of noscapine. Although earlier work indicated that the ZWF1 knockout leads to enhanced production of reticuline (11), our studies indicate that this deletion is not advantageous for production of noscapine. ZWF1 encodes a glucose-6-phosphate dehydrogenase, which catalyzes the oxidation of glucose-6-phosphate and synthesizes NADPH (18). Plant cytochrome P450s require NADPH for electron transfer, and the biosynthesis of noscapine includes five such enzymes, whereas the biosynthesis of reticuline employs only one. Thus, the NADPH requirement for a noscapine-producing strain is expected to be substantially greater than that for a reticuline-producing strain. We hypothesize that in our two constructed noscapine strains (CSY1149 and CSY1150), any positive effect on reticuline production resulting from deletion of ZWF1 in CSY1149 was small compared with the negative effect on the downstream pathway to noscapine caused by removal of this NADPH-generating enzyme. Therefore, CSY1150 was selected as the base noscapine-producing strain to further engineer for enhanced noscapine biosynthesis.
Increased Noscapine Production Through Genetic Modifications That Push Flux into the Pathway.
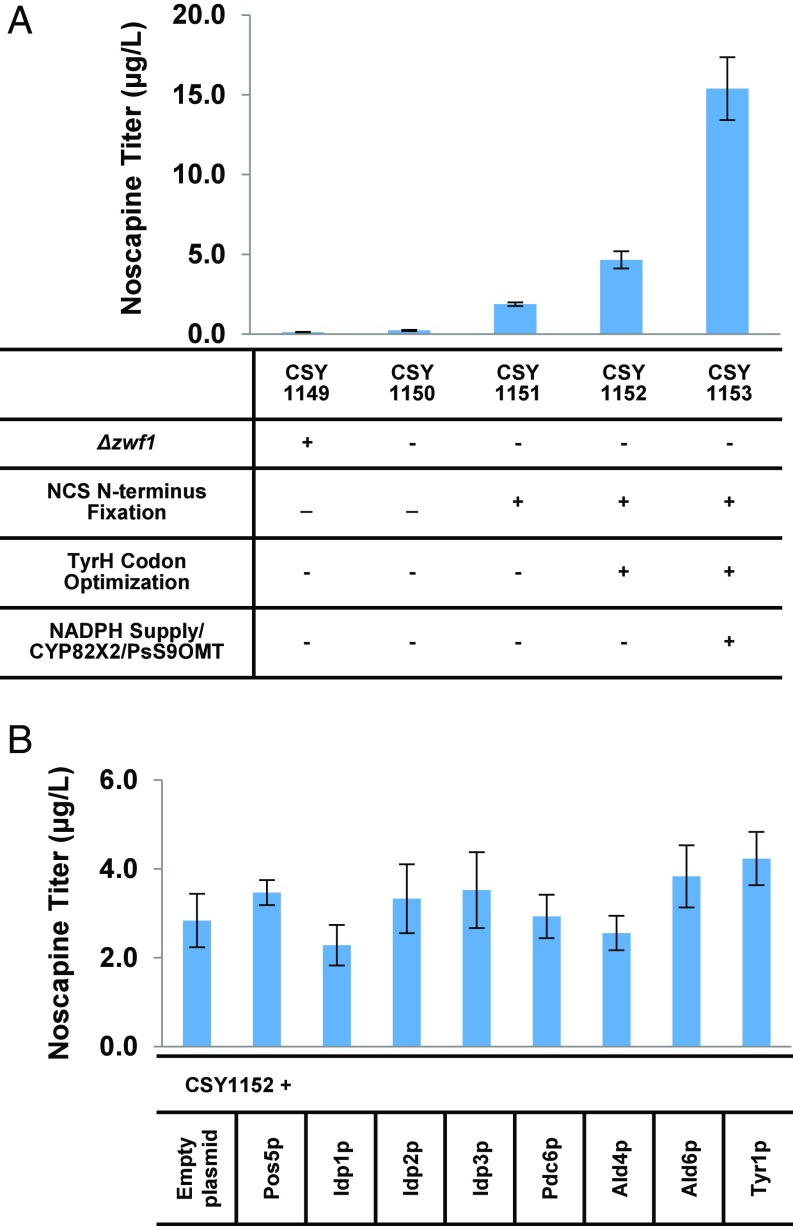
We first focused on increasing noscapine production by improving the activities of key bottleneck enzymes in the early part of the pathway to convert more precursor molecules into the 1-benzylisoquinoline backbone. NCS catalyzes the condensation reaction between dopamine and 4-HPAA to produce norcoclaurine. Earlier studies indicate substantial accumulation of dopamine in the reticuline-producing platform strain (11), suggesting that NCS activity is a key bottleneck in pushing more flux into the BIA pathway. Recent investigations on NCS indicate that the N-terminal signal peptide on some NCS variants may have a deleterious effect on enzyme function (19). Thus, we designed an NCS variant with the first 24 amino acids deleted and used a CRISPR/CAS9 genomic editing strategy to directly replace the original NCS variants in CSY1150 with this modified NCS variant (CSY1151; Table S1) (20). Production of noscapine from the strain harboring the modified NCS variant increased approximately eightfold to 1.9 µg/L (Fig. 2A).
Fig. 2.
Strain optimization for enhanced noscapine production in yeast. (A) Noscapine titers from yeast strains engineered with pathway and enzyme improvements. (B) Optimization of noscapine production through NADPH regeneration. S. cerevisiae endogenous genes were expressed from a low-copy plasmid in CSY1152. All strains were cultured in SD with 2% dextrose in the presence of 10 mM ascorbic acid at 25 °C for 72 h. Error bars represent SD of three biological replicates.
In addition, the tyrosine hydroxylase (TyrH) used in the reconstructed pathway is a variant from R. norvegicus and harbors mutations R37E, R38E, and W166Y (TyrHWR) to reduce substrate inhibition (11, 15). Tyrosine hydroxylase (TyrH) catalyzes the conversion of tyrosine to l-DOPA, which is one of the first steps in the early BIA biosynthetic pathway. Earlier studies observed nearly complete conversion of l-DOPA in the reticuline-producing platform strain, suggesting that TyrH activity also limits flux into the BIA pathway (11). We designed a codon-optimized version of this TyrH variant and used a CRISPR/CAS9 cloning strategy to directly replace the original TyrHWR variant in CSY1151 with this codon-optimized variant (CSY1152; Table S1). Production of noscapine from the strain harboring the codon-optimized TyrHWR variant increased a further ∼2.5-fold to 4.7 μg/L (Fig. 2A).
Increased Noscapine Production Through Genetic Modifications That Regenerate a Key Cofactor to Improve the Efficiency of Limiting Pathway Enzymes.
Noscapine biosynthesis requires five plant cytochrome P450s that each utilize NADPH for electron transfer. The decrease observed in noscapine production upon deletion of ZWF1 indicates that the supply of NADPH may be critical to the enzymatic efficiency of the plant cytochrome P450s. In addition to enhanced supply of NADPH, previous investigations also demonstrate that a higher NADPH/NADP+ ratio can improve the activity of cytochrome P450s (21). We thus attempted to increase the pool of available NADPH by introducing an extra copy of selected yeast enzymes that can regenerate NADPH (Fig. S2), specifically, NADH kinase (Pos5p), three isocitrate reductases (Idp1-3p), pyruvate decarboxylase (Pdc6p), two aldehyde dehydrogenases (Ald4p and Ald6p), and prephenate dehydrogenase (Tyr1p). The genes encoding each enzyme were cloned into a low-copy plasmid regulated by a strong GPD promoter. The noscapine-producing strain CSY1152 was transformed individually with each plasmid and noscapine production was measured. Noscapine production levels were assayed and compared with CSY1152 harboring an empty plasmid. All of the candidate NADPH-generating enzymes other than Idp1p and Ald4p resulted in more efficient synthesis of noscapine, whereas overexpression of Ald6p and Tyr1p led to the largest and statistically significant improvements in noscapine production, ∼1.4 fold (Fig. 2B). Tyr1p is also involved in the production of tyrosine and 4-hydroxyphenylacetaldehyde (4-HPAA) (Fig. S2), which are key precursors of the noscapine pathway (Fig. 1A). The noscapine titer of CSY1152 overexpressing both Ald6p and Tyr1p exhibits a higher noscapine titer than overexpression of Ald6p or Tyr1p alone in this strain (Fig. S2).
Earlier work indicated that the activities of CYP82X2 and S9OMT limit pathway flux to noscapine (14). Thus, we designed a ninth genetic module (IX) to enhance noscapine production. Module IX is designed to regenerate NADPH to improve the activity of cytochrome P450s along the pathway and pull more flux through the pathway and encodes four proteins—Ald6p and Tyr1p from yeast and CYP82X2 and S9OMT from P. somniferum. The introduction of this module into chromosomal loci in CSY1152 resulted in an enhanced noscapine-producing strain (CSY1153), from which noscapine production levels increased approximately threefold to 15.4 μg/L (Fig. 2A). Quantification of the pathway intermediates synthesized by CSY1152 and CSY1153 indicated that more scoulerine is converted to tetrahydrocolumbamine and 1-hydroxy-N-methylcanadine accumulates to a lower level in CSY1153 compared with CSY1152, consistent with the incorporation of additional copies of PsS9OMT and CYP82X2 in CSY1153 (Fig. S3).
Media Optimization Leads to Substantial Improvements in Noscapine Production.
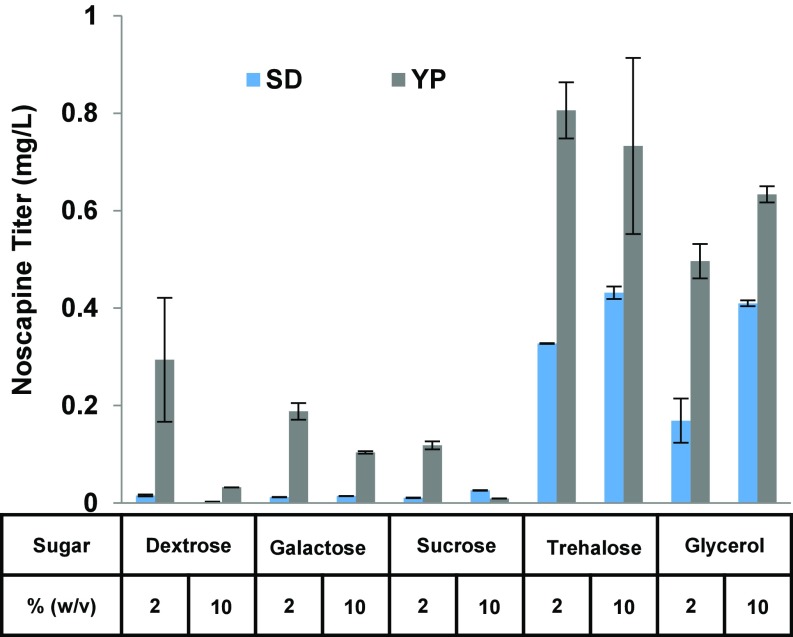
We next explored the effect of various base media and carbon sources on noscapine production from the highest-producing noscapine strain CSY1153. Specifically, synthetic defined medium (SD) and complex medium (Yeast extract and peptone, YP) were investigated with a number of carbon sources (2% wt/vol), including dextrose, galactose, sucrose, trehalose, and glycerol. Across all of the carbon sources examined, YP led to a pronounced increase in noscapine titers (SD with 2% dextrose, 15.4 μg/L; YP with 2% dextrose, 293.9 μg/L; SD with 2% galactose, 11.9 μg/L; YP with 2% galactose, 188.0 μg/L; SD with 2% sucrose, 10.5 μg/L; YP with 2% sucrose, 118.2 μg/L; SD with 2% trehalose, 32.7 μg/L; YP with 2% trehalose, 805.8 μg/L; SD with 2% glycerol, 43.2; YP with 2% glycerol, 496.3 μg/L) (Fig. 3). Under the best media condition (YP supplemented with 2% trehalose) production of noscapine was increased over 50-fold from 15.4 to 805.8 μg/L.
Fig. 3.
Media optimization for enhanced noscapine production in yeast. Noscapine titers were measured from CSY1153 in different medium and carbon sources. Base medium includes SD (synthetic complete) and YP (yeast extract and peptone). Carbon sources include 2% or 10% of dextrose, galactose, sucrose, trehalose, and glycerol. The strain was cultured at 25 °C for 72 h in the indicated media composition with 10 mM ascorbic acid. Error bars represent SD of at least three biological replicates.
The positive effect of YP on noscapine production relative to SD is likely to be a result of the higher concentrations of the noscapine precursor tyrosine and other nutrients. However, the effects of carbon sources varied; trehalose, a disaccharide composed of 2 α-glucose, afforded highest noscapine production levels in SD and YP. Galactose and sucrose, also disaccharide sugars, resulted in lower production of noscapine in SD and YP. Dextrose led to the intermediate noscapine production level in SD and in YP. Glycerol, the only nonsugar carbon source examined, afforded substantially higher noscapine titer in SD and YP compared with dextrose, sucrose, and galactose. Quantification of the pathway intermediates also implies more efficient conversion from reticuline, scoulerine, and tetrahydrocolumbamine to canadine and downstream pathway metabolites when utilizing glycerol as the carbon source (Fig. S4).
The effect of higher carbon concentration (10% wt/vol) was investigated with SD and YP as the base medium. The cells grew to a similar density across the various carbon sources in SD (OD600 ∼ 4.5–7) and YP (OD600 ∼ 23–32). In SD, higher concentration (10% wt/vol) of dextrose leads to a decrease in noscapine titer, whereas enhanced concentrations of other sugars generally lead to a slight increase in noscapine titer (dextrose, 1.7 μg/L; galactose, 14.3 μg/L; sucrose, 25.7 μg/L; trehalose, 43.2 μg/L) (Fig. 3). The data indicate that high concentration of dextrose in the defined SD media may inhibit the activities of certain enzymes associated with noscapine production, such as enzymes involved in aromatic amino acid biosynthesis (22), whereas high concentration of other sugars does not affect the production of noscapine as significantly. In YP, higher concentration of sugars (10% dextrose, galactose, sucrose, and trehalose) leads to reduced noscapine production (32.0, 103.8, 9.2, and 732.8 μg/L, respectively) (Fig. 3). The data also show that high concentration of trehalose and galactose in the complex YP media does not exhibit as deleterious an effect to noscapine production compared with the other sugars. The reduction in noscapine production under high concentrations of sugars (especially glucose and sucrose) is likely a result of (i) faster growth rates that may lead to misfolding of heterologous enzymes (or the relatively slower growth rate in high concentrations of nonfermentive carbon sources, compared with glucose and sucrose, may result in comparatively better enzyme folding) (23) and (ii) repressive effects on yeast metabolism leading to a redirection of metabolic production away from noscapine bioproduction related metabolism, such as aromatic amino acid biosynthesis (22, 24). In contrast, the higher concentrations of the nonsugar carbon source, glycerol, enhanced noscapine production by 243% from 168.9 to 409.8 μg/L in SD and 127% from 496.3 to 633.6 μg/L in YP (Fig. 3).
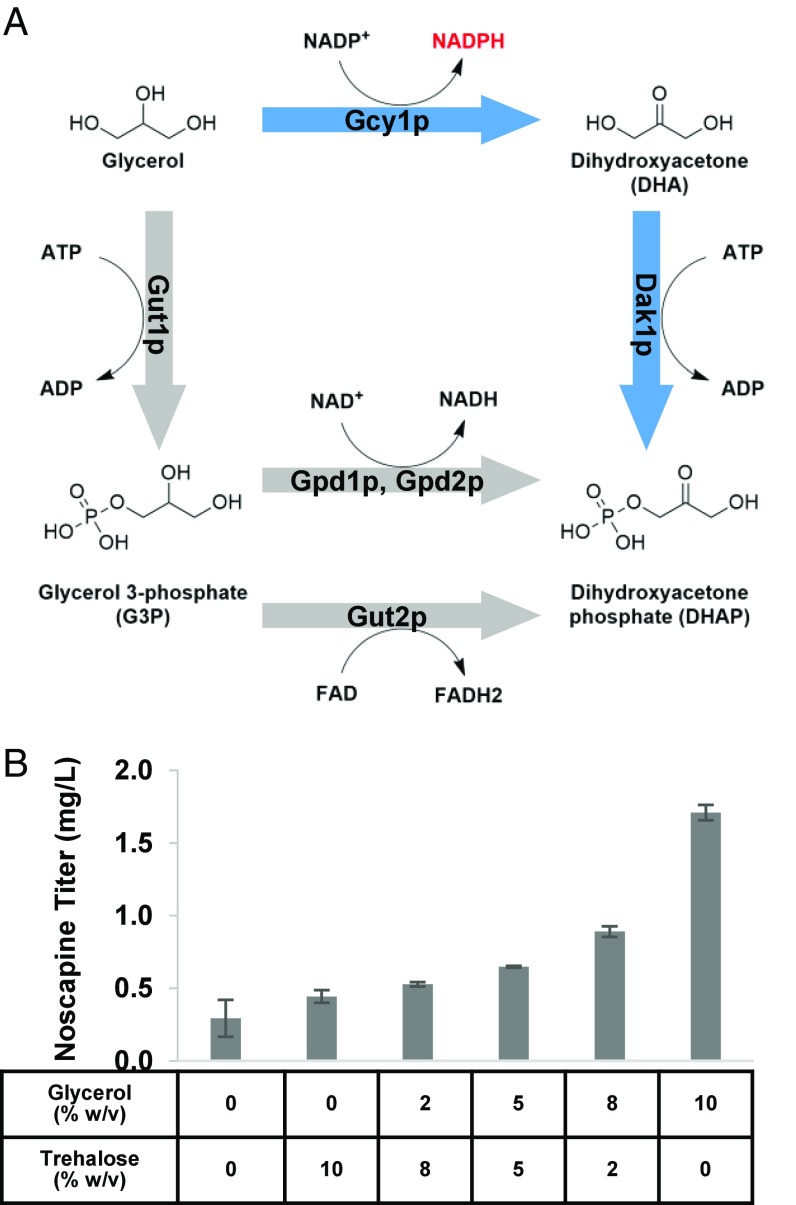
We continued to investigate the role glycerol plays in noscapine synthesis and further optimize medium composition. Glycerol may increase noscapine production through a number of mechanisms. For example, natural osmolytes (e.g., trehalose, sucrose, and glycerol) have been reported to promote stabilization of recombinant proteins (25) and cellular membranes (26–28). Additionally, glycerol and trehalose have also been reported to facilitate the correct folding of nascent polypeptides (29, 30) and thus have been utilized as chemical chaperones to reverse the misfolding or mislocalization of aggregated proteins both in vitro and in cells (31, 32). Apart from being able to stabilize the membrane and facilitate the correct folding of proteins, glycerol utilization in S. cerevisiae is also associated with the regeneration of NADPH (Fig. 4A): an NADP-dependent glycerol dehydrogenase (Gcy1p and Ypr1p) catalyzes the conversion of glycerol to dihydroxyacetone (DHA), which is subsequently phosphorylated to DHAP via a DHA kinase (Dak1p and Dak2p) (33). We compared noscapine production at different glycerol concentrations (0, 2, 5, 8, and 10%). The media in these experiments was maintained at 12% total carbon by including 2% dextrose (to support efficient cell growth in the early stages of fermentation), and the remaining carbon up to 12% was made up with trehalose. The cells grew to the same density under all media conditions (OD600 in the range of 22.83 ± 0.72–24.79 ± 0.86), and noscapine titer positively correlated with glycerol concentration in the media (Fig. 4B). The highest noscapine titer (1.71 ± 0.03 mg/L) was observed from YP media supplemented with 2% dextrose and 10% glycerol, likely resulting from availability of an efficiently utilized carbon source supporting early cell growth, protein stabilization, and NADPH regeneration. However, CSY1153 exhibits a lower noscapine titer in YP media supplemented with 2% dextrose and 10% trehalose (0.44 mg/L), compared with when cultured in YP media supplemented with only 10% trehalose (0.73 mg/L), which is likely due to glucose repression caused by the 2% dextrose (22). In contrast, CSY1153 exhibits a higher noscapine titer in YP media supplemented with 2% dextrose and 10% glycerol (1.71 mg/L), compared with when cultured in YP media supplemented with only 10% glycerol (0.63 mg/L). These data indicate that slower growth rate may not be the main reason for improved noscapine titers when using glycerol as the carbon source. The effect of higher concentration of glycerol (15, 20, and 25% glycerol supplemented to YP with 2% dextrose) was subsequently examined and revealed that concentrations of glycerol higher than 10% are not beneficial to noscapine production (Fig. S5).
Fig. 4.
Effect of glycerol on noscapine production. (A) Schematic illustrating the glycerol utilization mechanism. The pathway facilitating NADPH regeneration is highlighted in blue, and the pathway converting glycerol to dihydroxyacetone phosphate is highlighted in gray. (B) Noscapine titers from CSY1153 under different glycerol concentrations. Ten percent glycerol and trehalose at the indicated ratios were supplemented into YP with 2% dextrose. The strain was cultured at 25 °C for 72 h in the indicated media composition with 10 mM ascorbic acid. Error bars represent SD of three biological replicates.
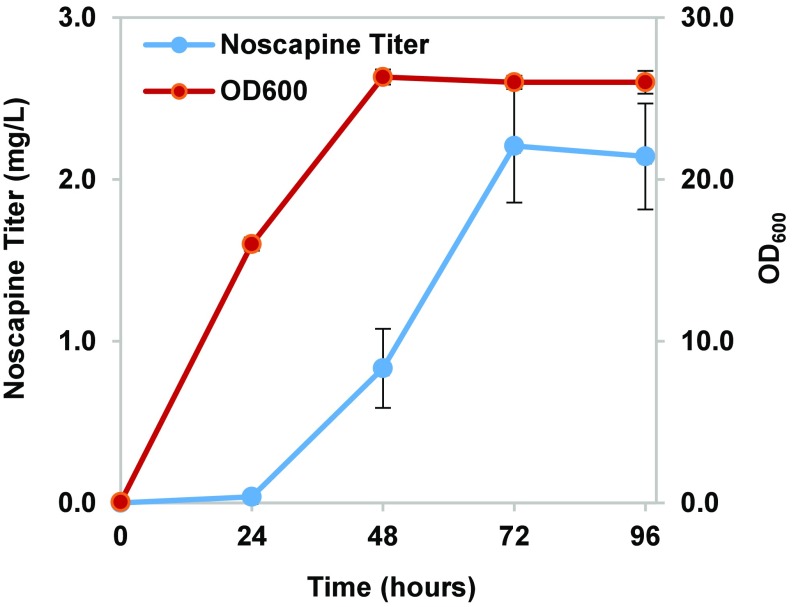
The optimized noscapine production media was utilized to grow the highest noscapine producing strain (CSY1153) in a 250-mL shake flask to model enhanced oxygen transfer in a larger culture vessel. The maximum noscapine titer was further increased to 2.21 ± 0.35 mg/L after 72 h of fermentation and remained stable after an additional 24 h of growth. The increase in noscapine titer observed in the shake flask experiment may be due to enhanced oxygen transfer in this culture vessel flask compared with the 96-deep well plate. The results from the shake flask experiments reveal that noscapine production is not coupled with biomass production, because the maximum cell density was reached on day 2, whereas over 50% of the accumulated noscapine was produced on day 3.
Our work highlights the central role of optimizing media and fermentation conditions to improve bioprocesses for complex plant natural products. Our media optimization efforts resulted in an ∼300-fold increase in noscapine titers. Our work also demonstrates that glycerol substantially enhances noscapine production, possibly by regenerating the essential cofactor NADPH for cytochrome-P450–catalyzed reactions and by stabilizing heterologous proteins localized to subcellular membranes.
Biosynthesis of Halogenated BIA Derivatives Through Feeding Tyrosine Derivatives.
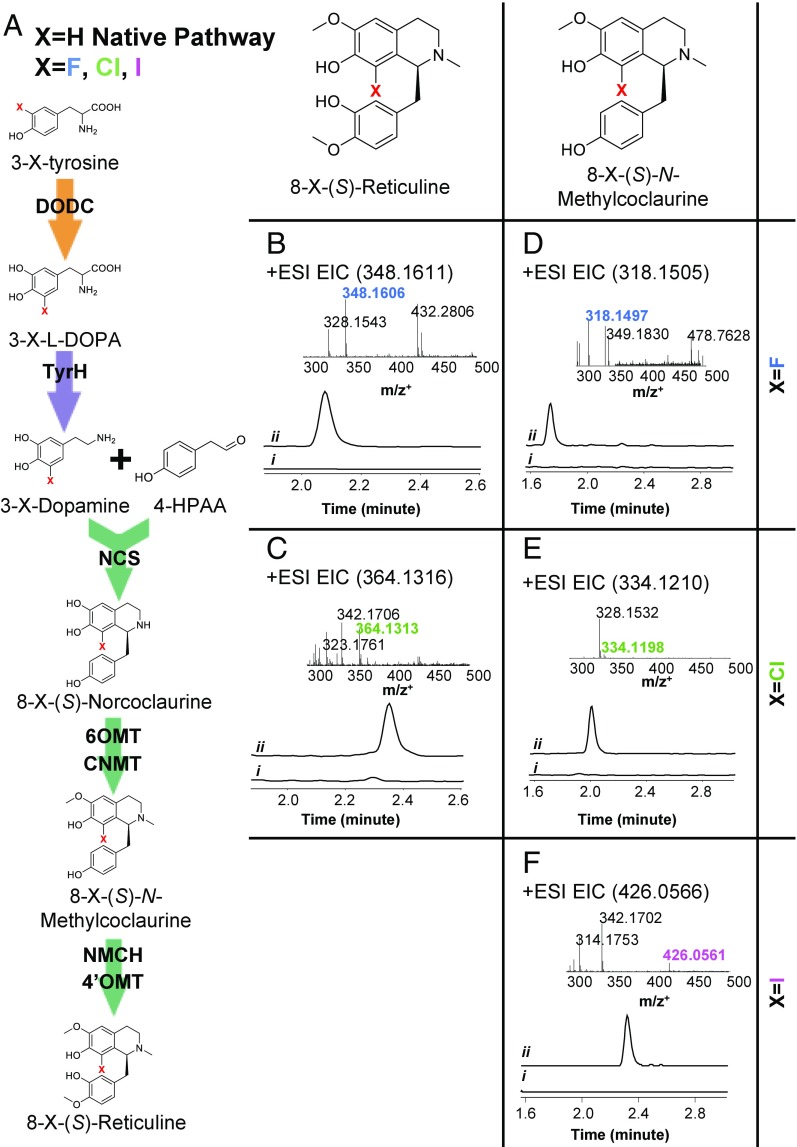
Chemical transformation, such as functional group interconversion and addition (e.g., acylation, alkylation, and halogenation), plays a critical role in drug candidate preparation in pharmaceutical synthesis (34). Noscapine derivatives have shown higher therapeutic potential toward certain cancer cell lines (5, 35), indicating a promising approach to enhance the bioactivity of noscapine-derived anticancer agents. However, the number and chemical diversity of noscapine derivatives are limited, due to the lack of commercially available noscapine synthetic intermediates and the structural complexity and chemical reactivity of noscapine (5). The reconstruction of the complete biosynthetic pathway to noscapine in yeast offers a versatile platform to provision previously inaccessible noscapine intermediates and to enable the de novo biosynthesis of unique noscapine derivatives (Fig. 5A).
Fig. 5.
Proposed tyrosine derivative incorporation mechanism and LC–MS analyses of noscapine producing strain fed tyrosine derivatives. (A) Proposed biosynthetic pathway of 8-halo-(S)-reticuline from 3-halo-tyrosine when X = F, Cl, or I and native biosynthetic pathway of reticuline from typrosine when X = H. The high-resolution EIC of CSY1153 fed with (i) tyrosine and (ii) the corresponding 3-halo-tyrosine. Mass spectra of eight-substituted-(S)-N-Methylcoclaurine synthesized from (B) 3-fluro-tyrosine, (C) 3-chloro-tyrosine, and (D) 3-iodo-tyrosine and eight-substituted-(S)-reticuline synthesized from (E) 3-fluro-tyrosine and (F) 3-chloro-tyrosine. The EIC was extracted using the calculated m/z+ of target intermediate and derivatives, with a mass accuracy below 100 ppm. The EIC and mass spectra were obtained from CSY1153 cultured at 25 °C for 72 h in YP supplemented with 2% dextrose and 10% glycerol with 10 mM ascorbic acid, fed with tyrosine or the indicated derivative. Error bars represent SD of three biological replicates.
We probed the flexibility of the noscapine biosynthetic pathway to convert tyrosine analogs fed to the engineered yeast during fermentation. Sixteen tyrosine derivatives were supplemented into a tyrosine-free SD growth media (Table S4). No significant difference was observed in the growth rates of the highest-producing noscapine strain (CSY1153) in SD when supplemented with tyrosine or tyrosine analogs at the same concentration. The growth media was analyzed for derivatized BIA molecules by high-resolution liquid chromatography quadrupole time-of-flight mass spectrometry (LC-QTOF). The exact masses of the expected products with the incorporation of tyrosine derivatives were utilized to filter and analyze the LC-QTOF chromatographs.
We observed peaks that match the exact masses of 8-fluoro-reticuline (Fig. 5B) and 8-chloro-reticuline (Fig. 5C), when feeding 3-fluoro-tyrosine and 3-chloro-tyrosine, respectively. The detection of peaks matching the exact mass of 8-fluoro- and 8-choloro-reticuline implies that these molecules were synthesized from fed 3-fluoro- and 3-chloro-tyrosine, respectively, via oxidation by TyrH and subsequent incorporation into the biosynthetic pathway by DODC, NCS, 6OMT, CNMT, NMCH, and 4′OMT toward reticuline (Fig. 5A). To confirm that the eight-substituted reticuline molecules were formed via uptake and transformation of tyrosine derivatives by our noscapine strains, we searched for additional pathway intermediates and observed 8-halogen substituted (S)-N-methylcoclaurine (Fig. 5 D and E). In addition, when fed with a third tyrosine derivative, 3-iodo-tyrosine, a peak that matches the exact mass of 8-iodo substituted (S)-N-methylcoclaurine (Fig. 5F) was observed. Notably, peaks that match the exact mass of 8-iodo substituted reticuline were not detected, suggesting that flux of this derivatized tyrosine molecule into the reticuline pathway is incomplete. Furthermore, conversion of the derivatized 1-benzylisoquinoline intermediates to the downstream berberine, secoberberine, and phthalideisoquinoline alkaloids was not detected. The results highlight that the reconstructed noscapine pathway is capable of incorporating tyrosine derivatives to generate reticuline derivatives, demonstrating the possibility of the substrate-feeding approach in synthesizing novel BIA derivatives. The lack of conversion to the downstream BIAs in the noscapine pathway may be due to the limited promiscuity of the native enzymes, low reaction efficiency, and low abundance of the substrate. Thus, the biosynthesis of halogenated noscapinoids could be achieved through two routes: (i) engineering of tailoring enzymes, such as halogenases, to accept noscapinoids as substrates and (ii) engineering limiting enzymes in the noscapine pathway, such as BBE, to exhibit increased activity on halogenated substrates. In addition, further optimization of the yeast biosynthetic platform will allow for increased synthesis efficiency of the halogenated intermediates, which could ultimately allow more flux to be directed to the downstream part of the noscapine pathway. Therefore, future efforts directed to enzyme engineering and further pathway improvements may address these limitations and enable the production of a broad array of novel noscapinoid derivatives.
Discussion
We have demonstrated yeast-based fermentation of noscapine and related pathway intermediates from simple carbon and nitrogen sources. Our pathway engineering and optimization efforts resulted in a yeast strain engineered to express 31 enzymes, including 25 heterologous pathway enzymes taken from plants, mammals, and bacteria and 6 mutant or overexpressed yeast enzymes, that produced noscapine titers up to 15.4 μg/L. Further highlighting yeast’s ability to serve as a platform host for complex plant natural product pathways, 7 of the 25 heterologous enzymes require proper anchoring and folding within the endoplasm reticulum membrane for function, including five plant cytochrome P450s, a plant cytochrome P450 reductase, and berberine bridge enzyme (BBE). Media and growth optimization efforts further enhanced noscapine titers by ∼140-fold to ∼2.2 mg/L. Although titers will need to be further increased by ∼100–1,000 fold to realize a commercially viable manufacturing process at scale, our initial work highlights the feasibility of using yeast fermentation for supplying complex plant alkaloids for global demand with further improvement of the process. We anticipate that such improvements will be realized through a combination of enzyme and strain engineering and fermentation process optimization. We also show the value of such fermentation processes for realizing alkaloid derivatives through feeding modified substrates to the engineered cells.
Our results indicate that NADPH balance is likely an important factor in optimizing noscapine production in yeast, likely due to the relatively large number of pathway enzymes requiring this cofactor (e.g., cytochrome P450s). Although prior work had shown that a ZWF1 knockout strain results in higher reticuline production (15), we observed higher noscapine titers in strains harboring the intact ZWF1 gene. This observation is consistent with prior work describing production of strictosidine in yeast, which involves five NADPH-dependent enzymes, where product titer was enhanced by the overexpression of ZWF1 to increase NADPH availability (10). We achieved enhanced noscapine titers by overexpressing two native genes, TYR1 and ALD6, which generate NADPH through their native activities. The overexpression of ZWF1, TYR1, and ALD6 is likely to result in a higher NADPH/NADP+ ratio, which is kinetically preferred by the cytochrome P450s. Future engineering efforts directed to improving noscapine production should focus on further optimizing NADPH levels or NADPH/NADP+ ratios and enhancing the activities of the cytochrome P450s. For example, protein engineering can be performed on the rate limiting P450s (e.g., CYP82X2 and CAS) to enhance their kinetic properties, and strain engineering can be directed to achieving higher ER capacity to support the overexpression of the multiple membrane-bound pathway enzymes.
The pattern of pathway intermediate accumulation indicates bottlenecks in the pathway; however, these patterns vary with media composition. When the strains are cultured in SD, (S)-reticuline, scoulerine, tetrahydrocolumbamine, and canadine accumulated to the highest relative levels and the intermediates downstream of N-methylcanadine (except for noscapine) are relatively low (Fig. S3). These results are consistent with a prior study indicating that S9OMT and CAS are rate-limiting steps and CYP82X2 exhibits low activity in yeast (16). In contrast, when strains are cultured in YP media, relative accumulation of the early stage intermediates is reduced, and N-methylcanadine, 1-hydroxy-N-methylcanadine, 1-hydroxy-13-O-acetyl-N-methylcanadine, and narcotinehemiacetal accumulate to relatively higher levels. This trend is more prominent when the strain is cultured in the presence of trehalose and glycerol (Fig. S4). The data imply that complex medium may help to enhance the activity of the enzymes responsible for conversion of the early stage intermediates (e.g., reticuline, scoulerine, and tetrahydrocolumbamine) to downstream intermediates (e.g., canadine, N-methylcanadine, and 1-hydroxy-N-methylcanadine), such as BBE, Ps9OMT, and CAS. Because YP is a complex medium, the specific components responsible for driving the metabolic flux farther downstream the pathway are unclear from these results alone. However, future experiments may focus on elucidating the mechanism underlying the effects of media components, including nitrogen, on pathway flux.
Results of carbon source optimization revealed that high sugar concentration and fermentative carbon sources that can be beneficial in the optimization of central metabolism (36) are not optimal for noscapine production, which may be due to the higher growth rate leading to protein misfolding and the redirection of metabolite production away from noscapine bioproduction-related metabolism. We observed that trehalose and glycerol enhanced plant natural product biosynthesis encoded by a cytochrome P450-rich, complex biosynthetic pathway. Trehalose and glycerol may simultaneously serve as carbon source and protein stabilizer, whereas glycerol also functions as a NADPH regeneration agent (Fig. 4A). In addition, we observed that adding low concentrations of dextrose (2%) to the YP medium supplemented with 10% glycerol further enhanced noscapine production. Because noscapine production is not fully correlated with cell growth (Fig. 6), the beneficial effect may be due to the accelerated cell growth before stationary phase. The effect of media and fermentation conditions observed in these preliminary studies indicates that the precise control afforded by transferring the process to a bioreactor may lead to even higher noscapine production.
Fig. 6.
Noscapine production in shake flasks. Noscapine titers from CSY1153 at different time points in a shake flask experiment. The strain was cultured in a 250-mL shake flask at 25 °C for 96 h. Medium contains YP with 2% dextrose and 10% glycerol with 10 mM ascorbic acid. Noscapine concentration, blue; cell density at OD600, red. Error bars represent SD of three biological replicates.
The de novo biosynthesis of noscapine in yeast provides a unique opportunity to generate novel natural product derivatives synthesized from precursor derivatives. As proof of concept, we demonstrate the capability of de novo noscapine-producing strain to generate various reticuline derivatives via the incorporation of tyrosine derivatives in the pathway, which illustrates the potential of a biological approach to produce novel drug leads. Although we did not observe the accumulation of BIAs downstream of reticuline, such as noscapine, this may be addressed by a combination of further improvement of pathway efficiency; engineering downstream enzymes, such as BBE, for higher activity on nonnatural substrates; and engineering tailoring enzymes for broader substrate specificity toward noscapinoids. The successful reconstruction of this complex plant natural product pathway and synthesis of benzylisoquinoline alkaloid derivatives via precursor feeding demonstrates the capacity of yeast to serve as a scalable production platform for complex plant natural products and highlights the transformative effect synthetic biology may have in drug development and production.
Methods
Materials and Culture Conditions.
Reticuline was purchased from Santa Cruz Biotechnology and Specs, and noscapine (97% purity) was purchased from Sigma–Aldrich. For DNA manipulation and amplification, we used chemically competent or electrocompetent Escherichia coli TOP10 strains purchased from Life Technologies. E. coli strains were grown at 37 °C in lysogeny broth medium obtained from Fisher Scientific supplemented with 100 mg/mL ampicillin (EMD Chemicals) or 50 mg/mL kanamycin (Fisher Scientific) for plasmid maintenance. All engineered yeast strains described in this work, as listed in Table S1, were constructed with the reticuline-producing yeast strain CSY1060 and CSY1061 as a starting point (11). Unless specified, yeast strains were cultured at 25 °C in complex yeast extract peptone dextrose (YPD; all components from BD Diagnostics) medium, SD [containing yeast nitrogen base (YNB) without amino acids (BD Diagnostics), ammonium sulfate (Fisher Scientific), 2% dextrose, and synthetic complete or the appropriate dropout solution for plasmid maintenance]. Selection in YPD medium used 200 mg/L G418 sulfate (Calbiochem) or 200 mg/L hygromycin B (Life Technologies).
Plasmid and Yeast Strain Construction.
Custom oligonucleotides were synthesized by Integrated DNA Technologies (IDT) and the Stanford Protein and Nucleic Acid Facility. Heterologous gene sequences were codon-optimized for expression in S. cerevisiae using the GeneArt GeneOptimizer program (Life Technologies) and synthesized by GeneArt (Life Technologies) or IDT. Expand High Fidelity PCR system (Roche Diagnostics) was used for PCR amplifications according to manufacturer’s instructions. PCR products were purified by agarose gel extraction with Zymoclean gel DNA recovery kit (Zymo Research) according to manufacturer’s instructions. Restriction enzymes, T4 DNA ligase, and deoxynucleotides were purchased from New England Biolabs. Plasmids were prepared from E. coli using QIAprep columns (Qiagen) and Econospin columns (Epoch Life Science) according to manufacturer's instructions. Sequencing was performed by Elim Biopharmaceuticals, Inc.
pos5, idp1, idp2, idp3, pdc6, ald4, ald6, and tyr1 were PCR amplified from S. cerevisiae genomic DNA and introduced into Gateway pDONR221 using BP Clonase II Enzyme Mix to yield pCS3636, pCS3642, pCS3643, pCS3637, pCS3663, pCS3640, pCS3639, and pCS3644, respectively (Table S2). The genes were subsequently recombined into selected pAG416GPD-ccdB expression vectors from the S. cerevisiae Advanced Gateway Destination Vector Kit (37) using LR Clonase II to generate the corresponding yeast expression constructs pCS3645, pCS3649, pCS3652, pCS3646, pCS3657, pCS3647, pCS3653, and pCS3655, respectively (Table S1).
S. cerevisiae CSY1060 and CSY1061 strains were used as the base strain for construction of the de novo noscapine biosynthetic strains (11). tyr1 and ald6 were each inserted into pCS3125 and pCS3131, respectively, using Gibson assembly to yield pCS3659 and 3661 (14), respectively. For chromosome-based DNA assembly, 500–1,000 bp flanking the target loci (from genomic DNA), certain expression cassettes (from pCS3146, pCS3064, pCS3060, pCS3075, pCS3145, pCS3143, pCS3134, pCS3141, pCS3147, pCS3140, pCS3142, pCS3244, pCS3659, pCS3136, pCS3064, and pCS3661; Table S2), and selection markers [hygR and trp1 from pCS2922 and pCS2923 (38)] were PCR amplified and incorporated into selected loci to generate CSY1049, CSY1050, and CSY1053 (Table S1). The chromosome-based DNA assemblies were conducted through transformation of yeast with a mixture of 100 ng of each DNA fragment using electroporation, and the correctly assembled constructs were verified through PCR analysis of the junctions between each adjacent DNA fragment (39). The integration selection marker was then rescued by heterologous expression of Cre recombinase (40), except for CSY1053.
CSY1051 was constructed by transforming CSY1050 with pCS3638 and the linear double strand DNA fragment (5′-gatcatcaaggaagtaattatctactttttacaacaaatatatacaatgaacggtagaccttt gttgcacagagtcaccaaagaagaaactgttatgttg-3′). pCS3638 is composed of a Cas9 expression cassette and two transcription cassettes for the two guide RNAs (5′-acaaatatatacaatgagaa-3′ and 5′-tgcga aagattgatctttaa-3′). Similarly, CSY1052 was constructed by transforming CSY1051 with pCS3658 and the linear double strand DNA fragment of codon-optimized tyrHWR (Table S3) flanked by 5′-cttagtttcgacggattctagaactagtggatcctataca-3′ on the N terminus and 5′-gggcccttcgaatgacgcgccacttctaaataagcgaatt-3′ on the C terminus. pCS3658 is composed of a Cas9 expression cassette and two transcription cassettes for the two guide RNAs (5′-ggcgagggg gcgctgggggt-3′ and 5′-gcgcggccgctcgagtctta-3′).
Culture and Fermentation Conditions.
To assay for noscapine production and for media optimization, yeast strains were initially inoculated in 96-well plates (BD Falcon) with 0.5 mL YP (1% yeast extract, 2% peptone) with 2% dextrose per well and incubated in a Kuhner LT-X plate shaker (Kuhner AG) at 25 °C, 480 rpm agitation, 1.24 cm orbital diameter, with 80% humidity in triplicates overnight. Cultures were then back-diluted 50× into 0.5 mL SD (0.17% yeast nitrogen base, 0.5% ammonium sulfate, and 1× amino acid drop-out mixture if necessary) or YP growth medium supplemented with corresponding carbon sources and 10 mM ascorbic acid per well, and incubated in a Kuhner LT-X plate shaker at 25 °C, 480 rpm agitation, 1.24 cm orbital diameter, with 80% humidity in triplicates for 72 h. To determine cell density, the final OD600 (after 10× or 50× dilution) was measured in a cuvette on a Nanodrop 2000c spectrophotometer (Thermo Scientific).
For enhanced batch culture in shake flasks, strains were initially inoculated into YP medium with 2% dextrose and grown at 30 °C overnight. Cultures were then back-diluted 100× into YP medium supplemented with 2% dextrose and 10% glycerol, with 10 mM ascorbic acid, and incubated in a 250-mL shake flask with 25 mL starting volume at 30 °C for 96 h in triplicates. At appropriate time points, 0.5 mL samples were taken for metabolite analysis, and cell density was recorded from diluted samples measured in a cuvette on a Nanodrop 2000c spectrophotometer.
Analysis of Noscapine and Intermediate Production.
Noscapine and other BIA intermediates secreted into the culture medium by the engineered yeast strains were identified and quantified by HPLCy mass spectrometry (HPLC–MS). Yeast culture medium was analyzed by reverse phase LC–MS/MS on an Agilent 6420 triple quad LC–MS (Agilent EclipsePlus C18, 2.1 × 50 mm, 1.8 μm) using positive ionization. Metabolites in the medium were separated on a linear gradient of 10% CH3CN (vol/vol in water, 0.1% formic acid) to 40% CH3CN (vol/vol in water, 0.1% formic acid) over 5 min with a flow rate of 0.4 mL/min using multiple reaction monitoring (MRM) mode. The identification of each compound was verified by MRM using the highest characteristic precursor ion/product ion transition (Table S5), and the quantification of noscapine was measured by comparing the integrated peak area to a standard curve of standard of noscapine using precursor ion/product ion transition 414 → 220.
Tyrosine Derivative Feeding Assay and Analysis.
Sixteen tyrosine derivatives were fed to CSY1153 (Table S4): 3-fluoro-l-tyrosine, 3-chloro-l-tyrosine, 3-iodo-l-tyrosine, 3-amino-l-tyrosine, 3-nitro-l-tyrosine, and α-methyl-l-tyrosine (all >95% purity) were purchased from Sigma–Aldrich; 3-O-methyl-DOPA (PubChem CID 13206354), 3-hydroxymethyl-l-tyrosine (PubChem CID 10081872), 2-amino-3-hydroxy-3-(4-hydroxyphenyl)propanoic acid (PubChem CID 13309269), 2-amino-3-(3-tert-butyl-4-hydroxyphenyl)propanoic acid (PubChem CID 18352476), (2S)-2-amino-3-[4-hydroxy-3-(phosphonomethyl)phenyl]propanoic acid (PubChem CID 15045708), 2-amino-3-(3-ethyl-4-hydroxyphenyl)-2-methylpropanoic acid (PubChem CID 18544337), D-α-methyl DOPA (PubChem CID 721860), 3-(2-Amino-2-carboxyethyl)benzoic acid (PubChem CID 265274), l-m-tyrosine (PubChem CID 6950578), 4-aminophenylalanine (PubChem CID 95174), 2-amino-3-(3,4,5-trihydroxyphenyl)propanoic acid (PubChem CID 22283619), 3-carboxytyrosine (PubChem CID 583884), and 4-amino-3-hydroxyphenylalanine (PubChem CID 23253805) were kindly gifted by Novartis Institutes for Biomedical Research. CSY1153 was initially inoculated into SD with 2% dextrose and grown at 30 °C overnight. Cultures were then back-diluted 50× into 0.5 mL tyrosine-free SD (0.17% yeast nitrogen base, 0.5% ammonium sulfate, and 1× tyrosine drop-out amino acid mixture) medium supplemented with 2% dextrose and 10 mM ascorbic acid per well. Tyrosine derivatives were supplemented into the medium at a concentration of 100 mg/L. Yeasts were cultured at 25 °C for 72 h in triplicates, and 2 μL of the yeast culture medium was analyzed by reverse phase LC–MS on an Agilent 6545 QTOF LC–MS using dual AJS positive ionization on a Zorbax SB-Aq column (3.0 mm × 50 mm, 1.8 μM particle size). Metabolites in the medium were separated on a linear gradient of 5% CH3CN (vol/vol in water, 0.1% formic acid) to 95% CH3CN (vol/vol in water, 0.1% formic acid) over 8 min with a flow rate at 0.6 mL/min. Exact mass was obtained at the Stanford ChEM-H (Chemistry, Engineering & Medicine for Human Health) Metabolic Chemistry Analysis Center and was extracted using the calculated m/z+ of target reticuline derivatives, with a mass accuracy below 10 ppm.
Supplementary Material
Acknowledgments
We thank the Stanford ChEM-H Metabolic Chemistry Analysis Center and Dr. Curt Fischer for instrument access and training and C. Schmidt for valuable feedback in the preparation of the manuscript. This work was supported by National Institutes of Health Grant AT007886 (to C.D.S.) and Novartis Institutes for Biomedical Research Grant IC2013-1373 (to C.D.S.).
Footnotes
Conflict of interest statement: C.D.S. and Y.L. report a patent application published on May 26, 2016, “Noscapinoid-Producing Microbes and Methods of Making and Using the Same,” application WO 2016/081371.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721469115/-/DCSupplemental.
References
- 1.Ye K, et al. Opium alkaloid noscapine is an antitumor agent that arrests metaphase and induces apoptosis in dividing cells. Proc Natl Acad Sci USA. 1998;95:1601–1606. doi: 10.1073/pnas.95.4.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barken I, Geller J, Rogosnitzky M. Noscapine inhibits human prostate cancer progression and metastasis in a mouse model. Anticancer Res. 2008;28:3701–3704. [PubMed] [Google Scholar]
- 3.Joshi HC, Salil A, Bughani U, Naik P. Medicinal Plant Biotechnology. Centre for Agriculture and Bioscience International South Asia Edition; Wallingford, UK: 2010. Noscapinoids: A new class of anticancer drugs demand biotechnological intervention; pp. 303–320. [Google Scholar]
- 4.Mahmoudian M, Rahimi-Moghaddam P. The anti-cancer activity of noscapine: A review. Recent Patents Anticancer Drug Discov. 2009;4:92–97. doi: 10.2174/157489209787002524. [DOI] [PubMed] [Google Scholar]
- 5.DeBono A, Capuano B, Scammells PJ. Progress toward the development of noscapine and derivatives as anticancer agents. J Med Chem. 2015;58:5699–5727. doi: 10.1021/jm501180v. [DOI] [PubMed] [Google Scholar]
- 6.Ke Y, et al. Noscapine inhibits tumor growth with little toxicity to normal tissues or inhibition of immune responses. Cancer Immunol Immunother. 2000;49:217–225. doi: 10.1007/s002620000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pushpangadan P, George V, Singh SP. 2012. Poppy. Handbook of Herbs and Spices, ed Peter KV (Woodhead Publishing, Cambridge, England), A2, pp 437–448. [Google Scholar]
- 8.Bradsher K. July 20, 2014 Shake-up on opium island. New York Times. Available at https://www.nytimes.com/2014/07/20/business/international/tasmania-big-supplier-to-drug-companies-faces-changes.html. Accessed on March 19, 2018.
- 9.Chen X, Dang TT, Facchini PJ. Noscapine comes of age. Phytochemistry. 2015;111:7–13. doi: 10.1016/j.phytochem.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 10.Brown S, Clastre M, Courdavault V, O’Connor SE. De novo production of the plant-derived alkaloid strictosidine in yeast. Proc Natl Acad Sci USA. 2015;112:3205–3210. doi: 10.1073/pnas.1423555112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galanie S, Thodey K, Trenchard IJ, Filsinger Interrante M, Smolke CD. Complete biosynthesis of opioids in yeast. Science. 2015;349:1095–1100. doi: 10.1126/science.aac9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winzer T, et al. A Papaver somniferum 10-gene cluster for synthesis of the anticancer alkaloid noscapine. Science. 2012;336:1704–1708. doi: 10.1126/science.1220757. [DOI] [PubMed] [Google Scholar]
- 13.Dang T-TT, Chen X, Facchini PJ. Acetylation serves as a protective group in noscapine biosynthesis in opium poppy. Nat Chem Biol. 2015;11:104–106. doi: 10.1038/nchembio.1717. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Smolke CD. Engineering biosynthesis of the anticancer alkaloid noscapine in yeast. Nat Commun. 2016;7:12137. doi: 10.1038/ncomms12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trenchard IJ, Siddiqui MS, Thodey K, Smolke CD. De novo production of the key branch point benzylisoquinoline alkaloid reticuline in yeast. Metab Eng. 2015;31:74–83. doi: 10.1016/j.ymben.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galanie S, Smolke CD. Optimization of yeast-based production of medicinal protoberberine alkaloids. Microb Cell Fact. 2015;14:144. doi: 10.1186/s12934-015-0332-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Urlacher VB, Girhard M. Cytochrome P450 monooxygenases: An update on perspectives for synthetic application. Trends Biotechnol. 2012;30:26–36. doi: 10.1016/j.tibtech.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 18.Nogae I, Johnston M. Isolation and characterization of the ZWF1 gene of Saccharomyces cerevisiae, encoding glucose-6-phosphate dehydrogenase. Gene. 1990;96:161–169. doi: 10.1016/0378-1119(90)90248-p. [DOI] [PubMed] [Google Scholar]
- 19.Li J, Lee EJ, Chang L, Facchini PJ. Genes encoding norcoclaurine synthase occur as tandem fusions in the Papaveraceae. Sci Rep. 2016;6:39256. doi: 10.1038/srep39256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryan OW, Poddar S, Cate JH. CRISPR-Cas9 genome engineering in Saccharomyces cerevisiae cells. Cold Spring Harb Protoc. 2016;2016:pdb.prot086827. doi: 10.1101/pdb.prot086827. [DOI] [PubMed] [Google Scholar]
- 21.Wu Y, et al. Improvement of NADPH-dependent P450-mediated biotransformation of 7α,15α-diOH-DHEA from DHEA by a dual cosubstrate-coupled system. Steroids. 2015;101:15–20. doi: 10.1016/j.steroids.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 22.Kayikci Ö, Nielsen J. Glucose repression in Saccharomyces cerevisiae. FEMS Yeast Res. 2015;15:fov068. doi: 10.1093/femsyr/fov068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baneyx F, Mujacic M. Recombinant protein folding and misfolding in Escherichia coli. Nat Biotechnol. 2004;22:1399–1408. doi: 10.1038/nbt1029. [DOI] [PubMed] [Google Scholar]
- 24.Marques WL, Raghavendran V, Stambuk BU, Gombert AK. Sucrose and Saccharomyces cerevisiae: A relationship most sweet. FEMS Yeast Res. 2016;16:fov107. doi: 10.1093/femsyr/fov107. [DOI] [PubMed] [Google Scholar]
- 25.Bolen DW. Protein stabilization by naturally occurring osmolytes. In: Murphy KP, editor. Protein Structure, Stability, and Folding. Humana Press; Totowa, NJ: 2001. pp. 17–36. [DOI] [PubMed] [Google Scholar]
- 26.Gekko K, Timasheff SN. Mechanism of protein stabilization by glycerol: Preferential hydration in glycerol-water mixtures. Biochemistry. 1981;20:4667–4676. doi: 10.1021/bi00519a023. [DOI] [PubMed] [Google Scholar]
- 27.Crowe JH, et al. Interactions of sugars with membranes. Biochim Biophys Acta. 1988;947:367–384. doi: 10.1016/0304-4157(88)90015-9. [DOI] [PubMed] [Google Scholar]
- 28.Crowe LM, Mouradian R, Crowe JH, Jackson SA, Womersley C. Effects of carbohydrates on membrane stability at low water activities. Biochim Biophys Acta. 1984;769:141–150. doi: 10.1016/0005-2736(84)90017-8. [DOI] [PubMed] [Google Scholar]
- 29.Sato S, Ward CL, Krouse ME, Wine JJ, Kopito RR. Glycerol reverses the misfolding phenotype of the misfolding phenotype of the most common cystic fibrosis mutation. Mol Biol Cell. 1996;7:1534. doi: 10.1074/jbc.271.2.635. [DOI] [PubMed] [Google Scholar]
- 30.Meng F, Park Y, Zhou H. Role of proline, glycerol, and heparin as protein folding aids during refolding of rabbit muscle creatine kinase. Int J Biochem Cell Biol. 2001;33:701–709. doi: 10.1016/s1357-2725(01)00048-6. [DOI] [PubMed] [Google Scholar]
- 31.Perlmutter DH. Chemical chaperones: A pharmacological strategy for disorders of protein folding and trafficking. Pediatr Res. 2002;52:832–836. doi: 10.1203/00006450-200212000-00004. [DOI] [PubMed] [Google Scholar]
- 32.Diamant S, Eliahu N, Rosenthal D, Goloubinoff P. Chemical chaperones regulate molecular chaperones in vitro and in cells under combined salt and heat stresses. J Biol Chem. 2001;276:39586–39591. doi: 10.1074/jbc.M103081200. [DOI] [PubMed] [Google Scholar]
- 33.Costenoble R, Valadi H, Gustafsson L, Niklasson C, Franzén CJ. Microaerobic glycerol formation in Saccharomyces cerevisiae. Yeast. 2000;16:1483–1495. doi: 10.1002/1097-0061(200012)16:16<1483::AID-YEA642>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 34.Carey JS, Laffan D, Thomson C, Williams MT. Analysis of the reactions used for the preparation of drug candidate molecules. Org Biomol Chem. 2006;4:2337–2347. doi: 10.1039/b602413k. [DOI] [PubMed] [Google Scholar]
- 35.Tomar V, Kukreti S, Prakash S, Madan J, Chandra R. Noscapine and its analogs as chemotherapeutic agent: Current updates. Curr Top Med Chem. 2017;17:174–188. doi: 10.2174/1568026616666160530153518. [DOI] [PubMed] [Google Scholar]
- 36.Borodina I, Nielsen J. Advances in metabolic engineering of yeast Saccharomyces cerevisiae for production of chemicals. Biotechnol J. 2014;9:609–620. doi: 10.1002/biot.201300445. [DOI] [PubMed] [Google Scholar]
- 37.Alberti S, Gitler AD, Lindquist S. A suite of gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast. 2007;24:913–919. doi: 10.1002/yea.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siddiqui MS, Choksi A, Smolke CD. A system for multilocus chromosomal integration and transformation-free selection marker rescue. FEMS Yeast Res. 2014;14:1171–1185. doi: 10.1111/1567-1364.12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shao Z, Zhao H, Zhao H. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 2009;37:e16. doi: 10.1093/nar/gkn991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Güldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.