Significance
Regulators essential for facilitating gene silencing are interesting targets of epigenetic studies. Our work describes a regulator, CSE1L, that is essential for the silencing of many endogenous methylated genes. Depletion of CSE1L reactivates these genes without causing DNA demethylation. Interestingly, such reactivation is not due to a direct chromatin role of CSE1L. Instead, it depends on the role of CSE1L in importin-mediated protein nuclear transportation, which is confirmed by similar effects observed in cells depleted of other players in the same protein transportation pathway. Intriguingly, importin-mediated protein nuclear transportation preferentially facilitates gene silencing with specificity for a subset of genes, suggesting that the cargo specificity of protein nuclear import systems may impact the selectivity of gene regulation.
Keywords: CSE1L, DNA methylation, gene silencing, nuclear import pathway
Abstract
Epigenetic silencing can be mediated by various mechanisms, and many regulators remain to be identified. Here, we report a genome-wide siRNA screening to identify regulators essential for maintaining gene repression of a CMV promoter silenced by DNA methylation. We identified CSE1L (chromosome segregation 1 like) as an essential factor for the silencing of the reporter gene and many endogenous methylated genes. CSE1L depletion did not cause DNA demethylation. On the other hand, the methylated genes derepressed by CSE1L depletion largely overlapped with methylated genes that were also reactivated by treatment with histone deacetylase inhibitors (HDACi). Gene silencing defects observed upon CSE1L depletion were linked to its nuclear import function for certain protein cargos because depletion of other factors involved in the same nuclear import pathway, including KPNAs and KPNB1 proteins, displayed similar derepression profiles at the genome-wide level. Therefore, CSE1L appears to be critical for the nuclear import of certain key repressive proteins. Indeed, NOVA1, HDAC1, HDAC2, and HDAC8, genes known as silencing factors, became delocalized into cytosol upon CSE1L depletion. This study suggests that the cargo specificity of the protein nuclear import system may impact the selectivity of gene silencing.
DNA methylation is a well-characterized epigenetic modification that plays important roles in various biological processes, including transcription regulation, genomic imprinting, X chromosome inactivation, and transposon silencing (1–4). The correlation between DNA methylation and gene silencing has been extensively documented by a large body of literature.
DNA methylation directly inhibits the DNA-binding capacity of certain transcription factors that are sensitive to methylated CpG moieties within their target DNA motifs, and such direct inhibition contributes to the silencing of some methylated genes (5–8). On the other hand, DNA methylation on naked DNA often does not directly inhibit gene transcription (9, 10). Thus, DNA methylation-mediated silencing can also function in the chromatin context that involves the recognition of methylated CpG by methylated DNA-binding proteins, such as MeCP2 and other methyl-CpG–binding domain (MBD) proteins and subsequent recruitment of HDACs (11–19).
In addition to the above-mentioned histone deacetylation-mediated silencing for methylated genes, various studies have also revealed the presence of alternative mechanisms (20–23). These alternative mechanisms potentially involve site-specific repressors, histone H3K9 methylation, and other chromatin factors responsible for condensed chromatin formation (24–26). It remains to be determined whether unknown factors are essential for methylated gene silencing and, if so, how they execute their function.
To advance our understanding of DNA methylation-related gene silencing, we developed an unbiased RNAi screening to identify regulators involved in this process. We identified CSE1L, a key player in the nuclear import pathway, as an essential factor for maintaining the repression of many methylated genes. Mechanistically, CSE1L functions by facilitating the nuclear import of certain cargo proteins that are essential for gene repression.
Results
A Genome-Wide siRNA Screening for Factors Required for Silencing a Methylated CMV Promoter.
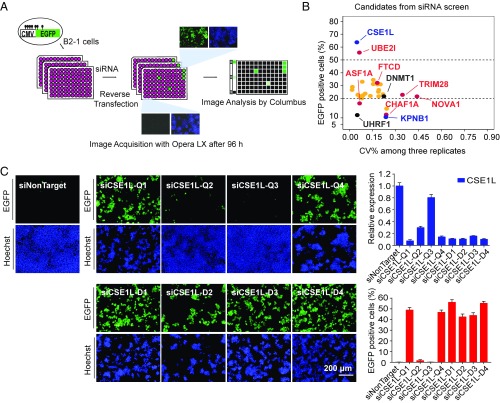
To identify regulators involved in DNA methylation-mediated gene silencing, we devised a cell-based genome-wide RNAi screening system. To generate the reporter cell line, a plasmid containing an EGFP reporter driven by the CMV promoter was first methylated in vitro with SssI methyltransferase. Then we cotransfected this reporter construct with a plasmid that contains a hygromycin-resistant selection marker into HEK293F cells (Fig. S1A). A stable reporter cell line, B2-1, in which the EGFP gene promoter remained fully methylated (Fig. S1B) and showed little basal EGFP expression (Fig. S1C), was chosen from individual hygromycin-resistant clones. To validate this reporter cell line, we performed siRNA-mediated knockdown for DNMT1 and UHRF1, two key players required for the maintenance of DNA methylation; EGFP was robustly reactivated (Fig. S1C).
Using this reporter cell line, we carried out image-based siRNA screening targeting 19,121 human genes (Fig. 1A). The cells were imaged for GFP and Hoechst fluorescence, and the mean percentage of GFP-expressing cells (RGFP) was calculated for each gene. The RGFP value in the negative control was less than 0.5. Genes with RGFP above 5 were considered as primary hits. Given that siRNAs often display off-target effects and each sample in the siRNA library was a pool of four pairs of siRNAs, we synthesized individual siRNA pairs for these primary hits and performed a secondary screening. After the secondary screening, 28 genes with at least two independent functional siRNA pairs were shortlisted as confirmed hits for further study (Fig. 1B and Table S1). DNMT1 (RGFP = 21.73) and UHRF1 (RGFP = 7.13), two genes initially used as positive controls, were among the confirmed hits. In addition, several factors previously implicated in gene silencing, including UBE2I, CHAF1A, NOVA1, ASF1A, and TRIM28 (27–29), were also on the list of confirmed hits (Fig. S1D and Table S1). These results indicate the robustness of our screening.
Fig. 1.
Genome-wide RNAi screening identifies a role for CSE1L in EGFP reporter silencing. (A) Experimental scheme of the high-content RNAi screening. (Magnification: 10×.) (B) Plot of the coefficients of variation (CV%) among three biological replicates and the mean RGFP for each of the confirmed hits, verified with at least two independent siRNA pairs. The scale of the y axis below 20 was adjusted for clarity. (C) Validation with eight independent siRNA pairs for CSE1L. Cell images (Left), knockdown efficiency (Upper Right), and RGFP values (Lower Right) of each siRNA pair are shown. Error bars represent SD.
CSE1L Is a Candidate Required for DNA Methylated EGFP Reporter Silence.
CSE1L, a candidate with an RGFP >60, was the top hit in our list (Fig. 1B). For further validation, eight independent siRNA pairs predicted to target CSE1L were tested (Table S2). Six of the eight siRNA pairs targeting CSE1L robustly activated EGFP expression in B2-1 cells, and only two siRNA pairs (siCSE1L-Q2 and siCSE1L-Q3) showed little or no EGFP activation (Fig. 1C), consistent with the knockdown efficiency of these siRNAs (Fig. 1C and Fig. S1E). These results suggest that CSE1L is likely a real hit.
CSE1L is an essential gene and cannot be knocked out (30). To obtain a cell line with tunable CSE1L expression, we established a stable cell line based on B2-1 that ectopically expressed a Flag-tagged CSE1L so that the expression of Flag-CSE1L could be turned off by the addition of doxycycline (Dox). Then, we knocked out the endogenous CSE1L in this cell line to generate a CSE1L-tunable cell line which lost CSE1L expression upon the addition of Dox (Fig. S1F). We named this cell line “DTO” (for “Dox-induced CSE1L turn off”). EGFP expression in DTO cells can be robustly induced by the addition of Dox (Fig. S1G). These experiments further confirm that CSE1L is required for the silencing of the EGFP reporter driven by a methylated CMV promoter and that CSE1L is likely a regulator of DNA methylation-mediated gene silencing.
Loss of CSE1L Expression Activates a Fraction of Endogenous Genes Silenced by DNA Methylation.
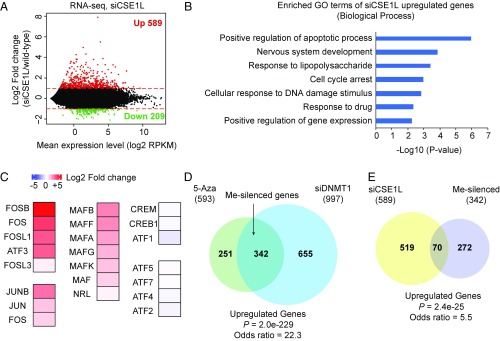
To study the role of CSE1L in the transcriptional regulation of endogenous genes, we performed RNA-sequencing (RNA-seq) in B2-1 cells with or without CSE1L. We identified 798 differentially expressed genes (with a greater than twofold expression change). Among them, 589 genes were up-regulated (Fig. 2A), supporting the potential role of CSE1L in gene repression. Gene Ontology (GO)-term analysis of these 589 up-regulated genes showed an enrichment in the positive regulation of apoptotic process (Fig. 2B). A large body of literature has linked dysregulated CSE1L expression with tumor progression, thereby implicating a role for CSE1L in promoting tumorigenesis (31–39). Interestingly, the expression levels of several AP-1 transcription factors, including oncogenes such as FOS, MAF, and JUN, were elevated upon the loss of CSE1L expression (Fig. 2C and Fig. S2A), which is also consistent with a role for CSE1L in tumorigenesis.
Fig. 2.
Depletion of CSE1L activates a fraction of endogenous genes silenced by DNA methylation. (A) MA plot (log ratio vs. average) showing differential gene expression in CSE1L-knockdown cells. The log2 fold change in the expression of each gene is plotted against the mean gene expression (log2 RPKM). Red dots represent genes with significantly increased expression in siCSE1L vs. wild-type cells. Green dots represent genes with significantly decreased expression in siCSE1L vs. wild-type cells. The red lines indicate the log2 fold change at 1 and −1. (B) GO-term analysis of up-regulated genes in siCSE1L-treated cells. (C) Heat-map showing the change in the expression of AP1 superfamily transcription factors in siCSE1L-treated cells. (D) Venn diagram showing the overlap of up-regulated genes in 5-Aza– and siDNMT1-treated cells (P = 2.0e-229). Me-silenced genes are the genes commonly up-regulated in 5-Aza and siDNMT1 treatments. (E) Venn diagram showing the overlap of up-regulated genes in siCSE1L-, 5-Aza–, and siDNMT1-treated cells (P = 2.4e-25).
To determine the genes silenced by DNA methylation, we performed RNA-seq experiments with B2-1 cells and with B2-1 cells treated with siRNA targeting DNMT1 (siDNMT1) or 5 μM 5-aza-deoxycytidine (5-Aza), a compound that impairs DNA methylation maintenance. Genes (342) that were commonly up-regulated in siDNMT1 and 5-Aza treatments were defined as DNA methylation-silenced (Me-silenced) genes (Fig. 2D). Approximately 20% of DNA Me- silenced genes became activated upon CSE1L knockdown (Fig. 2E), which was significantly higher than random overlap (P = 2.4e-25), and the Fisher’s exact test also gave an odds ratio of 5.5, representing the strength of association (40).
To examine whether the above observation can be generalized in other cells, we performed CSE1L knockdown and 5-Aza treatment in human HCT116 cells and then performed RNA-seq experiments with these cells. After 5-Aza treatment, 194 genes were up-regulated in HCT116 cells (Fig. S2B). Strikingly, among these 194 genes, 106 (55%) were also activated after CSE1L knockdown (Fig. S2B). This was significantly higher than random overlap (odds ratio = 22.1, P = 5.5e-79) (Fig. S2B).
Taken together, these results show that CSE1L is required for maintaining the silencing of a subset of endogenous methylated genes.
CSE1L Down-Regulation Does Not Cause DNA Demethylation.
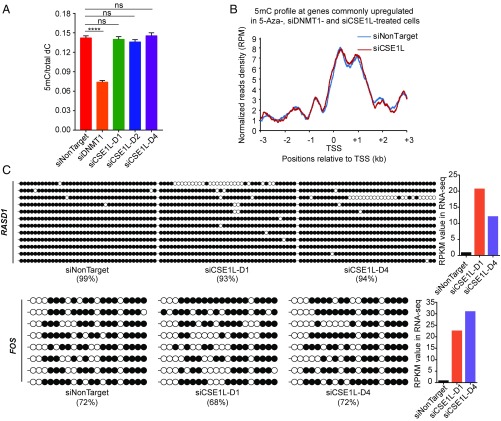
To determine if CSE1L down-regulation activates methylated genes by DNA demethylation, we measured the global 5-methylcytosine (5mC) level with ultra-high-pressure LC–multiple reaction monitoring (UHPLC-MRM) MS/MS using B2-1 cells with or without CSE1L knockdown. We did not observe a global change in 5mC level with three independent siRNA pairs targeting CSE1L, but DNMT1 knockdown clearly reduced the 5mC level in a control experiment (Fig. 3A). These results indicate that CSE1L is not required for the maintenance of global DNA methylation.
Fig. 3.
CSE1L down-regulation does not cause DNA demethylation. (A) UHPLC-MRM MS/MS quantitation of 5mC in genomic DNA from B2-1 cells treated with siNonTarget, siDNMT1, siCSE1L-D1, siCSE1L-D2, or siCSE1L-D4. 5mC represents 5-methyl-2′-deoxycytidine, and dC represents 2’-deoxycytidine. Error bars represent the SD of three replicates. An unpaired two-tailed t test was used for statistical calculation. ****P < 0.0001; ns, not significant. (B) Metagene analysis showing 5mC profiles around the TSS of genes commonly up-regulated in siCSE1L-, 5-Aza–, and siDNMT1-treated cells. (C, Left) Bisulfite sequencing results of the CpG island of RASD1 (Upper Left) and FOS (Lower Left). Black and white circles indicate methylated and unmethylated CpG sites. The percentages of methylated CpG sites are stated. (Right) Gene-expression levels of RASD1 (Upper Right) and FOS (Lower Right) in B2-1 cells treated with siNonTarget, siCSE1L-D1, or siCSE1L-D4.
We next asked whether gene activation induced by CSE1L knockdown was caused by locus-specific DNA demethylation. We performed methylated DNA immunoprecipitation followed by deep sequencing (MeDIP-seq) using B2-1 cells with or without CSE1L knockdown. DNA methylation levels around the transcription start site (TSS) of genes that were up-regulated by both CSE1L knockdown and 5-Aza treatment were evaluated, and no significant alteration in 5mC level was observed in these regions when CSE1L was knocked down (Fig. 3B).
We also examined the methylation status of the CMV promoter of the reporter gene, which remained heavily methylated after CSE1L knockdown with two independent siRNA pairs (Fig. S3A). Likewise, no apparent change in DNA methylation level was observed at the CpG islands of two endogenous genes (RASD1 and FOS) which were up-regulated upon CSE1L knockdown (Fig. 3C).
These data collectively indicate that CSE1L is not involved in DNA methylation maintenance and that genes activated by CSE1L knockdown did not undergo DNA demethylation.
Knockdown of CSE1L Preferentially Activates Genes That Can Be Activated by Histone Deacetylase Inhibitors.
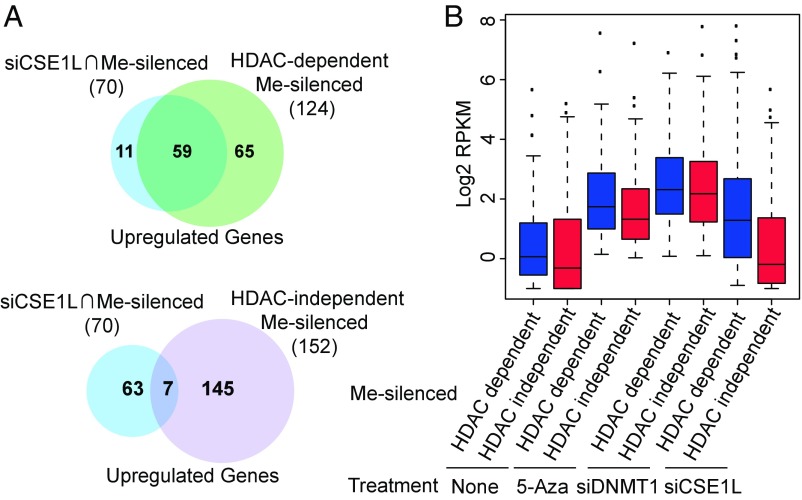
Some, but not all, DNA methylation-silenced genes can be effectively reactivated by treatment with histone deacetylase inhibitors (HDACi) (18, 20). To gain insight into the categorization of methylated genes derepressed by CSE1L knockdown, we treated B2-1 cells with two different HDACi, trichostatin A (TSA) and suberoylanilide hydroxamic acid (SAHA), and then performed RNA-seq experiments. Among the 342 DNA methylation-silenced genes defined in Fig. 2D, 124 were also activated by both TSA and SAHA treatments [categorized as the “histone deacetylase (HDAC)-dependent Me-silenced group”], while 152 genes were not activated in either TSA- or SAHA-treated B2-1 cells [categorized as the “HDAC-independent Me-silenced group” (Fig. S3B)]. Interestingly, approximately half of the genes in the HDAC-dependent Me-silenced group (59 of 124) were activated by CSE1L knockdown. In contrast, only ∼5% of the HDAC-independent Me-silenced group genes (7 of 124) were activated by CSE1L knockdown (Fig. 4A). This suggests that CSE1L knockdown preferentially activates genes that can be activated by HDACi.
Fig. 4.
CSE1L depletion preferentially activates genes that can be activated by HDACi. (A, Upper) Venn diagram showing the overlap between genes commonly up-regulated by siCSE1L, 5-Aza, and siDNMT1 treatments and genes commonly activated by 5-Aza and siDNMT1, TSA, and SAHA treatments. (Lower) Venn diagram showing the overlap between genes commonly up-regulated by siCSE1L and 5-Aza and siDNMT1 treatments and genes up-regulated by 5-Aza and siDNMT1 treatments but not by TSA or SAHA treatment. Me-silenced genes are the genes commonly up-regulated in 5-Aza and siDNMT1 treatments. HDAC-dependent Me-silenced genes are the genes commonly activated by 5-Aza and siDNMT1, TSA, and SAHA treatment. HDAC-independent Me-silenced genes are the genes activated only by 5-Aza and siDNMT1 treatment but not by TSA or SAHA treatment. (B) Box plots showing the gene-expression levels of the indicated gene sets in control and 5-Aza–, siDNMT1-, and siCSE1L-treated cells.
We then plotted the distribution of the reads per kilobase of transcript per million reads mapped (RPKM) values of these two groups of genes in B2-1 cells with various treatments. The expression levels of HDAC-dependent methylation-silenced genes were preferentially activated by CSE1L knockdown (Fig. 4B). These results further support the idea that CSE1L knockdown preferentially activates genes that can be activated by HDACi.
Dysregulated Nuclear Transportation Induced by CSE1L Depletion Contributes to Gene Activation.
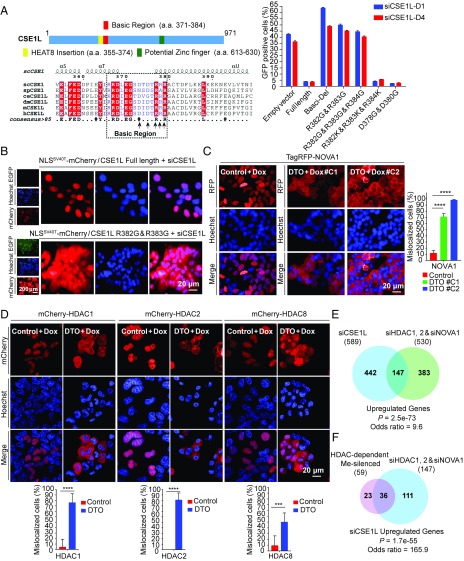
CSE1L consists of 20 HEAT (Huntingtin, elongation factor 3, a subunit of PP2A, and TOR) repeats and is conserved from yeast to mammals (41). To explore the mechanistic role of CSE1L in gene silencing, we constructed a series of siRNA-resistant CSE1L-mutant expression vectors and transfected them into B2-1 cells to generate corresponding stable cell lines (Fig. S4A). Then, we knocked down endogenous CSE1L and quantified the percentage of EGFP-positive cells using FACS. Both the N-terminal and C-terminal deletions impaired the silencing effect of CSE1L to some degree, suggesting the importance of the overall structure of CSE1L (Fig. S4B). In addition, the replacement of a small basic region of CSE1L (amino acids 371–384) (42) with a flexible linker also impaired the silencing effect of CSE1L, whereas a potential zinc finger motif (amino acids 613–630) (42) and the HEAT8 insertion region (amino acids 355–374) (41) were dispensable for the silencing function of CSE1L (Fig. S4B).
We performed further mutagenesis studies within the basic region and found that an R382G/R383G double mutation fully impaired the silencing effect of CSE1L (Fig. 5A). Interestingly, this region was previously reported to be a potential DNA-binding module (42), and CSE1L is indeed a nuclear protein (Fig. S4C) (42, 43). However, ChIP-sequencing (ChIP-seq) experiments using B2-1 cells with an antibody against CSE1L revealed no enrichment at all (Fig. S5), although this antibody detected a single major band in a nuclear extract of B2-1 cells and was able to immunoprecipitate endogenous CSE1L (Fig. S4D). To exclude the possibility that this antibody was not suited for ChIP-seq experiments, we also introduced the BirA-mediated site-specific biotinylation system (44–46) in 293F cells to overexpress a biotinylated CSE1L (Fig. S4E) and then performed streptavidin-mediated ChIP-seq for biotinylated CSE1L; again, no enrichment was observed (Fig. S5). In total, we were able to call only 118 peaks of bio-CSE1L under a fairly loose threshold (q <1e-2), but most of them ( 102/118; 86%) were located within ENCODE-blacklisted genomic regions (47) that tend to show artificially high signal (probably caused by unassembled repeats that are mistakenly uniquely mapped). Therefore, we wondered whether CSE1L was a chromatin-binding protein or just a protein in the nuclear plasma, and we performed fractionation experiments to clarify this. It is quite clear that nearly all CSE1L was present in the nuclear soluble fraction instead of in the chromatin fraction (Fig. S4F). This suggests that CSE1L does not function as a repressor or corepressor protein that directly associates with the target genes for silencing. Instead, CSE1L more likely exerts its silencing role in an indirect manner.
Fig. 5.
Dysregulated nuclear transportation induced by CSE1L depletion contributes to gene activation. (A, Left) Domain structure of CSE1L. The basic region of CSE1L, shown in the dashed box, is conserved from yeast to mammals. Mutation sites in the rescue assays are indicated by arrows. (Right) Percentages of EGFP-positive cells in various rescue experiments using CSE1L with the indicated deletions or point mutations in the basic regions. Error bars represent the SD of three biological replicates. (B) Fluorescence images demonstrating that the CSE1L R382G/R383G mutant that fails to rescue EGFP silencing in CSE1L-knockdown cells could not maintain the nuclear localization of NLSSV40T-mCherry. (C, Left) Fluorescence images showing that nuclear localization of TagRFP-NOVA1 is disrupted when expression of CSE1L is turned off. (Right) The percentage of cells with mislocalized TagRFP-NOVA1. Error bars represent SD. An unpaired two-tailed t test was used for statistical calculation. ****P < 0.0001. (D, Upper) Fluorescence images showing that the nuclear localization of mCherry-HDAC1, mCherry-HDAC2, and mCherry-HDAC8 are disrupted when expression of CSE1L is turned off. (Lower) The percentage of cells with mislocalized mCherry-HDACs. Error bars represent the SD. An unpaired two-tailed t test was used for statistical calculation. ****P < 0.0001; ***P < 0.001. (E) A Venn diagram showing the overlap between genes up-regulated by CSE1L knockdown and genes commonly up-regulated by knockdown of HDAC1 and HDAC2 and NOVA1 (P = 2.5e-73). (F) A Venn diagram showing the overlap between genes in the HDAC-dependent Me-silenced group and genes commonly up-regulated by CSE1L knockdown and HDAC1 and 2 and NOVA1 knockdown (P = 1.7e-55).
Although CSE1L was reported to associate with chromatin (42), it was also reported to function as an exportin, which re-exports importin α from the nucleus to the cytoplasm to facilitate the nuclear transportation of certain cargo proteins (41, 48). We then wondered whether CSE1L-mediated nuclear transportation is linked to its gene-silencing function. Interestingly, we observed that the CSE1L R382G/R383G mutant protein, which failed to maintain EGFP reporter silencing (Fig. 5A), also failed to maintain the nuclear localization of a mCherry protein fused with a nuclear localization signal (NLS) of SV40 large T protein (NLSSV40T-mCherry) (Fig. 5B). This suggests a connection between the two functions of CSE1L described above: gene silencing and nuclear transportation.
Given that the disruption of a nuclear transport pathway by CSE1L knockdown seemed to contribute to gene derepression, certain proteins required for gene silencing should be mislocalized into the cytosol upon CSE1L knockdown. To identify such protein(s), we established a system using DTO cells by transfecting candidate proteins fused with an TagRFP/mCherry tag under the control of a DOX-induced promoter. In these cells, the addition of DOX induced the expression of the TagRFP/mCherry-tagged candidate proteins and simultaneously turned off CSE1L expression. After screening with several genes in our confirmed hit list, we identified one protein, NOVA1, that was required for reporter gene silencing and became mislocalized in the cytosol upon CSE1L depletion. In B2-1 cells, TagRFP-tagged NOVA1 (TagRFP-NOVA1) showed a strict nuclear localization pattern, which was completely disrupted after CSE1L depletion (Fig. 5C). In contrast, the other hit proteins we tested, such as UHRF1 and TRIM28, showed no localization change in CSE1L-depleted cells (Fig. S6A).
Considering that a large fraction of genes activated by CSE1L knockdown were also activated by HDACi treatment (Fig. 4A), we wondered if any of the HDAC proteins were also cargo proteins of CSE1L-dependent nuclear transport pathway. Therefore we systematically tested all the HDAC proteins using the same strategy and found that HDAC1, HDAC2, and HDAC8 were significantly mislocalized into cytoplasm when the expression of CSE1L was turned off (Fig. 5D). Interestingly, the subcellular localization of the other HDAC proteins displayed little change, with HDAC3, HDAC5, and HDAC10 remaining localized in both cytoplasm and nuclei and all the other HDACs remaining enriched in the cytoplasm (Fig. S6B). Consistently, we also stained the endogenous HDAC1 and HDAC2 in DTO cells before and after turning off CSE1L and observed similar results (Fig. S6C).
To examine whether the mislocalization of NOVA1, HDAC1, and HDAC2 contributed to silencing defects in CSE1L-knockdown cells, we simultaneously knocked down NOVA1, HDAC1, and HDAC2 expression in B2-1 cells. RNA-seq experiments revealed up-regulation of 530 genes. Among them, 28% (147) overlapped with genes that were up-regulated upon CSE1L knockdown. This was significantly higher than random overlap (odds ratio = 9.6, P = 2.5e-73) (Fig. 5E). Moreover, of the 59 HDAC-dependent methylation-silenced genes that were up-regulated upon CSE1L knockdown (defined in Fig. 4A), 61% (36 of 59) were up-regulated in B2-1 cells, in which NOVA1, HDAC1, and HDAC2 were knocked down simultaneously (Fig. 5F). These results sufficiently support a partial contribution of NOVA1, HDAC1, and HDAC2 in CSE1L-mediated methylated gene silencing.
The cargo transportation selectivity may help explain the selectivity observed in gene activation upon CSE1L depletion, because the nuclear localization of some, but not all, repressors was impaired by CSE1L depletion, and some, but not all, silenced genes became derepressed after CSE1L depletion.
Disruption of Related Nuclear Import Pathway Components Leads to Similar Gene Activation.
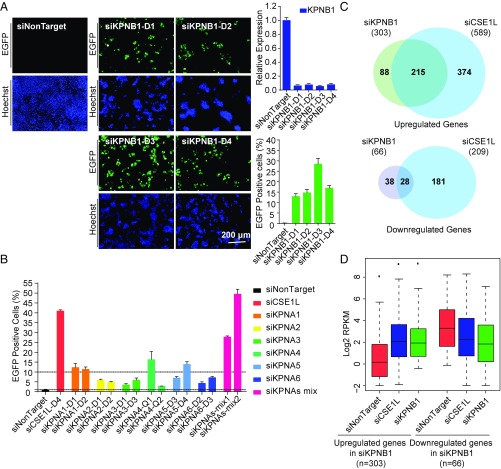
To further confirm the above conclusions, we tested whether disruption of the nuclear import pathway by knockdown of other factors related to CSE1L may also activate the silenced genes. In the nuclear import pathway, importin α recognizes and binds to classic NLS-containing proteins and then links them to importin β, which interacts with the nuclear pore complexes and mediates cargo translocation into the nucleus (49). Notably, humans have seven importin α proteins (KPNA1–7) and only one importin β (KPNB1) known to interact with importin α (50, 51). Given that KPNA7 was not expressed in B2-1 cells, we synthesized siRNAs targeting KPNA1–6 and KPNB1 and performed knockdown experiments. The knockdown efficiency was validated with qRT-PCR (Fig. 6A and Fig. S7A). Robust activation of methylated, silenced EGFP was observed in cells treated with four independent siRNA pairs targeting KPNB1 for knockdown (Fig. 6A). Knockdown of individual KPNA1–6 proteins caused derepression of the silenced EGFP gene to various degrees (Fig. 6B). We also used two sets of independent siRNA mixtures containing six siRNA pairs targeting all KPNAs, and both displayed robust derepression of the reporter gene (Fig. 6B). In addition, we treated B2-1 with 50 μM importazole and 40 μM ivermectin, two small molecules that block importin-β or importin α/β-mediated nuclear import (52, 53), and both robustly activated silenced EGFP (Fig. S7B). These results clearly indicate that the importin-mediated nuclear transportation pathway is indeed crucial for gene silencing.
Fig. 6.
Disruption of other components of the importin-mediated nuclear transportation pathway leads to similar transcription changes. (A) Four independent siRNA pairs targeting the KPNB1 activate reporter gene in B2-1 cells. Cell images (Left), knockdown efficiency (Upper Right), and RGFP values (Lower Right) of each siRNA pair are shown. Error bars represent the SD. (B) Knockdown of KPNAs activates silenced EGFP in B2-1 cells. The percentage of EGFP-expressing cells was measured with FACS. Error bars represent the SD of three biological replicates. (C) Venn diagrams showing the overlap of up-regulated genes (Upper) or down-regulated genes (Lower) in KPNB1- and CSE1L-knockdown cells. (D) Box plot showing the expression level of KPNB1-regulated genes in B2-1 cells treated with siNonTarget, siCSE1L, or siKPNB1.
To examine whether the derepression described above occurred at the genome-wide level, we performed RNA-seq experiments with KPNB1-knockdown B2-1 cells. After KPNB1 knockdown, 303 genes were up-regulated, and only 66 genes were down-regulated, supporting an overall role of KPNB1 in facilitating gene repression. Importantly, among the 303 up-regulated genes, 71% (215 genes) were also up-regulated after CSE1L depletion, and among the 66 down-regulated genes, 42% (28 genes) were also down-regulated after CSE1L depletion (Fig. 6C). A box plot analysis showed that the 303 genes up-regulated after KPNB1 depletion also displayed much higher expression upon CSE1L depletion (Fig. 6D). In addition, the expression of AP-1 superfamily members also displayed similar changes in KPNB1 and CSE1L knockdown cells (Fig. S7C). Collectively, these results support the idea that CSE1L and KPNB1 likely function in the same pathway in gene regulation.
Discussion
DNA methylation is a repressive marker critical for epigenetic gene silencing and often involves HDACs recruited by methylated DNA-binding proteins (11–19). However, not all methylated genes are necessarily silenced through the same mechanism. Approximately half the genes that are commonly up-regulated in siDNMT1 and 5-Aza treatments and are presumably silenced by DNA methylation fail to respond to HDACi treatment (Fig. S3B). Similar observations have been reported previously (20–22); a number of unbiased screenings have been performed, and several important regulators have been identified (27–29, 54–56). Some of these previously identified factors, including TRIM28, UBE2I, and CHAF1A, were also in the hit list of our screening. On the other hand, some of the previously identified factors were not in our hit list, and we also identified several silencing factors, including CSE1L. This can be explained by the specific features of the individual screening design and the efficiency of each screening. The specific feature of this study is the use of an integrated reporter gene with a methylated promoter. This may help identify factors related to DNA methylation-mediated silencing. In our results, the identification of CSE1L and its partner proteins (KPNAs and KPNB1) in the importin-mediated nuclear transportation system as indirect regulators of a fraction of methylated genes is interesting. However, their target genes largely overlap with the target genes of HDACi (Fig. 4A), suggesting that additional mechanisms controlling the silencing of other methylated genes remain to be uncovered. Further characterization of the other hits identified in this study may help resolve this question.
The discovery of a role for the importin-mediated nuclear transportation pathway in gene silencing is unexpected but in retrospect is not surprising. Here, we would like to emphasize an intriguing aspect of this discovery. The first expectation upon identifying the involvement of a nuclear transportation system in gene regulation is that it is unlikely to be specific. Interestingly, this expectation was incorrect. First, RNA-seq results from CSE1L knockdown cells showed that among the genes with altered expression (798 genes with more than twofold up- or down-regulation), 74% were up-regulated (Fig. 2A), clearly indicating a biased role for CSE1L in facilitating silencing. Second, importin-mediated nuclear transportation requires that a specific NLS be present in the cargo protein (48–50, 57). Therefore, the nuclear localization of some, but not all, repressive proteins was affected by CSE1L depletion (Fig. 5 C and D and Fig. S6 A and B). Due to the target-gene selectivity of these repressive proteins and their selectivity for the nuclear import machinery, the activation of a specific set of genes can be achieved by targeting the nuclear import machinery, similar to what we did for the importin pathway (Figs. 2 and 6). This could be of potential interest, especially when some DNA methyltransferase and HDACi have been approved as anticancer drugs. This point might be very relevant to the reported function of CSE1L in cancer progression. Dysregulated CSE1L expression and localization has been reported to correlate with cancer progression, and its abnormal distribution has been proposed to be a biomarker for prognosis of carcinomas (31, 32, 39, 58–62). We observed that CSE1L depletion robustly activated the expression of AP-1 superfamily transcription factors, including oncogenes such as FOS, JUN, and MAF (Fig. 2C). It would be interesting to dissect whether CSE1L promotes oncogenic metastasis in part through its regulation of AP-1 family transcription factors. Although these are speculations at this stage, our study provides a starting point for an alternative understanding and intervention strategy.
Another interesting observation is that we were able to activate methylated genes without affecting DNA methylation (Fig. 3 and Fig. S3A). An interesting future direction would be to test whether prolonged activation of these initially methylated genes would cause eventual demethylation and, if so, whether such a phenomenon would transform a short-term intervention into a cellular epigenetic memory.
The observations that we failed to immunoprecipitate CSE1L to chromatin (Fig. S5) and that CSE1L was predominantly localized in the nuclear plasma (Fig. S4 C and F) are also worthy of further investigation. CSE1L has been reported to associate with chromatin and regulates the expression of p53 target genes (42). In addition, CSE1L shows distinct localization patterns in different cell lines that have been previously studied (43, 63). Whether CSE1L can directly associate with chromatin and exert a regulatory function remains an open question. Nevertheless, the largely overlapping alteration to global transcription profiles in CSE1L- or KPNB1-depleted cells indicates that the silencing effect of CSE1L observed in this study is largely an indirect event.
In conclusion, our work reveals a role for CSE1L and the importin-mediated nuclear transportation pathway in gene silencing, suggesting that the cargo specificity of protein nuclear import systems may impact the selectivity of gene silencing.
Materials and Methods
High-Content RNAi Screening.
The Human Whole Genome siRNA Set V4.0 from Qiagen targeting 19,121 human genes was used for the genome-wide RNAi screening. The libraries consist of pools of four siRNA pairs per gene arrayed in 96-well plates. siRNA pairs targeting DNMT1 (siDNMT1) were included as the positive control, and a nontargeting siRNA pair (siNonTarget) was included as the negative control in every 96-well plate for screening. The addition of siRNA was carried out using a versatile pipetting robot, a Biomek FX workstation (Beckman Coulter), and the addition of transfection reagent and cells was performed with Matrix WellMate microplate dispenser (Thermo Scientific). The images were collected with the Opera LX high-content screening system (PerkinElmer) and analyzed with the Columbus Image Data Storage and Analysis System. Screenings were routinely performed in triplicate. The mean percentage of EGFP-positive cells in each well was determined from a total of seven image fields.
In brief, siRNA samples from the libraries were dispensed into 96-well plates containing Opti-MEM reduced serum medium. Then diluted siRNA solution was equally distributed into three CellCarrier 96-well plates (6005558; PerkinElmer). INTERFERin-HTS (410-060; Polyplus-transfection) diluted with tridistilled water (0.07 μL in 10 μL) was added into each well, and the plates were incubated at room temperature for 30 min. B2-1 cells were seeded in 96-well plates (4,500 cells per well) containing siRNAs with INTERFERin-HTS to give a final siRNA concentration of 40 nM. The transfected cells were incubated for 96 h for siRNA knockdown, and Hoechst 33342 was added to stain the nucleus. Then the cells were imaged with Opera LX.
Image Analysis with Columbus Image Analysis System.
Image acquisition was performed with seven fields per well using a 10× (air) objective lens for RNAi screening and 90 fields per well using a 60× (water) objective lens for protein cellular localization analysis.
The percentage of EGFP-positive cells was obtained using object identification modules in Columbus Image Analysis System to enumerate the number of Hoechst-stained nuclei and EGFP-expressing cells. In brief, images were submitted to Columbus to identify the nuclei based on Hoechst signal. Cells were then defined based on the identified nuclei, and the cytoplasm region around nuclei was determined. To reduce misdetection, morphology properties were also taken into consideration. The intensity properties of the EGFP signal in the cytoplasm region were calculated, and cells with an intensity value greater than 50 were defined as EGFP-positive cells. Then the percentage of EGFP-positive cells was calculated as the readout.
For calculating the percentage of mislocalized cells, images were imported into Columbus. The nucleus boundary was identified through the Hoechst signal, and regions that expanded or shrank from the nucleus boundary by 15 pixels were defined as the cytoplasm fraction or nuclear fraction, respectively. Next, the mean fluorescence density of TagRFP/mCherry in these two regions was measured, and cells with a ratio of fluorescence density between the cytoplasm fraction and the nuclear fraction higher than 0.75 were defined as “mislocalized cells.” The percentage of mislocalized cells was calculated as the readout.
RNAi.
siRNAs were chemically synthesized by the Biological Resource Center, National Institute of Biological Sciences (NIBS). siRNAs were dissolved in tridistilled water to a final concentration of 20 μM. siRNA transfection for hits validation or functional study was performed with Lipofectamine RNAiMax (13778150; Thermo); the final concentration of siRNA used was 10 nM. All siRNAs used in this study are listed in Table S1.
Generation of the DTO Cell Line.
The CSE1L DTO line was constructed in two steps. First, a Dox-induced CSE1L tetracycline (Tet)-off cell line was established using the Tet-Off Advanced system (Clontech). Guide RNA-resistant CSE1L cDNA was introduced into pLenti-TRE-puromycin for construction of the Tet-off vectors. Lentivirus was produced in 293FT cells by cotransfection of pLenti-TRE-CSE1L-puromycin or pLenti-tTA-neomycin with the lentivirus package vectors pMD2.g and psPAX2. These two lentiviruses were mixed (CSE1L:tTA = 1:10) to coinfect B2-1 cells. The infected population was selected in culture medium containing 1 μg/mL puromycin and 1 mg/mL G418. Individual surviving clones were screened for protein expression with or without 2 μg/mL Dox treatment. Then the endogenous CSE1L genes were knocked out using the CRISPR/Cas9 system, and cells for tests were grown in 96-well plates. Two single-guide RNA (sgRNA) sequences were used for CSE1L gene targeting: sgRNA-1-CSE1L (AATTTGTGAAGCCGATCGAGTGG) and sgRNA-2-CSE1L (GGCTTTAATGGCCACTCGATCGG).
RNA-Seq and Data Analysis.
Total RNA was directly extracted from cells cultured in 6-cm plates or six-well plates using TRIzol reagent (15596018; Thermo Fisher) with two biological replicates for each experiment. For RNA-seq, polyA-containing mRNA molecules were captured using attached oligo-dT and were subjected to library preparation according to the manufacturer’s instructions. RNA-seq reads were generated by the BGISEQ-500 platform (BGI, Ltd.) with the policy of single-ended 50 bp (SE50). The average RNA-seq depth was 24 million reads, ranging from 23.9–24.1 million reads. Clean reads were mapped to the hg19 genome with TopHat software (v1.4.1, cole-trapnell-lab.github.io/projects/tophat/). The transcription levels (RPKM) were quantified with Cufflinks software (v2.0.2, cole-trapnell-lab.github.io/projects/cufflinks/). Fold changes of gene expression were calculated with Cuffdiff software (v2.2.1, cole-trapnell-lab.github.io/cufflinks/cuffdiff/). Up-regulated/down-regulated genes were defined by log2(treatment/control) ≥1 or log2(treatment/control) equal to or less than −1, respectively, in which both RPKM values add a pseudo value of 0.5 to avoid being divided by zero.
Enrichment analysis of GO terms for gene sets was performed by Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resources 6.7 with default settings (64, 65).
Statistics.
All data with statistics are from at least three biological replicates. For image analysis, images with a total of at least 50 cells were used; the exact sample sizes are shown in Tables S3 and S4. Statistical analysis was performed with GraphPad Prism 7.00, and an unpaired two-tailed Student’s t test was used to determine the statistical significance between two groups. Data shown in bar graphs represent the mean ± SD, as indicated in figure legends. The statistical significance of the overlap between two groups of genes was calculated with the R package GeneOverlap (40).
Supplementary Material
Acknowledgments
We thank Dr. Jiemin Wong for providing human HDAC2 and HDAC11 cDNAs; the imaging facility at the Beijing NIBS for help with high-content screening; the Biological Resource Center at the Beijing NIBS for the synthesis of siRNAs used in this study; Junying Jia & Shuang Sun from FACS center, Yan Teng from the Center for Bio-Imaging, and Yihui Xu from the Key Laboratory of Infection and Immunity at the Institute of Biophysics, Chinese Academy of Sciences, for technical support. This work was primarily supported by China Natural Science Foundation Grant 31730047 and also by Chinese Ministry of Science and Technology Grants 2015CB856200 and 2017YFA0504200, Key Research Program of Frontier Sciences Grant QYZDY-SSW-SMC031, Chinese Academy of Sciences Strategic Priority Research Program Grant XDB08010103, and China Natural Science Foundation Grants 31425013 and 31521002. Z.Z. is sponsored by the Youth Innovation Promotion Association (Grant 2017133) of the Chinese Academy of Sciences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All datasets in this study have been deposited in the Gene Expression Omnibus databank (https://www.ncbi.nlm.nih.gov/geo/) (accession no. GSE96852).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800505115/-/DCSupplemental.
References
- 1.Cedar H. DNA methylation and gene activity. Cell. 1988;53:3–4. doi: 10.1016/0092-8674(88)90479-5. [DOI] [PubMed] [Google Scholar]
- 2.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 3.Li E, Zhang Y. DNA methylation in mammals. Cold Spring Harb Perspect Biol. 2014;6:a019133. doi: 10.1101/cshperspect.a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bestor TH, Edwards JR, Boulard M. Notes on the role of dynamic DNA methylation in mammalian development. Proc Natl Acad Sci USA. 2015;112:6796–6799. doi: 10.1073/pnas.1415301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iguchi-Ariga SM, Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989;3:612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- 6.Ben-Hattar J, Beard P, Jiricny J. Cytosine methylation in CTF and Sp1 recognition sites of an HSV tk promoter: Effects on transcription in vivo and on factor binding in vitro. Nucleic Acids Res. 1989;17:10179–10190. doi: 10.1093/nar/17.24.10179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu S, et al. DNA methylation presents distinct binding sites for human transcription factors. eLife. 2013;2:e00726. doi: 10.7554/eLife.00726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin Y, et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science. 2017;356:eaaj2239. doi: 10.1126/science.aaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kass SU, Landsberger N, Wolffe AP. DNA methylation directs a time-dependent repression of transcription initiation. Curr Biol. 1997;7:157–165. doi: 10.1016/s0960-9822(97)70086-1. [DOI] [PubMed] [Google Scholar]
- 10.Kass SU, Pruss D, Wolffe AP. How does DNA methylation repress transcription? Trends Genet. 1997;13:444–449. doi: 10.1016/s0168-9525(97)01268-7. [DOI] [PubMed] [Google Scholar]
- 11.Solage A, Cedar H. Organization of 5-methylcytosine in chromosomal DNA. Biochemistry. 1978;17:2934–2938. doi: 10.1021/bi00607a036. [DOI] [PubMed] [Google Scholar]
- 12.Keshet I, Lieman-Hurwitz J, Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986;44:535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- 13.Boyes J, Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991;64:1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- 14.Meehan RR, Lewis JD, Bird AP. Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 1992;20:5085–5092. doi: 10.1093/nar/20.19.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis JD, et al. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 16.Eden S, Hashimshony T, Keshet I, Cedar H, Thorne AW. DNA methylation models histone acetylation. Nature. 1998;394:842. doi: 10.1038/29680. [DOI] [PubMed] [Google Scholar]
- 17.Jones PL, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 18.Nan X, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, et al. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cameron EE, Bachman KE, Myöhänen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 21.Coffee B, Zhang F, Warren ST, Reines D. Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nat Genet. 1999;22:98–101. doi: 10.1038/8807. [DOI] [PubMed] [Google Scholar]
- 22.Benjamin D, Jost JP. Reversal of methylation-mediated repression with short-chain fatty acids: Evidence for an additional mechanism to histone deacetylation. Nucleic Acids Res. 2001;29:3603–3610. doi: 10.1093/nar/29.17.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lande-Diner L, et al. Role of DNA methylation in stable gene repression. J Biol Chem. 2007;282:12194–12200. doi: 10.1074/jbc.M607838200. [DOI] [PubMed] [Google Scholar]
- 24.Blattler A, Farnham PJ. Cross-talk between site-specific transcription factors and DNA methylation states. J Biol Chem. 2013;288:34287–34294. doi: 10.1074/jbc.R113.512517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuks F, et al. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J Biol Chem. 2003;278:4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- 26.Smallwood A, Estève PO, Pradhan S, Carey M. Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev. 2007;21:1169–1178. doi: 10.1101/gad.1536807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poleshko A, et al. Identification of a functional network of human epigenetic silencing factors. J Biol Chem. 2010;285:422–433. doi: 10.1074/jbc.M109.064667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poleshko A, et al. Human factors and pathways essential for mediating epigenetic gene silencing. Epigenetics. 2014;9:1280–1289. doi: 10.4161/epi.32088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang BX, et al. Systematic identification of factors for provirus silencing in embryonic stem cells. Cell. 2015;163:230–245. doi: 10.1016/j.cell.2015.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bera TK, Bera J, Brinkmann U, Tessarollo L, Pastan I. Cse1l is essential for early embryonic growth and development. Mol Cell Biol. 2001;21:7020–7024. doi: 10.1128/MCB.21.20.7020-7024.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wellmann A, et al. High expression of the proliferation and apoptosis associated CSE1L/CAS gene in hepatitis and liver neoplasms: Correlation with tumor progression. Int J Mol Med. 2001;7:489–494. doi: 10.3892/ijmm.7.5.489. [DOI] [PubMed] [Google Scholar]
- 32.Liao CF, et al. CSE1L/CAS, the cellular apoptosis susceptibility protein, enhances invasion and metastasis but not proliferation of cancer cells. J Exp Clin Cancer Res. 2008;27:15. doi: 10.1186/1756-9966-27-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tai CJ, et al. Correlations between cytoplasmic CSE1L in neoplastic colorectal glands and depth of tumor penetration and cancer stage. J Transl Med. 2013;11:29. doi: 10.1186/1479-5876-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winkler J, et al. Cellular apoptosis susceptibility (CAS) is linked to integrin β1 and required for tumor cell migration and invasion in hepatocellular carcinoma (HCC) Oncotarget. 2016;7:22883–22892. doi: 10.18632/oncotarget.8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holzer K, et al. Cellular apoptosis susceptibility (CAS) is overexpressed in thyroid carcinoma and maintains tumor cell growth: A potential link to the BRAFV600E mutation. Int J Oncol. 2016;48:1679–1687. doi: 10.3892/ijo.2016.3388. [DOI] [PubMed] [Google Scholar]
- 36.Behrens P, Brinkmann U, Wellmann A. CSE1L/CAS: Its role in proliferation and apoptosis. Apoptosis. 2003;8:39–44. doi: 10.1023/a:1021644918117. [DOI] [PubMed] [Google Scholar]
- 37.Pimiento JM, et al. Knockdown of CSE1L gene in colorectal cancer reduces tumorigenesis in vitro. Am J Pathol. 2016;186:2761–2768. doi: 10.1016/j.ajpath.2016.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu J-H, et al. Suppression of cellular apoptosis susceptibility (CSE1L) inhibits proliferation and induces apoptosis in colorectal cancer cells. Asian Pac J Cancer Prev. 2013;14:1017–1021. doi: 10.7314/apjcp.2013.14.2.1017. [DOI] [PubMed] [Google Scholar]
- 39.Tai CJ, Hsu CH, Shen SC, Lee WR, Jiang MC. Cellular apoptosis susceptibility (CSE1L/CAS) protein in cancer metastasis and chemotherapeutic drug-induced apoptosis. J Exp Clin Cancer Res. 2010;29:110. doi: 10.1186/1756-9966-29-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen L, Sinai M. 2013 GeneOverlap: An R Package to Test and Visualize Gene Overlaps, R Package Version 1.14.0. Available at shenlab-sinai.github.io/shenlab-sinai/. Accessed March 1, 2018.
- 41.Cook A, et al. The structure of the nuclear export receptor Cse1 in its cytosolic state reveals a closed conformation incompatible with cargo binding. Mol Cell. 2005;18:355–367. doi: 10.1016/j.molcel.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 42.Tanaka T, Ohkubo S, Tatsuno I, Prives C. hCAS/CSE1L associates with chromatin and regulates expression of select p53 target genes. Cell. 2007;130:638–650. doi: 10.1016/j.cell.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 43.Lorenzato A, et al. AKT activation drives the nuclear localization of CSE1L and a pro-oncogenic transcriptional activation in ovarian cancer cells. Exp Cell Res. 2013;319:2627–2636. doi: 10.1016/j.yexcr.2013.07.030. [DOI] [PubMed] [Google Scholar]
- 44.Wang J, et al. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 45.Kim J, Cantor AB, Orkin SH, Wang J. Use of in vivo biotinylation to study protein-protein and protein-DNA interactions in mouse embryonic stem cells. Nat Protoc. 2009;4:506–517. doi: 10.1038/nprot.2009.23. [DOI] [PubMed] [Google Scholar]
- 46.He A, Pu WT. Genome-wide location analysis by pull down of in vivo biotinylated transcription factors. Curr Protoc Mol Biol. 2010;92:21.20.1–21.20.15. doi: 10.1002/0471142727.mb2120s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dunham I, et al. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kutay U, Bischoff FR, Kostka S, Kraft R, Görlich D. Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- 49.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 50.Miyamoto Y, Yamada K, Yoneda Y. Importin α: A key molecule in nuclear transport and non-transport functions. J Biochem. 2016;160:69–75. doi: 10.1093/jb/mvw036. [DOI] [PubMed] [Google Scholar]
- 51.Friedrich B, Quensel C, Sommer T, Hartmann E, Köhler M. Nuclear localization signal and protein context both mediate importin alpha specificity of nuclear import substrates. Mol Cell Biol. 2006;26:8697–8709. doi: 10.1128/MCB.00708-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soderholm JF, et al. Importazole, a small molecule inhibitor of the transport receptor importin-β. ACS Chem Biol. 2011;6:700–708. doi: 10.1021/cb2000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wagstaff KM, Sivakumaran H, Heaton SM, Harrich D, Jans DA. Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem J. 2012;443:851–856. doi: 10.1042/BJ20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poleshko A, et al. Identification of cellular proteins that maintain retroviral epigenetic silencing: Evidence for an antiviral response. J Virol. 2008;82:2313–2323. doi: 10.1128/JVI.01882-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fang M, Ou J, Hutchinson L, Green MR. The BRAF oncoprotein functions through the transcriptional repressor MAFG to mediate the CpG island methylator phenotype. Mol Cell. 2014;55:904–915. doi: 10.1016/j.molcel.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Serra RW, Fang M, Park SM, Hutchinson L, Green MR. A KRAS-directed transcriptional silencing pathway that mediates the CpG island methylator phenotype. eLife. 2014;3:e02313. doi: 10.7554/eLife.02313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pemberton LF, Paschal BM. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187–198. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- 58.Alnabulsi A, et al. Cellular apoptosis susceptibility (chromosome segregation 1-like, CSE1L) gene is a key regulator of apoptosis, migration and invasion in colorectal cancer. J Pathol. 2012;228:471–481. doi: 10.1002/path.4031. [DOI] [PubMed] [Google Scholar]
- 59.Chang CC, et al. The prognostic significance of nuclear CSE1L in urinary bladder urothelial carcinomas. Ann Diagn Pathol. 2012;16:362–368. doi: 10.1016/j.anndiagpath.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 60.Yuksel UM, et al. Does CSE1L overexpression affect distant metastasis development in breast cancer? Oncol Res Treat. 2015;38:431–434. doi: 10.1159/000438501. [DOI] [PubMed] [Google Scholar]
- 61.Zhan C, Zhang XY, Pang D. High expression of CSE1L is associated with poor prognosis in breast cancer. Int J Clin Exp Pathol. 2016;9:11788–11794. [Google Scholar]
- 62.Cheng DD, et al. CSE1L interaction with MSH6 promotes osteosarcoma progression and predicts poor patient survival. Sci Rep. 2017;7:46238. doi: 10.1038/srep46238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Behrens P, Brinkmann U, Fogt F, Wernert N, Wellmann A. Implication of the proliferation and apoptosis associated CSE1L/CAS gene for breast cancer development. Anticancer Res. 2001;21:2413–2417. [PubMed] [Google Scholar]
- 64.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 65.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.