Significance
β-Lactam antibiotics are currently the most broadly used class of antibiotics. These antibiotics prevent bacterial cell wall from cross-linking, which leads to the accumulation of long non–cross-linked strands of peptidoglycan. Pseudomonas aeruginosa, a nefarious bacterial pathogen, attempts to repair this aberrantly formed peptidoglycan by the function of the lytic transglycosylase Slt. We document in the present report that Slt turns over the peptidoglycan by both scission of the glycosidic bonds from a terminus or in the middle of the peptidoglycan. In a series of seven X-ray crystal structures, we provide structural context to how these two reactions take place. These results disclose the details of bacterial response to the β-lactam antibiotic challenge.
Keywords: cell wall, cell-wall recycling, peptidoglycan, lytic transglycosylases
Abstract
β-Lactam antibiotics inhibit cell-wall transpeptidases, preventing the peptidoglycan, the major constituent of the bacterial cell wall, from cross-linking. This causes accumulation of long non–cross-linked strands of peptidoglycan, which leads to bacterial death. Pseudomonas aeruginosa, a nefarious bacterial pathogen, attempts to repair this aberrantly formed peptidoglycan by the function of the lytic transglycosylase Slt. We document in this report that Slt turns over the peptidoglycan by both exolytic and endolytic reactions, which cause glycosidic bond scission from a terminus or in the middle of the peptidoglycan, respectively. These reactions were characterized with complex synthetic peptidoglycan fragments that ranged in size from tetrasaccharides to octasaccharides. The X-ray structure of the wild-type apo Slt revealed it to be a doughnut-shaped protein. In a series of six additional X-ray crystal structures, we provide insights with authentic substrates into how Slt is enabled for catalysis for both the endolytic and exolytic reactions. The substrate for the exolytic reaction binds Slt in a canonical arrangement and reveals how both the glycan chain and the peptide stems are recognized by the Slt. We document that the apo enzyme does not have a fully formed active site for the endolytic reaction. However, binding of the peptidoglycan at the existing subsites within the catalytic domain causes a conformational change in the protein that assembles the surface for binding of a more expansive peptidoglycan between the catalytic domain and an adjacent domain. The complexes of Slt with synthetic peptidoglycan substrates provide an unprecedented snapshot of the endolytic reaction.
Transpeptidases, members of the superfamily of penicillin-binding proteins (PBPs), are inhibited by β-lactam antibiotics. These enzymes cross-link the peptidoglycan, the major constituent of the bacterial cell wall (Fig. 1) (1–3). This inhibition results in longer non–cross-linked strands of peptidoglycan, which are aberrant structures. The bacterium attempts to repair the damage to the cell wall by initiating turnover of these longer non–cross-linked strands by the functions of the superfamily of lytic transglycosylases (LTs) (Fig. 1) (4). Multiple LTs are known in various bacteria, exemplified by eight in Escherichia coli and 11 in Pseudomonas aeruginosa (5). It is of interest that the activities of these enzymes are overlapping to safeguard in large measure the important functions that the various LTs play (6, 7).
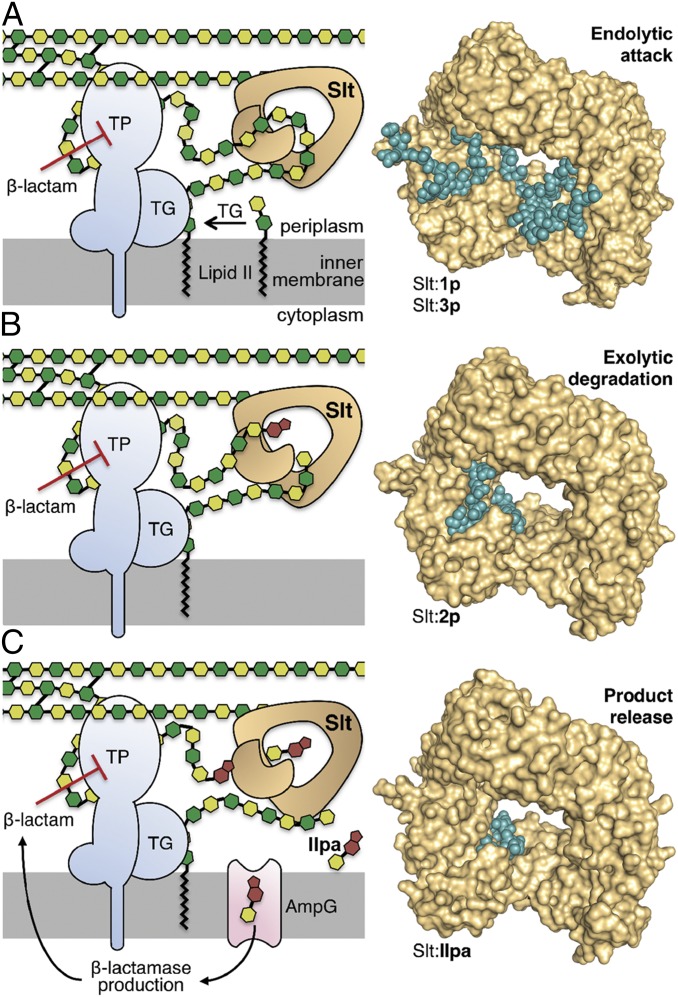
Fig. 1.
The interplay among transpeptidase (TP), transglycosylase (TG), and Slt is depicted by the cartoons to the Left of each panel, explaining the mechanistic concept that emerged from the crystal structures shown to the Right of each panel. The NAM and NAG are depicted as dark and light green hexagons, respectively. The bicyclic anhNAM is in brown. The peptide stems are not shown, except for cross-links (black lines). The undecaprenyl membrane anchor of Lipid II is represented by the zigzagging black lines. (A) Lipid II is polymerized by a TG to generate the nascent peptidoglycan, which experiences cross-linking to the cell wall by a TP. On inhibition of TP by a β-lactam antibiotic, aberrant longer nascent peptidoglycan is formed, which serves as substrate for the endolytic reaction of Slt. (B) The product of endolytic reaction serves as substrate for the exolytic reaction of Slt, (C) which, in turn, generates the ultimate product, compound IIpa. Internalization of IIpa by the permease AmpG initiated the cascade of events that leads to production of β-lactamase as an antibiotic-resistance determinant against β-lactam antibiotics. The conceptual framework for the schematic was adapted from Cho et al. (4).
Notwithstanding the aforementioned overlapping enzymatic activities, the principal LT implicated in turnover of the linear peptidoglycan that results from exposure to the antibiotic in E. coli is Slt70 (4) and its counterpart in P. aeruginosa Slt (8, 9). Both enzymes are members of the family 1 LTs (subfamily 1A) (5). We describe here our elucidation of the reactions of Slt and report the X-ray structures of the wild-type apo Slt, of a catalytically impaired mutant (E503Q), and of five complexes with synthetic substrates/products and an inhibitor. The collective data reveal that the enzyme catalyzes both exolytic (cleavage from one end of the peptidoglycan) and endolytic (cleavage in the middle) reactions and shed structural insights into these processes.
Results and Discussion
The peptidoglycan is composed of a repeating backbone of N-acetylglucosamine (NAG)-N-acetylmuramic acid (NAM), connected by β-1→4 glycosidic linkages. A pentapeptide, l-Ala-γ-d-Glu-m-DAP-d-Ala-d-Ala (DAP is diaminopimelate), is appended to the NAM subunit, which serves as the site of cross-linking of two neighboring peptidoglycan strands (Fig. 1). The NAM moiety is transformed to 1,6-anhydro-N-acetylmuramyl moiety (anhNAM) by the reactions of LTs via a transition state that goes through an oxocarbenium species (5). Hence, some strands of peptidoglycan terminate in anhNAM.
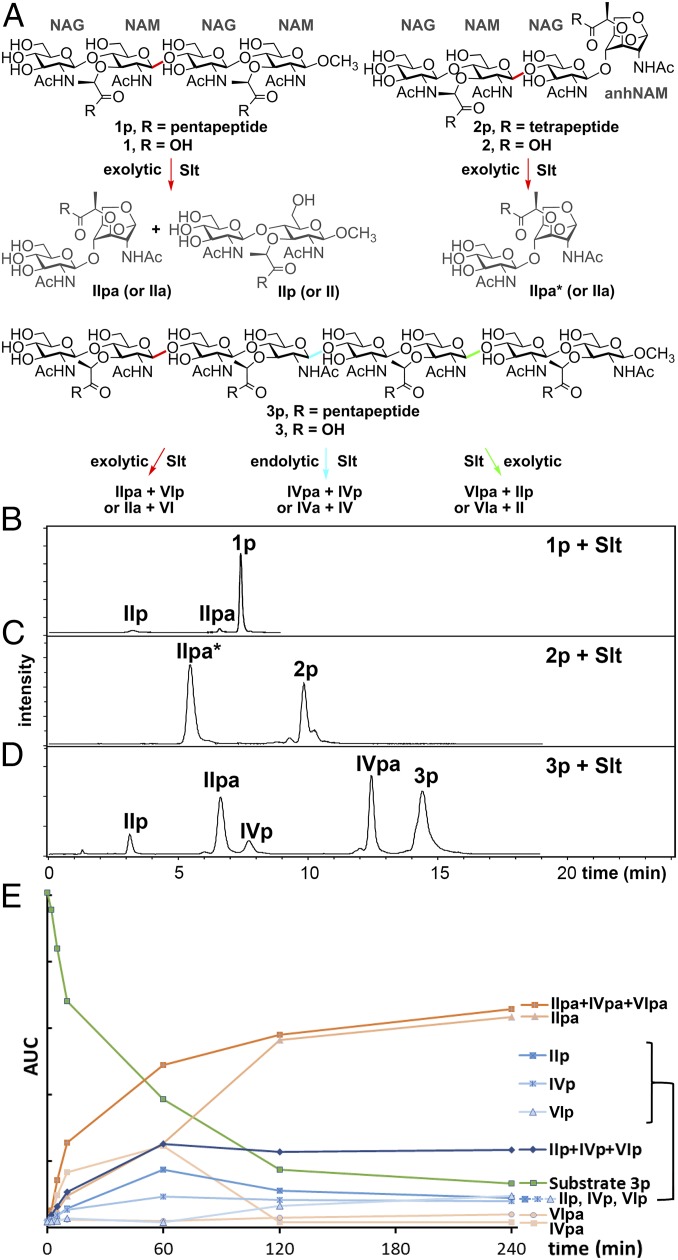
The cloning of the gene for Slt (locus tag PA3020) minus the segment corresponding to the signal peptide (the first 24 aa), its expression, and purification of the 73-kDa Slt were reported previously (6). We had documented that Slt from P. aeruginosa (PAO1) was able to turn over the sacculus—the purified bacterial cell wall—however, the amounts of the liberated muropeptides (products of the LT reactions) were relatively small (6). We carefully analyzed the incubation mixtures of Slt with four peptidoglycan-based synthetic tetrasaccharides (compounds 1p, 1, 2p, and 2; “p” for peptide stem; Fig. 2A) from our laboratory (6, 10, 11). Compounds 1 and 2, those without peptide stems, were not substrates. We noted marginal turnover of 1p (Fig. 2B), but 2p was a good substrate (Fig. 2C). These observations seemingly indicate that Slt requires in its minimal substrate the presence of the stem peptide(s) and the anhNAM (Fig. 2A). As a critical next experiment with more realistic potential substrates, we prepared by reported methodologies (11, 12) the two octasaccharides 3p and 3 in 58 and 36 synthetic steps, respectively. Both compounds have a long sugar backbone to closely mimic the non–cross-linked peptidoglycan that results from exposure of the organism to the β-lactam antibiotics. If both the presence of the stem peptides and anhNAM were indeed required, neither 3p nor 3 should serve as substrate. Consistent with the results with the tetrasacchrides 1 and 2, compound 3 is not a substrate. On the contrary, 3p was turned over by Slt. Hence, the presence of the peptide stems in peptidoglycan, but not that of the anhNAM moiety, is a requirement for Slt. The reaction of 3p by Slt produced all six possible products (IIpa + VIp, IVpa + IVp, and VIpa + IIp; roman numerals specify the number of sugars, “p” for peptide stem and “a” for anhNAM) by cleavage at any of the three colored glycosidic bonds [Fig. 2 A and D; the nature of the six distinct products were confirmed by liquid chromatography/mass spectrometry (LC/MS) and by comparison with authentic synthetic samples for three of them]. Therefore, Slt is capable of cleaving 3p in the middle glycosidic bond (shown in blue) and the flanking glycosidic bonds (colored in red and green; Fig. 2A). These competing reactions (Fig. 2D) result in both the endolytic and exolytic outcomes. The temporal analysis for the turnover flux reveals intermediates IVpa, VIpa, IVp, and VIp undergoing further reactions (Fig. 2E). However, we did observe preference for the presence of anhNAM, based on the rate of processing of products IVpa and VIpa to IIpa > IVp, VIp to IIpa and IIp.
Fig. 2.
Turnover of synthetic peptidoglycans by Slt. (A) The six synthetic peptidoglycans and the turnover chemistry by Slt. The reactive glycosidic sites are colored in red, blue, and green. (B–D) LC/MS traces of the reactions of three peptide-stem–containing substrates (1p, 2p, and 3p) with Slt. 1p, 3p, and their turnover products containing pentapeptide. 2p and IIpa* contains tetrapeptide instead of pentapeptide. (E) Time-course of reaction profile (area under the curve vs. time) of Slt with substrate 3p.
The same experiments and the set of substrates were also used with the E. coli homolog Slt70 (33%/96%, sequence identity/query coverage, respectively). The findings were very similar (Fig. S1). Overall, Slt requires the presence of peptide stems in its substrates. The minimum length of the glycan is four. Slt is capable of catalyzing both the endolytic and exolytic reactions with the octasaccharide substrate. Furthermore, as stated above, Slt requires the presence of the peptide stems in the substrates, but it could tolerate a missing peptide stem, as documented for an additional octasaccharide substrate (Fig. S2).
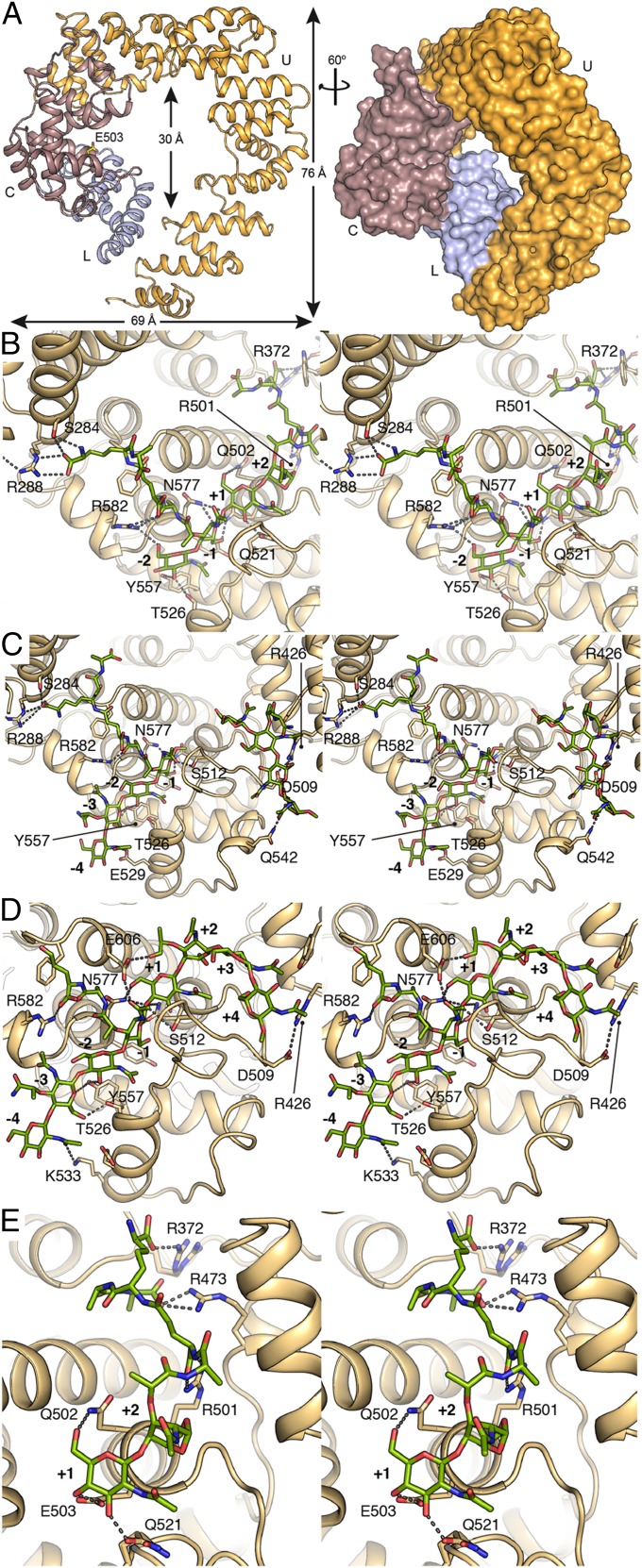
We undertook to correlate the analyses of the Slt reactions, described above, with its structure. The crystal structure of wild-type apo Slt was solved ab initio at 2.2-Å resolution (Fig. 3A, SI Materials and Methods, and Table S1) using the program Arcimboldo (13) to combine partial phase sets. It is important to note that all attempts to use the existing structure of the homologous Slt70 from E. coli as the initial model by the molecular-replacement method failed. Indeed, the structural homologs Slt70 (14) and LtgA (from Neisseria gonorrhoeae) (15) present large differences in the spatial arrangement of the domains as reflected by high rmsd values (4.6 Å for 528 Cα atoms for Slt70 and 6.0 Å for 439 Cα atoms for LtgA) (Fig. S3), while their catalytic domains are very similar (rmsd of 0.6 and 1.1 Å for Slt70 and LtgA, respectively) (Fig. S3C). This helix-rich protein (Fig. 3A) is composed of 612 aa (residues 29–641), and similar to the related Slt70 from E. coli (16), consists of three distinct domains: the N-terminal domain (also called the U domain; residues 29–387), the L domain (residues 388–478), and the C-terminal catalytic domain (residues 479–641). The overall fold of the catalytic domain is similar to that of goose-type lysozyme (family GH23 from www.cazy.org/). The functions of the U and L domains have not been known, but the L domain has been proposed as a potential peptidoglycan-binding domain (14). The three domains organize in a doughnut-shaped arrangement with a diameter for the central opening of ∼30 Å and outside maximum dimensions of 69 and 76 Å (Fig. 3A). The catalytic domain is composed of seven α-helices distributed across two lobes around the groove where the catalytic E503 is located. E503 is the source of the critical proton for the nonhydrolytic glycosidic bond cleavage in the substrates, en route to the high-energy oxocarbenium species (Fig. 4A). E503 is straddling subsites −1 and +1, where the scissile glycosidic bond of the peptidoglycan binds (17). This is the catalytic role that has been attributed to the residue for bond scission in glycosidases. The transient oxocarbenium ion entraps the C6 hydroxyl of the NAM to result in 1,6-anhNAM as one product at subsite −1, with the freed NAG as the other in subsite +1 (Fig. 4A).
Fig. 3.
Crystal structure of apo Slt and of complexes with compounds 1p, 2p, 3p, and IIpa. (A) 3D structure of the apo Slt. The catalytic groove is located in the C-terminal domain (in brown) where the catalytic E503 (labeled) is located; the neighboring domains U (in orange) and L (in blue) close the annular structure around the central opening. Stereo images for the (B) Slt:2p complex, (C) Slt:1p complex, (D) Slt:3p complex, and (E) Slt:IIpa complex. Catalytic subsites are labeled, and all of the compounds are shown as capped sticks in green for the carbon atoms. Residues interacting with the ligand are also depicted as capped sticks.
Fig. 4.
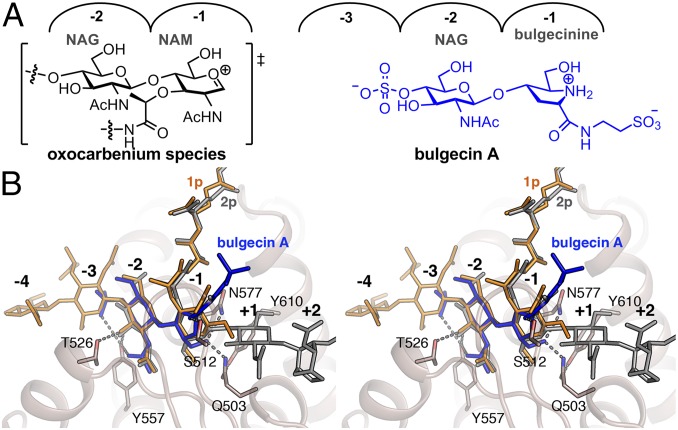
Inhibition of Slt by Bulgecin A. (A) The chemical structure of high-energy oxocarbenium species (shown in brackets), which is mimicked by bulgecine A. (B) Stereoview showing the Slt:bulgecin A complex superimposed with ligands 1p (orange) and 2p (gray) (as observed in the Slt:1p and the Slt:2p, respectively). Bulgecin A is represented in blue sticks. Residues interacting with the inhibitor are represented as capped sticks and labeled.
There does not exist any complexes of LTs with bona fide peptidoglycan substrates (5). In our efforts to get complexes of the peptidoglycan (substrate) with the enzyme (see below), we mutated the gene for Slt by PCR mutagenesis to generate the E503Q Slt variant. The gene was expressed and the protein was purified to homogeneity. We assayed the E503Q mutant protein with substrates 2p and 3p and did not observe any activity. The mutated enzyme was also inactive by the fluorescein-labeled sacculus assay (SI Materials and Methods). Crystals for the inactive E503Q mutant were obtained in different crystallization conditions than the wild-type enzyme (SI Materials and Methods), and its structure was solved at 3.05-Å resolution. As expected, the crystal structure of E503Q is essentially identical to that of the wild-type enzyme (rmsd of 0.5 Å for 612 Cα atoms). We used the crystals of the E503Q mutant in soaking experiments with the synthetic peptidoglycan substrates (1p, 2p, and 3p).
The structure of the E503Q Slt:2p complex was solved at 2.75-Å resolution, providing electron density for both the complete tetrasaccharide backbone and for the peptide stems attached to the NAM and the anhNAM (Fig. S4A). The substrate fully occupies the four active-site subsites −2 to +2 (Fig. 3B and Fig. S5A). The enzyme establishes a number of polar interactions with the sugars at subsites −2 to +1, but not with anhNAM at +2, which is ensconced in a relatively hydrophobic environment (Fig. 3B and Fig. S5A). It is worth recalling that 2p serves as substrate to furnish two molecules of NAG-anhNAM as products (Fig. 2A).
This structure gives an unprecedented view of the substrate 2p bound for the exolytic reaction, with the terminal NAG-anhNAM positioned at the threshold of the central opening of the annular shape. As indicated earlier, Slt exhibits strong preference for the presence of the peptide stem(s) in the substrate. Accordingly, the crystal structure of the Slt:2p complex provides information on how the peptide stems are recognized at the catalytic domain of an LT. (i) Notwithstanding that the entire peptide stem is seen in the structure at both −1 and +2 subsites, only polar interactions are established with Slt up to the m-DAP position (Fig. 3B and Fig. S5A). (ii) Five arginines are critical in peptide-stem recognition (R582 and R288 for peptide at −1, and R501, R473, and R372 for peptide at +2). Some of these arginines are held in place by a network of salt bridge interactions for their proper orientation (Fig. 3B and Fig. S5A). (iii) Peptide-stem recognition requires the intervention of the other domains (Fig. S5A). For instance, polar interactions stabilizing the m-DAP attached to NAM at −1 are made by S284 and R288 (both from the U domain), while that at +2 is made by R372 and R473 (from U and L domains, respectively). Residues from the L domain (such as W467, I458, D468, and R392) participate in van der Walls interactions or in the salt bridge network with the peptide stems at +2 subsite. Hence, both the U and L domains are involved in substrate recognition in conjunction with the catalytic domain, seen here because of the realism afforded by the authentic synthetic substrates. The observation of full density for a bona fide substrate is a first among LTs (5). Previously, only partial density for ligands was seen (18–20) or substrate analogs (chito-oligosaccharides) were employed (21, 22).
Furthermore, comparison of the structures of apo Slt to the Slt:2p complex reveals a significant structural tightening around the bound substrate, as seen also for the additional complexes that will be described below. These changes occur on one lobe of the catalytic domain involving helices (α1, α2, α3, and α4) and loops such as the clamping loop (residues 504–524) and the loop between α3 and α4 (residues 534–547) (Fig. S6A). The spatial rearrangement involves important movements (e.g., ∼3 Å for α3, ∼5 Å for α310, and ∼4 Å for the clamping loop) and results in repositioning of some critical residues for interaction with the peptidoglycan chain (among them Y557 and T526, which might trigger the movement by interactions within the subsite −2). Of special relevance is a 1.6-Å movement of the catalytic residue 503 toward the bound substrate, compared with the position in the apo structure. As indicated earlier, E503 is the source of the critical proton for initiation of the glycosidic bond scission. Finally, a set of conformational changes involve M507 and the formation of a strong bifurcated salt bridge interaction (with distances of 3.3 and 3.0 Å, respectively) between D509 from the catalytic domain and R426 from the L domain (Fig. S6A).
Next, we soaked the E503Q Slt crystals with substrate 1p and the structure of the complex was solved at 2.90-Å resolution (SI Materials and Methods and Table S1). We were intrigued to find two molecules of compound 1p bound to the enzyme (Fig. 3C and Fig. S4B). One tetrasaccharide was in the canonical active site spanning subsites −4 to −1 (Fig. S5B). Interactions that were observed in the Slt:2p complex for the sugar rings at subsites −2 and −1 are also conserved in the Slt:1p complex (Fig. 3C). Furthermore, Y557 and E529 extend polar interactions to the sugar rings at subsites −3 and −4. Interactions with the peptide stems in the Slt:2p complex are conserved in the Slt:1p complex. As with the Slt:2p complex, the terminal d-Ala-d-Ala in the stem peptides of the Slt:1p complex do not establish interactions with the protein. Interestingly, a second molecule of compound 1p was found in a newly constituted site at the interface between the catalytic and the L domains. This groove is not present in the apo form, but is created upon interaction with the substrate. The newly formed groove, arranged at a ∼99° angle to the first canonical substrate-binding site, intersects it at the +2 subsite. The +2 subsite was occupied by anhNAM in the Slt:2p complex for the exolytic reaction. However, in the Slt:1p complex, the active site has now been extended by the newly formed surface (subsites +3, +4, +5, and +6). The interactions of the second molecule of 1p within this extended section of the active site would appear to be mainly through shape complementarity via van der Waals interactions and by a few polar interactions (R426 and N542) with the protein. In retrospect, the absence of 1,6-anhNAM in the structure of the substrate 1p might have been a driving force in observing this alternative binding interaction with Slt. This creation of an extended active site upon exposure of the enzyme to the substrate is unusual and intriguing. Our synthetic octasaccharide substrate 3p provided the unique opportunity to test if indeed the expansion of the active site can be seen in binding to this more extended peptidoglycan molecule.
Soaking experiments were performed with the crystals of the E503Q Slt in the presence of compound 3p. The X-ray structure of the Slt:3p complex was solved at 3.20-Å resolution (SI Materials and Methods and Table S1). The octasaccharide binds Slt from subsites −4 to +4 (Fig. 3D and Fig. S5C), straddling equally the subsites on the two sides of the seat of the reaction (between subsites −1 and +1). This arrangement would be expected for an endolytic reaction, consistent with our turnover analyses with 3p. The interactions observed at subsites −4 to +1 in the complex with substrate 3p are essentially the same as those seen for complexes with 1p and 2p (Fig. 3). Importantly, the NAM ring at subsite +2 in the Slt:3p complex is oriented very differently than the anhNAM in the Slt:2p complex, bending the glycan chain sharply in the direction of the newly formed peptidoglycan-binding site seen in the complex with the tetrasaccharide 1p (Slt:1p), as described above. This arrangement positions the subsequent two NAG and NAM rings (located at subsites +3 and +4) close to the equivalent sugars in the Slt:1p complex (Fig. S5D). Therefore, reorganization of the active site to create the additional subsites for binding of the larger peptidoglycan substrate was also seen with 3p (Movie S1). This is an intriguing structure for the complex that can explain how the enzyme performs the endolytic reactions. The electron density for the peptidoglycan rings at subsites beyond +2 is poorer than for the other subsites (Fig. S4C), which we attribute to mobility of this segment of the substrate bound to these subsites. However, between the complexes for Slt:1p (for which the electron density is excellent) and for Slt:3p, there is clarity as to where substrate sugars bind.
We generated a hybrid computational model of the two X-ray structures, where we gave occupancy of subsites −4 to +6 to a large peptidoglycan substrate (Fig. S7A). The full occupancy complex for Slt was submitted to molecular-dynamics simulations for a duration of 200 ns according to the methodology reported previously (23) in the program AMBER16 program (24) (SI Materials and Methods). The simulation demonstrated a stable binding of the substrate within the extended substrate-binding groove. The occupancy of the peptidoglycan in the binding pocket was as per the X-ray structures for subsites −4 to +1 and +4 to +6 throughout the simulation. The conformation for both the unconstrained NAG and NAM rings is chair with all equatorial substituents. The X-ray structure for the NAM bound at subsite +2 assumes a half-chair conformation (2H1), which allows the peptidoglycan to bend for the occupancy of the rest of the active site in the endolytic reaction. The NAM at this subsite underwent a sampling of boat and skew conformations (3,OB and 3S1) during the simulation. In addition, sugars at subsites +2 and +3 made slight positional adjustments, but otherwise stable occupancy of the entire expanded active site was maintained throughout the simulation.
We add that we had thought initially that the doughnut-shaped catalyst could conceivably bind the long substrate at the canonical subsites −4 to +2 analogously to what we see in the Slt:2p complex, allowing for the subsequent segments of the substrate to thread through the opening of the protein. The notion was that as the longer substrate would thread through the annulus, it would undergo reaction at the seat of catalysis (subsites −1 to +1), and depending on which glycosidic bond would align with the seat of catalysis, a different reaction outcome could result. One drawback to this scheme is the fact that any segment of the substrate beyond subsite +2 could not have interactions with the protein. Furthermore, this model would stipulate that the strand of peptidoglycan that threads through the enzyme could be tethered only on one terminus (to membrane or to the cell wall). The complexes Slt:1p and Slt:3p firmly refute this possibility.
Beyond the work that we disclose in this report, there exists a paucity of structural information on binding of substrates to LTs. Furthermore, binding of the peptidoglycan for the endolytic reaction by Slt is important in several respects. The structure of the peptidoglycan in solution was solved by NMR to reveal it as a right-handed helix with a periodicity of three NAG-NAM repeats per turn of the helix (25). This solution structure has been seen in several X-ray structures for proteins that bind the peptidoglycan, where the 3D solution structure is recognized by the enzyme (10, 18, 26, 27). However, this is clearly not the conformation that is bound to Slt for the endolytic reaction. The extended binding site for the endolytic reaction begins at the −4 subsite moving diagonally toward the central opening of Slt, where it bends at 99° at the +2 subsite to move away from the annulus of the protein. This arrangement locates the entire active site on one side of the doughnut-shaped protein. The important implication is that the substrate need not be tethered only on one terminus, as the threading model would require. Indeed, the nascent peptidoglycan (Fig. 1A) initially would be tethered to the inner membrane by the undecaprenylpyrophosphate membrane anchor on one side and to the growing cell wall on the other. As the transpeptidase is inhibited by the β-lactam antibiotic, further cross-linking of the nascent peptidoglycan is prevented. However, the action of transglycosylase would continue lengthening the peptidoglycan in the absence of cross-linking (Fig. 1A), which accumulates in the periplasmic milieu. These peptidoglycan strands become candidate substrates for the endolytic reaction of Slt (4). An important observation is that the pentapeptide stem at the −1 subsite is bound at a “cul-de-sac.” This indicates that the space has evolved for binding to a pentapeptide stem, which is found in the nascent peptidoglycan. Once the peptidoglycan is processed by the endolytic reaction (Fig. 1A), the product strands would then become subjects to the exolytic activity of Slt (Fig. 1B). The exolytic reaction produces and releases the ultimate muropeptide product NAG-anhNAM-peptide (IIpa), which is internalized to the cytoplasm by the permease AmpG for the purpose of recycling (28) (Fig. 1C). As indicated earlier, the peptide stem in nascent peptidoglycan is the full-length pentapeptide. As the nascent peptidoglycan is turned over in the reactions of Slt, the reaction products, including NAG-anhNAM (IIpa), will retain the pentapeptide stem. NAG-anhNAM (IIpa) possessing the pentapeptide has been shown to be the inducer of the resistance response to β-lactam antibiotics in live P. aeruginosa (29). It serves as an effector for the AmpC β-lactamase transcriptional regulatory protein, AmpR (30, 31). As a corollary, our observations provide the mechanistic rationale for the increased susceptibility of P. aeruginosa to β-lactam antibiotics (8) in an slt-knockout strain, as in the absence of Slt, less effector molecules would be available to trigger production of AmpC β-lactamase.
One additional soaking experiment was made with the wild-type Slt crystals and the substrate 1p. As this is soaking of a substrate with the active enzyme, we were expecting a potential in crystallo transformation, which would result in IIpa (NAG-anhNAM) at subsites −2 and −1 and IIp at subsites +1 and +2. This experiment would validate the crystals as catalytically competent and would highlight the structural issues in the light of catalysis. A crystal structure was solved at 2.50-Å resolution (SI Materials and Methods and Table S1). Indeed, the structure revealed the complex for one IIpa bound to Slt, but it bound to subsites +1 and +2 (Fig. 3E and Fig. S4D). This indicates that the reaction took place in the crystal and that both products dissociated. However, because of the inherent affinity of IIpa for the +1 and +2 subsites, it bound back at these locations. This observation is consistent with recognition of 1,6-anhNAM at the +2 subsite, as we described for the exolytic reaction, and that substrates with the 1,6-anhNAM moiety were preferred. As observed in the complex of Slt:2p (with the E503Q mutant), interactions of the reaction product IIpa with the enzyme follows the same pattern. That is, the NAG at subsite +1 makes several polar interactions with the enzyme, whereas anhNAM participates in polar interactions through its peptide stem (Fig. 3E). The comparison of the Slt:IIpa complex with the structures of apo enzyme and of Slt:2p complex shows that, upon completion of bond-making and bond-breaking, the clamping loop is retracted in the direction of the loop position as seen in the apo structure (Fig. S6B). This movement results in (i) opening of the active site, (ii) retracting of the catalytic E503 by 1 Å (approaching the position found in the apo structure), and (iii) displacement of the sugar rings at subsites +1 and +2 (Fig. S6B). The dissociation of both products from the active site will bring the catalytic cycle to its completion.
Bulgecin A is a natural product inhibitor of lytic transglycosylases (Fig. 4A) (15, 16, 21). We soaked crystals of E503Q mutant with bulgecin A. The structure was solved for the complex at 3.10-Å resolution (SI Materials and Methods and Table S1). The inhibitor bound to the active site, occupying subsites −3 to −1 (Fig. 4B, Fig. S4E, and Fig. S8A). The 4-O-sulfate moiety is largely accessible to solvent within the subsite −3, and it interacts also with Y557. The bulgecin A NAG moiety occupies the −2 subsite and interacts with T526 and Y557. The hydroxymethyl segment on the modified proline (the bulgecinine moiety) at subsite −1 makes hydrogen bonds to N577 and to the catalytic residue at position 503. The carbonyl group of bulgecinine is hydrogen bonded to S512. Finally, the taurine amine interacts with E606 and one sulfonate oxygen interacts with the backbone of G513. While the complex of bulgecin A with Slt looks similar to those with Slt70 and LtgA (PDB ID codes 1SLY and 5MPQ, respectively) (Fig. S8B), we will make the following observations that have not been made previously (14, 15). The bulgecinine ring mimics the flatness of the oxocarbenium transition state species at the C1 (Fig. 4A). The bulgecinine hydroxymethyl moiety, anchored by residues N577 and Q503, would mimic the incoming C6 oxygen nucleophile of NAM to the C1 of the oxocarbenium species from one face. The Slt:bulgecin A complex shows broadly similar structural interactions to those found in the Slt:1p, Slt:2p, and Slt:3p complexes (Fig. 4B), consistent with it serving as a mimetic of the peptidoglycan and possibly of a transition state for the reaction that Slt catalyzes, based on the attributes that we described above.
In summary, the aberrant nascent peptidoglycan that results from the action of β-lactam antibiotics is turned over by Slt in an effort to repair the cell wall (4). Based on the arguments that we have presented, the endolytic reaction on the aberrant peptidoglycan generates products (Fig. 1A), which in turn serve as substrates for the exolytic reaction of Slt (Fig. 1B). Binding of the peptidoglycan to Slt entails a significant structural change in the entire protein (Movie S2), which tightens the active site around the bound substrate. This structural rearrangement of the protein around the substrate was documented in all Slt:1p, Slt:2p, and Slt:3p complexes, as well as in that for Slt:bulgecin A. The conformational change not only brings the protein to a catalytically competent state—as documented by the in crystallo reaction—it also extends the active site so as to enable endolytic reaction by physically binding the larger peptidoglycan. We hasten to add that our results shed light on why Slt and other members of family 1A are as large as they are, on the need for the annular structure, and on the functions of the additional U and L domains. Our findings reveal that the L and U domains are clearly involved in peptidoglycan recognition. Hence, the annular motif in its entirety serves to bind the non–cross-linked peptidoglycan to fulfill the needs of the endolytic (bent substrate) and exolytic (linear substrate) reactions. To accomplish this, the inner-ring regions are rich in basic patches (Fig. S7B) that would provide electrostatic attraction to the pentapeptide stems on the peptidoglycan, which we disclosed are required in Slt substrate(s). The bacterial physiological response to β-lactam antibiotics is complex indeed (2, 32). Upon encounter of bacteria to β-lactam antibiotics, inhibition of cell-wall cross-linking and the formation of the aberrant peptidoglycan lead to the demise of the organism (4). In the event of exposure of bacteria to subinhibitory concentrations of β-lactam antibiotics, the onset of recovery of the organism from the damage inflicted by the antibiotic is initiated by the processes that are disclosed in the present report.
Materials and Methods
Complete descriptions of the protein crystallization, structure determination, and refinement, of the computational methods, and of the reagent preparations are in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the Center for Research Computing of the University of Notre Dame for the computing resources. We also thank the staff from the ALBA synchrotron facility and European Synchrotron Radiation Facility for help during crystallographic data collection. The work in the United States was supported by NIH Grant GM61629 (to S.M.), and in Madrid, Spain, by Spanish Ministry of Economy and Competitiveness Grants BFU2014-59389-P and BFU2017-90030-P (to J.A.H.). D.A.D. is a Fellow of the Chemistry–Biochemistry–Biology Interface Program (NIH Training Grant T32GM075762) and a Fellow of the Eck Institute of Global Health at the University of Notre Dame. Grants BIO2015-64216-P (to C.M.), MDM2014-0435-01 (to I.U.), and BES-2015-071397 scholarship (to C.M.) supported the work in Barcelona, Spain.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 5OHU, 6FC4, 6FCS, 6FCR, 6FCU, 6FBT, and 6FCQ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1801298115/-/DCSupplemental.
References
- 1.Frère J-M, Page MGP. Penicillin-binding proteins: Evergreen drug targets. Curr Opin Pharmacol. 2014;18:112–119. doi: 10.1016/j.coph.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 2.Testero SA, Fisher JF, Mobashery S. β-Lactam antibiotics. In: Abraham DJ, Rotella DP, editors. Burger’s Medicinal Chemistry, Drug Discovery and Development (Antiinfectives) Vol 7. Wiley; Hoboken, NJ: 2010. pp. 259–404. [Google Scholar]
- 3.Lee W, et al. A 1.2-Å snapshot of the final step of bacterial cell wall biosynthesis. Proc Natl Acad Sci USA. 2001;98:1427–1431. doi: 10.1073/pnas.98.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho H, Uehara T, Bernhardt TG. β-Lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell. 2014;159:1300–1311. doi: 10.1016/j.cell.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dik DA, Marous DR, Fisher JF, Mobashery S. Lytic transglycosylases: Concinnity in concision of the bacterial cell wall. Crit Rev Biochem Mol Biol. 2017;52:503–542. doi: 10.1080/10409238.2017.1337705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee M, et al. From genome to proteome to elucidation of reactions for all eleven known lytic transglycosylases from Pseudomonas aeruginosa. Angew Chem Int Ed Engl. 2017;56:2735–2739. doi: 10.1002/anie.201611279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee M, et al. Reactions of all Escherichia coli lytic transglycosylases with bacterial cell wall. J Am Chem Soc. 2013;135:3311–3314. doi: 10.1021/ja309036q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavallari JF, Lamers RP, Scheurwater EM, Matos AL, Burrows LL. Changes to its peptidoglycan-remodeling enzyme repertoire modulate β-lactam resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2013;57:3078–3084. doi: 10.1128/AAC.00268-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamers RP, Nguyen UT, Nguyen Y, Buensuceso RN, Burrows LL. Loss of membrane-bound lytic transglycosylases increases outer membrane permeability and β-lactam sensitivity in Pseudomonas aeruginosa. Microbiologyopen. 2015;4:879–895. doi: 10.1002/mbo3.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee M, et al. Cell-wall remodeling by the zinc-protease AmpDh3 from Pseudomonas aeruginosa. J Am Chem Soc. 2013;135:12604–12607. doi: 10.1021/ja407445x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee M, et al. Synthetic peptidoglycan motifs for germination of bacterial spores. Chembiochem. 2010;11:2525–2529. doi: 10.1002/cbic.201000626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee M, et al. Deciphering the nature of enzymatic modifications of bacterial cell walls. Chembiochem. 2017;18:1696–1702. doi: 10.1002/cbic.201700293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sammito M, et al. Structure solution with ARCIMBOLDO using fragments derived from distant homology models. FEBS J. 2014;281:4029–4045. doi: 10.1111/febs.12897. [DOI] [PubMed] [Google Scholar]
- 14.van Asselt EJ, Thunnissen A-MWH, Dijkstra BW. High resolution crystal structures of the Escherichia coli lytic transglycosylase Slt70 and its complex with a peptidoglycan fragment. J Mol Biol. 1999;291:877–898. doi: 10.1006/jmbi.1999.3013. [DOI] [PubMed] [Google Scholar]
- 15.Williams AH, et al. Bulgecin A: The key to a broad-spectrum inhibitor that targets lytic transglycosylases. Antibiotics (Basel) 2017;6:8. [Google Scholar]
- 16.Thunnissen A-MWH, Rozeboom HJ, Kalk KH, Dijkstra BW. Structure of the 70-kDa soluble lytic transglycosylase complexed with bulgecin A. Implications for the enzymatic mechanism. Biochemistry. 1995;34:12729–12737. doi: 10.1021/bi00039a032. [DOI] [PubMed] [Google Scholar]
- 17.Davies GJ, Wilson KS, Henrissat B. Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem J. 1997;321:557–559. doi: 10.1042/bj3210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee M, et al. Turnover of bacterial cell wall by SltB3, a multidomain lytic transglycosylase of Pseudomonas aeruginosa. ACS Chem Biol. 2016;11:1525–1531. doi: 10.1021/acschembio.6b00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Asselt EJ, Kalk KH, Dijkstra BW. Crystallographic studies of the interactions of Escherichia coli lytic transglycosylase Slt35 with peptidoglycan. Biochemistry. 2000;39:1924–1934. doi: 10.1021/bi992161p. [DOI] [PubMed] [Google Scholar]
- 20.van Asselt EJ, et al. Crystal structure of Escherichia coli lytic transglycosylase Slt35 reveals a lysozyme-like catalytic domain with an EF-hand. Structure. 1999;7:1167–1180. doi: 10.1016/s0969-2126(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 21.Fibriansah G, Gliubich FI, Thunnissen A-MWH. On the mechanism of peptidoglycan binding and cleavage by the endo-specific lytic transglycosylase MltE from Escherichia coli. Biochemistry. 2012;51:9164–9177. doi: 10.1021/bi300900t. [DOI] [PubMed] [Google Scholar]
- 22.van Straaten KE, Barends TRM, Dijkstra BW, Thunnissen A-MWH. Structure of Escherichia coli lytic transglycosylase MltA with bound chitohexaose: Implications for peptidoglycan binding and cleavage. J Biol Chem. 2007;282:21197–21205. doi: 10.1074/jbc.M701818200. [DOI] [PubMed] [Google Scholar]
- 23.Mahasenan KV, et al. Conformational dynamics in penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus, allosteric communication network and enablement of catalysis. J Am Chem Soc. 2017;139:2102–2110. doi: 10.1021/jacs.6b12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Case DA, et al. 2017. AMBER16 (University of California, San Francisco), Version 16.
- 25.Meroueh SO, et al. Three-dimensional structure of the bacterial cell wall peptidoglycan. Proc Natl Acad Sci USA. 2006;103:4404–4409. doi: 10.1073/pnas.0510182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho S, et al. Structural insights into the bactericidal mechanism of human peptidoglycan recognition proteins. Proc Natl Acad Sci USA. 2007;104:8761–8766. doi: 10.1073/pnas.0701453104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pérez-Dorado I, et al. Insights into pneumococcal fratricide from the crystal structures of the modular killing factor LytC. Nat Struct Mol Biol. 2010;17:576–581. doi: 10.1038/nsmb.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson JW, Fisher JF, Mobashery S. Bacterial cell-wall recycling. Ann N Y Acad Sci. 2013;1277:54–75. doi: 10.1111/j.1749-6632.2012.06813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee M, et al. Muropeptides in Pseudomonas aeruginosa and their role as elicitors of β-lactam-antibiotic resistance. Angew Chem Int Ed Engl. 2016;55:6882–6886. doi: 10.1002/anie.201601693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dik DA, et al. Muropeptide binding and the X-ray structure of the effector domain of the transcriptional regulator AmpR of Pseudomonas aeruginosa. J Am Chem Soc. 2017;139:1448–1451. doi: 10.1021/jacs.6b12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vadlamani G, et al. The β-lactamase gene regulator AmpR is a tetramer that recognizes and binds the d-Ala-d-Ala motif of its repressor UDP-N-acetylmuramic acid (MurNAc)-pentapeptide. J Biol Chem. 2015;290:2630–2643. doi: 10.1074/jbc.M114.618199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fisher JF, Mobashery S. The sentinel role of peptidoglycan recycling in the β-lactam resistance of the Gram-negative Enterobacteriaceae and Pseudomonas aeruginosa. Bioorg Chem. 2014;56:41–48. doi: 10.1016/j.bioorg.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.