Significance
Spherical nucleic acids (SNAs) made thus far are inherently polydisperse due to variations in surface nucleic acid density, particle size, or both. In this article, we describe the synthesis and characterization of two types of molecular SNAs with precise numbers of surface DNA strands using T8 polyoctahedral silsesquioxane and buckminsterfullerene C60 scaffolds. The surface DNA densities for these molecular structures fall inside the range of typical SNAs, which allows them to exhibit SNA-related properties, including enhanced cellular uptake and the ability to function as a gene regulation agent. With a route to molecularly pure SNAs opened, it becomes possible to use them to unveil the molecular details of SNA interactions with complementary ligands and living systems.
Keywords: molecular nanoconjugates, spherical nucleic acids, oligonucleotides, gene regulation
Abstract
Herein, we report a class of molecular spherical nucleic acid (SNA) nanostructures. These nano-sized single molecules are synthesized from T8 polyoctahedral silsesquioxane and buckminsterfullerene C60 scaffolds, modified with 8 and 12 pendant DNA strands, respectively. These conjugates have different DNA surface densities and thus exhibit different levels of nuclease resistance, cellular uptake, and gene regulation capabilities; the properties displayed by the C60 SNA conjugate are closer to those of conventional and prototypical gold nanoparticle SNAs. Importantly, the C60 SNA can serve as a single entity (no transfection agent required) antisense agent to efficiently regulate gene expression. The realization of molecularly pure forms of SNAs will open the door for studying the interactions of such structures with ligands and living cells with a much greater degree of control than the conventional polydisperse forms of SNAs.
Spherical nucleic acids (SNAs) are polyvalent nanostructures made from particles chemically modified with a dense shell of highly oriented oligonucleotides (1). The first SNAs were made using gold nanoparticle (AuNP) cores (2), but the core compositions explored since then have included a wide variety of inorganic particles [silver (3), quantum dots (4), iron oxide (5), and silica (6)], organic materials [liposomes (7, 8), polymers (9), proteins (10), and other macromolecules (11, 12)], and hybrid structures [metal−organic frameworks (13) and infinite coordination polymers (14) in general]. In addition, core-less, hollow SNAs consisting of cross-linked oligonucleotides (15) have been developed, which exhibit many of the hallmark properties of the original AuNP structures. Indeed, all SNAs share several properties that substantially differentiate them from their linear counterparts. These include increased binding affinities for complementary sequences (16), resistance to nuclease degradation (17), and muted innate immune responses (18). Importantly, despite being highly negatively charged, SNAs rapidly enter cells [via scavenger receptor-mediated endocytosis in the case of endothelial cells (19)], making it possible to deliver large amounts of nucleic acids and other payloads into cells without the need for cocarriers (11, 12, 20, 21). These properties have made SNAs useful as probes for in vitro DNA and RNA detection (22–24) and lead compounds for gene regulation (25, 26), chemotherapy (11, 12, 20, 27), and immune system modulation (28). In addition, SNAs are the central building blocks for crystal engineering strategies based upon programmable assembly concepts (29, 30).
All SNAs studied thus far are inherently polydisperse, either by virtue of their core materials or the extent of surface modification, or both (31, 32) (Fig. 1). This limitation makes it impossible to utilize a variety of methods that would provide molecular-level information regarding such structures and their interactions with ligands and living systems. Herein, we address the challenge of synthesizing and characterizing molecularly well-defined SNA architectures from two different cores that allow for quantitative comparisons of structure−function relationships. In addition, in principle, they will provide the opportunity to create more chemically and biologically well-behaved and understood systems for both diagnostic and therapeutic uses.
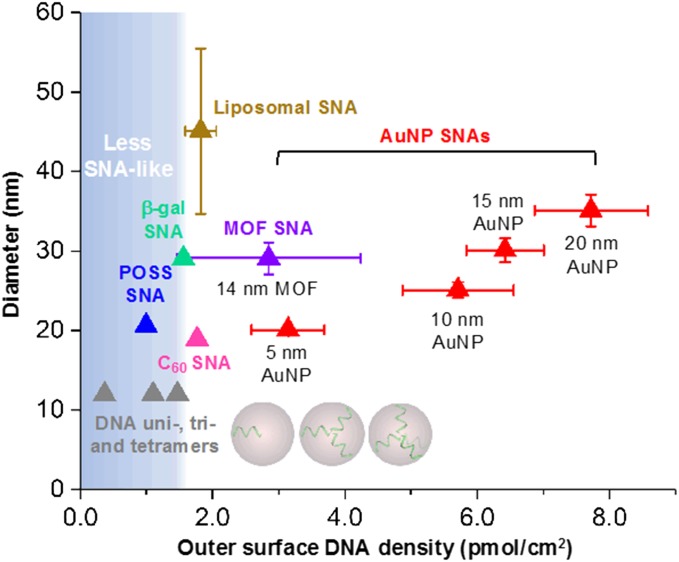
Fig. 1.
Size-density map of various types of SNAs showing potential polydispersity in size and nucleic acid density. Particle diameter is defined as the linear combination of core diameter and 2× the length of an 18-mer DNA (B-DNA form) with a 1.6-nm-long linker. Linkers are assumed to be in the fully stretched conformation. DNA density refers to the density at the outer surface of the particle as opposed to the footprint where DNA meets with the particle core. DNA loading and particle size data were obtained from the literature (7, 8, 10, 13, 32) (shown as mean ± σ). The values for DNA loading on AuNP surfaces represent typical SNA syntheses and may not indicate the highest achievable loading.
If one plots DNA surface density as a function of SNA diameter for all structures studied thus far and, within the same plot, compares the effective surface densities of unimer and multimer molecular DNA architectures (gray circles), two important observations can be made (Fig. 1). First, the SNA surface DNA density range is between 1.5 pmol/cm2 and 7.7 pmol/cm2, higher than the molecular univalent through tetravalent structures. Second, every nanoparticle structure studied thus far has inherent dispersity, hence the bars that define both diameter and DNA coverage; a molecularly pure system will be represented as a single dot on this graph. Note that, in this plot, surface coverage is defined based upon DNA density at the outer surface of the conjugate as opposed to on the particle template; this allows for a comparison with the nonparticle structures.
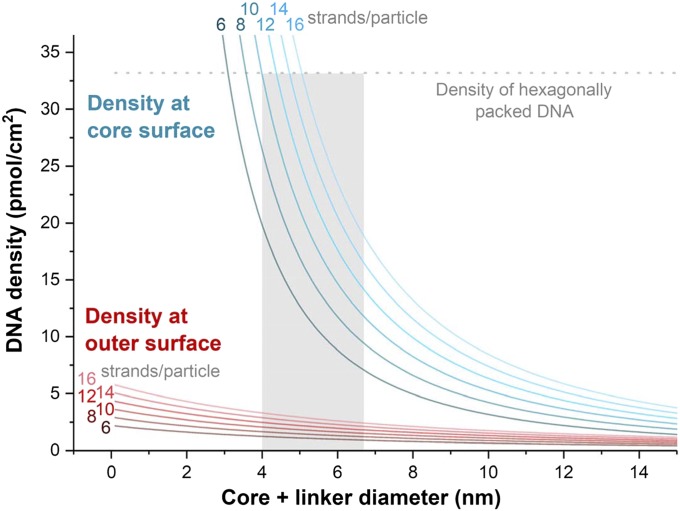
A survey of the literature suggests that, in developing a strategy for making molecularly well-defined SNAs, it is important to design an architecture where the oligonucleotide density and orientation can be highly controlled. Therefore, in addition to identifying appropriate candidate cores based upon the ability to make them in pure forms, the target structures need to have enough surface area to accommodate the large footprint of DNA ligands. In addition, the particle cores cannot possess a surface area that accommodates so many DNA strands that it becomes impossible to isolate one structure from a set of other structures with variable strand numbers. Finally, the targeted architectures should have a surface DNA density high enough to maintain the hallmark SNA properties that make them so useful and that are highly dependent on density and strand orientation. Considering that the typical length of an antisense DNA or siRNA strand (18- to 22-mer) is ∼6 nm to 7 nm (assuming a B-form conformation), one can plot the DNA densities at both the conjugate surface and the core/shell interface as a function of core size (Fig. 2). It can be seen that the combined core and linker size should be larger than 4.0 nm to accommodate a manageable number of DNA strands per particle (ref. 10, for example), but should be smaller than ∼6.7 nm to maintain a surface density of 1.5 pmol/cm2, which is the lowest density of previously studied SNAs. With these constraints and facts in hand, we have pursued T8 polyoctahedral silsesquioxanes (POSS) and C60 materials as cores to create molecular SNAs (Fig. 3).
Fig. 2.
DNA densities at the core surface (blue) or outer surface (red) as a function of core size for hypothetical SNAs with 6 to 16 strands per particle. The DNA is assumed to be 6 nm in length (18-mer in B-DNA form). To accommodate 10 strands on the particle while maintaining an outer surface density of >1.5 pmol/cm2, the combined core and linker size should be in a narrow window of ∼4.0 nm to 6.7 nm (indicated by a gray box).
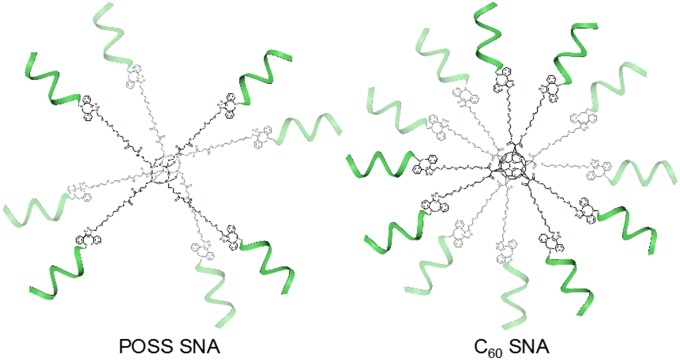
Fig. 3.
POSS- and C60-core SNAs. The DNA is drawn at 1/2 scale so that the core can be visualized. The linker molecules give the cores enough surface area to accommodate the large footprint of DNA.
Results and Discussion
Synthesis of SNA Cores.
To chemically attach DNA strands to the cores and avoid the incompatibility between Cu and DNA (33), we used strain-promoted click chemistry between azide-terminated linker molecules and dibenzocyclooctyne (DBCO)-modified DNA (34). The cubic T8 POSS has eight Si vertices for functionalization. To make clickable POSS, octaaminopropyl POSS was first reacted with an azide-terminated tetra (ethylene glycol) linker (N3-TEG) via amidation. The TEG was used to promote water solubility of the POSS, since the conjugation step is best conducted in the aqueous phase due to the poor solubility of DNA in other solvents. Fourier transform infrared spectroscopy (FT-IR) of the column chromatography-purified coupled product shows the presence of the highly diagnostic azide asymmetric stretching vibration (2,105 cm−1; SI Appendix, Fig. S20). The 1H NMR peak integrations (SI Appendix, Fig. S8) as well as matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS) analysis confirm the successful synthesis of the POSS core with eight azide sites ([M + H]+ calc: 3,290.3, found: 3,291.6; SI Appendix, Fig. S10).
In contrast, C60 can form a hexakis adduct with malonic esters to form a structure with a well-defined Th octahedral symmetry. However, if this is to be transformed into an adduct with only six DNA strands, the DNA surface density of the resulting SNA would be lower than all previously studied SNAs (∼0.9 pmol/cm2). Therefore, a symmetric malonic ester, bis(azidotetraethylene glycol) malonate, was synthesized and coupled to the C60 core in the presence of CBr4, 1,5-diaza-bicyclo[4.3.0]non-5-ene (DBU), and o-dichlorobenzene to yield a structure with 12 azide conjugation sites. Again, FT-IR confirms the presence of the azide groups, and 13C NMR spectroscopy shows only two peaks in the sp2 region of the spectra (141 and 145 ppm) due to the Th symmetry of the hexakis adduct (35). Importantly, the MALDI-TOF molecular weight measurement (3,746.1, [M + H]+) matches that of the calculated value (3,746.3, [M + H]+; SI Appendix, Fig. S17).
DNA Conjugation and Purification.
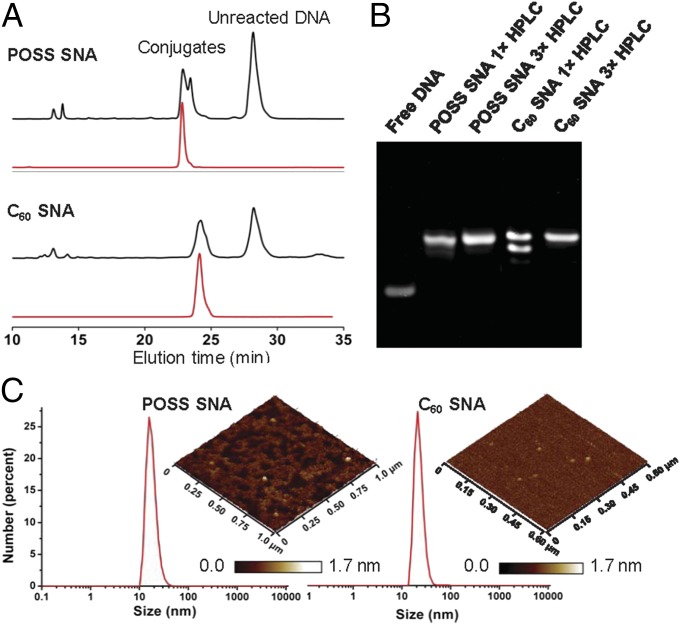
DNA sequences with a 5′ DBCO group were first synthesized following phosphoramidite chemistry protocols available in the literature (36). An 18-mer antisense sequence against the human epidermal growth factor receptor 2 (HER2) mRNA was selected as a proof-of-concept sequence to study the behaviors of the novel molecular SNA constructs. HER2 is involved in the signal transduction pathways leading to malignant cell growth and differentiation (37, 38). A scrambled sequence of equal length and base content was also prepared for comparative purposes. For the conjugation reaction, an azide:DBCO (m:m) ratio of 1:2 to 1:3 was used to ensure full chemical modification of the azide-terminated cores. The reaction was carried out in an aqueous solution containing 1.5 M NaCl, which screens the electrostatic repulsion between the DNA strands, allowing more strands to come into contact with the core. For the POSS core, a reaction temperature of 4 °C was used, since the core has a lower critical solution temperature (LCST) of ∼24 °C, a parameter experimentally determined (SI Appendix, Fig. S18). For the C60 core, 1 µL of dimethyl sulfoxide (DMSO) was used to deliver the core to ∼300 µL of an aqueous solution containing the DNA, since the fullerene-based core is water-insoluble. Initially, dynamic light scattering (DLS) shows aggregates of C60 with an average hydrodynamic diameter of ∼160 nm (number average), which gradually shifts to ∼20 nm as the reaction proceeds, reflecting the attachment of DNA and subsequent dissolution of the aggregates (Fig. 4C and SI Appendix, Fig. S19). RP-HPLC was employed to purify the POSS and the C60 conjugates using a step gradient of acetonitrile/ammonium acetate buffer (the more DNA grafting, the shorter the retention time; Fig. 4A). Although the baseline separation of the fully functionalized products from impurities missing one or two DNA strands (12 total expected for C60 SNA, 8 total for the POSS SNAs) was not achieved in a single HPLC run, repeated (3×) fractionation and reinjection of the product peak resulted in the isolation of the individual conjugates, as evidenced by single bands observed in the native polyacrylamide gel electrophoresis (PAGE, 15%, Fig. 4B). FT-IR spectra of the purified conjugates show the complete disappearance of the characteristic azide stretching vibration (2,105 cm−1), confirming that the cores have been fully derivatized (SI Appendix, Figs. S20 and S21). As one would expect for molecular entities, the POSS and C60 SNAs both exhibit narrow, unimodal size distributions by DLS, with number-average hydrodynamic diameters of 17.9 ± 4.9 nm and 22.1 ± 4.9 nm, respectively (Fig. 4C). Atomic force microscopy (AFM) shows very uniform dry-state diameters of ∼20 nm and a height of ∼1 nm for both SNAs, a consequence of the DNA collapsing around the core and onto the surface.
Fig. 4.
Characterization of molecular SNAs. (A) HPLC traces of the conjugation reaction mixture (black) and final purified products (red). (B) PAGE (15%) of free DNA and conjugates after 1× and 3× HPLC purification. (C) DLS size distribution by particle number. (Insets) AFM height images of the particles. The x-y scale is indicated on the sides of the images. Z-scale is shown in a scale bar below.
Nuclease Stability.
To determine if these conjugates exhibit properties characteristic of SNAs, we first examined their stability toward DNase I, an endonuclease that primarily cleaves dsDNA. Due to the dense oligonucleotide arrangement and the high local salt concentration associated with the nucleic acid corona, SNAs exhibit enhanced resistance to certain nucleases. A Förster resonance energy transfer (FRET)-based assay was utilized to analyze nuclease stability, which involves a fluorescein-modified strand hybridized to a dabcyl (quencher)-labeled complementary strand. In this assay, upon DNase I degradation, an increase of the fluorescence signal is observed due to the separation of the fluorophore−quencher pair, which can be used to measure degradation kinetics (Fig. 5A). Significantly, initial degradation half-lives of the POSS- and C60-based duplexes are ca. 1.4 and 2.7 times longer, respectively, than that of the free duplex of the same sequence (Fig. 5B). For other SNAs shown in the literature, the half-lives are typically between 2 and 5 times longer than the free duplex (11, 12, 15, 17).
Fig. 5.
(A) Scheme describing the FRET-based SNA stability assay in the presence of DNase I. (B) Percent duplex degraded as a function of time (0.5 units of DNase I were added to 1 mL of 0.5 µM duplex DNA solution).
Extent of Cellular Uptake.
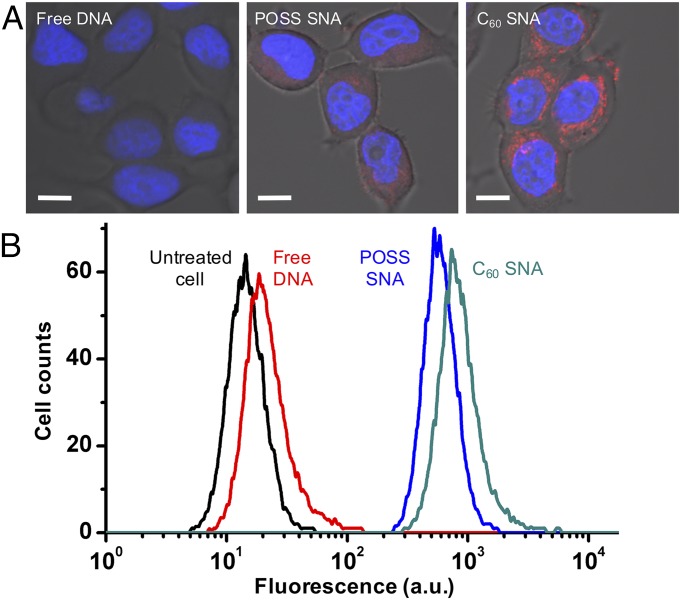
Next, we compared the extent of cellular uptake of the conjugates with that of free DNA. To study cellular uptake, MCF7 cells were incubated with either the conjugates or particle-free DNA bearing Cy3 labels (0.5 µM) in serum-free medium for 6 h (total DNA was equal in each experiment). Confocal microscopy shows that, although both SNA structures enter cells, the C60 conjugates enter them more effectively than the POSS conjugates, and the particle-free DNA exhibits no significant entry (Fig. 6A). These observations were corroborated by flow cytometry, which shows that the conjugates are taken up by the cells 400 to 1,000 times more effectively than free DNA (Fig. 6B). The C60-based conjugates were taken up ∼2 times faster than the POSS-based conjugates, likely due to having a higher-density DNA corona (12 strands vs. 8 strands). A very similar trend was also observed for SKOV-3 cells (SI Appendix, Fig. S22).
Fig. 6.
Cellular uptake of molecular SNAs. (A) Confocal microscopy of MCF7 cells treated with Cy3-labeled free DNA and conjugates (red). Cell nuclei are stained with Hoechst (blue). The cells were incubated for 6 h with samples with an equal DNA concentration of 0.5 µM in serum-free medium. (Scale bar: 10 μm.) (B) Flow cytometry measurement of MCF7 cells treated with the samples and the untreated cells.
Intracellular Gene Regulation.
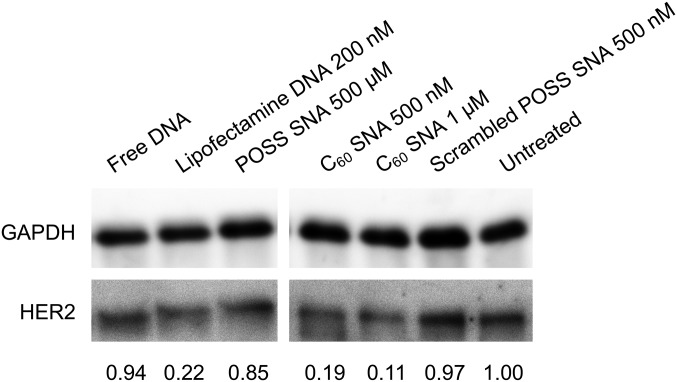
We evaluated the ability of the conjugates to regulate protein expression by antisense targeting of HER2 mRNA transcripts. The ovarian cell line, SKOV3, was chosen as a test case due to its overexpression of HER2. These cells were incubated with different concentrations of antisense or scrambled conjugates, as well as Lipofectamine-complexed antisense DNA in serum-free medium for 15 h, followed by additional culturing in full-serum media for another 48 h. The cells were harvested, and the HER2 expression was determined by Western blot analysis (Fig. 7). At equal DNA concentrations (500 nM), markedly reduced HER2 expression is found for the C60 conjugate (81% reduction), while the POSS conjugate showed only a slight decrease in HER2 expression (15% reduction). A positive control, Lipofectamine-complexed DNA, also showed significant knockdown using manufacturer-suggested protocols (78% reduction with 200 nM DNA). Increasing the antisense DNA concentration to 1 µM improves the antisense knockdown activity of the C60 conjugate to 89%. In contrast, when the cells were incubated with the scrambled conjugates, baseline-level HER2 expression was observed. These results indicate that, while both conjugates are multivalent, only the C60 conjugate can knock down target genes efficiently, resembling typical SNAs; the POSS conjugate may not cross a critical threshold in density for high antisense activity.
Fig. 7.
Western blot analysis of antisense gene silencing efficacy of HER2 using SNAs and controls in SKOV3 cells.
Conclusion.
These results are important because they outline two related, but distinct, strategies for making molecular SNAs. These structures exhibit properties that are both qualitatively and quantitatively consistent with their larger polydisperse counterparts. DNA density, which can be tailored with these two constructs, significantly affects cellular internalization and the subsequent ability of the conjugates to effect gene knockdown. These molecular SNAs herald a class of monodisperse systems that can be made with a variety of core materials and potentially higher DNA surface densities approaching that of AuNP-based systems. Significantly, with such structures now well characterized and in hand, we and others are poised to use them to begin to uncover the molecular details of the interactions between SNAs and complementary ligands as well as living systems.
Materials and Methods
Oligonucleotides were synthesized on an ABI 3400 DNA/RNA synthesizer (Applied Biosystems) using standard solid-phase phosphoramidite methodology (sequences for gene regulation and cellular uptake assays are included in SI Appendix, Materials and Instrumentation). Buckminsterfullerene C60 was supplied by Acros Organic. Ammonium chloride salt functionalized POSS (OctaAmmonium POSS) was purchased from Hybrid Plastics.
Synthesis of the Cores.
Synthesis of the POSS core.
A linker, butanedioic acid 2-{2-[2-(2-azidoethoxy)ethoxy]ethoxy}ethyl ester, was first synthesized in three steps (∼70% overall yield; see SI Appendix). To connect the linker to the octaammonium POSS hydrochloride core, N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC·HCl, 2.88 g, 15 mmol) and N-hydroxysuccinimide (NHS, 0.69 g, 6 mmol) were added to a solution of the linker (1.6 g, 5 mmol) in deionized water (5 mL). The reaction mixture was stirred at room temperature for 15 min, and a solution of octaammonium POSS hydrochloride (175 mg, 0.15 mmol) in PBS buffer (5 mL, pH 7.0) was added, followed by the addition of diisopropylethylamine (1.37 mL, 15 mmol). The reaction mixture was sonicated for 30 s and stirred overnight at room temperature, before being freeze-dried to remove water. The residue was dissolved in CH2Cl2, filtered, dried over anhydrous Na2SO4, and concentrated under reduced pressure. The residue was purified by flash chromatography with a silica gel (CH2Cl2:MeOH 98:2, v:v) to afford the POSS core as a clear oil (52 mg, 10%). The compound is stable in aqueous solution (neutral pH) for several months without apparent degradation.
Synthesis of the C60 core.
A symmetric malonic ester linker, bis(azidotetraethylene glycol) malonate, was first prepared in three steps (∼75% overall yield; see SI Appendix). To couple the linker to the C60 core, CBr4 (100 eq), the linker (10.0 eq), and DBU (20.0 eq) were added successively to a C60 solution (1.00 eq) in dry o-dichlorobenzene. The reaction mixture was stirred under argon for 72 h at room temperature. The crude reaction mixture was purified by silica gel column chromatography (CH2Cl2:MeOH 100:0 to 97:3 gradient, v:v) to yield a brown oil (∼15% yield).
Synthesis and Purification of Molecular SNAs.
Octakis DNA functionalization of the POSS core.
The POSS-based core shows an LCST of ∼24 °C. To maximize solubility, the coupling reaction was performed at 4 °C. In a typical reaction, DBCO-modified DNA strands (64 nmol to 100 nmol in 100 µL of 1.5 M NaCl aqueous solution) was placed in a 1.5-mL microcentrifuge tube and cooled to 4 °C on an Eppendorf Thermomixer C, and the POSS core (4 nmol in 2 µL of H2O, stored at 4 °C, DBCO being 2 to 3 equiv to azide) was added. The reaction mixture was gently shaken for 72 h before RP-HPLC purification.
Hexakis DNA functionalization of the C60 core.
In a typical reaction, DBCO-modified DNA strands (60 nmol to 90 nmol in 100 µL of 1.5 M NaCl aqueous solution) was placed in a 1.5-mL microcentrifuge tube at room temperature, and the C60-based core (2.5 nmol in 1 µL of DMSO, DBCO being 2 to 3 equiv to azide) was added. The reaction mixture was gently shaken for 72 h before RP-HPLC purification.
Purification of the SNAs.
The reaction mixtures containing the SNAs, free DNA, and partially derivatized cores were diluted to 1 mL and injected into a Waters RP-HPLC system equipped with a Waters Symmetry C18 column (3.5 μm, 4.6 × 150 mm). A gradient method was used, beginning with 95:5 vol/vol 0.05 M ammonium acetate (aq):MeCN, increasing to 65:35 vol/vol over 60 min (at a ramp of +0.5 vol% MeCN/min), with a flow rate of 1 mL/min. By comparing the peak integration of the conjugate vs. the unreacted DNA, it is estimated that >∼85% of the azide groups have been consumed. The conjugate fraction was collected, lyophilized, redissolved in H2O (0.2 mL), and subjected to further HPLC purification. The procedure was repeated until the product fraction was eluted as a single peak.
PAGE Analysis of the SNAs.
The purity of molecular SNAs was confirmed by native PAGE (15%). The gel (20 cm × 20 cm) was cast by polymerizing 19:1 acrylamide:bisacrylamide using 10% ammonium persulfate in the presence of N,N,N′,N′-tetramethylethylenediamine. The gel was clamped into a vertical electrophoresis chamber, filled with 1× Tris/Borate/EDTA buffer, and preheated at 350 V for 30 min. After the gel was loaded with samples, it was electrophoresed at 450 V for 120 min. Upon completion, the gel was stained using the Red Plus nucleic acid stain (Sangon Biotech) and imaged using a Fluochem Q imager (Proteinsimple).
Nuclease Degradation Kinetics Assay.
Free DNA, POSS SNA, and C60 SNA (1 µM DNA, fluorescein-labeled) were each mixed with a complementary, dabcyl-labeled DNA (2 µM) in 1× PBS buffer. The mixtures were heated to 80 °C and annealed by slow cooling to 25 °C in an Eppendorf PCR instrument for a period of 12 h. The mixtures were then diluted to 500 nM (DNA concentration) in assay buffer (10 mM Tris, 2.5 mM MgCl2, and 0.5 mM CaCl2, pH = 7.5), and 150 µL of each mixture was transferred to a 96-well black plate. DNase I (Beyotime) was then added and rapidly mixed to give a final concentration of 0.5 units per milliliter. The fluorescence of the samples (ex = 485 nm, em = 530 nm) was measured immediately and every 3 s thereafter for 2 h using a microplate reader (TECAN). The endpoint was determined by adding a large excess of DNase I (ca. 2 units per milliliter) to the mixture, and the fluorescence was monitored until no additional increase was observed.
Confocal Fluorescence Microscopy.
To evaluate the cellular uptake of POSS SNAs and C60 SNAs, MCF7 cells were seeded at a density of 5.0 × 105 cells/well in 3.5-cm glass bottom plates and were cultured for 24 h at 37 °C and 5% CO2. Serum-free DMEM containing Cy3-labeled free DNA, POSS SNA, and C60 SNA at equal doses of DNA (500 nM) were added to each well, followed by incubation for 6 h at 37 °C. Hoechest 33342 staining solution for live cells (100×) was then added to the medium for 10 min. The cells were gently washed with PBS 3× and imaged immediately on a Nikon confocal laser scanning microscope at excitation wavelengths of 408 nm (Hoechest) and 543 nm (Cy3). Imaging settings were kept identical for free DNA-, POSS SNA-, and C60 SNA-treated cells.
Western Blotting.
SKOV3 cells were plated in a 24-well plate at a density of 1.0 × 105 cells per well and cultured for 24 h. Then medium was replaced with serum-free DMEM immediately before treatment with free DNA (200 nM), POSS SNA (500 nM), C60 SNA (500 nM, 1,000 nM), and Lipofectamine-complexed DNA (200 nM, following manufacturer suggested protocol). After 15 h, the medium was replaced with fresh, full growth medium, and cells were cultured for another 48 h. Whole cell lysates were prepared in 75 µL of radioimmunoprecipitation assay (RIPA) Cell Lysis Buffer (Sangon Biotech). Protein concentrations were determined using a bicinchoninic acid assay (BCA) Protein Assay Kit (Sangon Biotech). Equal amounts (25 µg) of protein samples were fractionated by 4 to 20% precast gradient gels (Beyotime), transferred to polyvinylidene difluoride (PVDF) membrane, and blocked with 5% nonfat milk in tris-buffered saline with Tween 20 (TBST) for 1 h at room temperature (Bio-Rad). Proteins were analyzed by Western blotting with rabbit primary antibodies against HER2 (1,000:1) (Cell Signaling), rabbit primary antibodies against GAPDH (1,000:1) (Sangon Biotech), and HRP-conjugated goat anti-rabbit IgG secondary antibodies (2,000:1) (Sangon Biotech) using an ECL Western blotting substrate (Beyotime).
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Institutes of Health (National Cancer Institute Award U54CA199091 and National Institute of General Medical Sciences Award 1R01GM121612-01) and the National Science Foundation (CAREER Award 1453255). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the National Science Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1801836115/-/DCSupplemental.
References
- 1.Cutler JI, Auyeung E, Mirkin CA. Spherical nucleic acids. J Am Chem Soc. 2012;134:1376–1391. doi: 10.1021/ja209351u. [DOI] [PubMed] [Google Scholar]
- 2.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 3.Lee J-S, Lytton-Jean AKR, Hurst SJ, Mirkin CA. Silver nanoparticle-oligonucleotide conjugates based on DNA with triple cyclic disulfide moieties. Nano Lett. 2007;7:2112–2115. doi: 10.1021/nl071108g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell GP, Mirkin CA, Letsinger RL. Programmed assembly of DNA functionalized quantum dots. J Am Chem Soc. 1999;121:8122–8123. [Google Scholar]
- 5.Cutler JI, Zheng D, Xu X, Giljohann DA, Mirkin CA. Polyvalent oligonucleotide iron oxide nanoparticle “click” conjugates. Nano Lett. 2010;10:1477–1480. doi: 10.1021/nl100477m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young KL, et al. Hollow spherical nucleic acids for intracellular gene regulation based upon biocompatible silica shells. Nano Lett. 2012;12:3867–3871. doi: 10.1021/nl3020846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banga RJ, Chernyak N, Narayan SP, Nguyen ST, Mirkin CA. Liposomal spherical nucleic acids. J Am Chem Soc. 2014;136:9866–9869. doi: 10.1021/ja504845f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sprangers AJ, Hao L, Banga RJ, Mirkin CA. Liposomal spherical nucleic acids for regulating long noncoding RNAs in the nucleus. Small. 2017;13:1602753. doi: 10.1002/smll.201602753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, Zhang Y, Fullhart P, Mirkin CA. Reversible and chemically programmable micelle assembly with DNA block-copolymer amphiphiles. Nano Lett. 2004;4:1055–1058. [Google Scholar]
- 10.Brodin JD, Sprangers AJ, McMillan JR, Mirkin CA. DNA-mediated cellular delivery of functional enzymes. J Am Chem Soc. 2015;137:14838–14841. doi: 10.1021/jacs.5b09711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan X, et al. Light-triggered, self-immolative nucleic Acid-drug nanostructures. J Am Chem Soc. 2015;137:6112–6115. doi: 10.1021/jacs.5b00795. [DOI] [PubMed] [Google Scholar]
- 12.Tan X, et al. Blurring the role of oligonucleotides: Spherical nucleic acids as a drug delivery vehicle. J Am Chem Soc. 2016;138:10834–10837. doi: 10.1021/jacs.6b07554. [DOI] [PubMed] [Google Scholar]
- 13.Morris W, Briley WE, Auyeung E, Cabezas MD, Mirkin CA. Nucleic acid-metal organic framework (MOF) nanoparticle conjugates. J Am Chem Soc. 2014;136:7261–7264. doi: 10.1021/ja503215w. [DOI] [PubMed] [Google Scholar]
- 14.Calabrese CM, et al. Biocompatible infinite-coordination-polymer nanoparticle-nucleic-acid conjugates for antisense gene regulation. Angew Chem Int Ed Engl. 2015;54:476–480. doi: 10.1002/anie.201407946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cutler JI, et al. Polyvalent nucleic acid nanostructures. J Am Chem Soc. 2011;133:9254–9257. doi: 10.1021/ja203375n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lytton-Jean AKR, Mirkin CA. A thermodynamic investigation into the binding properties of DNA functionalized gold nanoparticle probes and molecular fluorophore probes. J Am Chem Soc. 2005;127:12754–12755. doi: 10.1021/ja052255o. [DOI] [PubMed] [Google Scholar]
- 17.Seferos DS, Prigodich AE, Giljohann DA, Patel PC, Mirkin CA. Polyvalent DNA nanoparticle conjugates stabilize nucleic acids. Nano Lett. 2009;9:308–311. doi: 10.1021/nl802958f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Massich MD, et al. Regulating immune response using polyvalent nucleic acid-gold nanoparticle conjugates. Mol Pharm. 2009;6:1934–1940. doi: 10.1021/mp900172m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel PC, et al. Scavenger receptors mediate cellular uptake of polyvalent oligonucleotide-functionalized gold nanoparticles. Bioconjug Chem. 2010;21:2250–2256. doi: 10.1021/bc1002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dhar S, Daniel WL, Giljohann DA, Mirkin CA, Lippard SJ. Polyvalent oligonucleotide gold nanoparticle conjugates as delivery vehicles for platinum(IV) warheads. J Am Chem Soc. 2009;131:14652–14653. doi: 10.1021/ja9071282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song Y, et al. Multimodal gadolinium-enriched DNA-gold nanoparticle conjugates for cellular imaging. Angew Chem Int Ed Engl. 2009;48:9143–9147. doi: 10.1002/anie.200904666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Briley WE, Bondy MH, Randeria PS, Dupper TJ, Mirkin CA. Quantification and real-time tracking of RNA in live cells using Sticky-flares. Proc Natl Acad Sci USA. 2015;112:9591–9595. doi: 10.1073/pnas.1510581112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halo TL, et al. NanoFlares for the detection, isolation, and culture of live tumor cells from human blood. Proc Natl Acad Sci USA. 2014;111:17104–17109. doi: 10.1073/pnas.1418637111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prigodich AE, et al. Multiplexed nanoflares: mRNA detection in live cells. Anal Chem. 2012;84:2062–2066. doi: 10.1021/ac202648w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosi NL, et al. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 26.Jensen SA, et al. Spherical nucleic acid nanoparticle conjugates as an RNAi-based therapy for glioblastoma. Sci Transl Med. 2013;5:209ra152. doi: 10.1126/scitranslmed.3006839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bousmail D, et al. Precision spherical nucleic acids for delivery of anticancer drugs. Chem Sci (Camb) 2017;8:6218–6229. doi: 10.1039/c7sc01619k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radovic-Moreno AF, et al. Immunomodulatory spherical nucleic acids. Proc Natl Acad Sci USA. 2015;112:3892–3897. doi: 10.1073/pnas.1502850112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park SY, et al. DNA-programmable nanoparticle crystallization. Nature. 2008;451:553–556. doi: 10.1038/nature06508. [DOI] [PubMed] [Google Scholar]
- 30.Jones MR, Seeman NC, Mirkin CA. Nanomaterials. Programmable materials and the nature of the DNA bond. Science. 2015;347:1260901. doi: 10.1126/science.1260901. [DOI] [PubMed] [Google Scholar]
- 31.Hill HD, Millstone JE, Banholzer MJ, Mirkin CA. The role radius of curvature plays in thiolated oligonucleotide loading on gold nanoparticles. ACS Nano. 2009;3:418–424. doi: 10.1021/nn800726e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hurst SJ, Lytton-Jean AKR, Mirkin CA. Maximizing DNA loading on a range of gold nanoparticle sizes. Anal Chem. 2006;78:8313–8318. doi: 10.1021/ac0613582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abel GR, Jr, Calabrese ZA, Ayco J, Hein JE, Ye T. Measuring and suppressing the oxidative damage to DNA during Cu(I)-catalyzed azide-alkyne cycloaddition. Bioconjug Chem. 2016;27:698–704. doi: 10.1021/acs.bioconjchem.5b00665. [DOI] [PubMed] [Google Scholar]
- 34.Jewett JC, Bertozzi CR. Cu-free click cycloaddition reactions in chemical biology. Chem Soc Rev. 2010;39:1272–1279. doi: 10.1039/b901970g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirsch A, Lamparth I, Grosser T, Karfunkel HR. Regiochemistry of multiple additions to the fullerene core: Synthesis of a Th-symmetric hexakis adduct of C60 with bis(ethoxycarbonyl)methylene. J Am Chem Soc. 1994;116:9385–9386. [Google Scholar]
- 36.Beaucage SL. Oligodeoxyribonucleotides synthesis. Phosphoramidite approach. Methods Mol Biol. 1993;20:33–61. doi: 10.1385/0-89603-281-7:33. [DOI] [PubMed] [Google Scholar]
- 37.Baselga J, Swain SM. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer. 2009;9:463–475. doi: 10.1038/nrc2656. [DOI] [PubMed] [Google Scholar]
- 38.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.