Significance
Carboxypeptidase O (CPO) is a membrane-anchored brush-border enzyme associated with the small intestinal phase of protein digestion with distinctive specificity toward acidic C-terminal (C-t) amino acids. The combined activity of human CPO (hCPO) and pancreatic carboxypeptidases enables the C-t proteolysis of the great majority of amino acids present in dietary proteins. Here we disclose mechanism and structures of hCPO, both ligand-free and -bound with a natural peptidic inhibitor ascribing the exquisite specificity toward C-t acidic residues to a single amino acid, Arg275, in the substrate-binding pocket. Mutations of this residue to Asp and Ala suffices to reverse the specificity to C-t basic and hydrophobic residues, respectively, and faithfully mirror the specificity variants (hCPB, hCPA1, hCPA2) in enzyme kinetic assays.
Keywords: carboxypeptidase, protein digestion, crystal structure, acidic protease
Abstract
Human metallocarboxypeptidase O (hCPO) is a recently discovered digestive enzyme localized to the apical membrane of intestinal epithelial cells. Unlike pancreatic metallocarboxypeptidases, hCPO is glycosylated and produced as an active enzyme with distinctive substrate specificity toward C-terminal (C-t) acidic residues. Here we present the crystal structure of hCPO at 1.85-Å resolution, both alone and in complex with a carboxypeptidase inhibitor (NvCI) from the marine snail Nerita versicolor. The structure provides detailed information regarding determinants of enzyme specificity, in particular Arg275, placed at the bottom of the substrate-binding pocket. This residue, located at “canonical” position 255, where it is Ile in human pancreatic carboxypeptidases A1 (hCPA1) and A2 (hCPA2) and Asp in B (hCPB), plays a dominant role in determining the preference of hCPO for acidic C-t residues. Site-directed mutagenesis to Asp and Ala changes the specificity to C-t basic and hydrophobic residues, respectively. The single-site mutants thus faithfully mimic the enzymatic properties of CPB and CPA, respectively. hCPO also shows a preference for Glu over Asp, probably as a consequence of a tighter fitting of the Glu side chain in its S1′ substrate-binding pocket. This unique preference of hCPO, together with hCPA1, hCPA2, and hCPB, completes the array of C-t cleavages enabling the digestion of the dietary proteins within the intestine. Finally, in addition to activity toward small synthetic substrates and peptides, hCPO can also trim C-t extensions of proteins, such as epidermal growth factor, suggesting a role in the maturation and degradation of growth factors and bioactive peptides.
Metallocarboxypeptidases (MCPs) are important zinc-dependent enzymes that cleave single amino acids from the C termini (C-t) of peptides and proteins, and participate in a wide range of physiological processes in humans, ranging from digestion of dietary proteins to the regulation of blood fibrinolysis and the maturation of neuropeptides and hormones, among others (1–3). According to the MEROPS database, the most thoroughly studied group of MCPs is the M14 family of proteases (4). Based on sequence homologies and structural features, members of this large family (about 25 members in humans) can be grouped into A, B, C, and D subfamilies. M14-A is the largest of these subfamilies and includes the human pancreatic digestive enzymes carboxypeptidase A1 (hCPA1), carboxypeptidase A2 (hCPA2), and carboxypeptidase B (hCPB). These three enzymes are ∼35-kDa nonglycosylated proteins synthesized with a signal peptide and secreted as stable and inactive zymogens by the exocrine pancreas. A distinctive feature of these inactive zymogens is the presence of an extra ∼100 residue prosegment or prodomain that partially blocks their catalytic activity (5–9). After their secretion into the intestinal lumen, trypsin-promoted limited proteolysis generates active enzymes with ∼300 residues (10–12), which actively contribute to digestion of dietary proteins through the hydrolysis of C-t amino acids from peptides that are end products of digestive endopeptidases (13). The active forms of hCPA1, hCPA2, and hCPB display distinct but complementary substrate specificities. While hCPA1 and hCPA2 show a higher preference for small aliphatic and bulky aromatic side chains, respectively (14), hCPB preferentially cleaves substrates with C-t basic amino acids (15–17). Although their combined action can release a wide range of amino acids that are absorbed in the intestinal tract, pancreatic MCPs are unable to release acidic C-t amino acids, such as Asp or Glu (13, 18, 19), even though these acidic residues are two of the most abundant amino acids in alimentary proteins (20–22).
Human carboxypeptidase O (hCPO) is a recently described brush-border (BB) digestive enzyme that belongs to the M14-A subfamily of MCPs (18, 23). Enzymes of the small intestinal BB are responsible for the final stage of luminal digestion before absorption (19). Unlike pancreatic MCPs, hCPO has strict specificity toward acidic C-t amino acids, a unique feature within the M14-A subfamily (18). In 2001, hCPO was first identified in a bioinformatic search for additional MCP members in the human genome (23). A decade later, the full-length CPO gene was shown to be present in a large number of vertebrate species, including mammals, birds, and fish (18). Whereas all other members of the M14-A subfamily are produced as inactive zymogens, hCPO does not contain a large prodomain and can be expressed as a constitutively active enzyme (18). hCPO is synthesized by cells of the intestinal epithelium, where it tethers to the apical plasma membrane through a covalently linked glycosylphosphatidylinositol (GPI) anchor (18). A large number of BB digestive enzymes have been found to contain anchors at the C-t, including alkaline phosphatase, trehalase, and dipeptidase 1, among others (24–26). In addition to GPI anchoring, posttranslational glycosylation is a feature common to BB digestive enzymes that presumably confers protection against proteolytic degradation (19). The glycosylation of hCPO probably accounts for its higher molecular mass (18).
The activity of MCPs can be specifically regulated by proteinaceous inhibitors (2). During the past 50 y, a number of such MCP inhibitors have been isolated and characterized so far from potato and tomato (PCI and MCPI, respectively) (27–30), intestinal parasites Ascaris suum and Ascaris lumbricoides (ACI) (31, 32), the medical leech Hirudo medicinalis (LCI) (33), ticks Rhipicephalus bursa and Haemaphysalis longicornis (TCI and H1TCI, respectively) (34, 35), mammals (latexin) (36, 37) and, more recently, from the marine annelid Sabellastarte magnifica (SmCI) (38) and sea snail Nerita versicolor (NvCI) (39). Among them, the latter inhibitor NvCI was found to be the most potent inhibitor discovered for members of the M14-A subfamily, displaying equilibrium dissociation constants in the picomolar range (39). During the last decades, several crystal structures of pancreatic and extrapancreatic MCPs in complex with protein inhibitors have been obtained (29, 39–45). Although these protease-inhibitor structures shed light on functional mechanisms of MCPs and inhibitory binding properties, the detailed structural features that define the substrate specificity for C-t acidic amino acids remain still unexplored.
In this study, we report the crystal structure of a soluble form of hCPO (hCPOΔC), unbound and in complex with NvCI, at 1.85-Å resolution; interestingly, both have been observed in the same asymmetric unit of a crystal. The structural analysis, functional characterization, and site-directed mutagenesis of hCPO reveal the molecular details by which this protease exhibits a preference toward acidic C-t amino acids and, hence, completes the analysis of the range of amino acid specificities found in digestive MCPs. In addition, structural changes in active site residues induced through NvCI binding and interactions between the protease and the inhibitor were investigated in detail and compared with those reported previously for other MCPs. We also report the substrate preference of hCPO for Glu over Asp residues, and describe the ability of this enzyme to cleave the C-t of endogenous bioactive proteinaceous molecules, such as the human epidermal growth factor (EGF).
Results
Expression and Purification of hCPO Using Mammalian Cells.
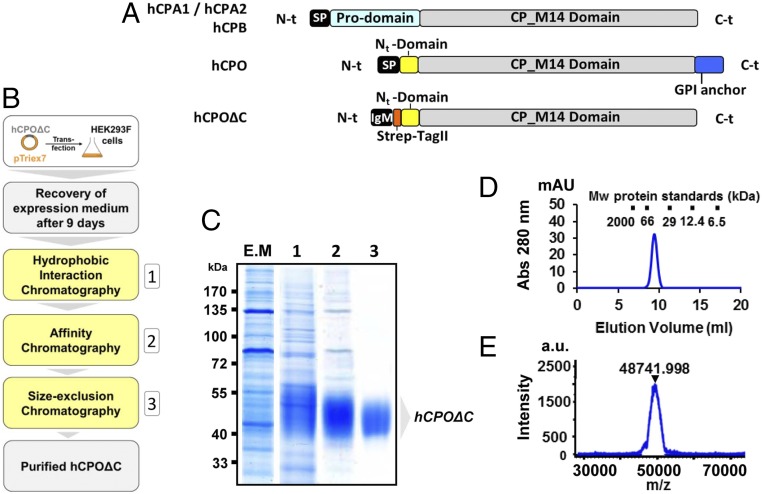
CPO is an N-glycosylated disulfide bond-containing protein with a GPI anchor located at the C-t (18). Such structural complexity represents a challenge for the expression and purification of the full-length hCPO in large amounts as soluble and functional enzyme for structural studies. Initial experiments showed that hCPO in which the C-t GPI signal sequence or the N-terminal (N-t) domain were removed retained full enzymatic activity (SI Appendix, Fig. S1). This suggested that the short N-t domain present within hCPO, with certain homology to the prodomain of related enzymes, did not function as an inhibitory prodomain, and that these truncated forms reflect the enzymatic functional properties of the wild-type enzyme (SI Appendix, Fig. S1E).
To overcome the difficulties of its production, we cloned a C-t truncated form of hCPO (residues Tyr1-Trp329, according to the numbering for mature hCPO, and termed hereafter as hCPOΔC) into the pTriEx-7 expression vector, where the endogenous N-t signal sequence was exchanged by an IgM exporting signal sequence followed by an N-t Strep-tag II fusion protein tag (Fig. 1A). The hCPOΔC–pTriEx-7 construct was transfected into HEK293 F mammalian cells for transient expression, leading to the secretion of the soluble recombinant protein into the culture media. At 9 d posttransfection, hCPOΔC was purified from the supernatant by a combination of three chromatographic steps (see Fig. 1B and SI Appendix, SI Materials and Methods for more details). Fig. 1C shows the SDS/PAGE analysis of the eluted samples after the different purification steps. After purification, hCPOΔC could be easily visualized on SDS/PAGE by Coomassie blue staining as a single broad band of ∼50 kDa. The identity and purity of the recombinant protein was additionally assessed by both size-exclusion chromatography and MALDI-TOF analysis, confirming its high purity and molecular mass of about 48.7 kDa (Fig. 1 D and E). The observed molecular mass of purified hCPOΔC is about ∼20% higher than its predicted molecular mass of 40.8 kDa. Digestion of the purified recombinant protein with PNGase F confirmed that the molecular size discrepancy is a consequence of N-glycosylation, which is in agreement with a previous report (18). To investigate whether hCPOΔC behaves as a functional carboxypeptidase, we determined the kinetic parameters for the purified hCPOΔC using the colorimetric substrate 3-(2-furyl)acryloyl-Glu-Glu-OH (FA-EE) at pH 7.5 (see SI Appendix, SI Materials and Methods for more details). The resultant kinetic parameters (SI Appendix, Table S1) were comparable to those obtained previously for the full-length hCPO produced in insect cells and purified in the presence of detergents (18).
Fig. 1.
Expression and purification of hCPOΔC. (A) Comparison of the domain structures of pancreatic MCPs (hCPA1, hCPA2, and hCPB) with hCPO and hCPOΔC. The latter corresponds to a C-t truncated form of hCPO that lacks the C-t tail and, therefore, the GPI membrane anchor. To improve protein expression and facilitate its purification, the endogenous N-t signal sequence (SP, in hCPO) of hCPOΔC was exchanged for an IgM signal sequence (IgM) plus an N-t Strep-tag II fusion protein tag (Strep-tag II). The tag was incorporated immediately before the N-t domain (Nt-domain). (B) Schematic diagram of the strategy followed for expression and purification of hCPOΔC. Protein expression was performed by transient transfection of HEK293 F mammalian cells grown in suspension. Medium was collected after 9-d incubation and the recombinant protein purified in three steps: 1) hydrophobic interaction chromatography using a Butyl 650-M resin, 2) affinity chromatography using anti–Strep-tag II resin, and 3) size-exclusion chromatography. (C) Coomassie-stained SDS/PAGE showing purity of hCPOΔC in the expression medium (E.M) and after each purification step (1, 2 or 3). (D) Representative size-exclusion chromatography and (E) MALDI-TOF MS of the purified hCPOΔC.
hCPOΔC Is Inhibited by a Wide Range of MCP Proteinaceous Inhibitors.
Inhibition kinetic constants (Ki) for hCPOΔC toward different proteinaceous MCPs inhibitors (PCI, LCI, TCI, ACI, and NvCI) were determined at pH 7.5. The results of such kinetics analyses are summarized in Table 1. As expected, all tested MCP proteinaceous inhibitors, except for NvCI, inhibit hCPOΔC with Ki values in the nanomolar range (PCI 3.2 ± 0.6 nM, LCI 12.8 ± 3.1 nM, TCI 16.7 ± 3.1 nM, and ACI 43.6 ± 6.3 nM), which are comparable to other members of the M14-A subfamily, such as hCPA1, hCPA2, and hCPB (28, 33, 34, 42).
Table 1.
Inhibitory constants (Ki) for different proteinaceous inhibitors against hCPOΔC
| Inhibitor | Ki (nM) |
| PCI | 3.20 ± 0.58 |
| LCI | 12.79 ± 3.06 |
| TCI | 16.66 ± 3.10 |
| ACI | 43.57 ± 6.30 |
| NvCI | 3,749 ± 521 |
Inhibitory measurements were performed in triplicate. Data are shown as mean ± SE. See SI Appendix, SI Materials and Methods for more details about the equations used for Ki determinations.
Interestingly, NvCI, which was recently reported as the strongest inhibitor discovered so far for some members of the M14-A subfamily (39), weakly inhibited hCPOΔC, showing a much higher Ki value (Table 1). In the Michaelis–Menten kinetic analysis (SI Appendix, Fig. S2), NvCI displays a typical competitive inhibition behavior toward hCPOΔC, with a Ki value of 3.8 ± 0.5 µM (Table 1). Preincubation of NvCI with the enzyme for various periods of time did not affect its inhibitory activity, suggesting that NvCI behaves as a fast binding and stable inhibitor of hCPOΔC.
Crystal Structure of hCPOΔC in Complex with NvCI.
After exploring a wide variety of protein combinations and crystallization conditions, crystals of hCPOΔC were only grown in complex with NvCI after incubation at 18 °C for several weeks. The complex of hCPOΔC with NvCI was formed in the crystallization drop by mixing the enzyme and the inhibitor in a molar ratio of 1:0.5 (hCPOΔC:NvCI). Crystals of hCPOΔC-NvCI belonged to the monoclinic space group C2 and diffracted beyond 1.85 Å resolution. The structure of the complex was solved by molecular replacement using hCPA4 as a starting model (PDB ID code 2PCU) (46) and was refined to a final R-factor and R-free of 18.6 and 20.8, respectively (SI Appendix, Table S2).
The crystals of the hCPOΔC–NvCI complex contained two hCPOΔC molecules in the asymmetric unit (Fig. 2A), chains A and B, but only one had NvCI bound. Each hCPOΔC polypeptide chain (chain A bound to NvCI and chain B unbound) could be clearly and completely traced in the electron density maps, from Glu23 to Trp329 (according to the numbering for mature hCPO without the signal peptide). In the initial unbiased electron density maps, NvCI (chain C) was clearly observed bound only to one of the two hCPOΔC molecules of the asymmetric unit, and could be completely traced from Phe1 to Ala53 (Fig. 2A). Despite the presence of two hCPOΔC molecules in the asymmetric unit, interface analysis with the PDBePISA software (47) and previous results obtained by gel-filtration chromatography indicate that the biological unit of hCPOΔC is the monomer (Fig. 1D).
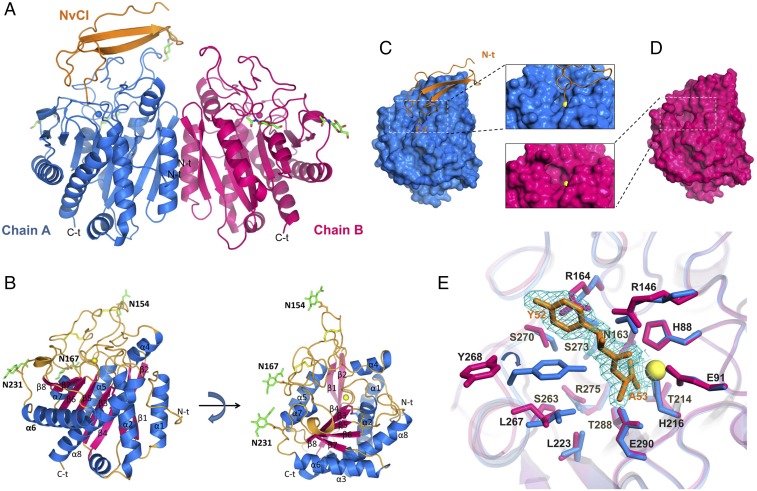
Fig. 2.
Three-dimensional structure of hCPOΔC in complex with NvCI. (A) The two monomers of hCPOΔC in the asymmetric unit (chains A and B, shown in blue and magenta, respectively) are stacked forming a homodimeric quaternary crystal structure. In contrast to chain B, chain A has a clear electron density in the active site region that accounts for a NvCI molecule (NvCI, in orange). In chain A, three N-glycan sites are indicated by an electron density of NAG at the Asn154, Asn167, and Asn231 consensus glycosylation sites as shown by the green stick models. In contrast, the electron density around the NAG at Asn154 in chain B is less complete, missing the N-glycan located in this position. (B) Two views of the 3D structure of recombinant hCPOΔC shown in ribbon representation. The catalytic carboxypeptidase domain of hCPOΔC displays a typical MCP-fold composed of eight α-helices and a mixed eight-stranded β-sheet that forms a globular α/β protein. The α-helices (α1–α8) and β-strands (β1–β8) are shown in blue and magenta, respectively. The three N-glycosylated residues (Asn154, Asn167, and Asn231) are depicted as green stick models. (C) Surface representation of hCPOΔC from chain A (blue model) bound with NvCI in a ribbon representation (orange model). (D) Surface representation of hCPOΔC from chain B in its unbound form. In C and D a magnification of the catalytic site cleft is shown. (E) Close-up view in stick representation of the active site of hCPOΔC in the absence (magenta) or in the presence of NvCI (blue). For clarity, only the last two C-t residues of NvCI (Tyr52 and Ala53) that enter the active site of hCPOΔC are shown in orange and superimposed with the final electron density corresponding to a 1.85-Å resolution F0–Fc Fourier map contoured at 1σ (mesh in cyan). The major change caused by the presence of NvCI in the active site is observed in the side chain of Tyr268, which rotates around its 1× angle and displaces the hydroxyl group 10.8 Å to further close the active site pocket. Other minor rearrangements at the active site are also observed, including minor movements of the side chains of Arg146, Arg164, Leu223, Ser270, Arg275, and Thr288. Residues are numbered according to the mature hCPO sequence without the signal peptide (Tyr1-Trp329). In B–E the catalytic zinc atom is shown as a yellow sphere. N-t and C-t correspond to the N and C termini in A–C, respectively.
Electron density maps clearly define three different N-linked glycosylation sites in the hCPOΔC crystal structure at Asn154, Asn167, and Asn231 in chain A; and Asn167 and Asn231 in chain B. In all cases the maps are well defined only for the N-acetylglucosamine (NAG) residue of the glycosyl moiety. Interestingly, NAG linked to Asn154 that is only observed in chain A is localized at contact distance to the NvCI inhibitor and displays the lowest B-factor of all observed NAGs in the structure (Fig. 2B).
The two different molecules of hCPOΔC in the asymmetric unit of the crystal, substrate-free (chain B) and complexed with the substrate-like inhibitor (chain A), allow us to compare the structural changes of residues forming the active-site pocket in closely similar environments (Fig. 2 C and D). The average main-chain rmsd deviation between chain A and chain B is 0.33 Å, increasing to 1.85 Å in the loop region between β-sheet β7 and α-helix α7 that contain the active site residue Tyr268 (Tyr248 in bCPA), which has been extensively reported in several MCP complex structures to move from the “up” to the “down” conformation upon binding of substrates or inhibitors (Fig. 2E) (12, 39, 42, 45). It marks the major structural difference observed between the “open” and “closed” conformations seen in chain B and chain A of hCPOΔC.
Structure of hCPOΔC.
The structure of the carboxypeptidase domain of hCPOΔC shows the classic compact α/β-hydrolase fold of the M14 family of MCPs, which is formed by a central twisted eight-stranded mixed β-sheet (strands β1 to β8) flanked by eight α-helices (Fig. 2B and SI Appendix, Fig. S3A). Structural alignment of the carboxypeptidase domains of hCPOΔC and pancreatic MCPs hCPA1, hCPA2, and hCPB confirms the structural similarities within the M14-A subfamily, displaying main-chain rmsd deviations of 0.85 Å for 299 aligned residues with a 40% sequence identity for hCPA1, of 0.83 Å for 297 aligned residues with a 39% sequence identity for hCPA2, and of 0.83 Å for 300 aligned residues with a 47% sequence identity for hCPB (SI Appendix, Fig. S3). All relevant M14 MCPs active site residues are shared by hCPOΔC and located in analogous positions, including: the Zn2+ coordinating residues His88, Glu91, and His216 (His69, Glu72, and His196 in bCPA); the catalytic Arg146 and Glu290 (Arg127 and Glu270 in bCPA); the C-t carboxylate substrate-binding Asn163 and Arg164 (Asn144 and Arg145 in bCPA); and Tyr268 (Tyr248 in bCPA), in its alternative conformations in liganded and unliganded hCPOΔC molecules (Fig. 3 A and B and SI Appendix, Fig. S3B).
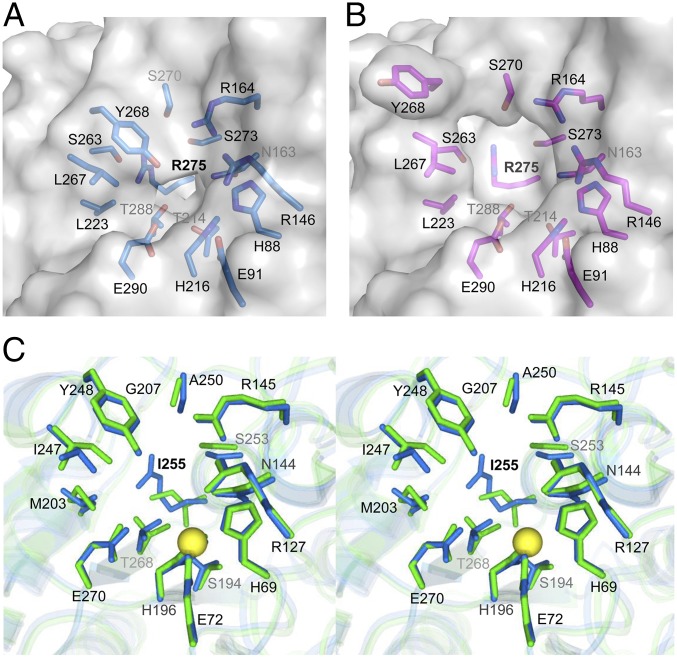
Fig. 3.
Structural features of the active site of hCPOΔC and comparison with the active sites of pancreatic MCPs. Detailed view of the active site groove of hCPOΔC in (A) the closed conformation observed in the complex with NvCI (chain A), and in (B) the open conformation observed in the substrate-free molecule (chain B). In A and B, the side chains of those residues important for catalysis and substrate binding are shown in stick representation (in blue and magenta, respectively), and shown together with the half-transparent surface (in gray). In both panels, residues were numbered according to the amino acid position in mature hCPO. The characteristic Arg275 amino acid that determines the substrate specificity for C-t acidic residues in hCPO is located at the bottom of the active site cleft. (C) Stereoview of the active site groove of hCPOΔC in the open conformation (blue sticks), superimposed with human hCPA1 (green sticks, PDB ID code 3FJU) (42). The same residues shown in A and B were labeled and numbered according to the reference bCPA nomenclature. For the sake of better visibility of Ile255, the side chain of Ser263 (Ile243 in bCPA) has been omitted in the figure. The catalytic zinc atom is shown as a yellow sphere.
A distinctive feature of hCPOΔC is its substrate-binding pocket, which endows it with the C-t processing specificity of Glu and Asp residues (18). This particular activity of hCPO depends on the presence of an arginine residue, Arg275, located at the bottom center of the substrate specificity pocket and which plays a central role by creating a positively charged surface optimal for interaction with amino acid side chains containing negatively charged carboxylate groups, such as glutamate or aspartate. Arg275 replaces Ile255 in hCPA1 and hCPA2, which prefer C-t aliphatic and aromatic amino acids, respectively, and Asp253 in hCPB, with a demonstrated specificity for C-t basic amino acids (Figs. 3C and 4). Only Pro C-t amino acids escape from the action of these enzymes; however, these are presumably cleaved by proline-hydrolyzing carboxypeptidases, such as angiotensin-converting enzyme and carboxypeptidase P, from the BB (19).
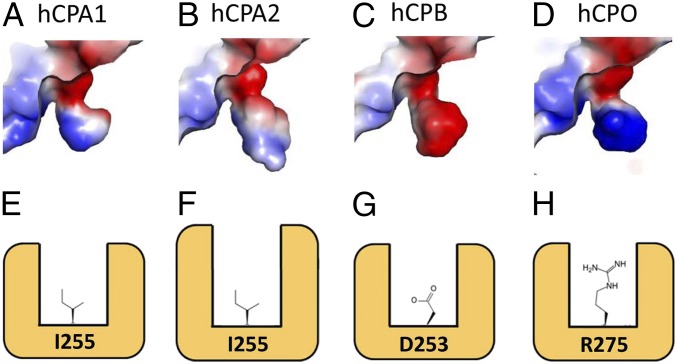
Fig. 4.
Characteristics of the binding pockets in digestive MCPs that determine the different substrate specificity. (A–D) Binding pockets of (A) hCPA1, (B) hCPA2, (C) hCPB, and (D) hCPO, displayed as a surface-rendered cavity. The electrostatic potential at the surface of the substrate-binding pocket is represented as a color gradient from red (−60 KBT/ec) to white (0 KBT/ec) to blue (+60 KBT/ec). Atomic coordinates of hCPA1 (PDB ID code 3FJU) (42), hCPA2 (PDB ID code 1DTD) (43), and hCPB (PDB ID code 1ZLI) (45) were obtained from the PDB. (E–H) Schematic representations of the substrate-binding pockets shown in A–D. (E) In hCPA1 a hydrophobic S1′ subsite binds small aliphatic residues such as Leu, Val, or Ala better. (F) In hCPA2 the presence of a much larger hydrophobic pocket than hCPA1 determines its stronger preference for bulkier hydrophobic amino acids such as Phe, Tyr, or Trp. However, in both cases, the specificity for hydrophobic/aliphatic amino acids is determined due to the presence of an Ile residue (I255, according to numbering in mature active hCPA1 and hCPA2) found in the base of a deep hydrophobic pocket. (G) hCPB specificity for basic residues is due to a negatively charged Asp residue (D253, according to numbering in mature active hCPB) located in the base of the binding pocket. (H) In hCPO the side chain of an Arg residue in position 275 (R275) is located in the base of the active site in an equivalent position to the I255 found in hCPA and hCPA2, and to D253 of hCPB. This contributes to its specificity for C-t acidic amino acids, such as Glu or Asp.
Site-Directed Mutagenesis.
To confirm the role of Arg275 for the substrate specificity of the enzyme, the positively charged Arg275 was replaced, in protein recombinant mutants, either by Asp (the hCPOΔC R275D mutant, in homology to hCPB) or by a small aliphatic residue Ala (the hCPOΔC R275A mutant). The substitution of the positive residue Arg275 for Asp drastically reduced the activity of the enzyme for the substrate FA-EE (Table 2 and SI Appendix, Fig. S4) and reversed the enzyme cleaving specificity: from glutamic to lysine C-t residues. As shown in Table 2, the Kcat/KM value of hCPOΔC R275D for 3-(2-furyl)acryloyl-Ala-Lys-OH (FA-AK) is 0.041 ± 0.004 µM−1 s−1 and is similar to CPB with 0.058 ± 0.018 µM−1 s−1 (Table 2). An earlier converse experiment by mutations of CPB (48) is in accord with these findings. The substitution by mutagenesis for Ala in the same position in hCPOΔC R275A reduced the activity of the enzyme toward FA-EE and led to a protease with preference for C-t hydrophobic residues, as shown by a Kcat/KM of 0.039 ± 0.006 µM−1 s−1 (Table 2 and SI Appendix, Fig. S4), comparable to type-A carboxypeptidase. It is remarkable that the substrate specificity of the different types of carboxypeptidases is determined by a single residue at the core of the specific pocket.
Table 2.
Effect of mutations of Arg275 on the substrate specificity of hCPO
| Substrate | |||
| Mutation | FA-EE | FA-AK | FA-FF |
| hCPOΔC WT | |||
| KM | 374.10 ± 48.36 | NM | NM |
| Vmax | 0.23 ± 0.01 | NM | NM |
| Kcat | 7.86 ± 0.40 | NM | NM |
| Kcat/KM | 0.021 ± 0.004 | NM | NM |
| hCPOΔC R275D | |||
| KM | NM | 671.50 ± 112.20 | NM |
| Vmax | NM | 0.81 ± 0.07 | NM |
| Kcat | NM | 27.77 ± 2.39 | NM |
| Kcat/KM | NM | 0.041 ± 0.010 | NM |
| hCPOΔC R275A | |||
| KM | NM | NM | 357.80 ± 40.34 |
| Vmax | NM | NM | 0.41 ± 0.02 |
| Kcat | NM | NM | 14.15 ± 0.64 |
| Kcat/KM | NM | NM | 0.039 ± 0.006 |
| CPB | |||
| KM | NM | 400.00 ± 86.98 | NM |
| Vmax | NM | 0.70 ± 0.006 | NM |
| Kcat | NM | 23.17 ± 2.05 | NM |
| Kcat/KM | NM | 0.058 ± 0.018 | NM |
| CPA | |||
| KM | NM | NM | 136.60 ± 15.94 |
| Vmax | NM | NM | 0.44 ± 0.02 |
| Kcat | NM | NM | 87.65 ± 4.04 |
| Kcat/KM | NM | NM | 0.64 ± 0.104 |
Enzymatic activity measurements were performed in triplicate. Data are shown as mean ± SE. See SI Appendix, SI Materials and Methods for more details about the equations used for kinetic constant determinations. Units are: KM: µM; Vmax: µM·s−1; Kcat: s−1; and Kcat/KM: µM−1·s−1. NM, not measurable levels of activity.
Besides the major role of Arg275, the cavity of the substrate-binding pocket in hCPO is also formed by Leu223, Gly227, Ser263, Leu267, Ser273, and Thr288 (Fig. 3 A and B and SI Appendix, Fig. S3B). These residues are mostly conserved in CPA- and CPB-like MCPs, shaping a cavity with enough space to accommodate all types of side chains of C-t residues. For example, Leu223 in hCPO is substituted by either Met or Ile in hCPAs and hCPB, respectively; and Gly227, Ser263, Ser273, and Thr288 in hCPO are substituted by analogous residues in hCPA1, hCPA2, and hCPB (Fig. 3C and SI Appendix, Fig. S3B).
Structural Determinants of hCPOΔC Inhibition by NvCI.
Only one of the hCPOΔC molecules in the asymmetric unit establishes a complex with NvCI, leaving the second hCPOΔC molecule unbound. This feature may be enforced by favorable crystal contacts and by the weak binding of NvCI to hCPO, as shown by an inhibition constant (Ki) value in the micromolar range (Table 1). Noteworthy, NvCI interacts extensively with the hCPO surface, with a total contact area of 1,734 A2, which is comparable to the previous structure of NvCI in complex with hCPA4 (39). NvCI is basically formed by a central two-stranded antiparallel β-sheet stabilized by three disulphide bonds (SI Appendix, Fig. S5A). The inhibitory mechanism is based on a competitive substrate-like interaction of its C-t tail, formed by Tyr52 and Ala53, with the active site of hCPO, occluding the active site subsites S2 and S1, respectively, as occurs in the other known carboxypeptidase proteinaceous inhibitors (SI Appendix, Fig. S5E). The C-t tail of NvCI constitutes the “primary” binding region, with the C-t carboxylate group of Ala53 coordinating the zinc atom in a bidentate form (Fig. 2E and SI Appendix, Fig. S5B and Table S3). Other relevant interactions of this primary binding region include a hydrogen bond between the amino group of Ala53 and the hydroxyl oxygen of Tyr268, which is involved in the aforementioned “up” to “down” conformational change of Tyr268 in the hCPOΔC structure (SI Appendix, Fig. S5B and Table S3), and the hydrogen bonds between the carbonyl oxygen of Tyr52 with side chains of Arg90 and Arg146 (SI Appendix, Fig. S5B and Table S3).
The interaction network of the primary binding region of NvCI in complex with hCPOΔC is quite similar to the previous complex with hCPA4 (39) and differences are mostly observed in contacts of the extended “secondary” binding region. The secondary binding region between NvCI and hCPOΔC is composed of contacts distant from the active site groove (SI Appendix, Fig. S5 C and D) and seems to determine the inhibitory capabilities toward MCPs (39).
NvCI displays micromolar Ki values for hCPOΔC, in contrast to the picomolar Ki values shown for hCPA4 (39). Whereas in NvCI the primary binding region between hCPA4 and hCPOΔC are quite similar, several changes are observed in the secondary binding region between both complexes (SI Appendix, Fig. S6 A and B). Perhaps the most significant difference in the hCPOΔC complex is the loss of hydrogen bonds between the amino groups of Cys51 and Tyr52 with the side chain of Gln182 (Glu163 in hCPA4), which was suggested to be a relevant interaction responsible for the low Ki value displayed toward hCPA4 (SI Appendix, Fig. S6 B and C) (39). Other differences include the loss of main-chain hydrogen bond contacts from Ile10 and Asn11, and from side chains of Lys28 and Gln39 (SI Appendix, Fig. S6 B, D, and E and Table S3). However, in the complex between NvCI and hCPOΔC, a few new contacts are observed, such as the side chain of Asn32 with the main-chain oxygen of Gly155, and of particular interest, the contact between Asp33 with the NAG-linked Asn154, contacting both the amino acid side chains and the sugar moieties (SI Appendix, Fig. S6D and Table S3).
hCPOΔC Cleaves C-t Acidic Residues, with a Clear Preference for Glu over Asp.
To evaluate the substrate preference of hCPOΔC for cleavage of Glu vs. Asp, we examined the activity of the purified enzyme toward a synthetic peptide with a Glu or Asp in the C-t position (ARLSQKFPKAE or ARLSQKFPKAD, respectively). Both synthetic peptides were incubated with purified hCPOΔC and reactions analyzed by MALDI-TOF MS (SI Appendix, Fig. S7 A and B). To obtain semiquantitative information about the cleavage, different incubation times for each peptide were analyzed. When tested with the synthetic peptide with a C-t Glu, the enzyme completely cleaved off the latter residue after 15 min of incubation (SI Appendix, Fig. S7A). Similarly, the enzyme was also able to cleave off the peptide containing a C-t Asp, but longer times were needed (>30 min) to observe a complete decrease in the peak intensity corresponding to the peptide substrate (SI Appendix, Fig. S7B). hCPOΔC was not able to process other peptides, such as Met-enkephalin peptides with C-t hydrophobic (Phe) or basic residues (Arg or Lys), even after much longer incubation times (SI Appendix, Fig. S7 C–E).
These results demonstrated a clear preference of the enzyme for Glu over Asp. Modeling analysis of either Asp or Glu substrate inside the substrate-binding pocket in our hCPOΔC structure, based on a previous model structure of hCPA4 in complex with a cleaved hexapeptide substrate (46), suggests that hCPO’s preference for C-t Glu substrates is facilitated by the contact between the carboxylate group of the substrate Glu with the side chain of Arg275 (SI Appendix, Fig. S8).
C-t Proteolysis of hEGF by hCPOΔC.
Human EGF (hEGF) is an important trophic factor produced mainly by salivary glands and the Brunner’s duodenal glands as a 53-amino acids peptide (EGF1–53), which is secreted in large amounts to the digestive tract (49–52). Although EGF1–53 is generally considered to be the “mature” form of EGF in humans, other circulating C-t truncated forms have been observed (EGF1–52, EGF1–51, EGF1–50, and so on), as a result of partial cleavage by unknown proteases from the gastrointestinal lumen (53–59). These relatively small changes in the C-t region of the molecule result in a marked effect on the EGF receptor binding (60, 61), thus affecting its biological activity (62).
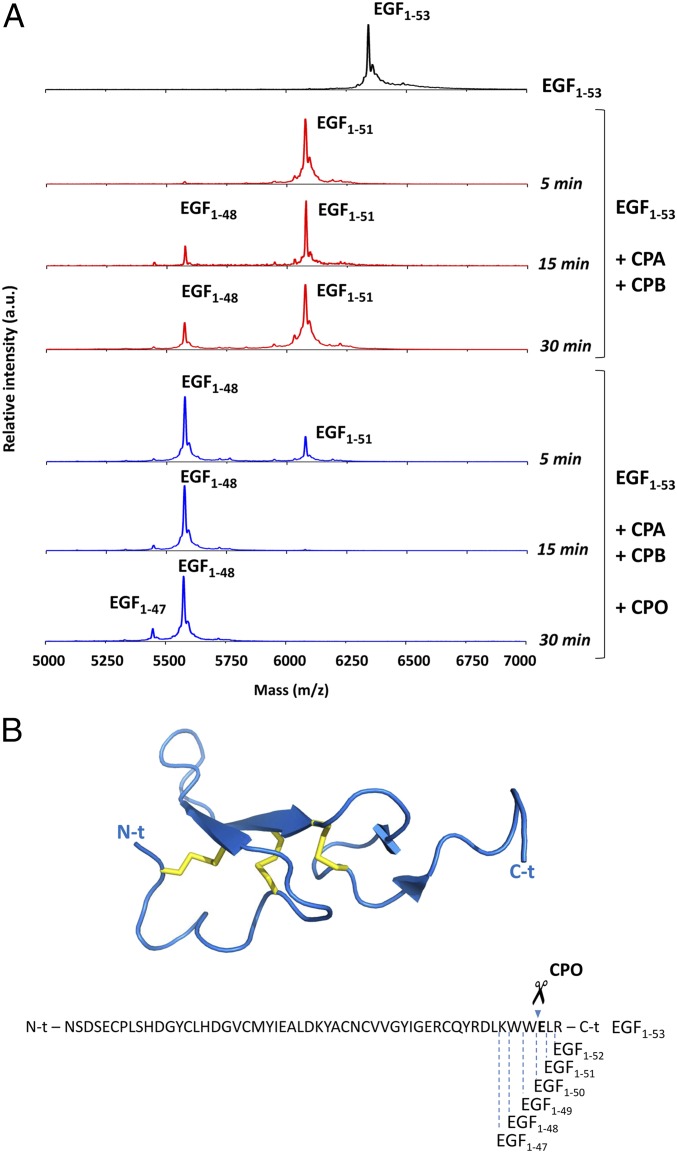
To determine whether hCPO processes EGF, we incubated the full-length EGF (EGF1–53) with both CPA and CPB in the absence or in the presence of purified hCPOΔC (see SI Appendix, SI Materials and Methods for more details). Fig. 5A depicts some points of the time course of EGF1–53 degradation as followed by MALDI-TOF MS analysis. The combination of CPA and CPB was rapidly able to excise Arg53 and Leu52 from EGF1–53 (red spectra, Fig. 5A). The presence of a Glu in position 51 of the EGF molecule limited the trimming action of CPA and CPB, which is to proceed further to generate smaller EGF forms (such as EGF1–50 or other shorter forms). In contrast, the addition of hCPOΔC to the reaction mixtures enhanced the C-t truncation of EGF, by cleaving Glu51 from EGF1–51 (blue spectra, Fig. 5). These results reveal the ability of hCPO to digest not only synthetic and small peptides, but also C-t residues of larger proteins and peptides, suggesting a potential role for this enzyme in the regulation of peptidic growth factors and bioactive peptides circulating in the BB of the intestinal tract.
Fig. 5.
Substrate specificity of hCPOΔC in the C-t proteolytic cleavage of synthetic peptides and hEGF. (A) Representative MALDI-TOF MS spectra of full-length recombinant human EGF (EGF1–53) incubated for different times at 37 °C with 30 nM bCPA and 50 nM pCPB in the absence (red spectra) or in the presence (blue spectra) of 30 nM hCPOΔC. (B) Ribbon representation and amino acid sequence of human EGF. The C-t peptide bonds cleaved by the proteolytic action of bCPA, pCPB, and hCPOΔC are indicated below the sequence. The peptide bond that is mainly cleaved by hCPOΔC is indicated above the sequence with scissors. Note that the full-length recombinant human EGF used in the experiment contains the EGF1−53 sequence plus an additional N-t Met residue. Accordingly, the monoisotopic molecular masses are: EGF1−53 = 6,348.77 Da; EGF1−51 = 6,078.85 Da; EGF1−48 = 5,577.32 Da; EGF1−47 = 5,449.15 Da. Atomic coordinates of hEGF (PDB ID code 1IVO) were obtained from the Protein Data Bank (www.rcsb.org).
Conclusions
For many decades, the pancreatic enzymes hCPA1, hCPA2, and hCPB were considered the only digestive carboxypeptidases of the M14 family present in humans. However, these enzymes are only able to release a fraction of all C-t residues present in dietary proteins. The discovery of the distinctive activity of hCPO for C-t acidic residues explains the cleavage of almost all types of amino acids and the nearly complete C-t degradation of proteins and peptides in the digestive tract. In this work we describe the structural details responsible for the specificity of hCPO for C-t Glu and Asp residues, and identify Arg275 located at the center of the substrate-binding pocket of the active site as the essential determinant. Structural comparison of the substrate-binding pockets of hCPA1, hCPA2, hCPB, and hCPO, the whole complement of digestive MCPs, reveals the essential roles of the equivalent residues in these enzymes in determining their cleavage specificities
Site-specific changes of Arg275 in hCPO to Asp and Ala reverses the specificity of these mutants, which cleave C-t basic and apolar residues, respectively, resembling the specificity of hCPB and hCPA1/2. In the case of hCPA1 and hCPA2, Ile255 provides the appropriate environment for binding aliphatic and aromatic substrate residues, whereas Asp253 endows hCPB with specificity for basic substrate residues (Fig. 4). Biochemical and kinetic analyses using hCPO also indicate a clear preference for processing C-t Glu over Asp substrates. Modeling suggests indeed a better accommodation of the longer side chain of Glu in the substrate-binding pocket by a salt linkage with Arg275. Finally, in addition to its role in the digestion of dietary peptides at the apical membrane of intestinal cells, hCPO accelerates C-t trimming of human EGF in vitro and might therefore have regulatory functions in the intestine.
Materials and Methods
A detailed description of procedures is provided in SI Appendix, SI Materials and Methods. Briefly, to facilitate production and crystallization, a recombinant C-t truncated form of hCPO (hCPOΔC) was produced and used instead of the intact/full form. Thus, it was overexpressed in HEK293 F mammalian cells by using the pTriEx-7 vector, which includes an N-t IgM exporting signal sequence and the Strep-Tag II affinity tag. The resultant recombinant protein hCPOΔC was purified to homogeneity by combining three chromatographic steps: hydrophobic interaction chromatography, affinity chromatography, and gel filtration. The catalytic activity of the purified hCPOΔC was tested against the typical colorimetric substrates FA-EE, FA-AK, and 3-(2-furyl)acryloyl-Phe-Phe-OH (FA-FF), and toward synthetic peptides and hEGF by MALDI-TOF MS. The same production procedure and assays apply to the mutants hCPOΔC R275D and hCPOΔC R275A. In addition, the inhibitory activities of a set of MCPs proteinaceous inhibitors were determined. Crystals of the NvCI–hCPOΔC complex were obtained by the sitting-drop vapor diffusion method by incubating a mix of the protease and the inhibitor (in a molar ratio of 1:0.5). The structure was solved by molecular replacement using the coordinates from hCPA4 in complex with NvCI as searching model. Crystallographic data are summarized in SI Appendix, SI Materials and Methods and Table S2.
Supplementary Material
Acknowledgments
The authors thank Lucía Díaz and Juan Fernandez-Recio (Center for Genomic Regulation-Institute for Research in Biomedicine Research Program in Computational Biology, Life Sciences Department, Barcelona Supercomputing Center) for their advice and experimental contribution in molecular docking; Silvia Bronsoms (Servei de Proteòmica i Biologia Estructural, Universitat Autònoma de Barcelona) for her valuable scientific advice and technical support in proteomics and mass spectrometry during the last years; and Carlos López Otin (Universidad de Oviedo) for the initial input to work with carboxypeptidase O. X-ray experiments were performed at the BL-13 beamline at the ALBA synchrotron (Cerdanyola, Barcelona, Spain), in collaboration with the ALBA staff. This work was funded by the Spanish Ministry of Innovation and Competitiveness Grants BFU2015-66417-P Ministry of Economy and Competitiveness (MINECO) and the European Fund for Regional Development (FEDER) (to D.R.) and BIO2016-78057-R (to F.X.A.); and by a Faculty Research grant from Andrews University (to P.J.L.). J.G.-P. and M.C.G.-G. were supported by PhD Fellowships BES-2011-044872 and FPU12/06137, respectively, from MINECO. J.G.-P. was supported by short-term Fellowship ASTF 603–2015 from the European Molecular Biology Organization.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 5MRV).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803685115/-/DCSupplemental.
References
- 1.Arolas JL, Vendrell J, Aviles FX, Fricker LD. Metallocarboxypeptidases: Emerging drug targets in biomedicine. Curr Pharm Des. 2007;13:349–366. doi: 10.2174/138161207780162980. [DOI] [PubMed] [Google Scholar]
- 2.Fernández D, Pallarès I, Covaleda G, Avilés FX, Vendrell J. Metallocarboxypeptidases and their inhibitors: Recent developments in biomedically relevant protein and organic ligands. Curr Med Chem. 2013;20:1595–1608. doi: 10.2174/0929867311320120009. [DOI] [PubMed] [Google Scholar]
- 3.Fernández D, Pallarès I, Vendrell J, Avilés FX. Progress in metallocarboxypeptidases and their small molecular weight inhibitors. Biochimie. 2010;92:1484–1500. doi: 10.1016/j.biochi.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Rawlings ND, Waller M, Barrett AJ, Bateman A. MEROPS: The database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2014;42:D503–D509. doi: 10.1093/nar/gkt953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waldschmidt-Leitz E, Purr A. Concering proteinase and carboxy-polypeptidase originating from the pancreas. (XVII. Communication concerning the specificity of animal proteinases.) Ber Dtsch Chem Ges. 1929;62:2217–2226. [Google Scholar]
- 6.Folk JE. A new pancreatic carboxypeptidase. J Am Chem Soc. 1956;78:3541–3542. [Google Scholar]
- 7.Gardell SJ, et al. A novel rat carboxypeptidase, CPA2: Characterization, molecular cloning, and evolutionary implications on substrate specificity in the carboxypeptidase gene family. J Biol Chem. 1988;263:17828–17836. [PubMed] [Google Scholar]
- 8.Aloy P, et al. Comparative analysis of the sequences and three-dimensional models of human procarboxypeptidases A1, A2 and B. Biol Chem. 1998;379:149–155. doi: 10.1515/bchm.1998.379.2.149. [DOI] [PubMed] [Google Scholar]
- 9.Coll M, Guasch A, Avilés FX, Huber R. Three-dimensional structure of porcine procarboxypeptidase B: A structural basis of its inactivity. EMBO J. 1991;10:1–9. doi: 10.1002/j.1460-2075.1991.tb07914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vendrell J, Cuchillo CM, Avilés FX. The tryptic activation pathway of monomeric procarboxypeptidase A. J Biol Chem. 1990;265:6949–6953. [PubMed] [Google Scholar]
- 11.Villegas V, Vendrell J, Avilés X. The activation pathway of procarboxypeptidase B from porcine pancreas: Participation of the active enzyme in the proteolytic processing. Protein Sci. 1995;4:1792–1800. doi: 10.1002/pro.5560040914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vendrell J, et al. Pancreatic procarboxypeptidases: Their activation processes related to the structural features of the zymogens and activation segments. Biol Chem Hoppe Seyler. 1992;373:387–392. doi: 10.1515/bchm3.1992.373.2.387. [DOI] [PubMed] [Google Scholar]
- 13.Beck IT. The role of pancreatic enzymes in digestion. Am J Clin Nutr. 1973;26:311–325. doi: 10.1093/ajcn/26.3.311. [DOI] [PubMed] [Google Scholar]
- 14.Tanco S, et al. Proteome-derived peptide libraries to study the substrate specificity profiles of carboxypeptidases. Mol Cell Proteomics. 2013;12:2096–2110. doi: 10.1074/mcp.M112.023234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Folk JE, Gladner JA. Carboxypeptidase B.I. Purification of the zymogen and specificity of the enzyme. J Biol Chem. 1958;231:379–391. [PubMed] [Google Scholar]
- 16.Aviles FX, Vendrell J. 2013. Carboxypeptidase B. Handbook of Proteolytic Enzymes (Academic, London), Vol 1, 3rd Ed, pp 1324–1329.
- 17.Bayés A, et al. Structural basis of the resistance of an insect carboxypeptidase to plant protease inhibitors. Proc Natl Acad Sci USA. 2005;102:16602–16607. doi: 10.1073/pnas.0505489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyons PJ, Fricker LD. Carboxypeptidase O is a glycosylphosphatidylinositol-anchored intestinal peptidase with acidic amino acid specificity. J Biol Chem. 2011;286:39023–39032. doi: 10.1074/jbc.M111.265819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hooton D, Lentle R, Monro J, Wickham M, Simpson R. The secretion and action of brush border enzymes in the mammalian small intestine. Rev Physiol Biochem Pharmacol. 2015;168:59–118. doi: 10.1007/112_2015_24. [DOI] [PubMed] [Google Scholar]
- 20.Blachier F, Boutry C, Bos C, Tomé D. Metabolism and functions of L-glutamate in the epithelial cells of the small and large intestines. Am J Clin Nutr. 2009;90:814S–821S. doi: 10.3945/ajcn.2009.27462S. [DOI] [PubMed] [Google Scholar]
- 21.Adibi SA, Mercer DW. Protein digestion in human intestine as reflected in luminal, mucosal, and plasma amino acid concentrations after meals. J Clin Invest. 1973;52:1586–1594. doi: 10.1172/JCI107335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burrin DG, Stoll B. Metabolic fate and function of dietary glutamate in the gut. Am J Clin Nutr. 2009;90:850S–856S. doi: 10.3945/ajcn.2009.27462Y. [DOI] [PubMed] [Google Scholar]
- 23.Wei S, et al. Identification and characterization of three members of the human metallocarboxypeptidase gene family. J Biol Chem. 2002;277:14954–14964. doi: 10.1074/jbc.M112254200. [DOI] [PubMed] [Google Scholar]
- 24.Low MG. Glycosyl-phosphatidylinositol: A versatile anchor for cell surface proteins. FASEB J. 1989;3:1600–1608. doi: 10.1096/fasebj.3.5.2522071. [DOI] [PubMed] [Google Scholar]
- 25.Hooper NM, Keen JN, Turner AJ. Characterization of the glycosyl-phosphatidylinositol-anchored human renal dipeptidase reveals that it is more extensively glycosylated than the pig enzyme. Biochem J. 1990;265:429–433. doi: 10.1042/bj2650429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerber LD, Kodukula K, Udenfriend S. Phosphatidylinositol glycan (PI-G) anchored membrane proteins. Amino acid requirements adjacent to the site of cleavage and PI-G attachment in the COOH-terminal signal peptide. J Biol Chem. 1992;267:12168–12173. [PubMed] [Google Scholar]
- 27.Hass GM, Hermodson MA. Amino acid sequence of a carboxypeptidase inhibitor from tomato fruit. Biochemistry. 1981;20:2256–2260. doi: 10.1021/bi00511a029. [DOI] [PubMed] [Google Scholar]
- 28.Lufrano D, et al. Biochemical characterization of a novel carboxypeptidase inhibitor from a variety of Andean potatoes. Phytochemistry. 2015;120:36–45. doi: 10.1016/j.phytochem.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 29.Rees DC, Lipscomb WN. Structure of the potato inhibitor complex of carboxypeptidase A at 2.5-A resolution. Proc Natl Acad Sci USA. 1980;77:4633–4637. doi: 10.1073/pnas.77.8.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hass GM, et al. The amino acid sequence of a carboxypeptidase inhibitor from potatoes. Biochemistry. 1975;14:1334–1342. doi: 10.1021/bi00677a036. [DOI] [PubMed] [Google Scholar]
- 31.Homandberg GA, Litwiller RD, Peanasky RJ. Carboxypeptidase inhibitors from Ascaris suum: The primary structure. Arch Biochem Biophys. 1989;270:153–161. doi: 10.1016/0003-9861(89)90017-9. [DOI] [PubMed] [Google Scholar]
- 32.Homandberg GA, Peanasky RJ. Characterization of proteins from Ascaris lumbricoides which bind specifically to carboxypeptidase. J Biol Chem. 1976;251:2226–2233. [PubMed] [Google Scholar]
- 33.Reverter D, et al. A carboxypeptidase inhibitor from the medical leech Hirudo medicinalis. Isolation, sequence analysis, cDNA cloning, recombinant expression, and characterization. J Biol Chem. 1998;273:32927–32933. doi: 10.1074/jbc.273.49.32927. [DOI] [PubMed] [Google Scholar]
- 34.Arolas JL, et al. A carboxypeptidase inhibitor from the tick Rhipicephalus bursa: Isolation, cDNA cloning, recombinant expression, and characterization. J Biol Chem. 2005;280:3441–3448. doi: 10.1074/jbc.M411086200. [DOI] [PubMed] [Google Scholar]
- 35.Gong H, et al. Characterization of a carboxypeptidase inhibitor from the tick Haemaphysalis longicornis. J Insect Physiol. 2007;53:1079–1087. doi: 10.1016/j.jinsphys.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 36.Normant E, Martres MP, Schwartz JC, Gros C. Purification, cDNA cloning, functional expression, and characterization of a 26-kDa endogenous mammalian carboxypeptidase inhibitor. Proc Natl Acad Sci USA. 1995;92:12225–12229. doi: 10.1073/pnas.92.26.12225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Q, et al. Cloning, tissue expression pattern and genomic organization of latexin, a human homologue of rat carboxypeptidase A inhibitor. Mol Biol Rep. 2000;27:241–246. doi: 10.1023/a:1010971219806. [DOI] [PubMed] [Google Scholar]
- 38.Del Rivero MA, et al. SmCI, a bifunctional inhibitor of metallo carboxypeptidase and serine proteinase isolated from the marine annelid Sabellastarte magnifica. Isolation, characterization, cDNA cloning and recombinant expression. FEBS J. 2008;275:157. [Google Scholar]
- 39.Covaleda G, del Rivero MA, Chávez MA, Avilés FX, Reverter D. Crystal structure of novel metallocarboxypeptidase inhibitor from marine mollusk Nerita versicolor in complex with human carboxypeptidase A4. J Biol Chem. 2012;287:9250–9258. doi: 10.1074/jbc.M111.330100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aloy P, et al. The crystal structure of the inhibitor-complexed carboxypeptidase D domain II and the modeling of regulatory carboxypeptidases. J Biol Chem. 2001;276:16177–16184. doi: 10.1074/jbc.M011457200. [DOI] [PubMed] [Google Scholar]
- 41.Pallarès I, et al. Structure of human carboxypeptidase A4 with its endogenous protein inhibitor, latexin. Proc Natl Acad Sci USA. 2005;102:3978–3983. doi: 10.1073/pnas.0500678102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanglas L, Aviles FX, Huber R, Gomis-Rüth FX, Arolas JL. Mammalian metallopeptidase inhibition at the defense barrier of Ascaris parasite. Proc Natl Acad Sci USA. 2009;106:1743–1747. doi: 10.1073/pnas.0812623106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reverter D, et al. Structure of a novel leech carboxypeptidase inhibitor determined free in solution and in complex with human carboxypeptidase A2. Nat Struct Biol. 2000;7:322–328. doi: 10.1038/74092. [DOI] [PubMed] [Google Scholar]
- 44.García-Castellanos R, et al. Detailed molecular comparison between the inhibition mode of A/B-type carboxypeptidases in the zymogen state and by the endogenous inhibitor latexin. Cell Mol Life Sci. 2005;62:1996–2014. doi: 10.1007/s00018-005-5174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arolas JL, et al. The three-dimensional structures of tick carboxypeptidase inhibitor in complex with A/B carboxypeptidases reveal a novel double-headed binding mode. J Mol Biol. 2005;350:489–498. doi: 10.1016/j.jmb.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 46.Bayés A, et al. Caught after the act: A human A-type metallocarboxypeptidase in a product complex with a cleaved hexapeptide. Biochemistry. 2007;46:6921–6930. doi: 10.1021/bi700480b. [DOI] [PubMed] [Google Scholar]
- 47.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 48.Edge M, et al. Engineered human carboxypeptidase B enzymes that hydrolyse hippuryl-L-glutamic acid: Reversed-polarity mutants. Protein Eng. 1998;11:1229–1234. doi: 10.1093/protein/11.12.1229. [DOI] [PubMed] [Google Scholar]
- 49.Playford RJ, Wright NA. Why is epidermal growth factor present in the gut lumen? Gut. 1996;38:303–305. doi: 10.1136/gut.38.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barnard JA, Beauchamp RD, Russell WE, Dubois RN, Coffey RJ. Epidermal growth factor-related peptides and their relevance to gastrointestinal pathophysiology. Gastroenterology. 1995;108:564–580. doi: 10.1016/0016-5085(95)90087-x. [DOI] [PubMed] [Google Scholar]
- 51.Heitz PU, et al. Immunohistochemical localisation of urogastrone to human duodenal and submandibular glands. Gut. 1978;19:408–413. doi: 10.1136/gut.19.5.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rowland KJ, Choi PM, Warner BW. The role of growth factors in intestinal regeneration and repair in necrotizing enterocolitis. Semin Pediatr Surg. 2013;22:101–111. doi: 10.1053/j.sempedsurg.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Araki F, Nakamura H, Nojima N, Tsukumo K, Sakamoto S. Stability of recombinant human epidermal growth factor in various solutions. Chem Pharm Bull (Tokyo) 1989;37:404–406. doi: 10.1248/cpb.37.404. [DOI] [PubMed] [Google Scholar]
- 54.Britton JR, George-Nascimento C, Udall JN, Koldovský O. Minimal hydrolysis of epidermal growth factor by gastric fluid of preterm infants. Gut. 1989;30:327–332. doi: 10.1136/gut.30.3.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Calnan DP, et al. Potency and stability of C terminal truncated human epidermal growth factor. Gut. 2000;47:622–627. doi: 10.1136/gut.47.5.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Playford RJ, et al. Epidermal growth factor is digested to smaller, less active forms in acidic gastric juice. Gastroenterology. 1995;108:92–101. doi: 10.1016/0016-5085(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 57.Rao RK, et al. Processing and transfer of epidermal growth factor in developing rat jejunum and ileum. Peptides. 1990;11:1093–1102. doi: 10.1016/0196-9781(90)90136-s. [DOI] [PubMed] [Google Scholar]
- 58.Tsukumo K, Nakamura H, Sakamoto S. Purification and characterization of high molecular weight human epidermal growth factor from human urine. Biochem Biophys Res Commun. 1987;145:126–133. doi: 10.1016/0006-291x(87)91296-4. [DOI] [PubMed] [Google Scholar]
- 59.Xian CJ, et al. Production of a human epidermal growth factor fusion protein and its degradation in rat gastrointestinal flushings. J Mol Endocrinol. 1996;16:89–97. doi: 10.1677/jme.0.0160089. [DOI] [PubMed] [Google Scholar]
- 60.Panosa C, et al. A comparison of non-biologically active truncated EGF (EGFt) and full-length hEGF for delivery of Auger electron-emitting 111In to EGFR-positive breast cancer cells and tumor xenografts in athymic mice. Nucl Med Biol. 2015;42:931–938. doi: 10.1016/j.nucmedbio.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 61.Gregory H, Thomas CE, Young JA, Willshire IR, Garner A. The contribution of the C-terminal undecapeptide sequence of urogastrone-epidermal growth factor to its biological action. Regul Pept. 1988;22:217–226. doi: 10.1016/0167-0115(88)90034-1. [DOI] [PubMed] [Google Scholar]
- 62.Goodlad RA, Boulton R, Playford RJ. Comparison of the mitogenic activity of human epidermal growth factor I-53 and epidermal growth factor I-48 in vitro and in vivo. Clin Sci (Lond) 1996;91:503–507. doi: 10.1042/cs0910503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.