Abstract
Aim:
We examined methylation patterns with aggressive tumor phenotypes and investigated demographic, socioeconomic and reproductive predictors of gene methylation.
Materials & methods:
Pyrosequencing quantified methylation of BRCA1, EGFR, GSTM2, RASSF1, TFF1 and Sat 2. We used quantile regression models to calculate adjusted median methylation values by estrogen and progesterone receptor (ER/PR) status. Bivariate associations between participant characteristics and methylation were examined.
Results:
Higher percent methylation of GSTM2 was observed in ER/PR-negative compared with ER/PR-positive tumors in ductal carcinoma in situ (14 vs 2%) and invasive (35 vs 3%) tissue components. Trends in aberrant GSTM2 methylation across tissue components were stronger among ER/PR-negative tumors (p-interaction <0.001). Black women were more likely to have ER/PR-negative tumors (p = 0.01) and show hypermethylation of GSTM2 compared with other women (p = 0.05).
Conclusion:
GSTM2 promoter hypermethylation may serve as a potential biomarker of aggressive tumor development and a mechanism for ER/PR-negative tumor progression.
Keywords: : breast cancer, breast cancer disparities, DNA methylation, estrogen/progesterone receptor, GSTM2, tumor progression
Background
Breast cancer is the most commonly diagnosed cancer in women worldwide [1]. The disease is highly heterogeneous and is commonly classified based on tumor receptor status for clinical purposes. Luminal A and B tumors (both estrogen and progesterone receptor [ER/PR]-positive) are among the most common subtypes and tend to have favorable prognoses [2–4]. In contrast, ER/PR-negative tumors including basal-like and triple-negative are less common, but are generally more aggressive and are associated with lower survival rates [4,5].
Previous research, including a study from our group, identified distinct patterns of DNA methylation between ER/PR-positive and ER/PR-negative breast tumors among adjacent normal and invasive tissue components [6–10]. Without altering the underlying genomic sequence, DNA methylation guides cellular differentiation by affecting gene expression; it may therefore serve as a potential mechanism for carcinogenesis and subtype formation [11]. In comparison to adjacent normal tissues, invasive tumor components exhibit reductions in global methylation [12–14] and increase in tumor suppressor gene promoter methylation [15–18]. Promoter methylation is often hypothesized to be associated with reduced gene expression [19].
To advance the knowledge of tumor etiology, a thorough understanding of the biological underpinnings of subtype development is necessary. Researchers have previously identified aberrant hypermethylation of SCGB3A1 among invasive ER/PR-positive tumors [20]. We sought to extend our previous work on aberrant methylation of genomic regions to a larger sample of breast cancer patients, and to explore associations with ductal carcinoma in situ (DCIS) tissue components [10]. Based on prior literature, we used an a priori approach to select genomic regions for analysis. We measured DNA methylation at five gene promoter regions (BRCA1, EGFR, GSTM2, RASSF1 and TFF1) as well as one marker of global methylation, Sat2, that have previously been associated with breast cancer incidence [21–28]. We hypothesized aberrant DNA methylation of these regions will be associated with aggressive breast cancer phenotypes and that participant characteristics will be predictors of aberrantly methylated genomic regions.
Methods
Study population
Breast Cancer Care in Chicago (BCCC) is a population-based sample of women who were diagnosed with breast cancer at one of 56 Chicago-area hospitals. The study population has been described in detail elsewhere [29,30]. Briefly, women who were eligible for enrollment were diagnosed with first primary DCIS or invasive breast cancer between 2005 and 2008; between the ages of 25 and 79; self-identified as non-Latina (nl) white, nl black or Latina; and resided in Chicago. Overall, 989 women were enrolled. Of these, 812 (82%) consented to allow samples of diagnostic tissue to be obtained by research staff. Breast tissue samples were obtained from 351 participants (43%) of whom ER/PR status was determined for 337 participants (42%). The University of Illinois at Chicago Institutional Review Board approved the protocol for this study and written informed consent was collected from all participants upon study enrollment.
Participant characteristics
Information on age at diagnosis, race/ethnicity, education, income, body mass index (BMI), age at first birth and menopause, and number of live births were collected via questionnaire upon enrollment in Breast Cancer Care in Chicago [29,30].
We constructed metrics of socioeconomic affluence and disadvantage at the neighborhood level, defined at the participant's census tract, using data from the 2000 United States Census. Neighborhood affluence was defined as the weighted contribution of the proportions of families with income of US$75,000 or more, adults with college education or more and civilian labor force employed in professional and managerial occupations. Neighborhood disadvantage was defined as the proportions of families with incomes below the poverty line, families receiving public assistance, persons unemployed and female-headed households with children. Both variables weighted the relevant variables equally, and standardized their sum to have a mean of 0 with a standard deviation of 1 [30]. Z-scores were used to determine low (<-1), intermediate (-1≤ z ≤1) and high levels (>1) of neighborhood affluence and disadvantage.
Tissue component collection procedures
Hematoxylin and eosin stained slides from formalin fixed, paraffin-embedded (FFPE) tumor blocks were examined to determine representative component areas of invasive, DCIS and histologically and morphologically normal-appearing breast tissue adjacent to the tumor (adjacent normal). We selected adjacent breast tissue from the same block as the tumor for women who received lumpectomies. However, when available we used a separate block containing breast tissue and no tumor as the nonmalignant, adjacent normal sample. Tissue core samples were cut precisely from the selected area using a semi-automated tissue arrayer (Beecher Instruments, Inc., Sun Prairie, WI, USA). Because the tissue was fixed and sealed by paraffin, cells from the invasive tissue could not become dislodged and contaminate the DCIS or adjacent normal tissue or vice versa. Overall, 714 individual tissue samples were collected: 109 women donated one sample from all three tissue components, 44 donated one sample from adjacent normal and DCIS tissues, 103 donated one sample from adjacent normal and invasive tissues, 12 donated one sample from DCIS and invasive tissues, 21 donated one sample from adjacent normal tissue, eight donated one sample from DCIS tissue and 40 donated one sample from invasive tissue.
Genomic region selection
We chose a diverse set of five genes and a DNA repeat to assay for DNA methylation in all tissue components [10]. Detailed information on the selected genomic regions is shown in Table 1. The DNA regions selected overlapped or were near regions previously reported to be aberrantly hypermethylated in breast cancer versus noncancerous breast tissue, namely BRCA1 [21,22], EGFR [23] and RASSF1 [24]; or aberrantly hypomethylated in breast cancer versus normal breast tissue, namely TFF1 [25] and DNA repeat, Sat2 [26,27]. We also examined a gene region from GSTM2 found to display hypermethylation in high-grade breast cancers [28]. Based on prior literature, we defined aberrant methylation as higher methylation of BRCA1, EGFR, GSTM2 and RASSF1 and lower methylation of TFF1 and Sat2.
Table 1. . List of studied DNA regions and number of CpG loci covered.
| Gene | Test region | Test region coordinates (hg19) | Distance from TSS (bp) | CGI | CpGs |
|---|---|---|---|---|---|
| BRCA1 | Exon 1 (extended promoter) | Chr17: 41277463–41277365 | +37 to +135 | No | 11 |
| EGFR | Intron 1 (extended promoter) | Chr7: 55088080–55088104 | +1355 to +1379 | Yes | 4 |
| GSTM2 | Promoter | Chr1: 110210582–110210641 | -62 to -3 | Yes | 8 |
| RASSF1 | Exon 1 (extended promoter) | Chr3: 50378294–50378232 | +74 to +134 | Yes | 9 |
| TFF1 | Promoter | Chr21: 43786664–43786628 | -20 to +16 | No | 5 |
| Sat2 | NA | DNA repeat | NA | NA | 2 |
CGI: CpG island overlapping the test region; NA: Not applicable; TSS: Transcription start site.
Sample processing & DNA methylation measurement
Dissolution of paraffin was accomplished by the addition of 1 ml of clearing agent (Histochoice) and incubation at 65°C for 30 min. Samples were digested by the addition of 100 μl of digestion buffer consisting of 10 μl 10× Target Retrieval Solution high pH (DAKO, Glostrup, Denmark), 75 μl of ATL Buffer (Qiagen, Hilden, Germany) and 15 μl of proteinase K and incubation at 65°C overnight. The sample volume was brought up to approximately 100 μl, and 20 μl of each sample was treated with bisulfite and purified using the Zymo EZ-96 DNA Methylation-Direct™ Kit (Irving, CA, USA), with a 15-min denaturation step at 98°C followed by a 3.5-h conversion at 64°C, an additional 15-min denaturation at 98°C and a 60-min incubation at 64°C. DNA was eluted in 40 μl of elution buffer. PCR was performed with 0.2 μM of each primer, one of which was biotinylated, and the final PCR product was purified (Streptavidin Sepharose HP, Amersham Biosciences, Uppsala, Sweden), washed, alkaline denatured and rewashed (Pyrosequencing Vacuum Prep Tool, Qiagen). The pyrosequencing primer (0.5 μM) was annealed to the purified single-stranded PCR product, and 10 μl of the PCR products were sequenced using the Pyrosequencing PSQ96 HS System (Biotage AB, Lund, Sweden) following the manufacturer's instructions. The methylation status of each locus was analyzed individually as a T/C SNP using Pyromark Q96 software (Qiagen).
Tumor hormone receptor assessment
ER/PR status was determined by immunohistochemical analysis. Copies of pathology reports were requested from the diagnosing institution. A single pathologist selected FFPE tumor blocks which generally represented the tumor. Recuts (4 μm each) of the selected tumor blocks were created for additional hematoxylin and eosin staining. The recuts were examined to identify invasive, DCIS and adjacent normal components of the tissue. Cores of the three tissue components were selected and tissue microarrays produced. Samples were stained using a monoclonal antibody for nuclear ER/PR status (manufacturer: Ventana [Oro Valley, AZ, USA], product catalogue number: 790–4324 and 790–2223 for ER and PR antibodies, respectively); stains were previously optimized on invasive breast tumor tissue before use in this study. ER and PR statuses were interpreted separately and given an H-score, which is the product of staining intensity (0, 1+, 2+, 3+) and the proportion of cells with the given intensity (possible range of H-score values: 0–300). A tumor sample was determined to be ER/PR-negative if it had an H-score less than ten for both receptor stains.
Statistical analysis
We compared individual-level demographic, socioeconomic and reproductive factors of ER/PR-positive and ER/PR-negative participants overall and stratified by race/ethnicity using χ2 tests.
For the methylation analyses, we averaged the percentage of methylated DNA across the tested CpG sites of each genomic region to compute a single methylation measurement for each gene. Nonparametric Wilcoxon rank sum tests were performed to test for differences in regional gene methylation by ER/PR status within tissue components. We calculated the median percentage methylated (and interquartile range) of the six genomic regions adjusted for individual-level characteristics using bootstrapped quantile regression models [31]. Quantile regression models were used to estimate adjusted median values (and 95% CIs), rather than adjusted means, of the DNA methylation variables. These models are more appropriate for our highly skewed DNA methylation outcomes. Models were adjusted for age at diagnosis, race/ethnicity, education, BMI, census tract affluence and disadvantage, age at first birth and number of live births. In order to model both number of live births and age at first birth together, we employed a method which combined information into one variable which generally represented ‘reproductive factors’ by assigning nulliparous women a value corresponding to an age at first birth equal to 40 [30]. For gene regions showing significant aberrant methylation associations with ER/PR-negative breast cancer, we used mixed-effect linear regression models to test for differences in methylation across tissue components by ER/PR status. Mixed-effect models were used to account for multiple tissue components donated from the same individual. Finally, we used the Kruskal–Wallis equality of population rank tests to examine associations between individual-level characteristics and gene DNA methylation. All analyses were conducted using Stata version 14.2 (Stata Corp., TX, USA). Given all hypotheses were prespecified, we considered p-values ≤0.05 to be statistically significant.
Results
Participants were between 25 and 78 years old (median: 56 years) at diagnosis. Most women were non-Latina black (40%), while non-Latina white and Latina women each comprised approximately 30% of the study sample. In general, the women were overweight/obese (76%), educated (74% completed high school/high school equivalency), and had annual household incomes of >US$25,000 (66%). A total of 86 (26%) women were diagnosed with ER/PR-negative tumors.
Race/ethnicity was significantly associated with tumor receptor status; nl black women were more likely to be diagnosed with ER/PR-negative tumors (p = 0.01) (Table 2). Participants’ neighborhood disadvantage was associated with ER/PR status (p = 0.01) such that women residing in census tracts with greater disadvantage were more likely to be diagnosed with ER/PR-negative tumors, although these associations did not persist after adjustment for race/ethnicity (p = 0.12). Obese and less educated women were marginally more likely to be diagnosed with ER/PR-negative tumors (p = 0.10 and 0.12, respectively). Women with an earlier age at first birth (p < 0.001) and a higher number of live births (p = 0.09) had a greater likelihood of being diagnosed with ER/PR-negative tumors. After adjustment for race/ethnicity, associations with earlier age at first birth and higher number of live births remained relatively unchanged (p = 0.001; p = 0.08). Notably, nl black women were significantly more likely to have earlier age at first birth and menopause as well as a higher number of births (data not shown).
Table 2. . Participant characteristic associations with tumor receptor status.
| Characteristic | ER/PR + | ER/PR - | p-value |
|---|---|---|---|
| n = 251 (%) | n = 86 (%) | ||
| Age at diagnosis (years): | 0.50 | ||
| – 18–49 | 67 (27) | 27 (31) | |
| – 50–59 | 79 (31) | 29 (34) | |
| – 60–79 | 105 (42) | 30 (35) | |
| Race/ethnicity: | 0.01 | ||
| – nl white | 87 (35) | 20 (23) | |
| – nl black | 90 (36) | 48 (56) | |
| – Latina | 74 (29) | 18 (21) | |
| Education: | 0.12 | ||
| – Less than high school | 59 (24) | 27 (31) | |
| – High school diploma/GED | 61 (24) | 25 (29) | |
| – Greater than high school | 131 (52) | 34 (40) | |
| Annual household income: | 0.43 | ||
| – <US$25,000 | 83 (34) | 32 (38) | |
| – US$25,000–US$87,499 | 112 (46) | 41 (48) | |
| – ≥US$85,000 | 50 (20) | 12 (14) | |
| Census tract affluence: | 0.83 | ||
| – Low | 21 (8) | 9 (11) | |
| – Intermediate | 206 (82) | 68 (80) | |
| – High | 24 (10) | 8 (9) | |
| Census tract disadvantage: | 0.01 | ||
| – Low | 37 (15) | 3 (4) | |
| – Intermediate | 168 (67) | 58 (68) | |
| – High | 46 (18) | 24 (28) | |
| BMI (kg/m2): | 0.10 | ||
| – ≤25 | 58 (23) | 21 (24) | |
| – 25–30 | 91 (37) | 21 (24) | |
| – >30 | 100 (40) | 44 (51) | |
| Age at first birth (years): | <0.01 | ||
| – <20 | 65 (26) | 39 (45) | |
| – 20–29 | 112 (45) | 37 (43) | |
| – 30+ | 74 (29) | 10 (12) | |
| Age at menopause (years): | 0.15 | ||
| – 20–39 | 20 (11) | 13 (20) | |
| – 40–45 | 50 (27) | 17 (26) | |
| – 46–50 | 74 (39) | 18 (27) | |
| – 51+ | 44 (23) | 18 (27) | |
| Live births (number): | 0.09 | ||
| – 0 | 46 (18) | 6 (7) | |
| – 1 | 40 (16) | 15 (17) | |
| – 2 | 61 (24) | 23 (27) | |
| – 3+ | 104 (42) | 42 (49) | |
ER/PR: Estrogen and progesterone receptor; GED: high school equivalency; nl: Non-Latina.
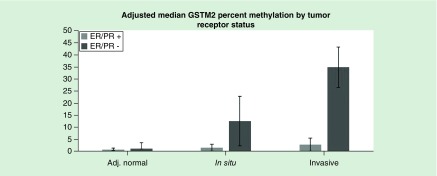
Table 3 presents the unadjusted median and interquartile ranges of methylation values for the six selected genomic regions by tissue component and tumor subtype. Aberrant hypermethylation of EGFR and RASSF1 was associated with less-aggressive, ER/PR-positive tumors in DCIS and invasive tissue components. Moreover, aberrant hypomethylation of TFF1 was associated with ER/PR-positive tumors in all three tissue components. Only GSTM2 showed significant aberrant hypermethylation with more aggressive, ER/PR-negative tumors in DCIS (p = 0.002) and invasive (p < 0.001) components. Less aggressive, ER/PR-positive tumors had percent methylation median values of 2 and 3% for the GSTM2 promoter region in DCIS and invasive tissue components, while more aggressive, ER/PR-negative tumors showed median values of 14 and 35% methylation in DCIS and invasive components. Adjustment for age at diagnosis, race/ethnicity, education, BMI, neighborhood affluence and disadvantage, and reproductive factors did not appreciably change the median methylation values for any of the tested genomic regions. Figure 1 displays the adjusted medians (with 95% CIs) for GSTM2 methylation by ER/PR status and tissue component. After accounting for paired samples and adjusting for covariates, we identified significant interaction between trends of GSTM2 methylation across all tissue components by ER/PR status (p-interaction <0.001). These interactions were present when restricting to adjacent normal and DCIS components (p-interaction = 0.02) or DCIS and invasive components (p-interaction = 0.008), with stronger trends observed for ER/PR-negative than ER/PR-positive tumors.
Table 3. . Bivariate associations examining DNA methylation by ER/PR status and tissue component.
| Gene region | ER/PR + | ER/PR - | p-value† | ||
|---|---|---|---|---|---|
| n | Median (IQR) | n | Median (IQR) | ||
| Aberrantly hypermethylated genes | |||||
| BRCA1: | |||||
| – Adj. normal | 163 | 0.3 (1.4) | 56 | 0.4 (1.0) | 0.73 |
| – DCIS | 123 | 0.8 (1.3) | 28 | 0.7 (0.7) | 0.32 |
| – Invasive | 175 | 0.6 (1.3) | 64 | 0.9 (1.5) | 0.06 |
| EGFR: | |||||
| – Adj. Normal | 207 | 6 (4) | 65 | 6 (4) | 0.60 |
| – DCIS | 140 | 18 (20) | 32 | 9 (12) | <0.01 |
| – Invasive | 195 | 25 (22) | 69 | 6 (12) | <0.01 |
| GSTM2: | |||||
| – Adj. Normal | 159 | 0.7 (3) | 54 | 0.8 (4) | 0.30 |
| – DCIS | 119 | 2 (6) | 29 | 14 (38) | <0.01 |
| – Invasive | 183 | 3 (18) | 64 | 35 (44) | <0.01 |
| RASSF1: | |||||
| – Adj. Normal | 171 | 7 (13) | 60 | 5 (10) | 0.11 |
| – DCIS | 128 | 53 (45) | 27 | 31 (39) | 0.01 |
| – Invasive | 184 | 50 (39) | 67 | 26 (47) | <0.01 |
| Aberrantly hypomethylated genes | |||||
| TFF1: | |||||
| – Adj. Normal | 191 | 70 (22) | 63 | 78 (18) | <0.01 |
| – DCIS | 134 | 35 (30) | 32 | 65 (33) | <0.01 |
| – Invasive | 193 | 36 (24) | 65 | 68 (39) | <0.01 |
| Sat2: | |||||
| – Adj. Normal | 187 | 59 (8) | 68 | 59 (12) | 0.40 |
| – DCIS | 133 | 53 (14) | 31 | 49 (14) | 0.45 |
| – Invasive | 189 | 52 (18) | 67 | 52 (14) | 0.72 |
†Wilcoxon rank sum test for differences between ER/PR status.
Adj.: Adjacent; DCIS: Ductal carcinoma in situ; ER/PR: Estrogen and progesterone receptor; IQR: Interquartile range.
Figure 1. . GSTM2 median values adjusted for age at diagnosis, race/ethnicity, education, body mass index, census tract affluence and disadvantage, age at first birth and number of live births by ER/PR status and tissue component.
Estimates calculated by bootstrapped quantile regression models.
Adj.: Adjacent; ER/PR: Estrogen and progesterone receptor.
We further examined participant characteristic associations with GSTM2 methylation as it was the only gene which showed aberrant methylation associations with more aggressive phenotypes (Table 4). In invasive tissue components, nl black women had nearly fivefold increased GSTM2 methylation compared with nl white and Latina women (p = 0.05). Similarly, women with earlier ages at menopause and first birth showed five- to eightfold higher GSTM2 methylation compared with women who were older at these life events (p = 0.04 for each); these associations did not persist after adjustment for race/ethnicity (p = 0.28; p = 0.22). In adjacent normal and DCIS components, we observed differences in GSTM2 methylation for neighborhood affluence (p = 0.07) and age at diagnosis (p = 0.05) although these differences were much smaller in magnitude. Associations between participant characteristics and DNA methylation of the five other genomic regions can be found in Supplementary Tables 1–10.
Table 4. . Participant characteristic associations with GSTM2 methylation by tissue component.
| Characteristic | Adjacent normal | DCIS | Invasive | ||||||
|---|---|---|---|---|---|---|---|---|---|
| n | Med. (IQR) | p-value† | n | Med. (IQR) | p-value† | n | Med. (IQR) | p-value† | |
| Age at diagnosis (years): | |||||||||
| – 18–49 | 63 | 0.8 (3) | 0.75 | 43 | 4 (21) | 0.05 | 73 | 7 (34) | 0.45 |
| – 50–59 | 64 | 0.5 (3) | 48 | 2 (22) | 77 | 4 (34) | |||
| – 60–79 | 86 | 0.9 (4) | 57 | 1 (5) | 97 | 4 (34) | |||
| Race/ethnicity: | |||||||||
| – nl white | 68 | 0.4 (2) | 0.14 | 44 | 1 (4) | 0.18 | 80 | 3 (29) | 0.05 |
| – nl black | 86 | 1 (4) | 64 | 4 (21) | 100 | 14 (41) | |||
| – Latina | 59 | 1 (4) | 40 | 2 (25) | 67 | 5 (35) | |||
| Education: | |||||||||
| – Less than high school | 49 | 0.5 (4) | 0.77 | 35 | 2 (34) | 0.94 | 61 | 9 (44) | 0.18 |
| – High school diploma/GED | 58 | 0.4 (3) | 38 | 3 (13) | 63 | 15 (41) | |||
| – Greater than high school | 106 | 0.9 (3) | 75 | 2 (12) | 123 | 3 (28) | |||
| Annual household income: | |||||||||
| – <US$25,000 | 65 | 0.9 (4) | 0.77 | 40 | 2 (21) | 0.70 | 85 | 8 (34) | 0.97 |
| – US$25,000–US$87,499 | 102 | 0.5 (3) | 73 | 2 (12) | 110 | 5 (35) | |||
| – ≥US$85,000 | 43 | 0.9 (2) | 31 | 3 (20) | 48 | 4 (28) | |||
| BMI (kg/m2): | |||||||||
| – ≤25 | 50 | 0.7 (4) | 0.89 | 37 | 2 (12) | 0.81 | 52 | 6 (28) | 0.95 |
| – 25–30 | 71 | 1 (3) | 47 | 2 (18) | 85 | 4 (35) | |||
| – >30 | 92 | 0.5 (4) | 64 | 2 (16) | 109 | 6 (35) | |||
| Census tract affluence | |||||||||
| – Low | 18 | 3 (4) | 0.07 | 8 | 2 (29) | 0.69 | 24 | 6 (15) | 0.77 |
| – Intermediate | 174 | 0.5 (3) | 124 | 2 (19) | 201 | 4 (34) | |||
| – High | 21 | 0.7 (2) | 16 | 1 (7) | 21 | 7 (47) | |||
| Census tract disadvantage: | |||||||||
| – Low | 25 | 0.8 (2) | 0.25 | 24 | 1 (4) | 0.51 | 26 | 3 (34) | 0.31 |
| – Intermediate | 142 | 0.5 (3) | 90 | 2 (20) | 172 | 4 (34) | |||
| – High | 46 | 2 (4) | 34 | 4 (22) | 48 | 15 (36) | |||
| Age at menopause (years): | |||||||||
| – 20–39 | 18 | 0.4 (2) | 0.78 | 14 | 1 (6) | 0.66 | 24 | 16 (53) | 0.04 |
| – 40–45 | 42 | 1 (4) | 28 | 0.9 (13) | 51 | 10 (45) | |||
| – 46–50 | 59 | 0.4 (3) | 39 | 2 (10) | 64 | 4 (27) | |||
| – 51+ | 35 | 0.9 (4) | 21 | 4 (12) | 49 | 2 (23) | |||
| Age at first birth (years): | |||||||||
| – <20 | 65 | 0.5 (3) | 0.76 | 45 | 3 (9) | 0.73 | 74 | 15 (44) | 0.04 |
| – 20–29 | 92 | 0.9 (4) | 69 | 2 (23) | 107 | 5 (31) | |||
| – 30+ | 56 | 0.4 (2) | 34 | 2 (5) | 66 | 3 (35) | |||
| Live births (number): | |||||||||
| – 0 | 34 | 0.3 (3) | 0.46 | 24 | 1 (3) | 0.80 | 38 | 2 (28) | 0.23 |
| – 1 | 33 | 0.7 (2) | 20 | 2 (23) | 41 | 4 (27) | |||
| – 2 | 52 | 0.9 (4) | 38 | 2 (33) | 57 | 16 (44) | |||
| – 3+ | 94 | 1 (4) | 66 | 3 (12) | 111 | 2 (33) | |||
†p-values determined by Kruskal–Wallis equality of populations rank test to examine differences between characteristic groups.
DCIS: Ductal carcinoma in situ; GED: High school equivalency; IQR: Interquartile range; Med.: Median.
Discussion
Our results suggest there are marked differences in DNA methylation by ER/PR status both within and across tissue components. Hypermethylation of GSTM2 was associated with ER/PR-negative tumors in DCIS and invasive tissue components. Aberrant methylation of the other gene regions, namely EGFR, RASSF1 and TFF1 were associated with less aggressive, ER/PR-positive tumors. Moreover, we observed greater methylation of GSTM2 among nl black women, as well among women who were younger when giving birth for the first time and women who undergo menopause earlier. Racial disparities in tumor subtype diagnosis have been identified in other populations [32–35]. While additional studies will be required, our results offer evidence that GSTM2 promoter hypermethylation may be one factor predisposing nl black women to the development of more aggressive tumors.
We observed a relationship between aberrant hypermethylation of the GSTM2 promoter region and ER/PR-negative tumors. GSTM2 encodes glutathione S-transferase Mu 2 which functions in the detoxification of electrophilic compounds including carcinogens, therapeutic drugs and environmental toxins [36]. GSTM2 methylation has previously been associated with aggressive, high-grade tumors [28]. To our knowledge, our group is the first to explicitly test the association between GSTM2 promoter methylation with ER/PR status by tissue component. We previously reported hypermethylation of this gene region in invasive tissue was associated with ER/PR-negative tumors using The Cancer Genome Atlas (TCGA) [10]. Notably, we also identified associations between DNA methylation of EGFR, TFF1 and RASSF1 with ER/PR status, although greater aberrant methylation was associated with less aggressive, ER/PR-positive tumors. Our findings for EGFR also suggest aggressive, ER/PR-negative breast cancer phenotypes show similar methylation levels across all tissue components.
We additionally identified associations between participant characteristics and GSTM2 hypermethylation. nl black women were more likely to have hypermethylated GSTM2 promoter regions in invasive tumor components compared with nl white and Latina women. These findings are unsurprising as we previously observed nl black women and women with hypermethylated GSTM2 promoter regions are more likely to be diagnosed with ER/PR-negative tumors. We similarly showed women with earlier age at menopause and age at first birth had increased GSTM2 methylation compared with women who experienced these events later in life, although race/ethnicity is likely to be driving these relationships. In our study sample, nl black women had significantly earlier ages at first birth and menopause compared with other women.
Although we identified increasing aberrant methylation of GSTM2 across all tissue components, particularly among ER/PR-negative tumors, this region was not identified in larger epigenome-wide studies examining tumor progression [37,38]. When testing across DCIS and invasive samples, we observed greater aberrant DNA methylation of GSTM2 among ER/PR-negative tumors compared with ER/PR-positive tumors (median percent methylation: 14% in DCIS vs 35% in invasive among ER/PR-negative, 2% in DCIS vs 3% in invasive among ER/PR-positive; p-interaction = 0.008). Fleisher et al. (2014) examined differences in DCIS and invasive methylation independent of ER/PR status and only reported sites with large aberrant methylation differences (change in median percent methylation >10%). Based on our sample, the strongest effects were only present among less prevalent, ER/PR-negative tumors and therefore may have been missed in analyses combining tumor subtypes. Moreover, Johnson et al. (2015) investigated differentially methylated regions between DCIS and invasive components, but only among ER/PR-positive tissue samples. Breast cancer is a heterogeneous disease; it is therefore plausible differentially methylated regions associated tumor progression from DCIS to invasive characteristics may be subtype-specific.
While our findings suggest GSTM2 promoter hypermethylation may predispose nl black women to the development of ER/PR-negative breast cancer, studies have suggested aberrant hypermethylation patterns among ER/PR-negative tumors are unlikely to drive tumor progression [39]. Holm et al. (2016) identified seven distinct epitypes, or patterns of epigenetic regulation across breast cancer subtypes; one of which was strongly associated with basal-like (ER/PR-negative) phenotypes. This epitype was defined by promoter gene hypermethylation, particularly among loci characterized by polycomb-repressed chromatin states. As such, these regions generally did not show correlations with gene expression. The authors therefore suggested promoter methylation in basal-like tumors is unlikely to be associated with DCIS to invasive tumor progression. Notably, our tested GSTM2 region does not have the characteristics of the basal-like epitype identified by Holm et al. (2016). In our previous study, we observed significant inverse correlations between GSTM2 hypermethylation and gene expression in invasive breast cancer samples from TCGA [10]. Moreover, using chromatin state segmentation data of human mammary epithelial tissue (HMEC) from the Encyclopedia of DNA elements (ENCODE), our tested region is characterized as a ‘weak/poised enhancer’, rather than polycomb repressed [40]. While our ER/PR-negative tumor samples are likely a mixture of basal-like and other hormone receptor-negative molecular subtypes, our tested region does not fit the characteristics and interpretations of Holm et al.'s (2016) basal-like epitype. It is therefore plausible hypermethylation of our tested GSTM2 region may influence tumor progression from DCIS to invasive components among ER/PR-negative tumors.
While this study observed associations between aberrant GSTM2 promoter methylation and ER/PR-negative phenotype, there are limitations worth noting. Our study was cross-sectional in the sense that both the tumor characteristics and DNA methylation were measured in the same set of tissue samples; we therefore cannot claim a temporal association. It is possible that aberrant methylation of GSTM2 is a consequence of ER/PR-negative status, rather than a cause of it. It is additionally possible that there exists a common underlying cause of both aberrant GSTM2 methylation and aggressive tumor characteristics which might result in associations that are not causal in nature. We also did not have gene expression data available to examine whether regional DNA methylation was associated with decreased gene expression. However, we previously examined associations between DNA methylation of these regions with gene expression using data from TCGA; in that study, we found significant inverse correlations with gene expression [10]. Finally, we used FFPE-derived DNA to examine tumor methylation. This type of DNA tends to be degraded and has greater issues with crosslinking compared with frozen tissue-derived DNA. We were therefore unable to use array-based assays to test for differentially methylated regions across the genome. Notably, the use of pyrosequencing is the gold-standard method for the accurate quantification of methylation in FFPE-derived DNA [41].
Conclusion & future perspective
This study identified aberrant hypermethylation of the promoter region of GSTM2 is associated with ER/PR-negative status among DCIS and invasive tissue components. The role of GSTM2 is understudied in relation to breast cancer but hypermethylation has previously been associated with aggressive, high-grade breast tumors. As GSTM2 functions in the detoxification of carcinogens and environmental toxins, future research may examine whether aberrant methylation results in toxins having prolonged cellular effects contributing to tumor progression among aggressive breast cancer phenotypes. Findings from these studies may offer evidence for a functional role for GSTM2 hypermethylation in the formation of ER/PR-negative breast cancer and may offer evidence that promoter GSTM2 methylation mediates racial disparities in aggressive tumor formation. Additional studies using novel populations are needed to confirm our findings as we examined gene regions not tested on array-based assays. Moreover, studies testing these relationships in peripheral blood would be required to use GSTM2 methylation as a potential risk assessment tool for aggressive breast cancer development. We conclude closer examination of GSTM2 methylation as a potential biomarker and mechanism of aggressive tumor development is warranted.
Summary points.
Aberrant DNA methylation has been linked to development and progression in human ductal carcinomas; less is known about how these biological mechanisms affect tumor characteristics.
We first screened for important genomic locations associated with tumor receptor status and followed up by examining methylation associations with participant characteristics.
We used quantile regression models to calculate adjusted median methylation values by estrogen and progesterone receptors (ER/PR) subtype and tissue component.
Non-Latina (nl) black women were more likely to be diagnosed with ER/PR-negative tumors compared with nl white and Latina women.
Hypermethylation of GSTM2 in ductal carcinoma in situ and invasive tissue components was associated with ER/PR-negative tumors.
nl black race/ethnicity was associated with hypermethylation of GSTM2 in invasive components.
GSTM2 promoter methylation may be one factor influencing racial disparities in aggressive tumor formation.
Studies testing relationships between GSTM2 methylation and breast cancer development in peripheral blood would be required to use GSTM2 methylation as a potential risk assessment tool for aggressive breast cancer development.
Closer examination of GSTM2 as a potential biomarker and mechanism of aggressive tumor development is warranted.
Supplementary Material
Footnotes
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.futuremedicine.com/doi/suppl/10.2217/epi-2017-0119
Financial & competing interests disclosure
The Breast Cancer Care in Chicago study is supported by the NIH (NIH P50 2CA106743). J Kresovich received additional support from the National Cancer Institute Cancer Education and Career Development Program (NIH R25 CA057699). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Desantis CE, Fedewa SA, Goding Sauer A, Kramer JL, Smith RA, Jemal A. Breast cancer statistics, 2015: convergence of incidence rates between black and white women. CA Cancer J. Clin. 2016;66(1):31–42. doi: 10.3322/caac.21320. [DOI] [PubMed] [Google Scholar]
- 2.Howlader N, Altekruse SF, Li CI, et al. US incidence of breast cancer subtypes defined by joint hormone receptor and HER2 status. J. Natl Cancer Inst. 2014;106(5) doi: 10.1093/jnci/dju055. pii:dju055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Metzger-Filho O, Sun Z, Viale G, et al. Patterns of recurrence and outcome according to breast cancer subtypes in lymph node-negative disease: results from international breast cancer study group trials VIII and IX. J. Clin. Oncol. 2013;31(25):3083–3090. doi: 10.1200/JCO.2012.46.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voduc KD, Cheang MC, Tyldesley S, Gelmon K, Nielsen TO, Kennecke H. Breast cancer subtypes and the risk of local and regional relapse. J. Clin. Oncol. 2010;28(10):1684–1691. doi: 10.1200/JCO.2009.24.9284. [DOI] [PubMed] [Google Scholar]
- 5.Millar EK, Graham PH, O'Toole SA, et al. Prediction of local recurrence, distant metastases, and death after breast-conserving therapy in early-stage invasive breast cancer using a five-biomarker panel. J. Clin. Oncol. 2009;27(28):4701–4708. doi: 10.1200/JCO.2008.21.7075. [DOI] [PubMed] [Google Scholar]
- 6.Fackler MJ, Umbricht CB, Williams D, et al. Genome-wide methylation analysis identifies genes specific to breast cancer hormone receptor status and risk of recurrence. Cancer Res. 2011;71(19):6195–6207. doi: 10.1158/0008-5472.CAN-11-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li L, Lee KM, Han W, et al. Estrogen and progesterone receptor status affect genome-wide DNA methylation profile in breast cancer. Hum. Mol. Genet. 2010;19(21):4273–4277. doi: 10.1093/hmg/ddq351. [DOI] [PubMed] [Google Scholar]
- 8.Rønneberg JA, Fleischer T, Solvang HK, et al. Methylation profiling with a panel of cancer related genes: association with estrogen receptor, TP53 mutation status and expression subtypes in sporadic breast cancer. Mol. Oncol. 2011;5(1):61–76. doi: 10.1016/j.molonc.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benevolenskaya EV, Islam AB, Ahsan H, et al. DNA methylation and hormone receptor status in breast cancer. Clin. Epigenetics. 2016;8:17. doi: 10.1186/s13148-016-0184-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rauscher GH, Kresovich JK, Poulin M, et al. Exploring DNA methylation changes in promoter, intragenic, and intergenic regions as early and late events in breast cancer formation. BMC Cancer. 2015;15:816. doi: 10.1186/s12885-015-1777-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Interesting paper examining field effects of DNA methylation across tumor subtypes in promoter and nonpromoter gene regions.
- 11.Ehrlich M, Lacey M. DNA methylation and differentiation: silencing, upregulation and modulation of gene expression. Epigenomics. 2013;5(5):553–568. doi: 10.2217/epi.13.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gama-Sosa MA, Slagel VA, Trewyn RW, et al. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 1983;11(19):6883–6894. doi: 10.1093/nar/11.19.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301(5895):89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 14.Varley KE, Gertz J, Bowling KM, et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23(3):555–567. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berman BP, Weisenberger DJ, Aman JF, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat. Genet. 2012;44(1):40–46. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costello JF, Frühwald MC, Smiraglia DJ, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat. Genet. 2000;24(2):132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 17.Ehrlich M, Jiang G, Fiala E, et al. Hypomethylation and hypermethylation of DNA in Wilms tumors. Oncogene. 2002;21(43):6694–6702. doi: 10.1038/sj.onc.1205890. [DOI] [PubMed] [Google Scholar]
- 18.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61(8):3225–3229. [PubMed] [Google Scholar]
- 19.Baylin SB, Esteller M, Rountree MR, Bachman KE, Schuebel K, Herman JG. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum. Mol. Genet. 2001;10(7):687–692. doi: 10.1093/hmg/10.7.687. [DOI] [PubMed] [Google Scholar]
- 20.Callahan CL, Wang Y, Marian C, et al. DNA methylation and breast tumor clinicopathological features: the Western New York Exposures and Breast Cancer (WEB) study. Epigenetics. 2016;11(9):643–652. doi: 10.1080/15592294.2016.1192735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dobrovic A, Simpfendorfer D. Methylation of the BRCA1 gene in sporadic breast cancer. Cancer Res. 1997;57(16):3347–3350. [PubMed] [Google Scholar]
- 22.Esteller M, Silva JM, Dominguez G, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J. Natl Cancer Inst. 2000;92(7):564–569. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 23.Montero AJ, Díaz-Montero CM, Mao L, et al. Epigenetic inactivation of EGFR by CpG island hypermethylation in cancer. Cancer Biol. Ther. 2006;5(11):1494–1501. doi: 10.4161/cbt.5.11.3299. [DOI] [PubMed] [Google Scholar]
- 24.Pasquali L, Bedeir A, Ringquist S, Styche A, Bhargava R, Trucco G. Quantification of CpG island methylation in progressive breast lesions from normal to invasive carcinoma. Cancer Lett. 2007;257(1):136–144. doi: 10.1016/j.canlet.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 25.Martin V, Ribieras S, Song-Wang XG, et al. Involvement of DNA methylation in the control of the expression of an estrogen-induced breast-cancer-associated protein (pS2) in human breast cancers. J. Cell. Biochem. 1997;65(1):95–106. doi: 10.1002/(sici)1097-4644(199704)65:1<95::aid-jcb10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 26.Jackson K, Yu MC, Arakawa K, et al. DNA hypomethylation is prevalent even in low-grade breast cancers. Cancer Biol. Ther. 2004;3(12):1225–1231. doi: 10.4161/cbt.3.12.1222. [DOI] [PubMed] [Google Scholar]
- 27.Cho YH, Yazici H, Wu HC, et al. Aberrant promoter hypermethylation and genomic hypomethylation in tumor, adjacent normal tissues and blood from breast cancer patients. Anticancer Res. 2010;30(7):2489–2496. [PMC free article] [PubMed] [Google Scholar]
- 28.Christensen BC, Kelsey KT, Zheng S, et al. Breast cancer DNA methylation profiles are associated with tumor size and alcohol and folate intake. PLoS Genet. 2010;6(7):e1001043. doi: 10.1371/journal.pgen.1001043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dookeran KA, Silva A, Warnecke RB, Rauscher GH. Race/ethnicity and disparities in mastectomy practice in the Breast Cancer Care in Chicago study. Ann. Surg. Oncol. 2015;22(1):66–74. doi: 10.1245/s10434-014-3945-6. [DOI] [PubMed] [Google Scholar]
- 30.Rauscher GH, Campbell RT, Wiley EL, Hoskins K, Stolley MR, Warnecke RB. Mediation of racial and ethnic disparities in estrogen/progesterone receptor-negative breast cancer by socioeconomic position and reproductive factors. Am. J. Epidemiol. 2016;183(10):884–893. doi: 10.1093/aje/kwv226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGreevy KM, Lipsitz SR, Linder JA, Rimm E, Hoel DG. Using median regression to obtain adjusted estimates of central tendency for skewed laboratory and epidemiologic data. Clin. Chem. 2009;55(1):165–169. doi: 10.1373/clinchem.2008.106260. [DOI] [PubMed] [Google Scholar]
- 32.Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California Cancer Registry. Cancer. 2007;109(9):1721–1728. doi: 10.1002/cncr.22618. [DOI] [PubMed] [Google Scholar]
- 33.Morris GJ, Naidu S, Topham AK, et al. Differences in breast carcinoma characteristics in newly diagnosed African–American and Caucasian patients: a single-institution compilation compared with the National Cancer Institute's Surveillance, Epidemiology, and End Results database. Cancer. 2007;110(4):876–884. doi: 10.1002/cncr.22836. [DOI] [PubMed] [Google Scholar]
- 34.Ooi SL, Martinez ME, Li CI. Disparities in breast cancer characteristics and outcomes by race/ethnicity. Breast Cancer Res. Treat. 2011;127(3):729–738. doi: 10.1007/s10549-010-1191-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295(21):2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 36.Tew KD, Ronai Z. GST function in drug and stress response. Drug Resist. Updat. 1999;2(3):143–147. doi: 10.1054/drup.1999.0086. [DOI] [PubMed] [Google Scholar]
- 37.Fleischer T, Frigessi A, Johnson KC, et al. Genome-wide DNA methylation profiles in progression to in situ and invasive carcinoma of the breast with impact on gene transcription and prognosis. Genome Biol. 2014;15(8):435. doi: 10.1186/s13059-014-0435-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Important paper of epigenetic mechanisms of breast cancer tumor progression.
- 38.Johnson KC, Koestler DC, Fleischer T, et al. DNA methylation in ductal carcinoma in situ related with future development of invasive breast cancer. Clin. Epigenetics. 2015;7:75. doi: 10.1186/s13148-015-0094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Important paper of estrogen and progesterone receptor-positive breast cancer tumor progression.
- 39.Holm K, Staaf J, Lauss M, et al. An integrated genomics analysis of epigenetic subtypes in human breast tumors links DNA methylation patterns to chromatin states in normal mammary cells. Breast Cancer Res. 2016;18(1):27. doi: 10.1186/s13058-016-0685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Interesting paper of epigenetic patterns of regulation across breast cancer subtypes.
- 40.Myers RM, Stamatoyannopoulos J, Snyder M, et al. A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9(4):e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat. Protoc. 2007;2(9):2265–2275. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.