Abstract
Enhancers are short noncoding segments of DNA (100–1000 bp) that control the temporal and spatial activity of genes in an orientation-independent manner. They can be separated from their target genes by large distances and are thus known as distal regulatory elements. One consequence of the variability in the distance separating enhancers and their target promoters is that it is difficult to determine which elements are involved in the regulation of a particular gene. Moreover, enhancers can be found in clusters in which multiple regulatory elements control expression of the same target gene. However, little is known about how the individual elements contribute to gene expression. Here, we describe how chromatin conformation promotes and constraints enhancer activity. Further, we discuss enhancer clusters and what is known about the contribution of individual elements to the regulation of target genes. Finally, we examine the reliability of different methods used to identify enhancers.
Keywords: : 3D conformation, chromatin contacts, cohesin, CTCF, enhancers, LCRs, stretch enhancers, super enhancers, TADs
Enhancers play a fundamental role in ensuring precise control of transcriptional patterns during development and differentiation. Since the original description of enhancers driving transcription of a reporter gene in specific constructs [1], many labs have focused on trying to understand how enhancers function, and in particular, how to identify which regulatory elements are involved in the control of a gene. Teasing apart the mode of action of enhancers has been difficult because only 7% of distal regulatory elements control the closest promoter as judged by a screen of transcription start sites and distal elements covering 1% of the human genome [2]. Indeed, enhancers can be located anywhere between 1 kb to tens of Mbs away from their target genes. An example of enhancer–promoter separation is the conserved Shh enhancer, ZRS, which when mutated is associated with polydactyly. Here, the enhancer and promoter are divided by almost a megabase of DNA along the linear chromosome [3]. Enhancers can also exert control over an even larger distance as in the case of the antigen receptor loci where Igk enhancers influence both the Tcrb locus, which is 30 MB away on mouse chromosome 6 and the Igh locus found on a different chromosome [4,5].
To exert control it is thought that enhancers and promoters need to be in physical contact [6]. The advent of chromosome conformation capture (3C) has made it possible to analyze interactions between enhancers and promoters at the molecular level and thereby determine the relevance of interaction [7]. 3C based techniques come in different flavors that can measure all possible interactions in the nucleus (Hi-C), specific interactions across a defined region (5C), or interactions from one or more viewpoints across the genome (4C-seq, Capture C and Capture Hi-C) [8]. 4C-seq, Capture C and Capture Hi-C are ideally suited for identifying enhancer–promoter contacts at high resolution and low cost.
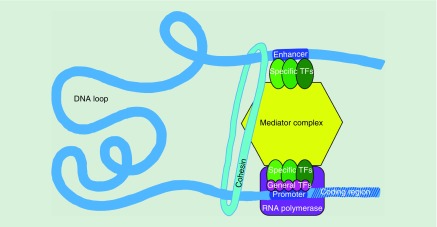
Enhancer–promoter contact via chromatin looping allows transcription factors (TFs) bound at enhancers to activate transcription of target genes [6]. Moreover, binding of TFs to both enhancers and promoters and recruitment of co-activators and chromatin remodelers, potentiates interaction between these regulatory elements (Figure 1) [9]. Identifying enhancer–promoter contacts provides a means of determining which genes are potentially regulated by a particular enhancer, and identifying new enhancers that could be involved in the regulation of a particular gene. Nonetheless, it is not clear whether interactions are predictive of functional regulation, so identifying enhancer–promoter contacts does not definitively determine the regulatory targets of individual enhancers and the extent to which they contribute to gene expression. Additionally, enhancer–enhancer contacts within enhancer clusters do not provide insight into whether interactions contribute to gene regulation and the extent to which they do so. Finally, genome-wide studies typically define enhancers as DNA stretches enriched for histone modifications or factors associated with active chromatin (H3K27Ac, p300, etc.) [7]. While this type of analysis provides a feasible way to determine enhancer candidates, it does not establish functional significance. To determine this, it is necessary to repress or delete these elements and characterize changes in interactions coupled with gene activity and binding of TFs. In this review, we discuss advances in the field that address the issues outlined above and provide new insight into the way enhancers contribute to the regulation of their target genes.
Figure 1. . Cell-type-specific transcription factors bind to both enhancers and promoters, recruiting co-activators, mediator complex and chromatin remodelers, all of which potentiate interaction between the regulatory elements and induce transcription.
TF: Transcripton factor.
Enhancer–promoter communication
The most convincing and detailed evidence to support the idea that enhancer–promoter contacts are important for transcriptional regulation comes from studies on the globin locus control region (LCR) and its target genes. The LCR activates distinct globin genes throughout erythroid development in a stage-specific manner starting with embryonic βh1-globin and fetal γ-globin and ending with two adult β-globin genes (β-major and β-minor). Under physiological conditions, activation of the individual globin genes is dependent on which transcription factors are expressed at the different stages of development. In adults, the transcription factor, GATA1 recruits the LDB1 factor to the β-globin locus to activate its transcription. LDB1 mediates looping via its self-association domain. To investigate the importance of chromatin contacts on gene expression, the Blobel group tethered the adult β-globin promoters to the LCR by fusing a zinc-finger DNA binding protein to LDB1 in GATA1 deficient erythroblasts [10]. LDB1 homodimerization mediates an interaction between the β-globin promoters and the LCR with the embryonic and fetal globin genes looped out. Forced physical contact between β-globin and its LCR was shown to be sufficient to recruit RNA polymerase to the β-globin promoters to activate expression in the absence of GATA1.
In subsequent experiments, LDB1 was tethered to the embryonic βh1-globin or fetal γ-globin promoters in primary erythroid cells where embryonic and fetal β-globin genes are normally silent [11]. Rewiring was again sufficient to trigger transcriptional activation. Furthermore, increasing the frequency of interactions between the enhancer and embryonic βh1-globin or fetal γ-globin promoters led to reduced contacts between the LCR and adult β-globin promoters, lowering adult globin expression. This strategy is now being applied to the treatment of hemoglobin-related disorders like sickle cell anemia or thalassemia, which arise from mutations in adult globin genes. These findings not only demonstrate that contacts between enhancers and promoters are important for control of gene expression, they also indicate that an enhancer can interact with more than one promoter.
There are numerous other examples of dynamic regulation involving tissue-specific enhancer–promoter contacts. At the Satb1 locus, which codes for a protein that functions as a scaffold for different chromatin remodeling enzymes, the promoter is not in contact with its enhancers in the brain, where the locus is silent. However, enhancer–promoter contacts form de novo in the thymus where Satb1 is highly expressed [12]. In another example of dynamic T-cell specific control, an enhancer of a master regulator gene, Bcl11b contacts its target promoter in early thymocyte development in a manner that is associated with repositioning of the entire region away from the nuclear lamina, production of enhancer RNAs (eRNAs) and deposition of activating epigenetic marks that allow for binding of chromatin architectural proteins, CTCF and cohesin, which establish an enhancer–promoter loop [13].
It is important to note that not all chromatin contacts are controlled in a cell type specific manner and not all interactions correlate with transcriptional activation. For example, in the Hoxd locus, Hoxd13 contacts five regulatory elements in the upstream gene desert in distal limb cells where it is expressed. However, Hoxd13 also contacts all these regulatory elements in proximal limb cells where the gene is inactive and it interacts with three out of five regulatory elements in the brain, where it is also silent [14–16]. Another example in which looping and expression are uncoupled comes from genes activated by the TNFα. Bing Ren's lab found that promoters were in contact with target enhancers prior to cytokine-mediated induction of gene expression in fibroblasts [17].
Genome-wide analysis of enhancer–promoter dynamics
A number of labs have now analyzed genome-wide stable versus dynamic looping in different contexts. The Furlong lab used 4C-seq to identify the interactomes of 103 enhancers in the Drosophila embryo, comparing mesodermal cells to the whole organism [18]. They found that most enhancer–promoter interactions were formed before gene activation and remained stable across development. In a mammalian-based study, the Mundlos lab used Capture-C to analyze contacts of the promoters of 446 loci in the mouse embryo [19]. With this approach, it was possible to annotate over a thousand putative enhancers for loci in forelimb, hindlimb and midbrain that exhibit both stable and dynamic (cell-type specific) interactions with promoters in different tissues. The stable contacts were found to be associated with CTCF and cohesin binding, while the dynamic contacts were linked with active chromatin modifications.
The Zhao lab examined the role of CTCF in enhancer–promoter interactions with Hi-C in mouse T helper (TH2) lymphocytes [20]. They found that CTCF binding is correlated with the activity of neighboring (<20 kb) enhancers and the strength of enhancer–promoter interactions. Furthermore, deletion of CTCF sites next to three TH2 specific genes led to increased cell-to-cell variation in their expression. These findings suggest that enhancer–promoter interactions associated with CTCF are more stable than those without CTCF and the former ensure robust gene expression by reducing cell-to-cell variability.
In another study, the Fraser group used Capture Hi-C to analyze promoter-associated genome architecture throughout 17 cell types of human hematopoietic lineage [21]. This analysis revealed the extent of 3D genome rewiring that occurs during differentiation in the hematopoietic lineage. The majority of enhancer–promoter interactions were found to be specific to the cell types where transcription of a promoter is activated. However, since progenitor cells were not included in the analysis, it remains unclear when cell-type specific contacts get established.
The Khavari lab used the same promoter Capture Hi-C approach to analyze chromosome interactions in human progenitor cells undergoing terminal epidermal differentiation in vitro. They identified two types of enhancer–promoter interactions: those that are pre-established in progenitor cells and those gained or newly formed during differentiation [22]. Some differentiation-induced genes participate in both types of interactions, with gained contacts strengthening their expression. However, the majority of interactions are stable and associated with the cohesin complex together with the constitutively expressed epidermal lineage-restricted ETS family transcription factor, EHF. Gained interactions lack cohesin but rely on the differentiation-induced transcription factors, KLF4 and ZNF750, which act as pioneer factors at a subset of enhancers established during development. In contrast to the constitutive interactions in developing Drosophila embryos discussed above, stable interactions identified in this study were not detected in human embryonic stem cells (ESCs), indicating that the interactomes of adult stem cells are not prewired.
Hi-C usually lacks sufficient resolution to detect enhancer–promoter interactions unless the sequencing depth is very high. Giacomo Cavalli's lab sequenced Hi-C libraries from an in vitro system of mouse neural differentiation to reach a resolution of 750 bp [23]. Ultra-deep sequencing revealed that active promoters contact each other even over a distance separation of 10–50 Mb along the linear chromosome. This is in contrast to CTCF-mediated interactions, which are confined to a much shorter range (<1 Mb). Active promoters also contact distal enhancer sites bound by the same transcription factors: NANOG in ESCs, PAX6 in neural progenitor cells (NPCs) and NEUROD2 and TBR1 in cortical neurons. It is of note that contacts associated with neural TFs were only significant in cells sorted ex vivo from the murine neurocortex, due to their lower expression in an in vitro system. Consistent with these findings, enhancer–promoter contacts bound by cell-type specific TFs were mostly established concomitantly with the start of gene expression and were disrupted when genes were silenced. These contacts were found to be independent of CTCF binding.
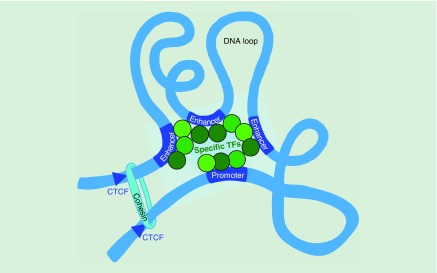
The role of cell type specific TFs was also highlighted by the Snyder lab's in situ Hi-C analysis of looping changes that occur during human monocyte to macrophage differentiation in vitro [24]. This group found both preformed stable and dynamic loops to be important for multiloop activation hubs at macrophage specific genes. The hubs incorporate enhancer–promoter interactions with a 3.4:1 ratio, respectively. Macrophage activation hubs are enriched for AP-1 bound long-range enhancer interactions. Together these findings support a model in which multiloop activation hubs and cell type specific transcription factors, drive changes in 3D interactions and transcription through regulatory loops (Figure 2).
Figure 2. . Multiloop activation hubs of regulatory elements controlled by cell-type-specific transcription factors drive changes in 3D interactions and gene expression.
TF: Transcription factor.
Multiloci interactions
Chromosome conformation capture methods typically identify pairwise interactions in a population of cells and cannot distinguish if interactions occur simultaneously between multiple loci in a single cell, or represent different 3D conformations that could be mutually exclusive. To investigate, the Tanay lab developed a technique called C-walk and analyzed multiloci contacts in human K562 cells and mouse ESCs. They found that contacts between regions within 1 Mb of each other do not show strong pairwise behavior indicating the potential formation of multiloci hubs. To investigate these at higher resolution, they used a modified 3way-4C procedure (Ay et al., 2015) that allows the detection of multiway contacts from designated viewpoints. Most of the interactions of the four highly expressed genes analyzed in K562 cells (EIF1B, GATA2, FTL, ANK1) represent independent pairwise contacts, but some synergistic contacts were detected particularly at the β-globin locus. However, in mouse ESCs the authors only detect what they term a ‘real’ synergistic hub with multiple elements interacting simultaneously at Hoxd loci that are repressed by Polycomb in these cells.
The De Laat lab used a method that relies on long reads from the minION sequencer to obtain information on simultaneous interactions between multiple partners from 4C data [25]. Focusing on the mouse β-globin super-enhancer (LCR) and its target genes they showed that in fetal liver, the β-globin super-enhancer represents an enhancer hub with concurrent contacts between individual enhancers and active adult β-globin genes. The interacting hub can even host both adult β-globin genes simultaneously. Chromatin hubs containing multiple genes and enhancers were also detected at the mouse protocadherin-alpha locus in fetal brain. Multicontact 4C provides a method to investigate the synergy of the multiway interactions in cells where individual regulatory elements within β-globin and protocadrein loci are deleted. The Skok lab performed this type of analysis on enhancers of the antigen receptor kappa locus (Igk) using a pipeline they developed for conventional Illumina sequencing reads (100 bps) from 4C data [26]. This is discussed in more detail in the section entitled ‘The impact of 3D organization on the function of enhancer clusters’.
Topologically associated domains
The finding that enhancers can act across large genomic distances begs the question about which features of chromatin preserve gene-target specificity? Emerging data indicate that sets of genes are organized into boundary-delimited territories and subterritories, within which there is a high level of coordination of epigenetic marks and transcriptional states. The larger domains defined as compartments A and B, are comprised of active and inactive chromatin, respectively. These compartments are subdivided into conserved megabase-sized topologically associated domains (TADs), encompassing highly self-interacting regions that are segregated by insulated boundaries [27,28]. At even higher resolution, gene expression is conferred by looping of cell context-specific gene enhancers to promoters within, and less frequently beyond TAD boundaries. CTCF (the only known vertebrate insulating protein) and cohesin are found at the loop bases of long-range interactions within TADs and are enriched at boundary regions. The current model explaining TAD formation and maintenance, involves a loop-extrusion mechanism [29–32], whereby cohesin rings organize the genome, creating loops by actively extruding DNA until the complex finds two CTCF binding sites in convergent orientation [33]. The main function of TADs is to limit the influence of regulatory elements to genes within the same domain. Indeed, most contacts between enhancers and promoters are restrained within the same domain. Global evidence for this comes from a study that integrated reporter genes into over a thousand sites in the mouse genome. These experiments reveal that enhancers function within regulatory domains that coincide with TADs [34].
The cooperation of CTCF and cohesin as chromatin architectural proteins became apparent from results of chromosome conformation experiments. A comparison of high-resolution 5C contact matrices encompassing seven genomic regions harboring pluripotency genes in ESCs versus NPCs demonstrate that different combinations of CTCF together with cohesin and mediator complex were enriched at chromatin interacting regions [35]. While CTCF and cohesin anchored invariant long-range contacts that formed TADs, cohesin and mediator clustered at the bases of short-range dynamic interactions within TADs, linking enhancers with promoters. In follow-up studies, the Phillips-Cremins lab showed that pluripotency gene-enhancer interactions anchored by CTCF are lost as the cells differentiate to NPCs. In contrast, CTCF binding sites in NPCs are already bound by CTCF in pluripotent stem cells. NPC-specific enhancer gene interactions are enriched for the ubiquitously expressed TF, YY1 only when they are engaged in 3D contacts and depletion of YY1 disrupts these interactions [36].
Insulating boundaries & their role in gene regulation
One of the first studies to identify TADs reported that deletion of an insulating boundary region at the X chromosome inactivation center led to a partial fusion of the neighboring domains [28] and long-range transcriptional misregulation. Further evidence for the importance of domains in transcriptional regulation comes from a growing list of reported examples of individual boundary disruptions. As an example, the Reinberg lab showed that deletion of CTCF binding sites at a boundary within the Hoxa cluster leads to the extension of active chromatin into repressive domains concomitant with increased interactions across the boundary [37,38].
In another elegant study, the Mundlos lab demonstrated the relevance of an intact domain boundary for the proper development of limb appendages in humans and mice [39]. Deletions, inversions and duplications across three TADs containing the WNT6, IHH, EPHA4 and PAX3 genes in human patient samples results in a distortion of boundaries. This in turn results in changes in chromatin contacts, bringing the promoters of WNT6, IHH and PAX3 under the control of a limb enhancer cluster that is normally associated with the EPHA4 locus, leading to their aberrant expression. The same genomic alterations were engineered in mice and shown to recapitulate the structural chromatin changes and limb malformations observed in humans. Boundary microdeletions can also result in misregulation of prominent proto-oncogene expression as shown in T-cell acute lymphoblastic leukemia [40]. The Young lab demonstrated that perturbation of two boundaries in the immortalized JURKAT T-cell line is sufficient to upregulate expression of the TAL1 and LMO2 oncogenes by hijacking enhancers across former borders.
Boundaries can additionally be altered under physiological conditions in response to signaling, as exemplified by the finding that border strength weakens in Drosophila cells that experience heat shock, leading to an increase in long-range inter-TAD interactions [41]. Boundary changes are also detected in primary human B lymphocytes as they transit from a naive to an activated state in the germinal center (GC). In this setting, numerous small domains become fused to form larger domains associated with a substantial increase in interactions at promoters and enhancers. Increased contacts go hand in hand with an elevated level of active histone marks and preferential binding of transcription factors with critical roles in GC formation such as PU.1, SPIB, IRF8 and upregulation of the Bcl6 gene, which drives the GC B cell phenotype [42].
Although CTCF and cohesin loss has been studied using conditional Cre-mediated deletion and shRNA knockdown [43–45], a more effective method of protein depletion has been achieved using the Auxin inducible degron system that enables rapid and acute protein degradation in a reversible manner (Nishimura et al., 2009). Use of this approach reveals that depletion of CTCF and cohesin results in loss of TAD insulation, however it does not affect compartmentalization of active (A) and inactive (B) regions, and cohesin loss even appears to strengthen interactions within each compartment [46–49]. Thus, the formation of domains and compartments occurs via different pathways. Somewhat surprisingly, removal of CTCF and cohesin has limited immediate transcriptional impact as measured by RNA-seq. Perhaps hijacking of regulatory elements caused by alterations in insulation may require time to manifest. Furthermore, ectopic exposure to enhancers may not be sufficient to alter transcriptional regulation as other factors such as histone modifiers and TFs required for determining the outcome of enhancer–promoter contacts may not be present.
A subset of active enhancers are found in clusters
In the past few years, attention has focused on clusters of enhancers that control the same target gene [50]. A paper from the Young lab described clusters of putative enhancers in mouse ESCs with an average size of 8 kb, which is in contrast to the 800 bp median length of regular enhancers [51]. The authors coined the term ‘super-enhancer’ to describe these clusters and defined them based on the level of enrichment of mediator, H3K27Ac, H3K4me1, p300 and master transcription factors. Datasets from four differentiated cell types show that super-enhancers, like LCRs are associated with cell identity genes. A catalog of super-enhancers in 86 different human cell and tissue samples was generated from extensive chromatin profiling analysis [52]. The Young lab showed that constituent enhancers of super-enhancer clusters tend to bind terminal transcription factors of multiple signaling pathways, which allows them to increase the sensitivity of their target genes to changes in these pathways [53]. In support of this idea, super-enhancers were shown to be acutely sensitive to pioneer master regulators in adult hair follicle stem cells [54]. This sensitivity allows super-enhancers in hair follicle stem cells to govern the dynamics of lineage commitment, following environmental changes like injury.
At around the same time, while studying chromatin signatures in ten different human cell types, the Collins’ lab identified 3 kb long DNA elements enriched with active enhancer marks and associated with cell-type-specific master regulator genes that they named ‘stretch enhancers’ [55]. The notion of enhancer clustering in large-scale regulatory domains is certainly not novel, but as more refined definitions emerge from ChIP-seq analysis, they come with new names. Nonetheless, there is clearly an overlap between the different groups of terms used to described enhancer clusters because the β-globin LCR, like other previously known LCR regions, is called as both a super-enhancer and a stretch enhancer in the K562 cell line.
Modes of cross talk between the individual elements in an enhancer cluster
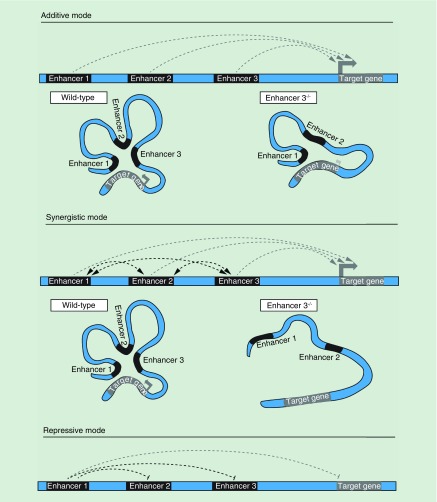
While the studies described above add new insight into the function of super-enhancers, they do not clarify whether clustering is important for activity. In this context, it is necessary to tease apart how the constituent elements of enhancer clusters contribute to gene regulation. For example, the individual elements within a cluster could act autonomously in a tissue specific manner or influence each other in an additive or synergistic manner (Figure 3). An additive mode of action is one in which individual neighboring enhancers act autonomously from each other such that the total transcriptional output is equal to the sum of the individual parts. Synergistic activity implies that the combined impact of the individual enhancers within the group is greater than the sum of the whole because neighboring enhancers influence each other's activity. To elucidate different modes of enhancer activity, it is necessary to delete the individual enhancer elements on their own or in combination.
Figure 3. . Modes of cross talk between the individual elements in an enhancer cluster.
Enhancers in additive mode act autonomously from each other so that deletion of one element does not impact other enhancers. In a synergistic mode, neighboring enhancers influence each other's activity, so that deletion of one enhancer has an impact on the way other enhancers in the cluster control a target gene. The impact of enhancer deletion on a target gene transcription is shown by the size and intensity of gray arrows. Enhancers in the cluster can also repress neighboring regulatory elements.
The eve locus [56] encodes a transcription factor responsible for the establishment of proper body patterning in the Drosophila embryo. It is expressed in odd numbered abdominal parasegments. In reporter assays, five enhancers separately drive eve expression in different parasegments at the same developmental stage of the fly embryo. In this example, modular control of target gene expression is exerted by the individual elements separately activating a target gene across different cell types or developmental time points.
Complex gene expression patterns of the Ihh gene, that encodes a master regulator of skeletal development, rely on a cluster of nine enhancers that act in a modular fashion [57]. Transgenic assays reveal that enhancers within the cluster show tissue specificity in the control of Ihh expression in digit anlagen, fingertips, growth plates and skull sutures with differences in activity occurring at different stages of development. A series of deletions and duplications of various parts of the Ihh enhancer cluster demonstrate that there is some redundancy in the activity of the individual elements. Interestingly, duplications in the Ihh enhancer cluster lead to Ihh misexpression highlighting the importance of the arrangement of duplicated elements relative to the enhancer cluster and Ihh gene.
The Fgf8 gene encodes a signaling molecule, whose precise expression patterns in brain, craniofacial structures, limbs and other tissues are crucial for embryo patterning. Transgenic analysis of DNA elements in the 220 kb region upstream of Fgf8 identified 19 enhancers interspersed throughout the region [58]. The majority of these enhancers drive transgene reporter expression in only one or two tissues overlapping Fgf8 expression in mouse embryo. However, multiple elements showed activity in almost every Fgf8 positive tissue, suggesting an additive mode of action. Nonetheless, there is some synergy among certain Fgf8 enhancers as the proximal enhancers fail to activate or maintain target gene expression when distal enhancers active in the same tissues are deleted.
Synergy between the individual elements of an enhancer cluster can manifest in a temporal order of activation, where the activity of one element is a prerequisite for the activity of neighboring elements [50]. A complex cis-regulatory network of enhancers governs expression of the endo16 locus in the endoderm of a developing sea urchin and its repression in ectoderm and mesenchymal lineages [59]. Endo16 encodes for a large extracellular calcium-binding protein with a possible role in cell adhesion [60]. There are six elements defined within the endo16 cis-regulatory network, of which the promoter proximal enhancer is required to initiate expression in the endoderm in early development. Two upstream elements act synergistically to enhance the activity of the promoter proximal enhancer and upregulate endo16 expression at later stages of development. The other three elements repress the endo16 gene in nonendodermal lineages in a manner that relies on the activity of the promoter proximal element.
Genome-wide analysis of super-enhancers that are progressively established in the mammary gland epithelium during pregnancy, reveal that individual super-enhancer components enriched for binding of STAT5, NFIB, ELF5 and GR transcription factors could sense hormonal cues separately, resulting in a temporal order of their activation [61]. The Wap gene, which is essential for nourishment of pups [62], is activated more than 1000-fold mid-pregnancy in mammary tissue. During pregnancy, three Wap enhancers establish a super-enhancer, which is activated by STAT5 binding to all three enhancers in response to hormonal signaling. Deletion of a STAT5 binding site at the most distal enhancer leads to a 91% reduction in Wap expression. However, deleting STAT5 binding sites at all three enhancers is necessary to completely abrogate target gene expression. Combined mutations of binding sites for STAT5, NFIB and ELF5 transcription factors in the proximal enhancer incapacitates the entire Wap locus preventing the super-enhancer from being established during pregnancy. Thus the proximal site acts as a seed enhancer.
Multiple enhancers can influence each other in a repressive manner to fine-tune the expression of their target gene. Three gap genes (hb, kr and kni) have precise spatial expression patterns in Drosophila and are responsible for proper segmentation of the embryo. Each of the three genes has a proximal and a distal enhancer that regulate transcriptional output. The distal and proximal enhancers influence each other to produce the correct expression patterns of the three gap genes [63]. Transgenes under the control of either distal or proximal enhancers alone exhibit abnormally broad expression patterns where transcription of the reporter gene is not restricted to the appropriate segments. In more recent studies the Levine group measured the transcriptional activity of transgenes carrying either proximal, distal or a combination of both hb, kni and sna enhancers with live-imaging [64]. In the anterior part of the embryo, the combined activity of proximal and distal Hb enhancers was shown to be less than the sum of the output from each individual enhancer, indicating sub-additive and potentially repressive behavior. In contrast, the two Hb enhancers act additively in the central region of the embryo. The authors attribute the increased output in this region to reduced levels of BICOID, a transcription factor which activates the two enhancers. The Kni enhancers on the other hand act synergistically as transgenes with both enhancers produce higher transcriptional activity than the combined activity of transgenes carrying either proximal or distal enhancer alone. Interestingly, the activity of a transgene with both Kni enhancers decreased at later stages of development, switching to additive behavior. The Sna enhancers also display a unique mode of action whereby the proximal enhancer attenuates the stronger distal enhancer. These studies demonstrate how distinct modes of cooperation between enhancers are necessary to establish precise spatial expression patterns of target genes.
Another example demonstrating that individual elements within clustered enhancers contribute unevenly to gene regulation comes from the Young lab. CRISPR/Cas9 deletion of the endogenous individual elements within the three super-enhancers of miR-290-295, Prdm14 and Sik1 loci have a different impact on their target gene expression in mouse ESCs as quantified by qPCR [53]. Indeed, knockout of one constituent of the Prdm14 super-enhancer increases Prdm14 expression.
Regulatory elements can also function redundantly and act as ‘shadow enhancers’. Michael Levine coined the term when he described the results of a genome-wide study of DORSAL transcription factor binding in a Drosophila embryo [65]. The results showed that nearly half of DORSAL's target genes can be regulated by secondary enhancers that are located further away from the target gene than primary enhancers, but which have overlapping activity profiles [66]. Shadow enhancers were shown to establish precise temporal and spatial transcription patterns of target genes under stress conditions [67,68].
The impact of 3D organization on the function of enhancer clusters
It is likely that cross talk between the neighboring regulatory elements of enhancer clusters described above will be influenced by their contact frequency and organization in 3D space. However, few enhancer clusters have been analyzed to determine how their interactions impact function. Given that chromatin is packaged within the nucleus in a manner that promotes or restricts contacts between regulatory elements, it is important to determine the functional significance of these associations. Although some of the five constituent enhancers of the Prdm14 super-enhancer described above were shown to contact each other in 3D nuclear space there were no genetic manipulations performed to determine whether interactions between elements are important for their activity [53]. Below, we describe in detail two studies that tackle this question. These studies focus on the super enhancers that regulate the Igk antigen receptor locus [4] and the LCR that controls the α-globin locus [69].
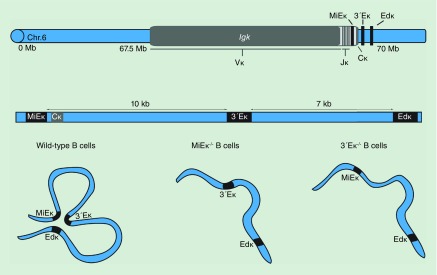
Immunoglobulin and T-cell receptor loci have served as a rich model system for analyzing the impact of nuclear organization and chromatin architecture on gene regulation. These antigen receptor loci consist of gene segments that span megabases of DNA and which come into close proximity in 3D nuclear space specifically to recombine [70–72]. Programmed recombination of the antigen receptor loci in developing lymphocytes is important for generating diversity of antigen receptors and for mounting an adaptive immune response to foreign antigens (for detailed review see [73]). As expected, enhancers of the loci play a major part in orchestrating antigen receptor recombination. The immunoglobulin Igk locus undergoes recombination of its variable (V) and joining (J) gene segments predominantly in pre-B cells to provide a rearranged immunoglobulin light chain. Productive rearrangement leads to surface expression of the IGK chain that forms part of the B-cell receptor, or antibody molecule secreted by plasma cells. At the Igk locus three enhancers MiEκ, 3′Eκ and Edκ are involved in controlling transcriptional output and recombination (Figure 4) [74–77]. The three enhancers have overlapping and distinct functions that contribute to Igk regulation. MiEκ and 3′Eκ are important for recombination and deletion of either one leads to a reduction in the frequency of IGK expressing B cells, while the double mutant is sufficient to abrogate the rearrangement of Igk altogether [78,79]. In contrast, deletion of Edκ leads to a reduction in expression of rearranged Igk in splenic B cells but it has no effect on recombination [80]. In the double 3′Eκ-/-Edκ-/- knockout mice however, there is negligible Igk expression and recombination [81]. These findings suggest that, in terms of their impact on Igk, the three enhancers act additively. However, in both MiEκ-/-3′Eκ-/- and 3′Eκ-/-Edκ-/- cells, the third remaining enhancer (Edκ or MiEκ, respectively) is not sufficient to rescue Igk expression and recombination.
Figure 4. . The immunoglobulin kappa locus, Igk on murine chromosome 6 consists of multiple variable and joining segments followed by a constant region gene.
Igk expression and recombination are controlled by three enhancers: MiEκ, 3′Eκ and Edκ, which are clustered at the 3′ end of the gene. The three enhancers form a hub in the developing B cells. Deletion of either MiEκ or 3′Eκ limits interactions between the two remaining enhancers.
MiEκ, 3′Eκ and Edκ comprise a super-enhancer in developing and mature B cells [4,82]. To determine whether the clustering and organization of enhancer elements in 3D space is functionally important, the Skok lab performed 4C-seq using different bait sequences within the Igk enhancer region in wild-type and enhancer deficient developing B cells [4]. These experiments revealed that the MiEκ, 3′Eκ and Edκ interact frequently in 3D nuclear space during recombination in wild-type pre-B cells. Furthermore, multiloci interaction analyses indicate that the interactions occur simultaneously in the same cell thereby forming an enhancer hub [26]. Deletion of either the MiEκ or 3′Eκ reduces enhancer interactions including pairwise interactions between the nondeleted enhancers, leading to the dissolution of the hub (Figure 4). Furthermore, RNA-seq analysis, demonstrates that a loss of either the MiEκ or the 3′Eκ results in reduced transcriptional output of the other two enhancers, which suggests that synergistic contacts between the individual components of a super-enhancer are important for their function. These findings highlight the interdependent nature of the individual enhancer components of the Igk super enhancer and the connections between transcriptional output and the contacts between them. Whether physical contact is important for the activity of other super-enhancers remains to be determined.
Capture-C analysis of the α-globin super-enhancer LCR organization from the viewpoint of all the α-globin promoters revealed that the five constituents of the α-globin super-enhancer form an interacting hub specifically in erythroid cells [69]. However, the individual elements were shown to have distinct contributions to target gene expression such that deletions of three out of five enhancers had no apparent phenotype. In contrast, combined deletion of the first and second enhancers resulted in a reduction in α-globin expression. The Higgs lab, who performed this analysis conclude that α-globin enhancers act in an additive manner, and that individual and combined enhancer deletions do not lead to the dissolution of interactions within the super-enhancer hub. However, one caveat to this study is that α-globin promoters were used as viewpoints in the chromatin conformation capture analysis of the α-globin super-enhancer hub and these could not reveal differences in interactions within the super-enhancer upon deletion of its individual elements. For this, it would be necessary to analyze interactions using the individual enhancers as viewpoints.
Predicting & validating enhancer function
Predictions about enhancers and their dysregulation in disease settings can be made using hierarchical analysis of recurring enhancer activation with altered levels of TF binding, function and gene expression. However, it is essential to validate these predictions by perturbing the system and examining the functional consequences. This point is nicely illustrated by a study focusing on the regulatory elements controlling the NEK6 gene, which is a mitosis-associated kinase that is commonly overexpressed in B-cell lymphoma [83]. The authors reveal that only a subset of enhancer sites, predicted by correlative algorithms to be involved in maintaining elevated NEK6 expression in B-cell lymphoma, are actually functional. Furthermore, an annotated super enhancer within the same TAD that was predicted to be involved in the regulation of NEK6 appears to be dispensable for its overexpression and for maintaining the architecture of a B-cell-specific enhancer hub. These findings, together with studies from other labs have shown that identifying candidate regulatory elements by chromatin marks (e.g., H3K27ac, H3K4me1, etc.) has limitations and can even produce unreliable results [4,83–85]. Indeed, the Skok lab have shown that a B-cell specific enhancer that regulates the Igk locus can also control Tcrb recombination in T cells, despite the absence of activating histone modifications and accessibility as measured by ATAC-seq in this lineage [4].
It is now possible to identify the full range of coding and noncoding mutations that contribute to gene regulation and clinically important outcomes. However, pairing noncoding regulatory elements such as enhancers with their target genes remains a challenge. Enhancers are typically identified by enrichment of marks such as H3K27Ac, H3K4me1 as well as binding of mediator, p300 and transcription factors [7], but, as pointed out above, this is not always reliable. Furthermore, assays that assess enhancer activity typically involve analyzing the effect of pieces of DNA outside of their normal chromatin environment in expression constructs [86–88]. To get around these limitations and to identify enhancers in a biologically relevant and unbiased manner, a number of studies described below used CRISPR-based mutagenesis or CRISPR effector noncoding screens.
Early CRISPR screens targeted small regions such as the 10 kb enhancer region of BCL11A implicated by genome-wide association studies to modulate fetal hemoglobin levels [89]. In this study, 500 single guide RNAs (sgRNAs) tiling the region, were used to identify which sequences were responsible for regulating BCL11A and fetal hemoglobin. With this approach, a narrow region that includes a GATA1 binding site essential for BCL11A expression was identified.
More recently the Zhang lab used a high-throughput CRISPR-based mutagenesis screen with 18,000 sgRNAs targeting 700 kb surrounding the NF1, NF2 and CUL3 genes (∼100 kb upstream and downstream of coding exons around the three genes) [90]. The screen relied on survival of tumor cells with CRISPR-mediated noncoding mutations in the regions surrounding the genes, NF1, NF2 and CUL3 that lead to loss of expression and resistance to the BRAF inhibitor, vemurafanib. Combined with other genome-wide analyses the screen identified regions where altered chromatin marks and abrogation of transcription factor binding was linked to a loss of transcriptional output. It is of note that the authors found enriched sgRNAs co-localized with regions of accessibility identified by assay for transposase-accessible chromatin using sequencing and DNase hypersensitivity, but the reverse was not true. Furthermore, they found that sgRNA enriched regions were more likely to interact frequently with the promoter as judged by 3C.
A similar CRISPR mutagenesis approach was used to target a 2 Mb region at the POU5F1 (OCT4) locus [91]. In this case, the screen was undertaken using POU5F1-GFP stem cells so that fluorescence-activated cell sorting could be used to sort positive and negative fluorescent cells. Fluorescence-activated cell sorting based screens with GFP knockins have also been used to identify enhancer elements controlling the programmed cell death locus, pcdh1, which is involved in regulating T-cell exhaustion and is a promising target for immunotherapy [92].
CRISPR effector screens can also be used to identify regulatory elements. As an example, dCas9-KRAB CRISPRi silencing was used to identify enhancers in regions around the MYC and GATA1 genes [93]. This screen relied on 98,000 sgRNAs to cover 1.3 Mbs of DNA in total. Decreased expression of these two genes in a leukemia cell line led to a reduction in proliferation and reduced representation of sgRNAs targeting the relevant elements.
CRISPR screens of this sort have a major advantage over other methods in that they provide an unbiased high-throughput method of identifying functional elements that act as enhancers. Nonetheless, not all of these screens identify distal regulatory elements as demonstrated by a deletion screen targeting a 206 kb region at the HPRT locus [94]. It is possible however that distal regulatory element/s lie outside the region targeted in this screen.
Future perspective
It is clear from the work described here that more functional studies are required to elucidate the contribution of individual enhancers to the expression of their target genes. From the evidence presented to date, it seems that enhancer clusters will need to be examined on a case-by-case basis as individual elements can act autonomously, additively, synergistically or in a repressive manner. To better understand the mode of action of the different enhancer clusters, it is important to analyze them in a physiological context at multiple stages of development. Understanding the temporal dynamics, by which the activities of enhancer clusters are established, can reveal insights into their modes of action. In addition, changes in the chromatin conformation of an enhancer cluster can provide a new perspective on the relationships between the constituents and provide insight into cause-and-effect relationships between nuclear organization and gene regulation. Finally, single cell approaches will be required to reveal the level of variability in enhancer promoter contacts and the significance this has on gene regulation.
Executive summary.
Enhancer–promoter communication: physical contact between enhancers and their target promoters is important for transcriptional activation. However, enhancer–promoter interactions are not predictive of transcriptional activation.
Genome-wide analysis of enhancer–promoter dynamics: enhancer–promoter contacts can be stable or cell-type specific, with dynamic contacts relying on cell-type specific transcription factors.
Multiloci interactions: approaches that identify multiloci interactions can distinguish if interactions occur simultaneously between multiple loci in a single cell, or represent different 3D conformations that could be mutually exclusive.
Topologically associated domains: contacts between enhancers and promoters are predominantly restrained within the same domain or topologically associated domain.
Insulating boundaries & their role in gene regulation: disruption of domain boundaries can alter gene regulation through changes in enhancer–promoter contacts.
A subset of active enhancers are found in clusters: super-enhancers are clusters of enhancers enriched for Mediator, H3K27Ac, H3K4me1, p300 and master transcription factors.
Modes of cross talk between the individual elements in an enhancer cluster: the individual elements within a cluster could act autonomously in a tissue specific manner or influence each other in an additive, synergistic or repressive manner.
The impact of 3D organization on the function of enhancer clusters: few super-enhancer clusters have been analyzed to determine how the individual elements and their interactions impact function.
Predicting & validating enhancer function: pairing noncoding regulatory elements such as enhancers with their target genes remains a challenge and it is important to validate predictions in a biologically relevant manner.
Acknowledgements
We would like to sincerely thank members of the Skok lab for their input and especially P Rocha for insightful comments and suggestions.
Footnotes
Financial & competing interests disclosure
JA Skok is supported by NIH grant R35GM122515, 4P30CA016087-36 Cancer Center Support Grant NIH/NCI (Neel) and 2R01CA140729-06A1 NIH/NCI (Carroll). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27(2 Pt 1):299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- 2.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489(7414):109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lettice LA, Heaney SJ, Purdie LA, et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 2003;12(14):1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 4.Proudhon C, Snetkova V, Raviram R, et al. Active and inactive enhancers cooperate to exert localized and long-range control of gene regulation. Cell Rep. 2016;15(10):2159–2169. doi: 10.1016/j.celrep.2016.04.087. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Investigates the contribution of enhancers to 3D organization of a multienhancer hub.
- 5.Hewitt SL, Farmer D, Marszalek K, et al. Association between the Igk and Igh immunoglobulin loci mediated by the 3′ Igk enhancer induces ‘decontraction’ of the Igh locus in pre-B cells. Nat. Immunol. 2008;9(4):396–404. doi: 10.1038/ni1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pennacchio LA, Bickmore W, Dean A, Nobrega MA, Bejerano G. Enhancers: five essential questions. Nat. Rev. Genet. 2013;14(4):288–295. doi: 10.1038/nrg3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buffry AD, Mendes CC, McGregor AP. The functionality and evolution of eukaryotic transcriptional enhancers. Adv. Genet. 2016;96:143–206. doi: 10.1016/bs.adgen.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Denker A, De Laat W. The second decade of 3C technologies: detailed insights into nuclear organization. Genes Dev. 2016;30(12):1357–1382. doi: 10.1101/gad.281964.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Laat W, Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 2013;502(7472):499–506. doi: 10.1038/nature12753. [DOI] [PubMed] [Google Scholar]
- 10.Deng WL, Lee J, Wang HX, et al. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149(6):1233–1244. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Forced physical contact between β-globin and its locus control region was shown to be sufficient to recruit RNA polymerase to the β-globin promoters in the absence of GATA1 to activate expression.
- 11.Deng WL, Rupon JW, Krivega I, et al. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell. 2014;158(4):849–860. doi: 10.1016/j.cell.2014.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van De Werken HJ, Landan G, Holwerda SJ, et al. Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nat. Methods. 2012;9(10):969–972. doi: 10.1038/nmeth.2173. [DOI] [PubMed] [Google Scholar]
- 13.Isoda T, Moore AJ, He Z, et al. Non-coding transcription instructs chromatin folding and compartmentalization to dictate enhancer–promoter communication and T cell fate. Cell. 2017;171(1):103–119. doi: 10.1016/j.cell.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fabre PJ, Leleu M, Mormann BH, et al. Large scale genomic reorganization of topological domains at the HoxD locus. Genome Biol. 2017;18(1):149. doi: 10.1186/s13059-017-1278-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montavon T, Soshnikova N, Mascrez B, et al. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147(5):1132–1145. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- 16.Andrey G, Montavon T, Mascrez B, et al. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science. 2013;340(6137):1234167. doi: 10.1126/science.1234167. [DOI] [PubMed] [Google Scholar]
- 17.Jin F, Li Y, Dixon JR, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503(7475):290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghavi-Helm Y, Klein FA, Pakozdi T, et al. Enhancer loops appear stable during development and are associated with paused polymerase. Nature. 2014;512(7512):96–100. doi: 10.1038/nature13417. [DOI] [PubMed] [Google Scholar]
- 19.Andrey G, Schopflin R, Jerkovic I, et al. Characterization of hundreds of regulatory landscapes in developing limbs reveals two regimes of chromatin folding. Genome Res. 2017;27(2):223–233. doi: 10.1101/gr.213066.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren G, Jin W, Cui K, et al. CTCF-Mediated enhancer–promoter interaction is a critical regulator of cell-to-cell variation of gene expression. Mol. Cell. 2017;67(6):1049–1058. doi: 10.1016/j.molcel.2017.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Javierre BM, Burren OS, Wilder SP, et al. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell. 2016;167(5):1369–1384. doi: 10.1016/j.cell.2016.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rubin AJ, Barajas BC, Furlan-Magaril M, et al. Lineage-specific dynamic and pre-established enhancer–promoter contacts cooperate in terminal differentiation. Nat. Genet. 2017;49(10):1522–1528. doi: 10.1038/ng.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonev B, Mendelson Cohen N, Szabo Q, et al. Multiscale 3D genome rewiring during mouse neural development. Cell. 2017;171(3):557–572. doi: 10.1016/j.cell.2017.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phanstiel DH, Van Bortle K, Spacek D, et al. Static and dynamic DNA loops form AP-1-bound activation hubs during macrophage development. Mol. Cell. 2017;67(6):1037–1048. doi: 10.1016/j.molcel.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allahyar A, Vermeulen C, Bouwman B, et al. Locus-specific enhancer hubs and architectural loop collisions uncovered from single allele DNA topologies. bioRxiv. 2017 [Google Scholar]
- 26.Jiang T, Raviram R, Snetkova V, et al. Identification of multi-loci hubs from 4C-seq demonstrates the functional importance of simultaneous interactions. Nucleic Acids Res. 2016;44(18):8714–8725. doi: 10.1093/nar/gkw568. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Employs 4C-seq to detect multiloci contacts that occur in the same cell as opposed to separate pairwise contacts observed in a cell population.
- 27.Dixon JR, Selvaraj S, Yue F, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nora EP, Lajoie BR, Schulz EG, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485(7398):381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nasmyth K. Cohesin: a catenase with separate entry and exit gates? Nat. Cell Biol. 2011;13(10):1170–1177. doi: 10.1038/ncb2349. [DOI] [PubMed] [Google Scholar]
- 30.Nichols MH, Corces VG. A CTCF code for 3D genome architecture. Cell. 2015;162(4):703–705. doi: 10.1016/j.cell.2015.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanborn AL, Rao SS, Huang SC, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl Acad. Sci. USA. 2015;112(47):E6456–E6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. Formation of chromosomal domains by loop extrusion. Cell Rep. 2016;15(9):2038–2049. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rao SS, Huntley MH, Durand NC, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159(7):1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Symmons O, Uslu VV, Tsujimura T, et al. Functional and topological characteristics of mammalian regulatory domains. Genome Res. 2014;24(3):390–400. doi: 10.1101/gr.163519.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phillips-Cremins JE, Sauria ME, Sanyal A, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153(6):1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beagan JA, Duong MT, Titus KR, et al. YY1 and CTCF orchestrate a 3D chromatin looping switch during early neural lineage commitment. Genome Res. 2017;27(7):1139–1152. doi: 10.1101/gr.215160.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Narendra V, Rocha PP, An D, et al. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 2015;347(6225):1017–1021. doi: 10.1126/science.1262088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Narendra V, Bulajic M, Dekker J, Mazzoni EO, Reinberg D. CTCF-mediated topological boundaries during development foster appropriate gene regulation. Genes Dev. 2016;30(24):2657–2662. doi: 10.1101/gad.288324.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lupianez DG, Kraft K, Heinrich V, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161(5):1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Demonstrates the importance of a topologically associated domain border for proper physiological development of a limb in both mice and humans.
- 40.Hnisz D, Weintraub AS, Day DS, et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016;351(6280):1454–1458. doi: 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li L, Lyu XW, Hou CH, et al. Widespread rearrangement of 3D chromatin organization underlies polycomb-mediated stress-induced silencing. Mol. Cell. 2015;58(2):216–231. doi: 10.1016/j.molcel.2015.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bunting KL, Soong TD, Singh R, et al. Multi-tiered reorganization of the genome during B cell affinity maturation anchored by a germinal center-specific locus control region. Immunity. 2016;45(3):497–512. doi: 10.1016/j.immuni.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seitan VC, Faure AJ, Zhan Y, et al. Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Res. 2013;23(12):2066–2077. doi: 10.1101/gr.161620.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sofueva S, Yaffe E, Chan WC, et al. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J. 2013;32(24):3119–3129. doi: 10.1038/emboj.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zuin J, Dixon JR, Van Der Reijden MI, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc. Natl Acad. Sci. USA. 2014;111(3):996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nora EP, Goloborodko A, Valton A-L, et al. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell. 2017;169(5):930–944. doi: 10.1016/j.cell.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwarzer W, Abdennur N, Goloborodko A, et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature. 2017;551(7678):51–56. doi: 10.1038/nature24281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rao SSP, Huang SC, Glenn St Hilaire B, et al. Cohesin loss eliminates all loop domains. Cell. 2017;171(2):305–320. doi: 10.1016/j.cell.2017.09.026. e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kubo N, Ishii H, Gorkin D, et al. Preservation of chromatin organization after acute loss of CTCF in mouse embryonic stem cells. bioRxiv. 2017 [Google Scholar]
- 50.Long HK, Prescott SL, Wysocka J. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell. 2016;167(5):1170–1187. doi: 10.1016/j.cell.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whyte WA, Orlando DA, Hnisz D, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153(2):307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hnisz D, Abraham BJ, Lee TI, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155(4):934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hnisz D, Schuijers J, Lin CY, et al. Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol. Cell. 2015;58(2):362–370. doi: 10.1016/j.molcel.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adam RC, Yang H, Rockowitz S, et al. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature. 2015;521(7552):366–370. doi: 10.1038/nature14289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parker SC, Stitzel ML, Taylor DL, et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc. Natl Acad. Sci. USA. 2013;110(44):17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maeda RK, Karch F. Gene expression in time and space: additive vs hierarchical organization of cis-regulatory regions. Curr. Opin. Genet. Dev. 2011;21(2):187–193. doi: 10.1016/j.gde.2011.01.021. [DOI] [PubMed] [Google Scholar]
- 57.Will AJ, Cova G, Osterwalder M, et al. Composition and dosage of a multipartite enhancer cluster control developmental expression of Ihh (Indian hedgehog) Nat. Genet. 2017;49(10):1539–1545. doi: 10.1038/ng.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marinic M, Aktas T, Ruf S, Spitz F. An integrated holo-enhancer unit defines tissue and gene specificity of the Fgf8 regulatory landscape. Dev. Cell. 2013;24(5):530–542. doi: 10.1016/j.devcel.2013.01.025. [DOI] [PubMed] [Google Scholar]
- 59.Yuh CH, Bolouri H, Davidson EH. Genomic cis-regulatory logic: experimental and computational analysis of a sea urchin gene. Science. 1998;279(5358):1896–1902. doi: 10.1126/science.279.5358.1896. [DOI] [PubMed] [Google Scholar]
- 60.Romano LA, Wray GA. Conservation of Endo16 expression in sea urchins despite evolutionary divergence in both cis and trans-acting components of transcriptional regulation. Development. 2003;130(17):4187–4199. doi: 10.1242/dev.00611. [DOI] [PubMed] [Google Scholar]
- 61.Shin HY, Willi M, Yoo KH, et al. Hierarchy within the mammary STAT5-driven Wap super-enhancer. Nat. Genet. 2016;48(8):904–911. doi: 10.1038/ng.3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Triplett AA, Sakamoto K, Matulka LA, Shen L, Smith GH, Wagner KU. Expression of the whey acidic protein (Wap) is necessary for adequate nourishment of the offspring but not functional differentiation of mammary epithelial cells. Genesis. 2005;43(1):1–11. doi: 10.1002/gene.20149. [DOI] [PubMed] [Google Scholar]
- 63.Perry MW, Boettiger AN, Levine M. Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proc. Natl Acad. Sci. USA. 2011;108(33):13570–13575. doi: 10.1073/pnas.1109873108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bothma JP, Garcia HG, Ng S, Perry MW, Gregor T, Levine M. Enhancer additivity and non-additivity are determined by enhancer strength in the Drosophila embryo. Elife. 2015;4:e07956. doi: 10.7554/eLife.07956. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Explores how pairs of enhancers controlling Gap gene expression in Drosophila embryo exhibit different modes of behavior ranging from additive to synergistic.
- 65.Hong JW, Hendrix DA, Levine MS. Shadow enhancers as a source of evolutionary novelty. Science. 2008;321(5894):1314–1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zeitlinger J, Zinzen RP, Stark A, et al. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Gene Dev. 2007;21(4):385–390. doi: 10.1101/gad.1509607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frankel N, Davis GK, Vargas D, Wang S, Payre F, Stern DL. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature. 2010;466(7305):490–493. doi: 10.1038/nature09158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perry MW, Boettiger AN, Bothma JP, Levine M. Shadow enhancers foster robustness of Drosophila gastrulation. Curr. Biol. 2010;20(17):1562–1567. doi: 10.1016/j.cub.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hay D, Hughes JR, Babbs C, et al. Genetic dissection of the alpha-globin super-enhancer in vivo . Nat. Genet. 2016;48(8):895–903. doi: 10.1038/ng.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Roldan E, Fuxa M, Chong W, et al. Locus ‘decontraction’ and centromeric recruitment contribute to allelic exclusion of the immunoglobulin heavy-chain gene. Nat. Immunol. 2005;6(1):31–41. doi: 10.1038/ni1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Skok JA, Gisler R, Novatchkova M, Farmer D, De Laat W, Busslinger M. Reversible contraction by looping of the Tcra and Tcrb loci in rearranging thymocytes. Nat. Immunol. 2007;8(4):378–387. doi: 10.1038/ni1448. [DOI] [PubMed] [Google Scholar]
- 72.Jhunjhunwala S, Van Zelm MC, Peak MM, et al. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell. 2008;133(2):265–279. doi: 10.1016/j.cell.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Proudhon C, Hao B, Raviram R, Chaumeil J, Skok JA. Long-range regulation of V(D)J recombination. Adv. Immunol. 2015;128:123–182. doi: 10.1016/bs.ai.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Queen C, Baltimore D. Immunoglobulin gene-transcription is activated by downstream sequence elements. Cell. 1983;33(3):741–748. doi: 10.1016/0092-8674(83)90016-8. [DOI] [PubMed] [Google Scholar]
- 75.Cockerill PN, Garrard WT. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986;44(2):273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- 76.Meyer KB, Neuberger MS. The immunoglobulin kappa-locus contains a second, stronger b-cell-specific enhancer which is located downstream of the constant region. Embo J. 1989;8(7):1959–1964. doi: 10.1002/j.1460-2075.1989.tb03601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu ZM, George-Raizen JB, Li S, Meyers KC, Chang MY, Garrard WT. Chromatin structural analyses of the mouse Igkappa gene locus reveal new hypersensitive sites specifying a transcriptional silencer and enhancer. J. Biol. Chem. 2002;277(36):32640–32649. doi: 10.1074/jbc.M204065200. [DOI] [PubMed] [Google Scholar]
- 78.Inlay M, Alt FW, Baltimore D, Xu Y. Essential roles of the kappa light chain intronic enhancer and 3′ enhancer in kappa rearrangement and demethylation. Nat. Immunol. 2002;3(5):463–468. doi: 10.1038/ni790. [DOI] [PubMed] [Google Scholar]
- 79.Inlay MA, Gao HH, Odegard VH, Lin T, Schatz DG, Xu Y. Roles of the Ig kappa light chain intronic and 3′ enhancers in Igk somatic hypermutation. J. Immunol. 2006;177(2):1146–1151. doi: 10.4049/jimmunol.177.2.1146. [DOI] [PubMed] [Google Scholar]
- 80.Xiang Y, Garrard WT. The downstream transcriptional enhancer, Ed, positively regulates mouse Ig kappa gene expression and somatic hypermutation. J. Immunol. 2008;180(10):6725–6732. doi: 10.4049/jimmunol.180.10.6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou X, Xiang Y, Garrard WT. The Igkappa gene enhancers, E3′ and Ed, are essential for triggering transcription. J. Immunol. 2010;185(12):7544–7552. doi: 10.4049/jimmunol.1002665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Qian J, Wang Q, Dose M, et al. B cell super-enhancers and regulatory clusters recruit AID tumorigenic activity. Cell. 2014;159(7):1524–1537. doi: 10.1016/j.cell.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang Y, Koues OI, Zhao JY, et al. cis-regulatory circuits regulating NEK6 kinase overexpression in transformed b cells are super-enhancer independent. Cell Rep. 2017;18(12):2918–2931. doi: 10.1016/j.celrep.2017.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Reveals that only a subset of enhancers predicted to affect NEK6 expression are actually functional, with an annotated neighboring super-enhancer being dispensable.
- 84.Engreitz JM, Haines JE, Perez EM, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539(7629):452–455. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Moorthy SD, Davidson S, Shchuka VM, et al. Enhancers and super-enhancers have an equivalent regulatory role in embryonic stem cells through regulation of single or multiple genes. Genome Res. 2017;27(2):246–258. doi: 10.1101/gr.210930.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Patwardhan RP, Hiatt JB, Witten DM, et al. Massively parallel functional dissection of mammalian enhancers in vivo . Nat. Biotechnol. 2012;30(3):265–270. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Arnold CD, Gerlach D, Stelzer C, Boryn LM, Rath M, Stark A. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339(6123):1074–1077. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- 88.Dickel DE, Zhu Y, Nord AS, et al. Function-based identification of mammalian enhancers using site-specific integration. Nat. Methods. 2014;11(5):566–571. doi: 10.1038/nmeth.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Canver MC, Smith EC, Sher F, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527(7577):192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sanjana NE, Wright J, Zheng K, et al. High-resolution interrogation of functional elements in the noncoding genome. Science. 2016;353(6307):1545–1549. doi: 10.1126/science.aaf7613. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Uses a high-throughput CRISPR-based mutagenesis screen to identify noncoding regulatory elements that control expression of genes responsible for tumor survival.
- 91.Diao Y, Fang R, Li B, et al. A tiling-deletion-based genetic screen for cis-regulatory element identification in mammalian cells. Nat. Methods. 2017;14(6):629–635. doi: 10.1038/nmeth.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sen DR, Kaminski J, Barnitz RA, et al. The epigenetic landscape of T cell exhaustion. Science. 2016;354(6316):1165–1169. doi: 10.1126/science.aae0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fulco CP, Munschauer M, Anyoha R, et al. Systematic mapping of functional enhancer–promoter connections with CRISPR interference. Science. 2016;354(6313):769–773. doi: 10.1126/science.aag2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gasperini M, Findlay GM, McKenna A, et al. CRISPR/Cas9-mediated scanning for regulatory elements required for HPRT1 expression via thousands of large, programmed genomic deletions. Am. J. Hum. Genet. 2017;101(2):192–205. doi: 10.1016/j.ajhg.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]