Abstract
Aim:
To investigate epigenomic changes in pregnancy and early postpartum in women with and without type 2 diabetes.
Methods:
Dimethylation of histones H3K4, H3K9, H3K27, H3K36 and H3K79 was measured in white blood cells of women at 30 weeks pregnancy, at 8–10 and 20 weeks postpartum and in never-pregnant women.
Results:
Dimethylation levels of all five histones were different between women in pregnancy and early postpartum compared with never-pregnant women and were different between women with and without type 2 diabetes.
Conclusion:
Histone methylation changes are transient in pregnancy and early postpartum and may represent normal physiological responses to hormones. Different epigenomic profiles in women with type 2 diabetes mellitus may correlate with hormonal responses, leading to high risk pregnancy outcomes.
Keywords: : epigenetics, histone methylation, pregnancy, type 2 diabetes mellitus
Pregnancy is a critical period where maternal and fetal physiology undergoes changes that can have significant implications for their future health. Studies have shown the importance of epigenetics in these processes. Considerable evidence exists that environmental pollutants known as endocrine disrupting agents can mimic naturally occurring hormones including pregnancy-related hormones, causing epigenetic changes [1]. Rat studies show endocrine disrupting agents, including diethylstilbestrol, act through the estrogen receptor causing a global decrease in H3K27 methylation [2]. Epigenetic changes mediated by environmental estrogens disrupt estrogen receptor signaling, and influence histone methylation leading to reprograming of the epigenome [3]. However, there is a paucity of clinical reports of epigenomic changes associated with the normal reproductive cycle and although epigenetic changes can be induced in cells following exposure to gestational hormones, there is little information about epigenomic changes associated with pregnancy.
The development of type 2 diabetes mellitus (T2DM) is strongly influenced by environmental factors, including diet, obesity and physical inactivity. These factors act through epigenetic mechanisms to alter gene expression, contributing to the onset of diabetes. Genes that show altered activity in T2DM include NF-κB-p65, PPAR-γ, Pdx1, PTEN and glut4 [4–8]. Although many epigenetic changes to diabetes-related genes have been identified, in particular to DNA methylation, no association between global gene methylation and T2DM, glucose, insulin or insulin resistance has been shown [9].
In addition to influencing DNA directly, diabetes risk factors also act through epigenetic mechanisms that alter histones. Methylation of lysine residues on histones is a key mechanism that regulates gene expression and is linked to lipid metabolism and diabetes [10–12]. Of the three methylation states of histones (mono, di and tri), dimethylation is most clearly linked to diabetes in human monocyte and lymphocyte studies [10]. High glucose levels can mediate histone modifications in vitro, establishing the causal mechanism by which gene expression patterns are altered [13].
In a previous study, we reported lower global histone lysine dimethylation of H3K27 and H3K4 in women with gestational diabetes who subsequently developed T2DM compared with women with gestational diabetes who did not, as well as differences in histone lysine dimethylation levels in women with gestational diabetes during pregnancy compared with the postpartum period [14]. This previous study compared groups of women, including those with gestational diabetes, during and after pregnancy. The current study is a further analysis comparing women who were not pregnant with those who were pregnant and did or did not have type 2 diabetes. These differences in histone dimethylation levels between pregnant and postpartum women suggest physiological changes occurring during pregnancy cause epigenetic effects, possibly through hormonal influences.
In this exploratory pilot study, to investigate epigenomic changes associated with pregnancy, we measured levels of lysine dimethylation in five histones: H3K4, H3K9, H3K27, H3K36 and H3K79 in never pregnant, pregnant and postpartum women. We also measured the histone dimethylation levels of women with and without T2DM in pregnancy and early postpartum to establish whether the epigenomic profile of women with T2DM were different from those of nondiabetic women.
Materials & methods
Participant recruitment & inclusion criteria
Participant recruitment was carried out at Sunshine Hospital and Deakin University, Melbourne, Australia. The experimental protocol received ethics approval from Melbourne Health Human Research Ethics Committee (HREC-12-MH-94), Western Health Governance (2012-093) and Deakin University Ethics committee (2012-254). All participants gave their written and informed consent.
Twenty-seven participants (six–seven in each group) were recruited over 12 months, representing four different groups of women: never-pregnant women with normal glucose metabolism; pregnant women with normal glucose metabolism; never-pregnant women with pre-existing T2DM; and pregnant women with pre-existing T2DM (Table 1). The women in the never-pregnant group, who were younger in age from the other groups, were volunteers from the university staff and different from the pregnant and postpartum group, so the study was a combination of cross-sectional and longitudinal.
Table 1. . Clinical and anthropometric data.
| Population characteristics | Nonpregnant nondiabetic | Pregnant nondiabetic | Nonpregnant pre-existing T2DM | Pregnant pre-existing T2DM |
|---|---|---|---|---|
| Number of participants | 7 | 7 | 7 | 6 |
| Age (mean ± SD) | 29.1 ± 3.1 | 36.7 ± 3.0 | 41.8 ± 7.1 | 38.2 ± 9.3 |
| Country of birth | Australia (6) Vietnam (1) |
Australia (4) New Zealand (1) Japan (1) India (1) |
Australia (6) South Korea (1) |
India (1) Kuwait (1) El Salvador (1) Philippines (1) Vietnam (1) Samoa (1) |
| Parity | N/A | P (4) M (3) |
N/A | P (1) M (5) |
| Family history of diabetes | Y (1) N (6) |
Y (0) N (7) |
Y (7) N (0) |
Y (5) N (1) |
M: Multipara; N: No; N/A: Not applicable; P: Primipara; SD: Standard deviation; T2DM: Type 2 diabetes mellitus; Y: Yes.
Selection criteria for the four groups were as follows: healthy women with no diabetic history; pregnant nondiabetic women with fasting glucose <5.1 mmol/l and a 2-h glucose <8.5 mmol/l following 75-g oral glucose tolerance test (OGTT); women with pre-existing T2DM diagnosed prior to the study.
Criteria for exclusion from participation were: women with gestational diabetes in pregnancy; women with severe mental illness or substance abuse in the 3 months prior to the study; women who had difficulty communicating in English or were <18 years of age; and heavy smokers (>20 cigarettes/day). It was estimated that 25% of the projected 32 women who would initially consent would drop out before the end of the study and thus the final sample size would be at least 24 (six per group).
Fasting blood samples were collected and analyzed to identify differences in levels of histone modification at three time points: 30 weeks gestation; 8–10 weeks postpartum; and 20 weeks postpartum for all women except for nonpregnant groups (A and C) where blood was collected only once. The time points relate to the times in and after pregnancy when women are generally tested for diabetes. Women are screened for the presence of gestational diabetes at around 30 weeks, then retested at 8–10 weeks postpartum to ascertain if their gestational diabetes mellitus (GDM) has resolved. We additionally tested at 20 weeks postpartum to ascertain if diabetes was present or not as we noted in our earlier study [14] that some women who had GDM were still diabetic at both 8–10 and 20 weeks postpartum.
Pregnant women participated in the study for 8 months and completed a brief survey to establish ethnicity, number of pregnancies, family history of diabetes and treatment, including medications that might affect glucose levels. Study nurses collected blood samples (10 ml) from each participant at their homes at the three time points.
Blood processing
Whole blood was separated by layering on Ficoll-Paque™ PLUS (GE Healthcare, Melbourne, Australia) and centrifuged at 400 × g for 40 min. Layers of plasma, buffy coat, granulocytes and red blood cells were collected separately and stored at -80°C. The buffy coat was washed in phosphate buffered saline, stored at -80°C and used for histone methylation analysis. All analyses were conducted in a PC2 lab and cell isolation was completed in a type 2 biohazard cabinet.
Sample storage & disposal
Samples were stored in a locked -80°C freezer.
Histone methylation analysis
The dimethylation status of histone 3 at five lysine sites was measured in white blood cells at all time points. Buffy coat lysates were prepared using 250 μl of 5% (w/v) SDS in 10 mM Tris-HCl (pH 7.5) lysis buffer (tris-buffered saline [TBS]) and homogenized by passing through a 21 gauge needle followed by sonication (15 pulses, 40% power output, 30% duty cycles, Microson XL2000 Ultrasonic Cell Disruptor, Misonix, Farmingdale, NY, USA) on ice in 1× EDTA-free inhibitor cocktail (Roche Diagnostics, Melbourne, Australia). The total protein content of lysates was measured using the BCA Protein Assay Kit (Pierce, IL, USA). Sixty μg lysates were fractionated by SDS-PAGE on a 12% gel and transferred to nitrocellulose membranes (Whatman, Dassal, Germany). Membranes were stained with 0.1% Ponceau S (Sigma-Aldrich, Sydney, Australia). Ponceau S stain was removed with 0.1% NaOH. After 1-h blocking in 5% (w/v) milk powder in TBS at room temperature, membranes were exposed overnight at 4°C to histone H3 antibodies (Methyl-Histone H3 Antibody Kit #9847, Cell Signalling Technology distributed by Australian Biosearch, Queensland, Australia) diluted in 1/1000 in 1% bovine serum albumin in TBS. Membranes were washed in TBS and antibodies were detected using 1/2000 dilution of horseradish peroxidase–conjugated goat antirabbit secondary antibody (Millipore, Melbourne, Australia) in 1% bovine serum albumin in TBS for 45 min at room temperature. Proteins were detected by enhanced chemiluminescence (Immobilon Western Chemiluminescent HRP Substrate, Life Science, Melbourne, Australia) and a LAS-3000 FujiFilm Lumino-Image Analyzer (Fuji Photo Film, Tokyo, Japan). Blots were stripped for 5–10 min using Reblot Plus Strong (Life Science) solution then reprobed with anti-β-actin monoclonal antibody (Sigma-Aldrich) diluted 1/5000 to determine protein loading. Densitometry was performed using Fuji Film Multi Gauge V3.0 computer software. Ratios for protein levels calculated relative to β-actin and total histone H3 expression. Western blots (triplicate experiments) were run and to allowed for accurate comparative analyses. One selected sample (from a nondiabetic pregnant participant) was loaded onto each of the gels. Densitometry readings were then normalized against this sample (blot control).
Statistical methods
One-way repeated measures analysis of variance was used to analyze differences in individual histone methylation at three time points (30 weeks pregnant, 8–10 weeks postpartum and 20 weeks postpartum) for women with and without T2DM mellitus. A Mauchly's test was used to assess sphericity. When the Greenhouse–Geisser epsilon was <0.75, a Greenhouse–Geisser correction was used; when the value was ≥0.75, a Huynh-Feldt correction was used. No correction was used if the assumption of sphericity was met. Post hoc tests were conducted using a Sidak adjustment. Epigenetic profiles are presented for each control group (nonpregnant women with and without T2DM) and for the pregnant and postpartum time points for women with and without T2DM mellitus. No statistical analyses were conducted on these data. All other data were analyzed using independent samples t-tests. Where the Levene's test for equality of variance was found to be significant; corrected values were used. Data were analyzed using SPSS versions 23 and 24; results are expressed as mean ± standard deviation and were deemed significant when p < 0.05.
Results
Recruitment outcomes
A total of 53 women were invited and 32 (60%) consented to participate in the study. Five women (18%) dropped out between the time of consent and final data collection. The total number of participants with full data collection was 27 (groups ranged from six–seven women). Their characteristics are shown in Table 1.
Epigenetic analyses
The dimethylation status of histone H3 at five different lysine sites (H3K4, H3K9, H3K27, H3K36 and H3K79) was measured in white blood cells from never-pregnant women, women at 30 weeks gestation, 8–10 weeks postpartum and 20 weeks postpartum.
We compared never-pregnant women with pregnant women, never-pregnant women with women at 20 weeks postpartum, and made comparisons between the same population of women at pregnancy and at 8–10 and 20 weeks postpartum, for nondiabetic groups (Figure 1). Similar comparisons were carried out for women with type 2 diabetes (Figure 2).
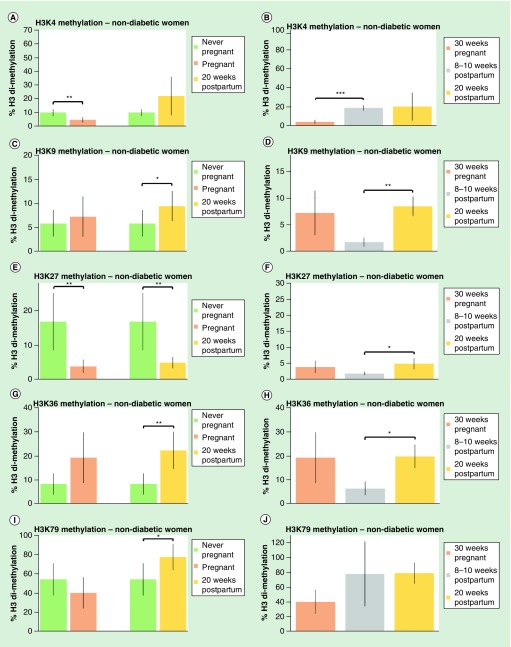
Figure 1. . Pregnancy affects histone methylation levels in nondiabetic women.
Dimethylation levels on five histones, expressed as a percentage of total histone H3 levels, were measured in peripheral blood mononuclear cells of never-pregnant women, women at 30 weeks pregnancy, women at 8–10 weeks postpartum and women at 20 weeks postpartum. (A) Dimethylation levels of H3K4 in never-pregnant women compared with women at 30 weeks pregnancy; and dimethylation levels in never-pregnant women compared with women at 20 weeks postpartum. (B) Dimethylation levels of H3K4 women at 30 weeks pregnancy, women at 8–10 weeks postpartum and women at 20 weeks postpartum. (C) Dimethylation levels of H3K9 in never-pregnant women compared with women at 30 weeks pregnancy; dimethylation levels in never-pregnant women compared with 20 weeks postpartum women. (D) Dimethylation levels of H3K9 women at 30 weeks pregnancy, women at 8–10 weeks postpartum and women at 20 weeks postpartum. (E) Dimethylation levels of H3K27 in never-pregnant women compared with women at 30 weeks pregnancy; dimethylation levels in never-pregnant women compared with 20 weeks postpartum women. (F) Dimethylation levels of H3K27 women at 30 weeks pregnancy, women at 8–10 weeks postpartum and women at 20 weeks postpartum. (G) Dimethylation levels of H3K36 in never-pregnant women compared with women at 30 weeks pregnancy; dimethylation levels in never-pregnant women compared with 20 weeks postpartum women. (H) Dimethylation levels of H3K36 women at 30 weeks pregnancy, women at 8–10 weeks postpartum and women at 20 weeks postpartum. (I) Dimethylation levels of H3K79 in never-pregnant women compared with women at 30 weeks pregnancy; dimethylation levels in never-pregnant women compared with 20 weeks postpartum women. (J) Dimethylation levels of H3K79 women at 30 weeks pregnancy, women at 8–10 weeks postpartum and women at 20 weeks postpartum. Values are expressed as a mean percentage of arbitrary units ± standard deviation normalized against total H3 levels.
*p < 0.05; **p < 0.01; ***p < 0.001.
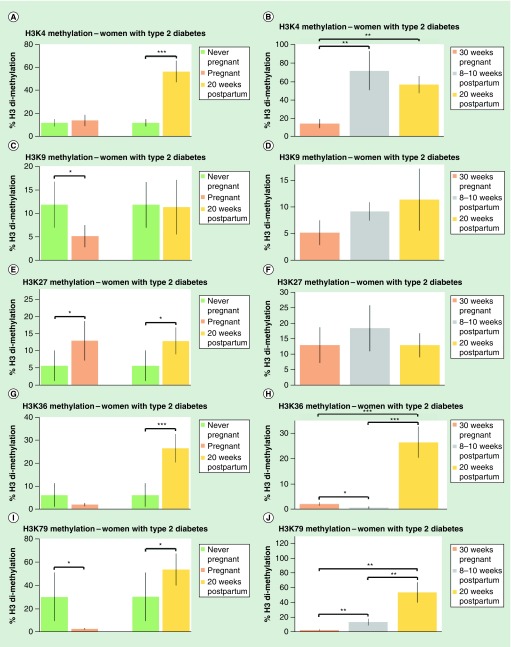
Figure 2. . Pregnancy affects histone methylation levels in women with type 2 diabetes mellitus.
Dimethylation levels on five histones, expressed as a percentage of total histone H3 levels, were measured in never-pregnant women, women with type 2 diabetes mellitus (T2DM), women with T2DM at 30 weeks pregnancy, women with T2DM at 8–10 weeks postpartum and women with T2DM at 20 weeks postpartum. (A) Dimethylation levels of H3K4 in never-pregnant women compared with women at 30 weeks pregnancy; dimethylation levels in never-pregnant women compared with women at 20 weeks postpartum. (B) Dimethylation levels of H3K4 women at 30 weeks pregnancy, 8–10 weeks postpartum women and 20 weeks postpartum women. (C) Dimethylation levels of H3K9 in never-pregnant women compared with women at 30 weeks pregnancy; dimethylation levels in never-pregnant women compared with 20 weeks postpartum women. (D) Dimethylation levels of H3K9 women at 30 weeks pregnancy, women at 8–10 weeks postpartum and women at 20 weeks postpartum. (E) Dimethylation levels of H3K27 in never-pregnant women compared with women at 30 weeks pregnancy; dimethylation levels in never-pregnant women compared with 20 weeks postpartum women. (F) Dimethylation levels of H3K27 women at 30 weeks pregnancy, women at 8–10 weeks postpartum and women at 20 weeks postpartum. (G) Dimethylation levels of H3K36 in never-pregnant women compared with women at 30 weeks pregnancy; dimethylation levels in never-pregnant women compared with 20 weeks postpartum women. (H) Dimethylation levels of H3K36 women at 30 weeks pregnancy, women at 8–10 weeks postpartum and women at 20 weeks postpartum. (I) Dimethylation levels of H3K79 in never-pregnant women compared with women at 30 weeks pregnancy; dimethylation levels in never-pregnant women compared with 20 weeks postpartum women. (J) Dimethylation levels of H3K79 women at 30 weeks pregnancy, women at 8–10 weeks postpartum and women at 20 weeks postpartum. Values are expressed as a mean percentage of arbitrary units ± standard deviation normalized against total H3 levels.
*p < 0.05; **p < 0.01; ***p < 0.001.
Overall, the levels of histone dimethylation in women with T2DM were different from nondiabetic women and varied over pregnancy and the postpartum period.
Women without diabetes
H3K4 dimethylation (H3K4me2) was 50% lower in pregnant women compared with never-pregnant women (Figure 1A). H3K4me2 was fourfold higher in women 8–10 weeks and 20 weeks postpartum compared with pregnant women (Figure 1B).
H3K9 dimethylation (H3K9me2) was 40% higher in women at 20 weeks postpartum compared with never-pregnant women (Figure 1C). H3K9me2 was higher in women 20 weeks postpartum compared with women at 8–10 weeks postpartum (Figure 1D).
H3K27 dimethylation (H3K27me2) was 75% lower in pregnant women compared with never-pregnant women and was 60% lower in women at 20 weeks postpartum compared with never-pregnant women (Figure 1E). H3K27me2 was higher in women 20 weeks postpartum compared with women at 8–10 weeks postpartum (Figure 1F).
H3K36 dimethylation (H3K36me2) was over twofold higher in women at 20 weeks postpartum compared with never-pregnant women (Figure 1G). H3K36me2 was higher in women 20 weeks postpartum compared with women at 8–10 weeks postpartum (Figure 1H).
H3K79 dimethylation (H3K79me2) was 20% higher in 20 weeks postpartum women compared with never-pregnant women (Figure 1I). H3K79me2 was not different in women at 8–10 weeks and 20 weeks postpartum compared with pregnant women (Figure 1J).
Women with type 2 diabetes
In women with T2DM, H3K4me2 was fivefold higher in women at 20 weeks postpartum (Figure 2A). H3K4me2 was three- to fourfold higher in women 8–10 weeks postpartum and 20 weeks postpartum compared with pregnant women (Figure 2B).
In women with T2DM, H3K9me2 was 50% lower in pregnant women compared with never-pregnant women (Figure 2C). H3K9me2 was not different between women 8–10 weeks and 20 weeks postpartum compared with pregnant women (Figure 2D).
In women with T2DM, H3K27me2 was twice as high in pregnant women and 20 weeks postpartum women compared with never-pregnant women (Figure 2E). No differences were observed in H3K27me2 in 8–10 or 20 weeks postpartum compared with pregnant women (Figure 2F).
In women with T2DM, H3K36me2 was over fourfold higher in 20 weeks postpartum women compared with never-pregnant women (Figure 2G). H3K36me2 in women with diabetes was 50% lower in 8–10 weeks postpartum women than it was in pregnant women and was over tenfold higher in women 20 weeks postpartum compared with pregnant women and 8–10 weeks postpartum women (Figure 2H).
In women with T2DM, H3K79me2 was tenfold higher in never-pregnant women than in pregnant women and was 40% higher in 20 weeks postpartum women compared with never-pregnant women (Figure 2I). H3K79me2 was fivefold higher in 8–10 weeks postpartum women and 20-times higher in 20 weeks postpartum women compared with pregnant women, it was also fourfold higher in women at 20 weeks postpartum compared with 8 weeks postpartum (Figure 2J).
Women with T2DM versus women without diabetes
Methylation of the five histones was compared between nonpregnant women, women with and without diabetes at 30 weeks of pregnancy, 8–10 weeks and 20 weeks postpartum.
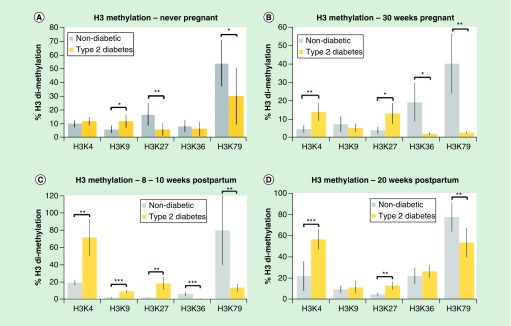
In never-pregnant women, H3K9me2 was twofold greater in women with T2DM compared with women without diabetes while H3K27me2 and H3K79me2 were lower in women with T2DM compared with women without diabetes (by 60 and 40%, respectively) (Figure 3A).
Figure 3. . Histone methylation is different between nondiabetic women and women with type 2 diabetes mellitus.
(A) Comparison between histone methylation levels of nondiabetic women and women with type 2 diabetes mellitus (T2DM) in never-pregnant women. (B) Comparison between histone methylation levels of nondiabetic women and women with T2DM in women at 30 weeks pregnancy. (C) Comparison between histone methylation levels of nondiabetic women and women with T2DM in women 8–10 weeks postpartum. (D) Comparison between histone methylation levels of nondiabetic women and women with T2DM in women at 20 weeks postpartum.
Values are expressed as a mean percentage of arbitrary units ± standard deviation normalized against total H3 levels.
*p < 0.05; **p < 0.01; ***p < 0.001.
In pregnant women at 30 weeks gestation, H3K4me and H3K27me2 were over threefold higher in women with T2DM compared with women without diabetes while H3K36 and H3K79 were ten- to 15-fold higher in women without diabetes compared with women with T2DM (Figure 3B).
In 8–10 weeks postpartum women, H3K4me2, H3K9me2 and H3K27me2 were three- to tenfold higher in women with T2DM compared with nondiabetic and H3K36me2 and H3K79me2 were six- to tenfold higher in women without diabetes compared with women with T2DM (Figure 3C).
In 20 weeks postpartum women, H3K4me2 and H3K27me2 were over twofold higher in women with T2DM compared with normal women and H3K79 was 30% lower in women with T2DM compared with nondiabetic women (Figure 3D).
Discussion
We measured levels of dimethylation of five histones: H3K4, H3K9, H3K27, H3K36 and H3K79 in white blood cells from women in pregnancy and in the early postpartum period in this pilot study. Although the number of participants in each group was relatively low (six–seven participants), significant differences in the levels of histone lysine dimethylation between the different groups of participants were found. These variations in histone dimethylation are consistent with our previous report showing differences in histone dimethylation between women with gestational diabetes mellitus during pregnancy and early postpartum [14]. Our data show that epigenomic changes occur as a consequence of pregnancy, as seen by individual histone dimethylation levels that were different between normal women at 30 weeks of pregnancy and those at 8–10 and 20 weeks postpartum. Epigenetic changes have frequently been associated with the concept of the developmental origins of disease [15] rather than a feature of normal physiological processes. Long-term epigenetic changes were found in the offspring of women with diabetes potentially leading to increased risk of the subsequent generation developing obesity, glucose intolerance and T2DM [16,17]. Long-term epigenetic changes were also associated with chronic vascular dysfunction [18]. Our results provide clear evidence that histone methylation changes are associated with normal physiology.
In contrast to many previous reports, our data show that histone methylation can be transient. Early methylation studies reported that histone methylation was not reversible [19] and generally histone methylation has been considered more stable than histone acetylation and phosphorylation [20,21]. It is now clear that short-term histone modifications are central to memory formation in the brain [22], suggesting that epigenetic changes may be more plastic than previously thought. Short-term changes in methylation of H3K4 have been detected in as little as 30 min in cultured 3T3–L1 preadipocytes exposed to insulin under hyperglycemic conditions [23].
Our data suggest that histone dimethylation levels remain altered for at least until 20 weeks postpartum. At that time, dimethylation levels of four of the five histones (H3K9, H3K27, H3K36 and H3K79) were different from the never pregnant levels, suggesting they had not reverted to prepregnancy levels by 20 weeks postpartum. As we did not have prepregnancy data for the same women whose methylation levels were measured in pregnancy and postpartum, we used a never pregnant population for comparison. Future studies with more time points would indicate the degree of plasticity of the epigenomic changes associated with pregnancy and would establish if over a longer time, the histone methylation profiles returned to the nonpregnant levels.
Given the key roles of hormones in the reproductive cycle, the labile nature of the histone methylation levels suggest that global histone changes may result from the action of hormones during pregnancy and the postpartum period. Such hormones include estrogen, progesterone, human chorionic gonadotropin and adrenocorticotrophic hormone [24].
Estrogen and progesterone levels increase during pregnancy and drop rapidly at the time of delivery [25,26]. Histone methylation is a key regulatory mechanism in estrogen receptor activation. In vitro studies show that demethylation H3K4me2 is associated with activation of the estrogen receptor-α, a transcription factor that mediates estrogen-dependent gene expression [27]. Our finding of reduced methylation of H3K4me2 in healthy pregnancy could be explained by the influence of estrogen on H3K4me2.
The progesterone receptor mediates the effects of progesterone through estrogen signaling pathway. A role for H3K4 methylation in controlling estrogen receptor-activated progesterone effects was demonstrated in cultured MCF7 human breast cancer cells. Demethylation of H3K4me2 occurred following estrogen-controlled progesterone receptor activation [28]. Our data showed a reduction in H3K4me2 in pregnancy, which could be a consequence of increasing levels of progesterone.
Glucocorticoids are essential for fetal development. Maternal adrenocorticotropin and cortisol blood levels increase during pregnancy with adrenocorticotropin levels rising dramatically at birth [29,30]. Glucocorticoids are essential late in pregnancy for mammary gland development and milk protein gene expression [31]. While glucocorticoids caused a decrease in H3K27 trimethylation and increase in H3K9 acetylation in sheep and guinea pig fetuses [32,33], effects on histone lysine dimethylation were not reported.
Analysis of histone methylation in women with T2DM showed differences in H3K4, H3K9, H3K27, H3K36 and H3K79 dimethylation between 30 weeks pregnancy, 8–10 weeks postpartum and 20 weeks postpartum. Women with T2DM had over 30-fold higher levels of H3K36m2 and up to 20-fold higher levels of K3K79me at 20 weeks postpartum relative to pregnancy and 8–10 weeks postpartum. This trend was not seen in nondiabetic women. Methylation of H3K36 and H4K79 are generally associated with active regions of chromatin [21] which may indicate an increase in expression of genes involved in the development of T2DM.
Histone dimethylation levels in women with T2DM were often considerably different from nondiabetic women. For example, H3K79me2 was lower in all groups of women with T2DM compared with nondiabetic women, in particular tenfold lower in women with T2DM in pregnancy compared with nondiabetic women. This may be related to differences in lipid metabolism as methylation of H3K79 is a component of PPAR-γ1-induced adipogenesis [11]. A rodent study demonstrated that high glucose levels induced vascular disease and were associated with a reduction of H3K9me [34], however, our human data did not show this trend. Methylation patterns are tissue-specific [10] and few studies have been carried out on PBMCs, however, one study [13] has shown that hyperglycemia induces changes in H3K4 and H3K9 dimethylation in human blood monocytes. These data are consistent with our study showing increases in H3K4 dimethylation in pregnancy and postpartum women with type 2 diabetes relative to nondiabetic women.
Histone modifications are critical determinants of gene expression. Differences in methylation profiles between nondiabetic and diabetic women during pregnancy and postpartum indicate different gene expression patterns that underlie the pathological features of T2DM. Women with T2DM may respond differently from women without diabetes to hormones during pregnancy. Abnormal hormonal responses in women with T2DM may have downstream effects that may contribute to insulin resistance. Evidence for this is that in animal experiments, altered H3K4 methylation causes insulin resistance, seen where knockout of the H3K4 methyl transferase gene, Mll2, caused hyperglycemia and hyperinsulinemia in fasting animals and impaired glucose tolerance in response to a glucose load [35]. Although our data showed increased methylation of H3K4 in women with T2DM compared with women without diabetes, it is clear that changes in histone methylation can influence cellular pathways involved in insulin regulation. Species differences may account for differences in results.
Higher blood glucose levels in women with T2DM might account for the differences in histone methylation levels between diabetic and nondiabetic women. This concept is supported by studies using cultured 3T3–1 preadipocytes, where glucose levels affected H3K4 methylation following treatment of cells with insulin. In the presence of high glucose, H3K4 methylation was greater compared with low glucose [23]. In our studies the higher levels of H3K4me found in women with T2DM relative to nondiabetic women could be related to their high blood glucose levels.
Over 100 genes, some of which regulate insulin secretion, are differentially expressed in human pancreatic islets, related to the presence of diabetes [36]. Epigenetic analyses, such as that involving histone methylation have the potential to lead to the discovery of the cellular pathways and downstream genes that contribute to the pathophysiology of T2DM as well as providing potential targets for pharmacological interventions.
This pilot study was limited by the small number of participants. With a larger study we could obtain data on other important parameters of the participants that could influence histone methylation, such as gestational age, preeclampsia, infection, birth defects and breast feeding, which were not obtained as part of this pilot study. It is possible that participant age also has an important role on H3 methylation levels in addition to T2DM.
Epigenomic fingerprint
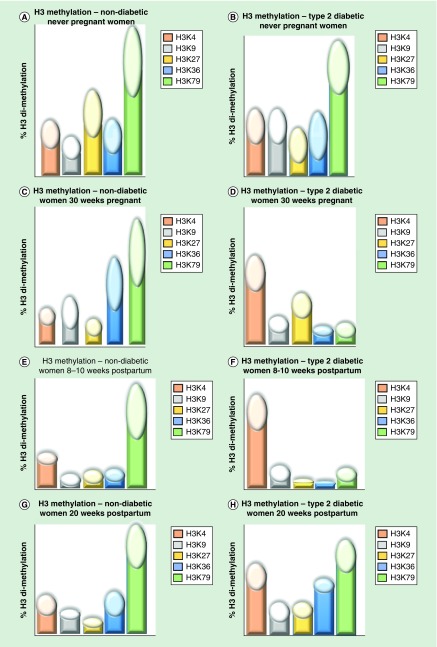
Although this study was short-term, substantial differences in global histone dimethylation were found between women with T2DM and nondiabetic women. These patterns can be represented an epigenetic fingerprint (Figure 4). Such fingerprints may correlate with disease severity, with potential as a tool to indicate different types of metabolic heterogeneity in women with T2DM. Epigenomic fingerprints might also have more global applications for prevention or treatment of T2DM in nonpregnant individuals, by indicating which pharmacological approaches are most promising [37], a form of ‘precision medicine’. For example, molecules influencing the activity of enzymes controlling epigenetic events including histone modifications are being trialed for cancer treatment [38] and may have applications to other diseases including diabetes, in which histone modifications occur.
Figure 4. . Epigenetic fingerprint that shows dimethylation levels for five histones in never pregnant, 30 weeks pregnant, 8–10 weeks postpartum and 20 weeks postpartum women with and without type 2 diabetes mellitus.
The dimethylation levels are shown as fingers and the fingernail lengths represent the standard deviation. (A) Dimethylation levels of H3 in nondiabetic never-pregnant women. (B) Dimethylation levels of H3 in type 2 diabetic never-pregnant women. (C) Dimethylation levels of H3 in nondiabetic women at 30 weeks pregnancy. (D) Dimethylation levels of H3 in type 2 diabetic women at 30 weeks pregnancy. (E) Dimethylation levels of H3 in nondiabetic women compared with women at 30 weeks pregnancy; dimethylation levels in 8–10 weeks postpartum women at 8 weeks postpartum. (F) Dimethylation levels of H3 in nondiabetic women at 8 weeks postpartum. (G) Dimethylation levels of H3 in nonpregnant women at 20 weeks postpartum. (H) Dimethylation levels of H3 in type 2 diabetic women at 20 weeks postpartum.
Conclusion
This study provides novel longitudinal data that epigenomic changes occur with normal pregnancy, most likely a consequence of hormonal changes, not yet fully elucidated. The changes in histone dimethylation levels observed in pregnancy and the early postpartum period were transient up to 20 weeks postpartum. Epigenetic fingerprints depicting methylation changes in five histones showed that women with type 2 diabetes had distinctly different epigenomic profiles relative to nondiabetic women. These changes could have important consequences for pregnancy outcomes and long-term metabolic changes as well as providing opportunities for personalized medicine. Further studies will determine whether the changes in histone dimethylation caused by pregnancy are maintained beyond 20 weeks postpartum and their role in pregnancy health and disease.
Summary points.
The epigenetic effects of endocrine disrupting agents suggest that hormones can mediate epigenetic changes.
Pregnancy is a critical period of hormonal changes, but little is known of epigenomic changes associated with the reproductive cycle.
To investigate epigenomic changes in pregnancy, we measured levels of lysine dimethylation in five histones: H3K4, H3K9, H3K27, H3K36 and H3K79 in white blood cells of never pregnant, 30 weeks pregnant and early postpartum women.
We also measured histone dimethylation in women with and without type 2 diabetes mellitus in pregnancy and early postpartum.
Dimethylation levels of all five histones varied across pregnancy and the postpartum period of normal women.
Dimethylation levels of all five histones were different between women with and without type 2 diabetes.
Histone methylation changes are transient in pregnancy and early postpartum.
Different epigenomic profiles in women with type 2 diabetes mellitus women may correlate with hormonal responses, leading to high risk pregnancy outcomes.
Footnotes
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Khan D, Ahmed SA. Epigenetic regulation of non-lymphoid cells by bisphenol A, a model endocrine disrupter: potential implications for immunoregulation. Front. Endocrinol. 2015;6:91. doi: 10.3389/fendo.2015.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ho SM, Cheong A, Adgent MA, et al. Environmental factors, epigenetics, and developmental origin of reproductive disorders. Reprod. Toxicol. 2016;68:85–104. doi: 10.1016/j.reprotox.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trevino LS, Wang Q, Walker CL. Hypothesis: activation of rapid signaling by environmental estrogens and epigenetic reprogramming in breast cancer. Reprod. Toxicol. 2015;54:136–140. doi: 10.1016/j.reprotox.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y, Reddy MA, Miao F, et al. Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-kappaB-dependent inflammatory genes. Relevance to diabetes and inflammation. J. Biol. Chem. 2008;283(39):26771–26781. doi: 10.1074/jbc.M802800200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L, Xu S, Lee JE, et al. Histone H3K9 methyltransferase G9a represses PPARgamma expression and adipogenesis. EMBO J. 2013;32(1):45–59. doi: 10.1038/emboj.2012.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park JH, Stoffers DA, Nicholls RD, Simmons RA. Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1. J. Clin. Invest. 2008;118(6):2316–2324. doi: 10.1172/JCI33655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bulger DA, Conley J, Conner SH, Majumdar G, Solomon SS. Role of PTEN in TNFalpha induced insulin resistance. Biochem. Biophysical. Res. Com. 2015;461(3):533–536. doi: 10.1016/j.bbrc.2015.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raychaudhuri N, Raychaudhuri S, Thamotharan M, Devaskar SU. Histone code modifications repress glucose transporter 4 expression in the intrauterine growth-restricted offspring. J. Biol. Chem. 2008;283(20):13611–13626. doi: 10.1074/jbc.M800128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muka T, Nano J, Voortman T, et al. The role of global and regional DNA methylation and histone modifications in glycemic traits and type 2 diabetes: a systematic review. Nutr. Metab. Cardiovasc. Dis. 2016;26(7):553–566. doi: 10.1016/j.numecd.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 10.Miao F, Wu X, Zhang L, Riggs AD, Natarajan R. Histone methylation patterns are cell-type specific in human monocytes and lymphocytes and well maintained at core genes. J. Immunol. 2008;180(4):2264–2269. doi: 10.4049/jimmunol.180.4.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Analysis of histone methylation in white blood cells.
- 11.Ge K. Epigenetic regulation of adipogenesis by histone methylation. Biochim. Biophys. Acta. 2012;1819(7):727–732. doi: 10.1016/j.bbagrm.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper ME, El-Osta A. Epigenetics: mechanisms and implications for diabetic complications. Circ. Res. 2010;107(12):1403–1413. doi: 10.1161/CIRCRESAHA.110.223552. [DOI] [PubMed] [Google Scholar]
- 13.Miao F, Wu X, Zhang L, Yuan YC, Riggs AD, Natarajan R. Genome-wide analysis of histone lysine methylation variations caused by diabetic conditions in human monocytes. J. Biol. Chem. 2007;282(18):13854–13863. doi: 10.1074/jbc.M609446200. [DOI] [PubMed] [Google Scholar]; •• Lysine methylation is linked to diabetes.
- 14.Michalczyk AA, Dunbar JA, Janus ED, et al. Epigenetic markers to predict conversion from gestational diabetes to type 2 diabetes. J. Clin. Endocrinol. Metab. 2016;101(6):2396–2404. doi: 10.1210/jc.2015-4206. [DOI] [PubMed] [Google Scholar]; •• Differences in epigenetic profiles between women with gestational diabetes and type 2 diabetes.
- 15.Ozanne SE, Constancia M. Mechanisms of disease: the developmental origins of disease and the role of the epigenotype. Nat. Clin. Pract. Endocrinol. Metab. 2007;3(7):539–546. doi: 10.1038/ncpendmet0531. [DOI] [PubMed] [Google Scholar]
- 16.Pinney SE, Simmons RA. Metabolic programming, epigenetics, and gestational diabetes mellitus. Curr. Diab. Rep. 2012;12(1):67–74. doi: 10.1007/s11892-011-0248-1. [DOI] [PubMed] [Google Scholar]
- 17.Holbrook JD. Catching diabetes. Epigenomics. 2016;8(9):1173–1177. doi: 10.2217/epi-2016-0079. [DOI] [PubMed] [Google Scholar]
- 18.Reddy MA, Natarajan R. Epigenetic mechanisms in diabetic vascular complications. Cardiovasc. Res. 2011;90(3):421–429. doi: 10.1093/cvr/cvr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bannister AJ, Schneider R, Kouzarides T. Histone methylation: dynamic or static? Cell. 2002;109(7):801–806. doi: 10.1016/s0092-8674(02)00798-5. [DOI] [PubMed] [Google Scholar]
- 20.Miao F, Natarajan R. Mapping global histone methylation patterns in the coding regions of human genes. Mol. Cell Biol. 2005;25(11):4650–4661. doi: 10.1128/MCB.25.11.4650-4661.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell Biol. 2007;8(4):307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 22.Halder R, Hennion M, Vidal RO, et al. DNA methylation changes in plasticity genes accompany the formation and maintenance of memory. Nat. Neurosci. 2016;19(1):102–110. doi: 10.1038/nn.4194. [DOI] [PubMed] [Google Scholar]
- 23.Gupta J, Tikoo K. Involvement of insulin-induced reversible chromatin remodeling in altering the expression of oxidative stress-responsive genes under hyperglycemia in 3T3-L1 preadipocytes. Gene. 2012;504(2):181–191. doi: 10.1016/j.gene.2012.05.027. [DOI] [PubMed] [Google Scholar]; • Methylation changes can be short-term.
- 24.Pepe G, Albrecht E. Steroid endocrinology of pregnancy. In: Arulkumaran S, editor. The Global Library of Women's Medicine. David GT Bloomer and Paula Bloomer; London, UK: 2008. pp. 1–33. [Google Scholar]
- 25.Levitz M, Young BK. Estrogens in pregnancy. Vitam. Horm. 1977;35:109–147. doi: 10.1016/s0083-6729(08)60522-1. [DOI] [PubMed] [Google Scholar]
- 26.Tulchinsky D, Hobel CJ, Yeager E, Marshall JR. Plasma estrone, estradiol, estriol, progesterone, and 17-hydroxyprogesterone in human pregnancy. I. Normal pregnancy. Am. J. Obstet. Gynecol. 1972;112(8):1095–1100. doi: 10.1016/0002-9378(72)90185-8. [DOI] [PubMed] [Google Scholar]
- 27.Mann M, Cortez V, Vadlamudi RK. Epigenetics of estrogen receptor signaling: role in hormonal cancer progression and therapy. Cancers. 2011;3(3):1691–1707. doi: 10.3390/cancers3021691. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Estrogen receptor signaling contributes to epigenetic changes.
- 28.Stratmann A, Haendler B. The histone demethylase JARID1A regulates progesterone receptor expression. FEBS J. 2011;278(9):1458–1469. doi: 10.1111/j.1742-4658.2011.08058.x. [DOI] [PubMed] [Google Scholar]
- 29.Carr BR, Parker CR, Jr, Madden JD, MacDonald PC, Porter JC. Maternal plasma adrenocorticotropin and cortisol relationships throughout human pregnancy. Am. J. Obstet. Gynecol. 1981;139(4):416–422. doi: 10.1016/0002-9378(81)90318-5. [DOI] [PubMed] [Google Scholar]
- 30.Lindsay JR, Nieman LK. The hypothalamic-pituitary-adrenal axis in pregnancy: challenges in disease detection and treatment. Endocr. Rev. 2005;26(6):775–799. doi: 10.1210/er.2004-0025. [DOI] [PubMed] [Google Scholar]
- 31.Chida D, Miyoshi K, Sato T, Yoda T, Kikusui T, Iwakura Y. The role of glucocorticoids in pregnancy, parturition, lactation, and nurturing in melanocortin receptor 2-deficient mice. J. Endocrinol. 2011;152(4):1652–1660. doi: 10.1210/en.2010-0935. [DOI] [PubMed] [Google Scholar]
- 32.Begum G, Davies A, Stevens A, et al. Maternal undernutrition programs tissue-specific epigenetic changes in the glucocorticoid receptor in adult offspring. J. Endocrinol. 2013;154(12):4560–4569. doi: 10.1210/en.2013-1693. [DOI] [PubMed] [Google Scholar]
- 33.Crudo A, Suderman M, Moisiadis VG, et al. Glucocorticoid programming of the fetal male hippocampal epigenome. J. Endocrinol. 2013;154(3):1168–1180. doi: 10.1210/en.2012-1980. [DOI] [PubMed] [Google Scholar]
- 34.Chen J, Zhang J, Yang J, et al. Histone demethylase KDM3a, a novel regulator of vascular smooth muscle cells, controls vascular neointimal hyperplasia in diabetic rats. Atherosclerosis. 2017;257:152–163. doi: 10.1016/j.atherosclerosis.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 35.Goldsworthy M, Absalom NL, Schroter D, et al. Mutations in Mll2, an H3K4 methyltransferase, result in insulin resistance and impaired glucose tolerance in mice. PLoS ONE. 2013;8(6):e61870. doi: 10.1371/journal.pone.0061870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dayeh T, Volkov P, Salo S, et al. Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. PLoS Genet. 2014;10(3):e1004160. doi: 10.1371/journal.pgen.1004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raciti GA, Nigro C, Longo M, et al. Personalized medicine and type 2 diabetes: lesson from epigenetics. Epigenomics. 2014;6(2):229–238. doi: 10.2217/epi.14.10. [DOI] [PubMed] [Google Scholar]
- 38.McGrath J, Trojer P. Targeting histone lysine methylation in cancer. Pharmacol. Ther. 2015;150:1–22. doi: 10.1016/j.pharmthera.2015.01.002. [DOI] [PubMed] [Google Scholar]