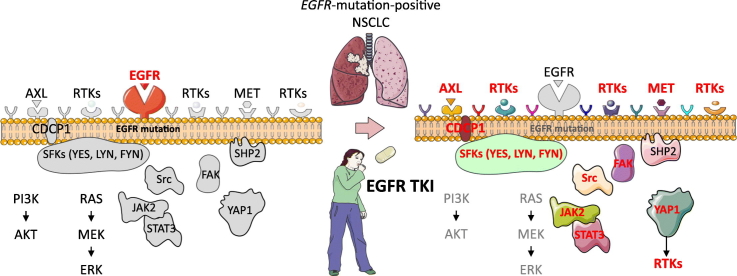
Abstract
Epidermal growth factor receptor (EGFR)-mutation-positive non-small cell lung cancer (NSCLC) is incurable, despite high rates of response to EGFR tyrosine kinase inhibitors (TKIs). We investigated receptor tyrosine kinases (RTKs), Src family kinases and focal adhesion kinase (FAK) as genetic modifiers of innate resistance in EGFR-mutation-positive NSCLC. We performed gene expression analysis in two cohorts (Cohort 1 and Cohort 2) of EGFR-mutation-positive NSCLC patients treated with EGFR TKI. We evaluated the efficacy of gefitinib or osimertinib with the Src/FAK/Janus kinase 2 (JAK2) inhibitor, TPX0005 in vitro and in vivo. In Cohort 1, CUB domain-containing protein-1 (CDCP1) was an independent negative prognostic factor for progression-free survival (hazard ratio of 1.79, p = 0.0407) and overall survival (hazard ratio of 2.23, p = 0.0192). A two-gene model based on AXL and CDCP1 expression was strongly associated with the clinical outcome to EGFR TKIs, in both cohorts of patients. Our preclinical experiments revealed that several RTKs and non-RTKs, were up-regulated at baseline or after treatment with gefitinib or osimertinib. TPX-0005 plus EGFR TKI suppressed expression and activation of RTKs and downstream signaling intermediates. Co-expression of CDCP1 and AXL is often observed in EGFR-mutation-positive tumors, limiting the efficacy of EGFR TKIs. Co-treatment with EGFR TKI and TPX-0005 warrants testing.
Keywords: Lung cancer, EGFR, Resistance, AXL, CDCP1, Combination therapies
Graphical Abstract
Highlights
-
•
AXL and CDCP1 are co-expressed in treatment-naïve EGFR-mutation-positive NSCLC patients.
-
•
AXL and CDCP1 are related to shorter progression-free survival with EGFR inhibitors and shorter overall survival.
-
•
Src family kinases and YAP1 are regulatory nodes for AXL and CDCP1 expression.
-
•
The combination of EGFR TKI with TPX-0005 is synergistic in EGFR-mutation-positive lung tumors in culture and in vivo.
We explore the molecular changes that occur after the application of an EGFR inhibitor in EGFR-mutation positive tumors. The tumors do not acquire secondary drivers to overcome a primary driver but, counter-regulatory nodes observable before treatment, are immediately made apparent by pathway-specific intervention. The expression of the receptor tyrosine kinase AXL and the transmembrane protein CDCP1 in baseline samples of EGFR-mutation positive NSCLC patients can provide us with information on the treatment outcome. The upfront combination of an EGFR inhibitor with a multikinase inhibitor, that controls the regulatory nodes for RTKs activation, is a therapeutic approach that deserves to be further explored.
1. Introduction
Epidermal growth factor receptor (EGFR) gene mutations were identified following the sequencing of 47 out of 58 receptor tyrosine kinase (RTKs) in tumors of non-small cell lung cancer (NSCLC) patients (Paez et al., 2004). EGFR mutations, mostly small in-frame exon 19 deletions and amino acid substitutions within exon 21, like Leu858Arg, are sensitive to EGFR tyrosine kinase inhibitors (TKIs) gefitinib (Paez et al., 2004; Lynch et al., 2004) and erlotinib (Rosell et al., 2009). Despite the higher response rate and longer progression-free survival, there is no survival benefit with erlotinib in patients with EGFR mutations (Tsao et al., 2005). EGFR-mutation-positive lung cancer cells undergo apoptosis following EGFR knockdown, or pharmacological inhibition of AKT and signal transducer and activator of transcription 3 (STAT3), while they are relatively resistant when treated with chemotherapy (Sordella et al., 2004). Despite these observations, studies have focused on comparing EGFR TKIs versus chemotherapy for EGFR-mutation-positive NSCLC. Phase 3 trials have compared gefitinib or erlotinib with chemotherapy, confirming the significantly longer progression-free survival with EGFR TKIs (Mok et al., 2009; Rosell et al., 2012). The second-generation irreversible EGFR TKIs, afatinib and dacomitinib, have been found to be superior to gefitinib (Park et al., 2016; Wu et al., 2017). Osimertinib, a third-generation irreversible EGFR-TKI that inhibits also the EGFR Thr790Met point resistant mutation, yields significantly longer progression-free survival in comparison with standard EGFR TKIs as first-line treatment (Soria et al., 2018). Complete responses are uncommon, regardless of the EGFR TKI, and patients ultimately succumb to the disease.
EGFR-mutation-positive NSCLC expresses multiple RTKs and/or non-RTKs, mainly focal adhesion kinase (FAK), encoded by the protein tyrosine kinase 2 (PTK2) gene and Src family kinases (SFK) (Rikova et al., 2007). EGFR inhibition increases the phosphorylation of the co-existing non-targeted RTKs, maintaining redundant downstream signaling (Stommel et al., 2007). Inter-receptor crosstalk of MET, integrin beta-4, erythropoietin-producing hepatocellular (EphA2), CUB domain-containing protein-1 (CDCP1) and AXL was described in EGFR-mutation-positive NSCLC (Gusenbauer et al., 2013). Numerous co-expressed RTKs and non-RTKs were observed in the EGFR-mutation-positive PC9 and PC9 gefitinib-resistant cells. The combination of afatinib or osimertinib with the Src inhibitor dasatinib caused tumor regression in EGFR-mutation-positive cells (Yoshida et al., 2014; Ichihara et al., 2017). SFK and FAK induce resistance to afatinib, erlotinib and osimertinib (Murakami et al., 2017). Moreover, RTK-driven cancers depend on Src-homology 2 domain-containing phosphatase 2 (SHP2), encoded by the protein tyrosine phosphatase, non-receptor type 11 (PTPN11) gene and SFKs for survival (Sausgruber et al., 2015). We reported that either gefitinib or osimertinib activates STAT3 and Src-YES-associated protein 1 (YAP1) in EGFR-mutation-positive lung cancer cells (Chaib et al., 2017). STAT3, when activated, translocates into the nucleus and promotes gene transcription (Gao et al., 2007). Combined EGFR, STAT3 and Src inhibition abrogated tumor growth more efficiently than single EGFR inhibition both in culture and in vivo (Chaib et al., 2017). AXL was identified as a downstream target of YAP1 in hepatocellular carcinoma (Xu et al., 2011). We noted that in EGFR TKI resistant cells, due to either EGFR Thr790Met point resistant mutation or AXL overexpression (Zhang et al., 2012), afatinib combined with a STAT3 blocker induced tumor growth regression (Codony-Servat et al., 2017).
Here, we show that the co-activation of RTKs and non-RTKs is a common trait in treatment-naïve EGFR-mutation-positive NSCLC cells and patients, with AXL and CDCP1 being commonly over-expressed. The combination of gefitinib or osimertinib with the Src/FAK/Janus kinase 2 (JAK2) inhibitor TPX-0005 (Zhai et al., 2016) abrogates STAT3, YAP1 and SFKs activation and down-regulates AXL and CDCP1 expression.
2. Methods
2.1. Study Oversight and Sample Collection
Clinical data were assessed in accordance with the protocol approved by the institutional review board of Germans Trias i Pujol Hospital, Badalona and de-identified for patient confidentiality. The first and last author wrote the first version of the manuscript. All authors contributed to subsequent drafts and vouch for the accuracy of the data. We studied pre-treatment tumors from 2 cohorts of EGFR-mutation-positive NSCLC patients. Cohort 1 was from hospitals in Spain, France, Italy and Colombia (Chaib et al., 2017). Cohort 2 was from the Clinica del country in Bogota, Colombia. The baseline characteristics of the patients are found in Table 1.
Table 1.
Patient characteristics in Cohort 1 and Cohort 2.
| Clinical characteristics | Cohort 1 (no = 64) | Cohort 2 (no = 53) | p Value test |
|---|---|---|---|
| Sex—no. (%) | |||
| Male | 22 (34.4) | 13 (24.5) | Chi-Square: 0.2469 |
| Female | 42 (65.6) | 40 (75.5) | |
| Age—yr | |||
| Median | 67 | 63 | Wilcoxon: 0.0494 |
| Range | 35–89 | 26–87 | |
| ECOGa performance status — no. (%) | |||
| 0 | 15 (23.4) | 10 (18.9) | Fisher: 0.2204 |
| 1 | 49 (76.6) | 43 (81.1) | |
| Smoking status — no. (%) | |||
| Never smoked | 42 (65.6) | 42 (79.2) | Fisher: <0.0001 |
| Former smoker | 17 (26.6) | 1 (1.9) | |
| Current smoker | 5 (7.8) | 0 (0.0) | |
| Unknown | 0 (0.0) | 10 (18.9) | |
| Disease stage — no. (%) | |||
| IIIB | 12 (18.7) | 0 (0.0) | |
| IV | 52 (81.3) | 53 (100.0) | |
| Brain metastasis — no. (%) | |||
| No | 43 (67.2) | 26 (49.1) | Chi-Square: 0.6585 |
| Yes | 21 (32.8) | 27 (50.9) | |
| Bone metastasis— no. (%) | |||
| No | 40 (62.5) | 31 (58.5) | Chi-Square: 0.0472 |
| Yes | 24 (37.5) | 22 (41.5) | |
| Type of EGFR mutation— no. (%) | |||
| Exon 19 deletion | 44 (68.8) | 26 (49.1) | Fisher: 0.0551 |
| Leu858Arg | 18 (28.1) | 21 (39.6) | |
| Otherb | 2 (3.1) | 6 (11.3) | |
| Type of EGFR TKI— no. (%) | |||
| Erlotinib | 37 (57.8) | 46 (86.8) | |
| Gefitinib | 25 (39.1) | 7 (13.2) | |
| Afatinib | 2 (3.1) | 0 (0.0) | |
| Progression-free survival | |||
| Median—mo (CI 95%) | 14.1 (8.8–16.3) | 13.0 (10.7–17.8) | |
| Overall survival | |||
| Median—mo (CI 95%) | 26.7 (17.9–37.1) | 27.1 (22.8–29.9) | |
| Best response— no. (%) | |||
| Complete response | 3 (4.7) | 12 (22.6) | |
| Partial response | 39 (60.1) | 20 (37.7) | |
| Stable disease | 16 (25.8) | 18 (34.0) | |
| Progressive disease | 6 (9.4) | 3 (5.7) |
ECOG, Eastern Cooperative Oncology Group.
Other, Cohort 1 Leu861Glu, Gly719X; Cohort 2 L861Q (no = 2), exon 20 insertion (no = 3), S768I (no = 1); no, number; mo, months.
2.2. Chemicals and Reagents
Gefitinib was purchased from Tocris Bioscience Company (Bristol, UK). Afatinib, dacomitinib, osimertinib and dasatinib were purchased from Selleck Chemicals (Houston, TX, U.S). TP Therapeutics Inc. (San Diego, CA, U.S) has developed the Src/FAK/JAK2 inhibitor TPX-0005 (Table 2). The compound was kindly provided to us by TP Therapeutics, Inc. under a material transfer agreement (MTA). TPX-0005 is a multikinase inhibitor (Zhai et al., 2016; Cui et al., 2017) with a rigid three-dimensional macrocyclic structure, currently in a phase I/II clinical trial (NCT03093116). Drugs were prepared in dimethyl sulfoxide (DMSO) at a concentration of 10-100 mmol/l stock solutions and stored at −20 °C. Further dilutions were made in culture medium to final concentration before use. Primary antibodies and secondary antibodies used are in Supplementary Table 1.
Table 2.
In vitro kinase activity of TPX-0005.
| Target | IC50 (nM) at 10 μM ATP | Target | IC50 (nM) at 10 μM ATP | Target | IC50 (nM) at 10 μM ATP |
|---|---|---|---|---|---|
| TRKB | 0.05 | SNARK | 13.0 | JAK3 | 50 |
| ROS1 | 0.07 | HCK | 16.4 | EPHA8 | 50.2 |
| TRKC | 0.1 | IRR | 18.1 | IGFR | 111 |
| TRKA | 0.83 | LCK | 18.6 | PLK4 | 126 |
| ALK | 1.04 | JAK1 | 19 | AXL | 149 |
| JAK2 | 1.04 | TYK2 | 21.6 | MARK3 | 512 |
| FYN | 1.05 | LTK | 21.8 | ||
| LYN | 1.66 | DDR2 | 23 | ||
| YES | 2.15 | BTK | 23.7 | ||
| FGR | 3.05 | TNK2 | 24.1 | ||
| TXK | 3.17 | EPHA1 | 25.0 | ||
| ARK5 | 4.46 | BLK | 32.3 | ||
| SRC | 5.3 | GRK7 | 35.2 | ||
| DDR1 | 5.7 | PYK2 | 39.9 | ||
| FAK | 6.96 | RET | 47.1 |
2.3. Cell Viability Assay
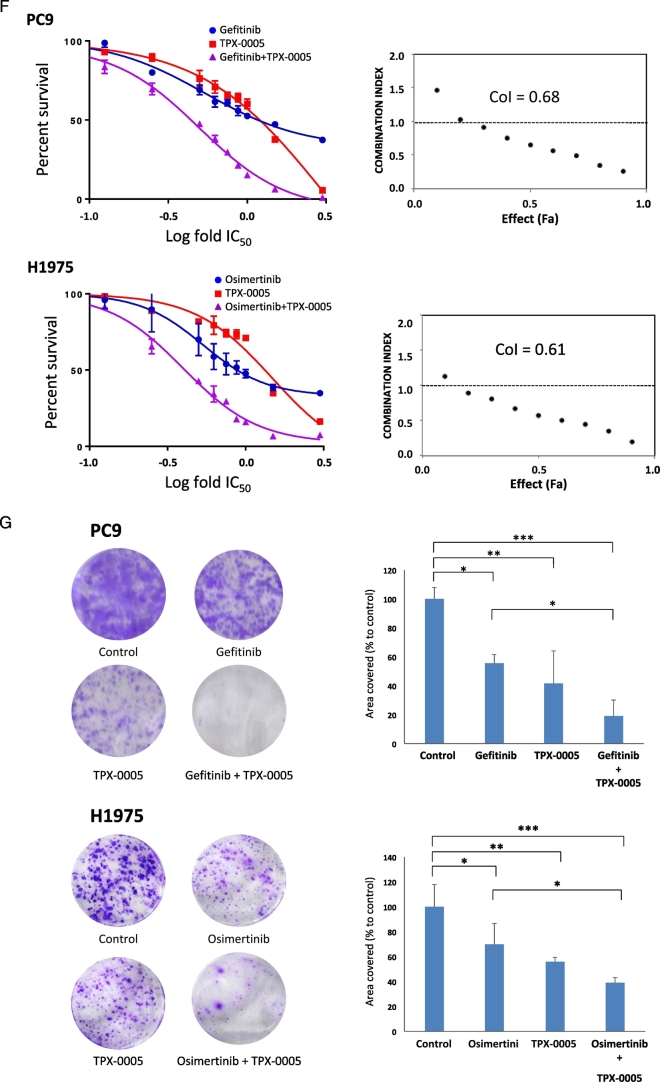
PC9 (exon 19 deletion) cells were provided by F. Hoffmann-La Roche Ltd. 11–18 (Leu858Arg) cells were provided by Dr. Mayumi Ono. H1975 (Leu858Arg and Thr790Met), HCC4006 (exon 19 deletion) and HCC827 (exon 19 deletion) cells were purchased from the American Type Culture Collection (ATCC). Cells were seeded on 96-well plates at the following densities: 2 × 103, 3 × 103 and 4 × 103 cells/well and incubated for 24 h, as previously described (Chaib et al., 2017). Cells were treated with serial dilutions of the drugs administrated at indicated doses. After 72 h of incubation, 0.5 mg/ml of MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) reagent (Sigma-Aldrich, St. Louis, MO, U.S) was added to the medium in the wells for 2 h at 37 °C and formazan crystals in viable cells were solubilized with 100 μl DMSO and spectrophotometrically quantified using a microplate reader (Varioskan Flash; Thermo Fisher Scientific, Waltham, MA, U.S) at 550 nm of absorbance. Fractional survival was then calculated as percentage to control cells. Data of combined drug effects were subsequently analyzed by the Chou and Talalay method (Chou, 2010; Narayan et al., 2017). Combination Index (CoI) values <1, =1 and >1 indicated synergism, additive effect and antagonism, respectively.
2.4. Colony Formation Assay
Cells were plated in six-well plates at 1000 cells/well in Roswell Park Memorial Institute (RPMI) 1640 Medium and 10% fetal bovine serum (FBS), as previously described (Chaib et al., 2017). The cells were cultured for 24 h and the media were then replaced with RPMI, 1% FBS with or without inhibitors. After 72 h the media were removed and replaced with fresh media without inhibitors for a total of 10 days. At the end of the experiment, the media were removed and the cells were washed with phosphate-buffered saline (PBS). The colonies were fixed and stained simultaneously with 0.5% crystal violet in 10% of ethanol for 15 min. The stain was aspirated and the wells were washed with deionized water until the background was clear. The wells were then photographed. As a semiquantitative measurement, the crystal violet was extracted from the colonies with Triton X-100 0.5% solution overnight and the absorbance was measured at 570 nm.
2.5. Western Blotting
Cells were washed with cold PBS and re-suspended in ice-cold radio-immunoprecipitation assay buffer (50 mM Tris- hydrochloric acid in pH 7.4, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 150 mM sodium chloride, 1 mM ethylenediaminetetraacetic acid, 1 mM sodium vanadate and 50 mM sodium fluoride) containing protease inhibitor mixture. Following cell lysis by sonication and centrifugation at 14000 rpm (revolutions per minute of rotor) for 15 min at 4 °C, the resulting supernatant was collected as the total cell lysate. Briefly, the lysates containing 30 μg proteins were electrophoresed on 10% SDS-polyacrylamide gel electrophoresis (Life Technologies, Carlsbad, CA, U.S) and transferred to polyvinylidene difluoride membranes (Bio-Rad laboratories Inc., Hercules, CA, U.S). Membranes were blocked in Odyssey blocking buffer (Li-Cor Biosciences, Lincoln, NE, U.S). All target proteins were immunoblotted with appropriate primary and either IRDye-conjugated or horseradish peroxidase (HPR)-conjugated secondary antibodies. Fluorescent bands (IRDye-conjugated) were detected with Odyssey CLx Imager (Li-Cor Biosciences), whereas chemiluminescent (HRP-conjugated) were detected in a ChemiDoc MP Imaging System (Bio-Rad laboratories Inc.). β-actin was used as an internal control to confirm equal gel loading.
2.6. Phospho-receptor Tyrosine Kinase and Phosphokinase Arrays
For phosphoproteomics analysis, 0.7 × 106 cells were seeded in T25 flasks (Starstedt, Newton, U.S). The next day, PC9 cells were treated with 50 nM gefitinib, 1 μM TPX-0005 or a combination of both. H1975 cells were treated with 10 nM osimertinib, 1 μM TPX-0005 or a combination of both. Untreated cells received an equivalent dose of vehicle (DMSO). After 24 h cells were lysed and hybridized in Proteome Profiler Human Phospho-RTK (ARY001B) or Phospho-Kinase (ARY003B) array membranes (R&D Systems Inc., Minneapolis, MN, U.S). Lysis, hybridization and development of the membranes were performed following manufacturer's instructions. Images were taken in a Chemidoc MP Imaging System and processed using Image Lab software (both from Bio-Rad laboratories Inc.). Spot intensity was determined using Quantity One software (Bio-Rad laboratories Inc.). Spot intensities were normalized to control (untreated) levels and a heatmap was generated using Microsoft Excel. The whole experiment was repeated twice with similar results.
2.7. Small Interfering RNA Transfections
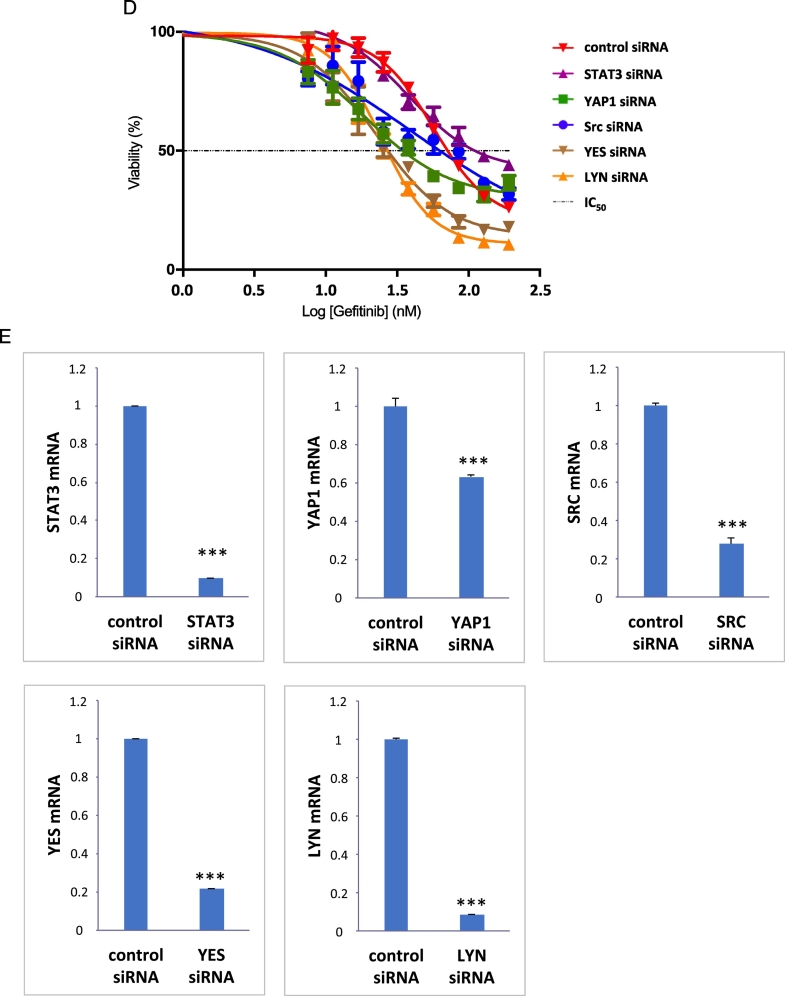
Flexi-tube small interfering RNAs (siRNA) (Qiagen Inc., Valencia, CA, U.S) were transfected using Lipofectamine 3000 (Thermo Fisher Scientific) following manufacturer instructions. The same transfection mix was used to transfect cells for viability assays and for western blotting to check knockdown. In parallel, PC9 and H1975 cells were transfected with Alexa Fluor 488-labeled Allstars siRNA (Qiagen Inc.) to verify transfection efficiency, which was higher than 95% for both cell lines. For Western blot, cells were lysed 48 h after transfection. For viability assays, cells were treated the day after transfection and drugs were maintained for 72 h before the MTT assay. The experiment was repeated three times with different set of siRNAs. siRNAs used were: SI03650325 for non-targeting control; SI02662898, SI02662338 and SI00048377 for STAT3; SI02664151, SI02223928 and SI02223921 for Src, SI02662954, SI04438651 and SI04438644 for YAP1, SI02223942, SI02223935 and SI00302218 for YES and SI00605577, SI00605570 and SI04713513 for LYN.
2.8. Immunocytochemistry and Imaging Flow Cytometry
For immunocytochemistry, 2 × 104 cells were seeded on coverslips placed on 24-well plates and grown overnight. The next day cells were treated with IC50 values of the indicated drugs. 24 h later, coverslips were washed with PBS, fixed with 4% paraformaldehyde (PFA) for 15 min and treated with cold methanol. After a wash in immunocytochemistry wash buffer (PBS 1×, 0.1% Troton-X-100, 0.05% Sodium Azide, 0.5% bovine serum albumine), coverslips were blocked in immunocytochemistry wash buffer containing 10% FBS for 20 min at room temperature and incubated overnight with primary antibodies against the indicated epitopes at 4 °C. After three wash steps, Alexa Fluor 488-coupled secondary antibodies (Thermo Fisher Scientific) were incubated at room temperature for 2 h. Coverslips were washed three times with immunocytochemistry wash buffer before 4′,6-diamidino-2-phenylindole (DAPI) counterstain and washed once more with H2O before being mounted with Vectashield (Vector Laboratories, Burlingame, CA, U.S) on glass slides. For imaging flow cytometry analysis, 1 × 106 cells were seeded on 21 cm2 cell-repellent plates. The next day, cells were treated with the indicated drugs with concentrations equivalent to their IC50 values for 24 h, stained with the Live/Dead Fixable Far Red Dead Cell Stain Kit (Thermo Fisher Scientific) to label dead cells and immunostained as indicated above. A minimum of 105 events were collected at 4000× in an ImagestreamX imaging flow cytometer (EMD Millipore, Billerica, MA, U.S) using Inspire software (EMD Millipore). Data was compensated and processed to render focused images of single cells and intensity and localization of the Alexa-488 signal was analyzed using IDEAS software (EMD Millipore).
2.9. Immunohistochemistry
All the immunohistochemistry experiments were performed on a Ventana Benchmark Ultra automated staining (Ventana Medical Systems Inc., Tuscon, AZ, U.S). Immunohistochemistry of the tumor samples was performed on 4 μm sections. The settings included pre-treatment with cell conditioner 1 (CC1) buffer for 56 min, incubation with a phosphorylated STAT3 (1:200) antibody for 40 min, and pre-treatment with CC1 buffer for 76 min, incubation with a phosphorylated YAP1 (1:200) antibody for 28 min. The detection was performed with 3,3-diaminobenzidine (DAB) detection kit (Ventana Medical Systems Inc.) according to the manufacturer's instructions. Slides were counterstained with hematoxylin and mounted. The evaluation of immunohistochemically labeled samples with these antibodies included the percentage of positively stained tumor cells and their intensity as well as the staining localization (cytoplasmic, nuclear). The semi-quantitative evaluation was performed using the H-score classification. The percentage of negative (0), weakly stained (1+), moderately stained (2+) and strongly stained (3+), tumor cells was estimated and the H-score was calculated as follows: H-score = 1 × (% of 1+ staining) + 2 × (% of 2+ staining) + 3 × (% of 3+ staining).
2.10. Quantitative Real-time Polymerase Chain Reaction (RT-PCR) Analyses
Paraffin-embedded samples and slides were obtained by standard procedures. If the tumor infiltration rate was <85%, the selected tumor area was captured by laser micro-dissection (Zeiss-Palm, Oberlensheim, Germany); otherwise, manual macro-dissection was performed. RNA was isolated from the tumor tissue specimens and the cell lines in accordance with a proprietary procedure (European patent number EP1945764-B1). Briefly, samples were lysed in a trischloride, ethylenediaminetetraacetic acid (EDTA), SDS and proteinase K containing buffer. Then, RNA was extracted with phenol-chloroform-isoamyl alcohol, followed by precipitation with isopropanol in the presence of glycogen and sodium acetate. RNA was re-suspended in water and treated with DNAse I to avoid DNA contamination. Complementary DNA (cDNA) was synthesized using M-MLV (Moloney Murine Leukaemia Virus Reverse Transcriptase) retro-transcriptase enzyme. cDNA was added to Taqman Universal Master Mix (Applied Biosystems, Carlsbad, CA, U.S) in a 12.5 μl reaction with specific primers and probe for each gene. The primer and probe sets were designed using Primer Express 3.0 Software (Applied Biosystems) according to their Ref Seq (http://www.ncbi.nlm.nih.gov/LocusLink). Gene-specific primers are provided in the Supplementary material. Quantification of gene expression was performed using the ABI Prism 7900HT Sequence Detection System (Applied Biosystems) and was calculated according to the comparative Ct method. Final results were determined as follows: 2-(ΔCt sample-ΔCt calibrator), where ΔCt values of the calibrator and sample are determined by subtracting the Ct value of the target gene from the value of the endogenous gene (β-actin). Commercial RNA controls were used as calibrators (Liver and Lung; Stratagene, La Jolla, CA, USA). In all quantitative experiments, a sample was considered not evaluable when the standard deviation of the Ct values was >0.30 in two independent analyses. As a result, the number of evaluable samples varied among the genes examined. All analyses of the cell lines and samples in the 2 cohorts were carried out at the ISO 15189-certified Pangaea Oncology laboratory located in the Quiron Dexeus University Hospital (Barcelona, Spain).
2.11. Animal Experiments
Nu/Nu mice of four to five weeks were obtained from Nanjing Biomedical Research Institute of Nanjing University (Nanjing, Jiangsu, China). All animals were maintained in a clean facility in Jiangsu Province Academy of Traditional Chinese Medicine (Nanjing, Jiangsu, China). Mice were kept in Individually Ventilated Cages (IVCs) (6 per cage) with free access of food and water, at 20 °C and 50 ± 20% relative humidity under a 12:12 h light:dark cycles and pathogen free conditions. All procedures were based on Guide for Care and Use of Laboratory Animals of National Institutes of Health and approved by Institutional Animal Care and Use Committee of Jiangsu Province Academy of Traditional Chinese Medicine (SYXK(SU) 2016-0017). PC9 and H1975 xenograft tumors were established by subcutaneously injecting each mouse in the right flank with 4 × 106 cells suspended in PBS mixed 1:1 with Corning Matrigel (Corning, Cat. No.356237). Tumor size was measured in two orthogonal directions using calipers every two days and weights were determined twice weekly. When established tumors become palpable (~100 mm3), mice were randomised into vehicle group and treated groups with osimertinib alone, TPX-0005 alone, osimertinib plus TPX-0005. Osimertinib was suspended in 0.5% [weight/volume (w/v)] methylcellulose and administered once daily by oral gavage (1 mg/kg osimertinib for the PC9 and H1975 model). TPX0005 was suspended in 0.5% (w/v) methylcellulose and administered once daily by oral gavage (30 mg/kg for the PC9 and H1975 model). Mice in the untreated group were given the same volumes of 0.5% (w/v) methylcellulose by oral gavage. The tumor volume (mm3) was estimated using the equation length × (width)2 × 0.5.
2.12. Statistical Analysis
The primary endpoint of the study was to examine the potential effects of gene messenger RNA (mRNA) expression levels on survival. Progression-free survival and overall survival were estimated by means of the Kaplan–Meier method and compared with a nonparametric log-rank test. In addition to analyzing gene expression as a continuous variable, expression levels were divided into two groups according to the median relative expression. A multivariate Cox proportional hazard model was applied with potential risk factors as covariates, obtaining Hazard Ratios (HR) and their 95% confidence intervals (CI). Each analysis was performed with the use of a two-sided 5% significance level and a 95%CI. Association between biomarkers was assessed using Spearman correlation analysis. The statistical analyses were performed using SAS version 9.4. Forest plots for subgroup analysis were generated using GraphPad Prism 7.0. The preclinical data were analyzed with GraphPad Prism statistical software using one-way or two-way ANOVA with ad hoc Bonferroni post-test (for comparison of treatment effects).
3. Results
3.1. Co-expression of RTKs and Non-RTKs and Clinical Outcome to EGFR Inhibition
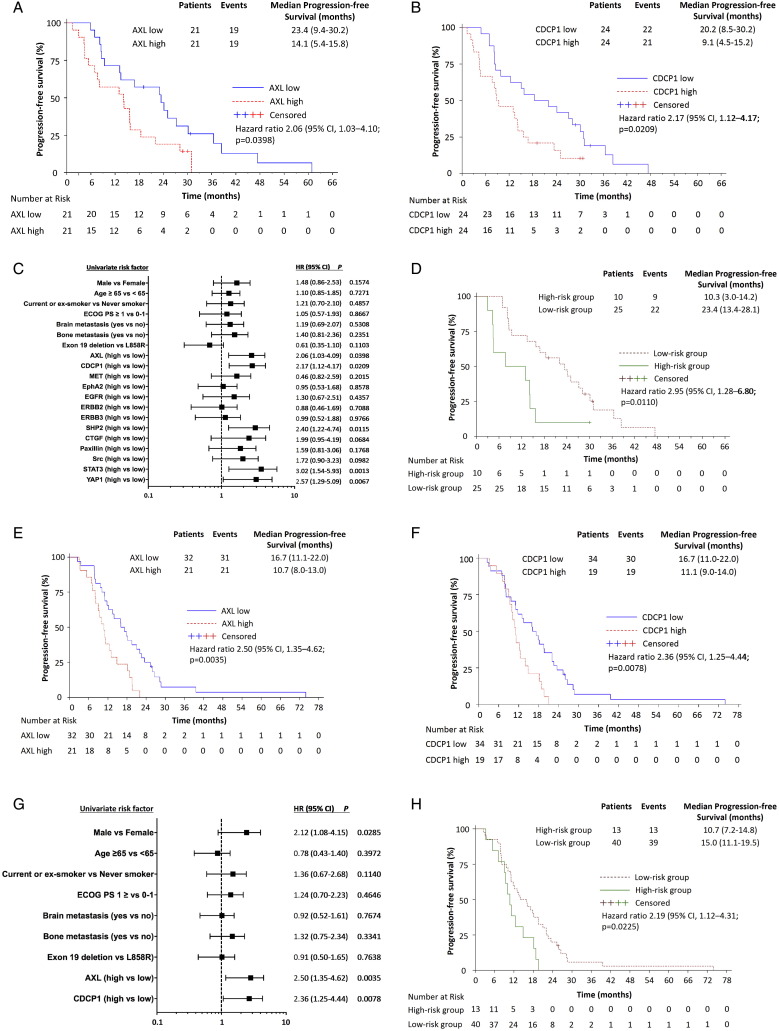
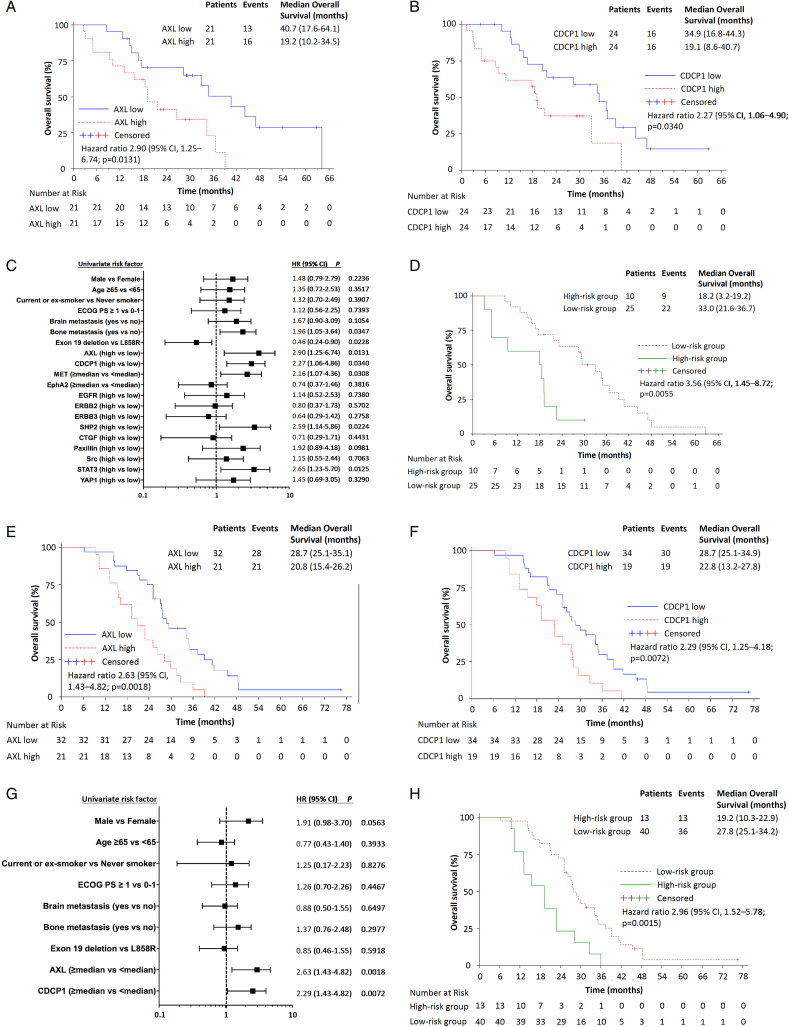
Extending our previous work (Chaib et al., 2017), we re-analyzed the baseline tissues of 64 EGFR-mutation-positive patients treated with an EGFR TKI (Cohort 1) for AXL, CDCP1 and other RTK and non-RTK mRNA expression. Gene expression levels were dichotomized at the median (Supplementary Table 2). The Spearman correlation coefficients among the biomarkers explored are presented in Supplementary Fig. 1. With a median follow-up of 26.7 months, median progression-free survival was 14.1 (95%CI, 5.4 to 15.8) and 23.4 months (95%CI, 9.4 to 30.2) for patients with high and low AXL mRNA, respectively (p < 0.001). Median progression-free survival was 9.1 (95%CI, 4.5 to 14.2) and 20.2 months (95%CI, 8.5 to 30.2) for patients with high and low CDCP1 mRNA, respectively (p = 0.0179) (Fig. 1A–C). 27% of the patients co-expressed high AXL and CDCP1 mRNA. Significant differences were observed in median overall survival according to AXL and CDCP1 mRNA expression (Fig. 2A–C). A multivariate Cox model suggested an independent association of AXL and CDCP1 mRNA expression and progression-free survival (hazard ratio [HR] for disease progression or death, 1.72; 95%CI, 1.50 to 2.94; p = 0.0461 and 1.79; 95%CI, 1.78 to 3.14; p = 0.0407) and CDCP1 mRNA expression and overall survival (HR for death, 2.23; 95%CI, 1.14 to 4.36; p =0.0192). In a two-gene model, patients were divided into two groups: a high-risk group with high both AXL and CDCP1 expression and a low-risk group with at least one of the two genes low. The model yielded a strong association between risk status and progression-free and overall survival (Figs. 1D and 2D).
Fig. 1.
Progression-free survival by the expression of biomarkers in 2 cohorts of EGFR-mutation positive NSCLC patients. A. Median progression-free survival was 23.4 months (95% CI, 9.4 to 30.2) for the 21 patients with low AXL and 14.1 months (95% CI, 5.4 to 15.8) for the 21 patients with high AXL mRNA expression; p < 0.001 (Cohort 1). B. Median progression-free survival was 20.2 months (95% CI, 8.5 to 30.2) for the 24 patients with low CDCP1 and 9.1 months (95% CI, 4.5 to 14.2) for the 24 patients with high CDCP1 mRNA expression; p = 0.0179 (Cohort 1). C. Univariate analysis was performed in Cohort 1. The bars correspond to 95% confidence intervals. D. Combined AXL and CDCP1 mRNA expression higher than the median denotes a high-risk group with a median progression-free survival of 10.3 months (95%CI, 3.0 to 14.2) and combined AXL or CDCP1 mRNA expression lower than the median denotes a low-risk group with a median progression-free survival of 23.4 months (95% CI, 13.4 to 28.1); p = 0.0079 (Cohort 1). E. Median progression-free survival was 16.7 (95% CI, 11.1 to 22.0) for the 32 patients with low AXL and 10.7 (95% CI, 8.0 to 13.0) for the 21 patients with high AXL mRNA expression; p = 0.0025 (Cohort 2). F. Median progression-free survival was 16.7 months (95% CI, 11.0 to 22.0) for the 34 patients with low CDCP1 and 11.1 months (95% CI, 9.0 to 14.0) for the 19 patients with high CDCP1 mRNA expression; p = 0.0062 (Cohort 2). G. Univariate analysis was performed in Cohort 2. The bars correspond to 95% confidence intervals. H. Combined AXL and CDCP1 mRNA expression higher than the median denotes a high-risk group with a median progression-free survival of 10.7 months (95% CI, 7.2 to 14.8) and combined AXL or CDCP1 mRNA expression lower than the median denotes a low-risk group with a median progression-free survival of 15.0 months (95% CI, 11.1 to 19.5); p = 0.0192 (Cohort 2).
Fig. 2.
Overall survival by the expression of biomarkers in 2 cohorts of EGFR-mutation-positive NSCLC patients. A. Median overall survival was 40.7 months (95% CI, 17.6 to 64.1) for the 21 patients with low AXL and 19.2 months (95% CI, 10.2 to 34.5) for the 21 patients with high AXL mRNA expression; p =0.0097 (Cohort 1). B. Median overall survival was 34.9 months (95% CI, 16.8 to 44.3) for the 24 patients with low CDCP1 and 19.1 months (95% CI, 8.6 to 40.7) for the 24 patients with high CDCP1 mRNA expression; p = 0.0300 (Cohort 1). C. Univariate analysis was performed in Cohort 1. The bars correspond to 95% confidence intervals. D. Combined AXL and CDCP1 mRNA expression higher than the median denotes a high-risk group with a median overall survival of 18.2 months (95%CI, 3.2 to 19.2) and combined AXL or CDCP1 mRNA expression lower than the median denotes a low-risk group with a median overall survival of 33.0 months (95% CI, 21.6 to 36.7); p =0.0031 (Cohort 1). E. Median overall survival was 28.7 months (95% CI, 25.1 to 35.1) for the 32 patients with low AXL and 20.8 months (95% CI, 15.4 to 26.2) for the 21 patients with high AXL mRNA expression; p = 0.0011 (Cohort 2). F. Median overall survival was 28.7 months (95% CI, 25.1 to 34.9) for the 34 patients with low CDCP1 and 22.8 months (95% CI, 13.2 to 27.8) for the 19 patients with high CDCP1 mRNA expression; p = 0.0056 (Cohort 2). G. Univariate analysis was performed in Cohort 2. The bars correspond to 95% confidence intervals. H. Combined AXL and CDCP1 mRNA expression higher than the median denotes a high-risk group with a median overall survival of 19.2 months (95% CI, 10.3 to 22.9) and combined AXL or CDCP1 mRNA expression lower than the median denotes a low-risk group with a median overall survival of 27.8 months (95% CI, 25.1 to 34.2); p < 0.001 (Cohort 2).
We next evaluated AXL and CDCP1 mRNA expression in a second cohort of 53 EGFR-mutation-positive NSCLC patients (Cohort 2). The patients were classified into high and low group according to the pre-specified in Cohort 1 median cut-off points for AXL (1.68) and CDCP1 (18.63) expression. 25% of the patients co-expressed high AXL and CDCP1 mRNA. The median follow-up time was 26.2 months. We confirmed the inverse relationship of high AXL or CDCP1 expression with progression-free and overall survival (Figs. 1E–G and 2E–G). The two-gene model defined according to AXL and CDCP1 expression of produced similar results like in Cohort 1 (Figs. 1H and 2H). Although we unable to find significant correlations between biomarkers and response to EGFR TKI in cohort 1, in Cohort 2, the objective response rate was 72% (95% CI, 53 to 86) in the low AXL mRNA expression group and 28% (95% CI, 53 to 86) in the high AXL mRNA expression group (odds ratio 2.03; 95%CI 1.04 to 3.96; p =0.0346). We finally used R2 genomics visualization tool (http://r2.amc.nl) (Santo et al., 2012) in a mixed lung tumor dataset (Bild dataset), to confirm the inverse relation between AXL and CDCP1 mRNA levels and overall survival probability (Supplementary Fig. 2).
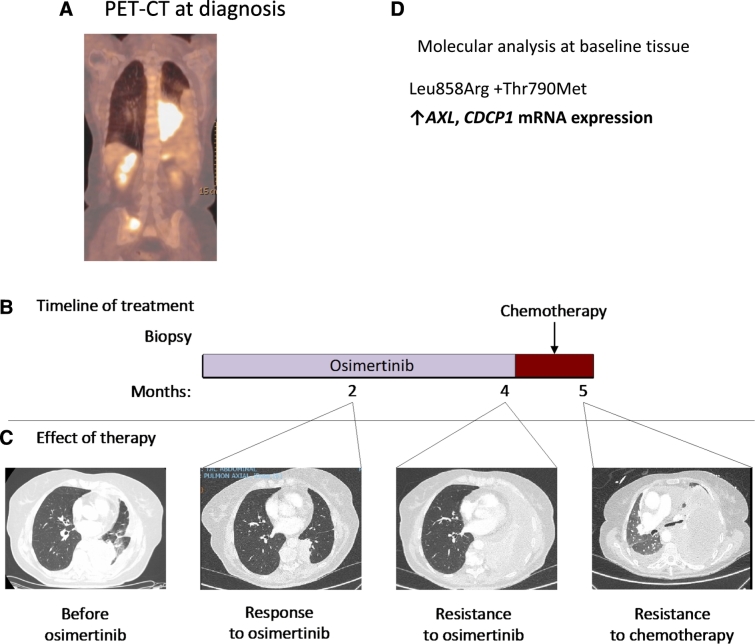
We report the case of a 78-year-old never smoker woman who was found to have advanced poorly differentiated adenocarcinoma of the lung in March, 2017 (Fig. 3). The diagnostic transbronchial tumor biopsy specimen showed a point Leu858Arg exon 21 and an EGFR Thr790Met point resistant mutation (allelic frequency of 0.009 ± 0%) mutation (Rosell et al., 2017). Further molecular testing performed in her tumor biopsy revealed high mRNA expression of AXL and CDCP1. The patient was included in the AZENT (AZD9291 in EGFR-mutation-positive NSCLC and concomitant Thr790Met pre-treatment) study (NCT02841579) and started first-line treatment with osimertinib. First restaging computed tomography (CT) after two months of treatment showed partial response (Fig. 3). However, she did well only until two months later when the second restaging CT showed worsening lung tumor and pleural infiltration. Osimertinib was discontinued and chemotherapy (carboplatin-pemetrexed) was administered. After a single dose of chemotherapy, the disease rapidly worsened, with multi-organ failure and finally death of the patient, only five months after the initial diagnosis (Fig. 3). Our data indicate that, AXL and CDCP1 can affect clinical outcome to EGFR TKIs.
Fig. 3.
Innate resistance to osimertinib therapy in a patient with high expression of AXL and CDCP1. Panel A shows the positron emission tomography-computed tomography (PET-CT) image of the patient at the time of diagnosis. Panel B shows the treatments the patient received for metastatic EGFR-mutation-positive NSCLC as well as the duration of each treatment. Panel C shows CT images of the patient's lung disease before she received osimertinib, during the time she had a response to osimertinib, when the disease subsequently relapsed at 4 months, and continue to progress after one cycle of chemotherapy. Panel D shows the results of the molecular analysis performed in the patient's baseline tissue biopsy.
3.2. Transcriptional and Proteomic Profiling of EGFR-mutation-positive NSCLC Cell Lines
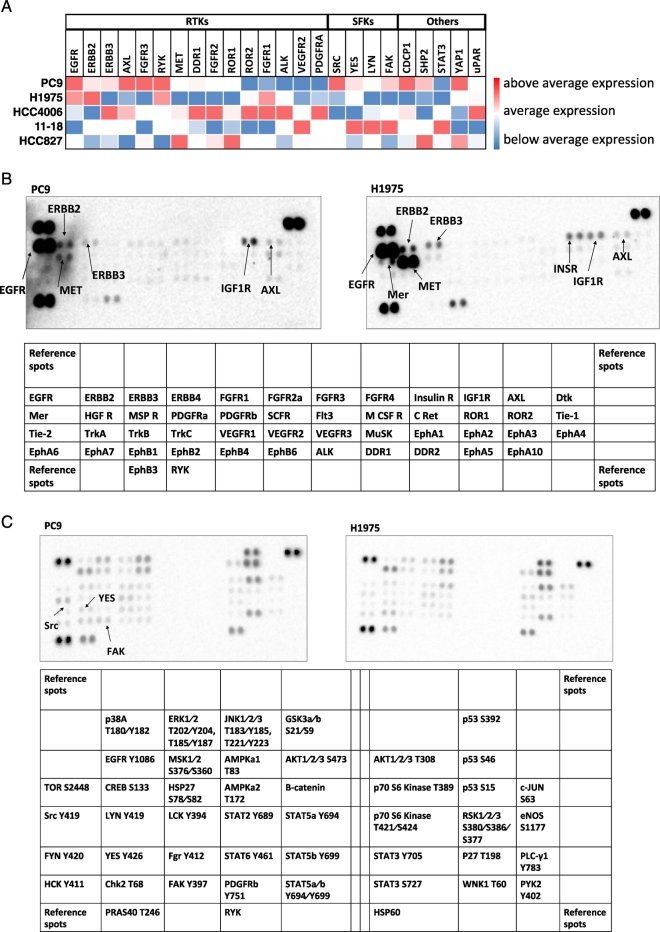
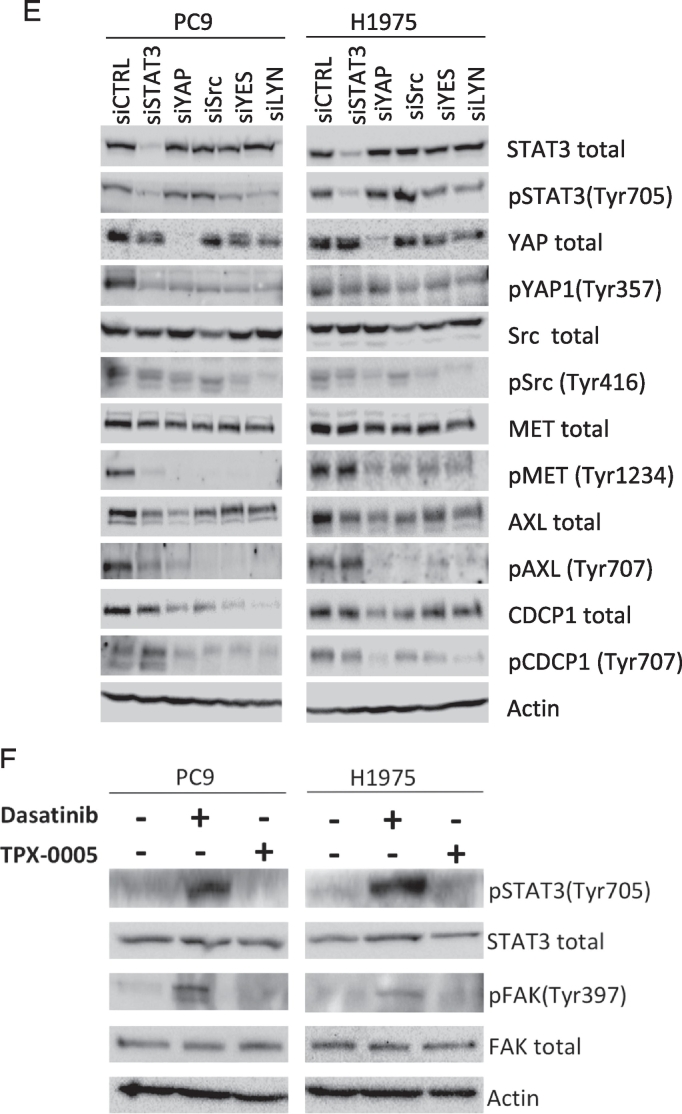
We then profiled five EGFR-mutation-positive NSCLC cell lines for the mRNA expression of RTK and non-RTK. PC9 cells showed high expression of most of the RTKs examined, including AXL and EGFR family members. SFKs, FAK and YAP1, as well as CDCP1 and SHP2, were also highly expressed in PC9 cells in comparison with the rest of the cell lines. The H1975 cell line had low mRNA expression of AXL, MET, and CDCP1, as well as SFKs, FAK SHP2 and YAP1, in comparison with the rest of the cell lines (Fig. 4A). HCC827 cells had moderate expression for most of the transcripts examined, with the exception of MET, SHP2 and YAP1. Compared to the rest of the cell lines, HCC827 had the highest mRNA expression of the receptor tyrosine kinase-like orphan receptor 1 (ROR1) (Karachaliou et al., 2014). HCC4006 had high expression of RTKs and low expression of non-RTKs, and 11–18 cells scored high expression levels only for SFKs and FAK (Fig. 4A). Based on these results we decided to use mainly the PC9 and H1975 cell lines for most of the rest of our preclinical experiments. Considering that mRNA does not predict functional activity, we then explored RTK and non-RTK activation using phospho-RTK and phosphokinase arrays in PC9 and H1975 cells. Both cell lines had several RTKs activated including AXL, MET, EGFR family members and insulin growth factor 1 receptor. Mer, a member of the TAM (Tyro3-AXL-Mer) family of RTKs (Linger et al., 2013), was also activated in the H1975 cell line (Fig. 4B). PC9 cells had higher activation of Src, YES and FAK in comparison with H1975 cells (Fig. 4C). Our findings indicate that coexpression of multiple RTKs and non-RTKs may occur in EGFR-mutation positive cells.
Fig. 4.
Quantitative RT-PCR analysis and phospho- RTK and non-RTK arrays in EGFR-mutation-positive NSCLC cell lines. Effects of genetic and pharmacologic inhibition of STAT3, YAP1 and SFKs in PC9 and H1975 cells treated with an EGFR TKI A. Expression of various genes in EGFR-mutation-positive NSCLC cell lines. Heatmap depicts gene mRNA expression (columns) in different EGFR- mutation-positive NSCLC cell lines (rows) compared to the average mRNA expression of each gene in all cell lines. B. Phospho-RTK profile in PC9 and H1975 cells obtained by human phospho-kinase array. Each membrane contains kinase specific and positive control antibodies spotted in duplicate. Template tables show the location of tyrosine kinase antibody spotted onto human phospho-RTK array. C. Phospho-kinase array profile in PC9 and H1975 cells obtained by human phospho-kinase array. Each membrane contains kinase specific and positive control antibodies spotted in duplicate. Template tables show the location of tyrosine kinase antibody spotted onto human phospho-kinase array. D. PC9 cells were transiently transfected with STAT3, Src, YAP1, YES, LYN or control siRNA (15 pmol/well). 24 h later cells were treated with serial dilutions of gefitinib. Cell viability was assessed by MTT assay after 72 h of treatment. Plots shown are representative of three independent experiments. E. Effects of negative control siRNA or STAT3, YAP1, SRC, YES and LYN siRNA on STAT3, YAP1, SRC, YES and LYN mRNA expression after 24 h of transfection. The control condition is set at 1 (arbitrary units). F. PC9 and H1975 cells were treated with serial dilutions of gefitinib or osimertinib, and TPX-0005 alone and with their double combinations for 72 h. The cell viability was measured by MTT and the synergy between the drugs was determined using the Chou and Talalay method (Chou and Talalay plot or Fraction affected [Fa] plot). The dotted horizontal line at 1 indicates the line of additive effect. Fa indicates the fractional inhibition for each CoI. The results represent the means of at least three independent experiments. Data are presented as the means ± standard deviation. G. PC9 and H1975 cells grown in six-well plates (1000 cells/well) for 24 h and then left untreated or treated with gefitinib (0.05 μM), osimertinib (0.02 μM), TPX-0005 (1 μM) alone and with their double combinations. After 72 h, media was replaced with fresh media without drugs. After seven more days cells were washed and stained with crystal violet and then photographed. The crystal violet was extracted and assayed by spectrophotometry. Data are means ± standard deviation of three independent experiments. One-way ANOVA test, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
3.3. Correlation of mRNA with Protein Expression or Gene Amplification
An important issue that is commonly raised when mRNA analysis data are reported is whether gene expression is correlated with protein expression or activation. With our real-time PCR analysis, as shown in Fig. 4A, high MET mRNA expression was detected in the HCC827 and HCC4006 cell lines that are previously described to be MET phosphorylated (Kubo et al., 2009). We have also recently shown that AXL mRNA expression is related to AXL protein expression, as defined by western blotting and immunohistochemistry in EGFR-mutation-positive NSCLC cell lines (Jacobsen et al., 2017). Finally, we have performed an exploratory analysis, where, in 14 tumor samples of the cohort 1 patients, we investigated whether the immunohistochemistry expression of both phosphorylated STAT3 at tyrosine 705 and phosphorylated YAP1 at tyrosine 357 are correlated with the mRNA expression of STAT3 and YAP1, respectively. For pSTAT3 immunohistochemistry analysis, any nuclear staining was considered positive (5 out of 14, 36%). The non-parametric Wilcoxon test was used to compare the mRNA and immunohistochemistry results. No significant correlation was found between parameters (p =0.6892). Furthermore, a median two-sample test was performed (p = 0.5909). For pYAP1 immunohistochemistry analysis, the results were considered positive when nuclear staining was observed with an histoscore ≥ 200 (5 out of 16, 31%). The non-parametric Wilcoxon test was used to compare the mRNA and immunohistochemistry results. Significant correlation was found between parameters (p = 0.034). Furthermore, a median two-sample test was performed and a significant correlation was found (p = 0.0027).
3.4. Effect of TPX-0005 with EGFR TKIs in Cell Culture
We have reported the combinatorial efficacy of gefitinib or osimertinib with TPCA-1 (STAT3 inhibitor) and AZD0530 (Src inhibitor) in EGFR-mutation-positive cells and the inverse relation of STAT3 and YAP1 mRNA expression with EGFR-TKI outcome (Chaib et al., 2017). To get insight into how STAT3 and Src-YAP1 contribute to innate resistance to EGFR TKI, STAT3, YAP1 and SFKs were knockdown by siRNA and the half-maximal inhibitory concentration (IC50) of gefitinib and osimertinib was calculated in PC9 and H1975 cells, respectively. Knockdown of Src, YAP1, YES, LYN or STAT3 enhances sensitivity to EGFR TKI (Fig. 4D, E and Supplementary Fig. 3). We next combined gefitinib or osimertinib with the Src/FAK/JAK2 inhibitor TPX-0005 (Table 2 and Supplementary Fig. 4), and examined cell viability. TPX-0005 was synergistic with gefitinib or osimertinib in PC9 and H1975 cells, respectively (Fig. 4F and G). Gefitinib with TPX-0005 suppressed PC9 cells migration (Supplementary Fig. 5). The combination of TPX-0005 was consistently synergistic or additive with all EGFR TKIs tested (Table 3). These results indicate that genetic or pharmacologic inhibition of SFKs and/or STAT3 enhances the anti-tumor effect of EGFR TKIs.
Table 3.
IC50 values and combination indexes of first-, second- and third-generation EGFR inhibitors in combination with TPX-0005 tested in EGFR-mutation-positive cell lines.
| IC50 (μM) |
Combination Index |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gefitinib | Afatinib | Dacomitinib | Osimertinib | TPX-0005 | Gefitinib + TPX-0005 | Afatinib + TPX-0005 | Dacomitinib + TPX-0005 | Osimertinib + TPX-0005 | |
| PC9 | 0.05 | <0.01 | <0.01 | 0.02 | 1.00 | 0.68 synergism | 0.80 synergism | 0.57 synergism | 0.60 synergism |
| H1975 | 10.00 | 0.04 | 0.15 | 0.02 | 1.00 | 0.84 additive | 1.12 additive | 0.84 additive | 0.61 synergism |
| 11–18 | 0.80 | 0.03 | 0.01 | 0.06 | 1.50 | 0.64 synergism | 0.76 synergism | 0.64 synergism | 0.62 synergism |
| HCC4006 | 0.03 | <0.01 | <0.01 | 0.01 | 1.48 | 0.92 additive | 0.93 additive | 0.66 synergism | 0.56 synergism |
| HCC827 | 0.02 | <0.01 | <0.01 | 0.01 | 0.43 | 0.75 synergism | 0.73 synergism | 0.63 synergism | 0.75 synergism |
3.5. Effect of TPX-0005 on STAT3, Src-YAP1 and RTK Signaling
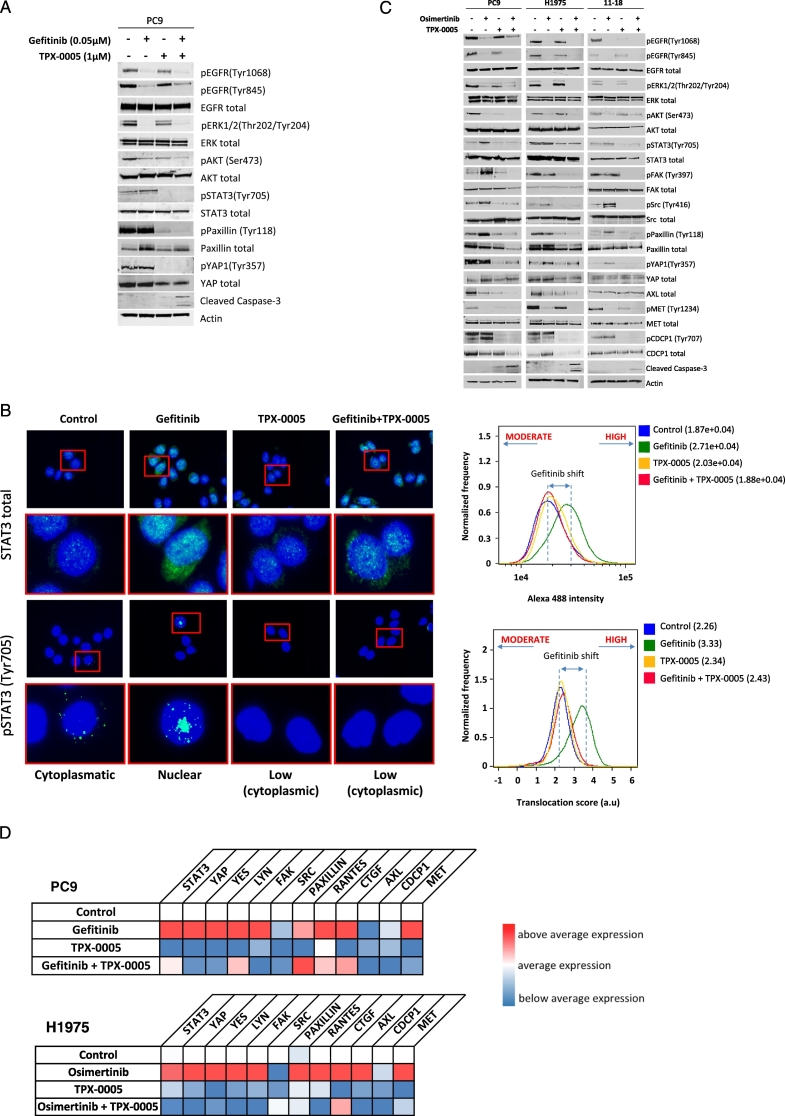
We evaluated whether EGFR TKIs combined with TPX-0005 have similar effect on EGFR downstream signal transduction pathways as we have reported with the triple combination (Chaib et al., 2017). Gefitinib suppressed EGFR, ERK1/2 and AKT phosphorylation but increased or not ablated STAT3, paxillin and YAP1 phosphorylation in PC9 cells (Fig. 5A). Gefitinib plus TPX-0005 abolished STAT3, paxillin and YAP1 phosphorylation and increased the expression of cleaved caspase-3. We adopted immunofluorescence to track STAT3 activation state by its intracellular localization. STAT3 was activated and translocated into the nucleus with gefitinib but this did not occur with TPX-0005 and the combination (Fig. 5B).
Fig. 5.
Effects of the double combination of gefitinib or osimertinib with TPX-0005 in EGFR-mutation-positive NSCLC cells. A. Extracts from the PC9 cell line treated with gefitinib (0.05 μM), TPX-0005 (1 μM), or the double combination for 24 h, were analyzed using the indicated antibodies. Similar results were obtained in three independent experiments. B. Representative immunofluorescence images showing total STAT3 and phospho-STAT3 expression and localization (green) in control, gefitinib, TPX-0005 and gefitinib plus TPX-0005 treated PC9 cells. Cell nuclei were stained with DAPI (blue). Right: expression levels and nuclear translocation of phospho-STAT3 were quantified using an ImageStreamX imaging flow cytometer. C. Extracts from PC9, 11–18 and H1975 cell lines treated with osimertinib (0.05, 0.8, and 0.02 μM respectively), TPX-0005 (1 μM), or osimertinib combined with TPX-0005 for 24 h, were analyzed by Western blotting using the indicated antibodies. β-Actin was used as a loading control. Similar results were obtained in three independent experiments. D. Heatmap depicts the mRNA expression of genes (columns) in control cells and after indicated treatments (rows) compared to the average mRNA expression of each gene in all cell lines. Gefitinib was used at 0.05 μM for PC9, osimertinib was used at 0.02 μM for H1975 and TPX-0005 was used at 1 μM for both cell lines. Quantitative RT-PCR time course experiments from two hours to seven days identified five days as the optimal time-point to compare pathway signaling after treatments (data not shown) and subsequent mRNA expression experiments are presented at this time point for consistency. Data were generated from a minimum of three replicates. β-Actin was used to normalize gene expression. E. Extracts from the PC9 and H1975 cell lines transfected with control siRNA or siRNA against STAT3, Src, YAP1, YES and LYN, were analyzed using the indicated antibodies. Similar results were obtained in three independent experiments. F. Extracts from the PC9 and H1975 cell lines treated with dasatinib (50 and 100 μM respectively) or TPX-0005 (1 μM) for 24 h, were analyzed by Western blotting using the indicated antibodies. Similar results were obtained in three independent experiments.
We then explored the effect of osimertinib with TPX-0005 on EGFR downstream signal transduction pathways and RTKs in three EGFR-mutation-positive NSCLC cell lines. Osimertinib not alone but combined with TPX-0005 ablated STAT3, paxillin and YAP1 phosphorylation (Fig. 5C). The osimertinib-induced Src and FAK phosphorylation was abrogated with TPX-0005 or the double combination. Similarly, CDCP1 phosphorylation was diminished with TPX-0005 and the double combination (Fig. 5C). TPX-0005 had a minor effect on AXL total protein expression (Fig. 5C). TPX-0005 alone or combined with an EGFR TKI abolished or attenuated the mRNA expression of most of the transcripts explored (Fig. 5D). We finally used siRNA knockdown to address the relationship of Src-YAP1 and RTKs in EGFR-mutation-positive NSCLC (Fig. 5E). Src, YES or LYN siRNA reduced YAP1 phosphorylation. AXL, CDCP1 and MET phosphorylation were diminished when YAP1 or SFKs were knocked-down, highlighting YAP1 and SFKs as regulatory nodes for RTKs activation (Fig. 5E).
We also explored the combination of EGFR TKIs with the Src inhibitor dasatinib. Dasatinib was highly synergistic with all EGFR TKIs in PC9 and H1975 cells (Supplementary Table 3). However in contrast with the Src/FAK/JAK2 inhibitor TPX-0005, the Src inhibitor dasatinib induced STAT3 and FAK phosphorylation (Fig. 5F).
The above data reconfirm our previous findings that EGFR inhibition alone increases STAT3 and YAP1 expression and activation in EGFR-mutation-positive cells. We also highlight the interplay between RTKs and SFKs-YAP1. TPX-0005 suppressed the increased expression or activation of CDCP1 and AXL observed at baseline or upon treatment with an EGFR TKI.
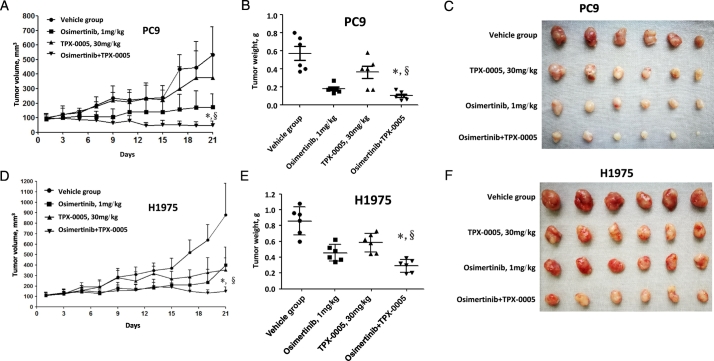
3.6. TPX-0005 and Osimertinib In Vivo
To determine the effect of EGFR TKI with TPX-0005 in vivo, nude mice bearing PC9 or H1975 cells, grown subcutaneously as tumor xenografts, were randomised to receive vehicle, osimertinib and TPX-0005 or the agents in monotherapy. TPX-0005 significantly potentiated the effect of osimertinib in the PC9 and H1975 models (Fig. 6). No substantial toxicity was noted with the combination (Supplementary Fig. 6) supporting the efficacy of osimertinib with TPX-0005 in inhibiting the growth of EGFR-mutation-positive NSCLC.
Fig. 6.
Effect of TPX-0005 and osimertinib in vivo. A. Mice with established PC9 tumors were treated with vehicle control, TPX-0005 (30 mg/kg), osimertinib (1 mg/kg) and TPX-0005 plus osimertinib. Each point represents the mean ± standard deviation of the tumor volume (n = 6 per group). Statistically significant differences on day 21 are shown for osimertinib plus TPX-0005 versus osimertinib alone (*p < 0.01). Statistically significant differences on day 21 are shown osimertinib plus TPX-0005 versus TPX-0005 alone (§p < 0.01). B. After 21 days, the mice were killed and the tumors were removed and weighed. Tumor weights were individually plotted, and comparisons between of TPX-0005 plus osimertinib group and single drug treatment groups were analyzed by Student's t-test. The tumor weight obtained with of TPX-0005 plus osimertinib in the PC9 xenograft model was statistically significantly different compared with osimertinib alone (*p < 0.05) and/or TPX-0005 alone (§p < 0.05). C. Representative PC9 tumors surgically removed. D. Mice with established H1975 tumors were treated with vehicle control, TPX-0005 (30 mg/kg), osimertinib (1 mg/kg) and TPX-0005 plus osimertinib. Each point represents the mean ± standard deviation of the tumor volume (n = 6 per group). Statistically significant differences on day 21 are shown for osimertinib plus TPX-0005 versus osimertinib alone (*p < 0.01). Statistically significant differences on day 21 are shown osimertinib plus TPX-0005 versus TPX-0005 alone (§p < 0.001). E. After 21 days, the mice were killed and the tumors were removed and weighed. Tumor weights were individually plotted, and comparisons between of TPX-0005 plus osimertinib group and single drug treatment groups were analyzed by Student's t-test. The tumor weight obtained with of TPX-0005 plus osimertinib in the H1975 xenograft model was statistically significantly different compared with osimertinib alone (*p < 0.05) and/or TPX-0005 alone (§p < 0.00001). F. Representative H1975 tumors surgically removed. The two-sided Student's t-test was used for the statistical analysis.
4. Discussion
Previously, we found that gefitinib or osimertinib activates STAT3 and Src-YAP1 in EGFR-mutation-positive NSCLC cells (Chaib et al., 2017). Here we find that the genetic or pharmacologic inhibition of Src or SFKs diminishes YAP1, AXL and CDCP1 phosphorylation or expression. In two cohorts of EGFR-mutation-positive NSCLC patients treated with EGFR TKI, we show that a risk-model combining AXL and CDCP1 mRNA expression was strongly associated with progression-free survival with hazard ratios of 2.95 and 2.19 and overall survival with hazard ratios of 3.56 and 2.96 between high-risk and low-risk group.
In accordance with earlier findings (Yoshida et al., 2014), we show that EGFR-mutation-positive NSCLC cells co-express more than just EGFR RTKs. Non-RTKs, especially, Src, YES and FAK, are also expressed and activated. Combined EGFR TKI with TPX-0005 is more effective than EGFR TKI alone both in culture and in vivo. At the time of our work, it was reported that the AKT and the mitogen-activated protein kinase (MAPK) pathways remain active, even in the presence of osimertinib (Ichihara et al., 2017). EGFR, SFK and FAK concomitant inhibition enhances osimertinib activity and suppresses resistance (Ichihara et al., 2017). Although co-targeting EGFR and MEK (known as mitogen-activated protein kinase kinase 1, or MAP2K1) (Tricker et al., 2015) or, from our experience, AKT (Jacobsen et al., 2017), causes growth inhibition in EGFR TKI resistant NSCLC models, the combination of osimertinib with dasatinib was superior to the combination of osimertinib with MEK or phosphoinositide 3-kinase (PI3K) inhibitor inhibitors (Ichihara et al., 2017). In summary, three studies show that SFK and FAK contribute to EGFR TKI resistance (Yoshida et al., 2014; Ichihara et al., 2017; Murakami et al., 2017).
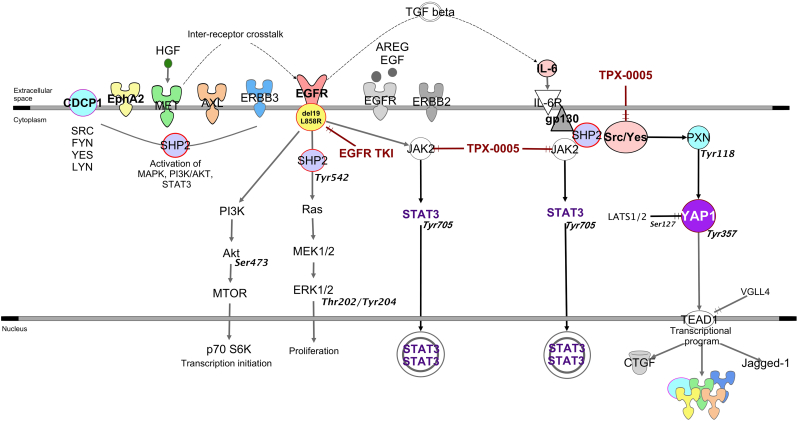
Tyrosine kinase inhibitors do not induce the activation of new RTKs, rather, only increase the phosphorylation of existing RTKs to compensate the reduced phosphorylation of ERK and AKT. A combination of TKIs could completely inactivate all the RTKs and their downstream signaling molecules (Sun et al., 2016). Paradoxically, inhibition of AXL or MET led to greater Src-induced AXL and MET phosphorylation in breast and lung cancer cell lines (Baumann et al., 2017). CDCP1 overexpression triggered a cascade of tyrosine phosphorylation events, leading to the activation of signaling networks, including SFKs, to promote tumor cell growth and survival (Leroy et al., 2015). Through the combination of genomic, biological, in vivo models and clinical cohorts of patients, our findings lead to the model in Fig. 7. Our data indicate that Src-YAP1 signaling leads to further activation of AXL, CDCP1 and MET (Fig. 5E). By targeting SFK and FAK, we can obviate the need to inhibit the dominant RTKs that can be activated with the paradoxical detrimental effect of causing more activation (Baumann et al., 2017). The combination of a TKI plus SFK and FAK inhibitor has been proposed (Yoshida et al., 2014; Ichihara et al., 2017; Murakami et al., 2017). However, in our work, dasatinib induced STAT3 and FAK phosphorylation (Fig. 5F). Erlotinib plus dasatinib was not effective in a clinical trial in EGFR-mutation-positive NSCLC patients with acquired resistance to erlotinib or gefitinib (Johnson et al., 2011). Clinical trials with EGFR TKI plus SFK and FAK inhibitors, such as the Src/FAK/JAK2 inhibitor TPX-0005, deserve to be carried out.
Fig. 7.
Our Signal transduction pathway model EGFR activating mutations located in the tyrosine kinase domains and mainly in the form of a base-pair deletion at exon 19 (ΔE746_A750) or a point mutation at exon 21 (Leu858Arg) enhance cell growth and invasion via tyrosine phosphorylation and lead to the activation of mitogen-activated protein kinase (MAPK), signal transducer and activator of transcription 3 (STAT3) and AKT pathways. SHP2, a widely expressed cytoplasmic tyrosine phosphatase with two src homology (SH)2 domains plays an essential role in most receptor-tyrosine kinase (RTK) signaling pathways. SHP2 function is required for MAPK pathway and activation of its downstream transcriptional targets. When phosphorylated on tyrosine 542, SHP2 increases the half-life of activated Ras (GTP-Ras) in the cell by interfering with the process of Ras inactivation catalyzed by Ras GTPase-activating protein (RasGAP). Raf activates MAPK/ERK Kinase 1 (MEK1) and MEK2. Extracellular signal-regulated kinase (ERK1 and ERK2) is activated upon phosphorylation by MEK1/2. Dual threonine 202 and tyrosine 204 residue phosphorylation activate ERK1. AKT is the major downstream target of phosphatidylinositol 3-kinase (PI3K), which is activated by EGFR. When PI3K is activated induces the recruitment of AKT to the cell membrane. The recruitment of AKT to the cell membrane drives a conformational change in the protein. This enables AKT phosphorylation on serine 473, which is required for full activation. Following activation, AKT translocates to the cytoplasm and nucleus, and phosphorylates various downstream substrates including mammalian target of rapamycin (mTOR). mTOR regulates the serine/threonine kinase, p70S6K, whose function regulates protein synthesis. STAT3 can be activated not only by growth factor receptors, like EGFR, but also by interleukins, like IL-6. The transforming growth factor-beta (TGF-β) drives the expression of IL-6. IL-6 is a glycoprotein which first binds to a-chain (IL-6R) and then recruits the b-chain (gp130) of the receptor. Subsequently, the IL-6/IL-6R complex initiates homo-dimerization of gp130, activates a cytoplasmic tyrosine kinase bound to gp130 and triggers signaling cascades through janus-like kinase (JAK) and Src kinase. JAK2 mediates STAT3 phosphorylation and activation. The major phosphorylation site in STAT3 is tyrosine at position 705. When activated, STAT3 undergoes phosphorylation-induced homo-dimerization, leading to nuclear translocation, DNA binding and gene transcription. Independently of STAT3, gp130 activates the Yes-Associated Protein (YAP1) oncoprotein through direct association with the Src family kinase (SFK) Yes. SHP2 activates several SFKs, including Src as top hit. Upon Src activation, several downstream Src binding partners are targeted for phosphorylation, including paxillin on tyrosine 118. YAP1 is normally kept inactive in the cytoplasm through serine phosphorylation by the Hippo effector large tumor suppressor kinases (LATS). One Hippo independent mechanism implicated in cancer involves phosphorylation of YAP1 on tyrosine 357 by Yes1. As a transcriptional co-activator, YAP1 has been reported to bind several DNA-binding transcription factors. Among the reported YAP1 partners, the TEA domain (TEAD) transcription factor is well characterized. Connective tissue growth factor (CTGF) is a prototypical target of the YAP1-TEAD complex and a commonly used marker of YAP1 activation. Several RTKs or their ligands have been described as mediators of YAP1-dependent oncogenic activities.
The tumor microenvironment-derived ligand hepatocyte growth factor (HGF) induces inter-receptor cross talk of MET with erythropoietin-producing hepatocellular receptor A2 (EphA2), the complement C1r/C1s, Uegf, Bmp1 (CUB) domain-containing protein-1 (CDCP1) or AXL. This provides an alternative signaling mechanism for EGFR, thereby circumventing TKI resistance. SHP2 modulates RTK signals at the level of the MAPK pathway.
EGFR-TKI combined with the Src/FAK/JAK2 inhibitor TPX-0005 may more efficiently block the EGFR downstream signaling pathways than EGFR-TKI alone.
A trial is foreseen with osimertinib plus TPX-0005 and, according to our knowledge, TPX-0005 is the only compound that inhibits Src, FAK and JAK2 at a similar affinity level. Finally, we advocate customizing the therapy of EGFR-mutation-positive NSCLC patients and the use of a tool kit to optimize EGFR mutational screening by incorporating CDCP1 and AXL mRNA assessment.
The following are the supplementary data related to this article.
Correlation among the most relevant biomarkers explored in the Cohort 1 of the study. Heat maps of Spearman's rank correlation coefficients.
Kaplan-Meier survival analysis of the selected genes (AXL and CDCP1) as indicated in the R2: Genomics Analysis and Visualization Platform database. A. Overall survival probability according to AXL expression levels. B. Overall survival probability according to CDCP1 expression levels.
Effects of genetic inhibition of STAT3, YAP1 and SFKs in H1975 cells treated with osimertinib. A. H1975 cells were transiently transfected with STAT3, Src, YAP1, YES, LYN or control siRNA. 24 h later cells were treated with serial dilutions of osimertinib. Cell viability was assessed by MTT assay after 72 h of treatment. Plots shown are representative of three independent experiments. B. Effects of negative control siRNA or STAT3, YAP1, SRC, YES and LYN siRNA on STAT3, YAP1, SRC, YES and LYN mRNA expression after 24 h of transfection. The control condition is set at 1.
Effect of TPX-0005 on STAT3 and paxillin phosphorylation. PC9 and H1975 cells were treated with TPX-0005 at indicated concentrations for 24 h. Extracts were analyzed using the indicated antibodies. Similar results were obtained in three independent experiments.
Effect of gefitinib, TPX-0005 and the double combination on the migration of PC9 cells. The cell migration of PC9 cells untreated and treated with gefitinib, TPX-0005 and gefitinib plus TPX-0005 was determined by wound healing assay A cross-shaped wound in a confluent monolayer of PC9 cells was created by scratching with a pipette tip and cells were incubated with or without the indicated drugs. Cell migration at the wound edge to the wound space was captured using an OLYMPUS CKX41 microscope equipped with an NIKON DXM1200C camera, after 12 h of incubation, and the migration distance was calculated using Image J analysis software. A. Representative images of wound healing/cell migration. B. Migration distance of cells treated with the indicated compounds. Data are presented as mean ± standard deviation of three independent experiments. One-way ANOVA test, *p <0.05, **p < 0.01 and ***P ≤ 0.001 versus values at t = 12 h.
Effect of TPX-0005 and osimertinib combination treatment on the body weight of xenograft models. A. Effect of TPX-0005 and osimertinib combination treatment on the body weight of xenograft PC9 tumor. Statistically significant differences on day 21 are shown for osimertinib plus TPX-0005 vs osimertinib alone alone (*p = 0.0309), Statistically significant differences on day 21 are shown osimertinib plus TPX-0005 versus TPX-0005 alone (§p = 0.1741). B. Effect of TPX-0005 and osimertinib combination treatment on the body weight of xenograft H1975 tumor. Statistically significant differences on day 21 are shown for osimertinib plus TPX-0005 vs osimertinib alone (*p = 0.0078), Statistically significant differences on day 21 are shown osimertinib plus TPX-0005 versus TPX-0005 alone (§p = 0.0019).
Supplementary material
Funding
Work in Dr. Rosell's laboratory is partially supported by a grant from La Caixa Foundation, and an Instituto de Salud Carlos III grant (RESPONSE, PIE16/00011). Work in Dr. Cao's laboratory is partially supported by the Major National Science and Technology Program of China for Innovative Drug (2017ZX09101002-002-006), the National Natural Science Foundation of China (No. 81573680 and 81403151) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (Integration of Chinese and Western Medicine) grant Work in Dr. Bivona's laboratory is partially supported by R01CA204302, R01CA211052, R01CA169338 National Institute of Health (NIH) grants. The funders did not have any role in study design, data collection, data analysis, interpretation and writing of the report.
Conflicts of Interest
The authors declare no conflicts of interest with respect to this work.
Author Contributions
R.R. and P.C. conceived the presented idea. N.K. and R.R. designed the experiments and wrote the manuscript. N.K. collected all the clinical data and created the database. A.D. performed the statistical analyses. N.K., E.A. and A.G.C. performed the experiments with the clinical tumor samples. A.F.C., J.R. and L.R. contributed in the collection of tumor samples with complete clinical data. I.C., J.B, J.W.P.B., C.C.S., J.C.S., M.I, I.A., M.A.M carried out the experiments in vitro. J.Y., X.C., Z.W. C.H. and P.C. carried out the experiments in vivo. S.V., SH.I., M.Ok., T.S.M., T.G.B., M.O. and S.R.C. helped supervise the project. All authors discussed the results and contributed to the final manuscript.
Contributor Information
Peng Cao, Email: pcao79@yahoo.com.
Rafael Rosell, Email: rrosell@iconcologia.net.
References
- Baumann C., Ullrich A., Torka R. GAS6-expressing and self-sustaining cancer cells in 3D spheroids activate the PDK-RSK-mTOR pathway for survival and drug resistance. Mol. Oncol. 2017;11(10):1430–1447. doi: 10.1002/1878-0261.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaib I., Karachaliou N., Pilotto S. Co-activation of STAT3 and YES-associated protein 1 (YAP1) pathway in EGFR-mutant NSCLC. J. Natl. Cancer Inst. 2017;109(9) doi: 10.1093/jnci/djx014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T.C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70(2):440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- Codony-Servat C., Codony-Servat J., Karachaliou N. Activation of signal transducer and activator of transcription 3 (STAT3) signaling in EGFR mutant non-small-cell lung cancer (NSCLC) Oncotarget. 2017;8(29):47305–47316. doi: 10.18632/oncotarget.17625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J.J., Li Y., Rogers E.W. 2017. Chiral Diaryl Macrocycles and Uses Thereof. (Google Patents) [Google Scholar]
- Gao S.P., Mark K.G., Leslie K. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J. Clin. Invest. 2007;117(12):3846–3856. doi: 10.1172/JCI31871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusenbauer S., Vlaicu P., Ullrich A. HGF induces novel EGFR functions involved in resistance formation to tyrosine kinase inhibitors. Oncogene. 2013;32(33):3846–3856. doi: 10.1038/onc.2012.396. [DOI] [PubMed] [Google Scholar]
- Ichihara E., Westover D., Meador C.B. SFK/FAK signaling attenuates osimertinib efficacy in both drug-sensitive and drug-resistant models of EGFR-mutant lung cancer. Cancer Res. 2017;77(11):2990–3000. doi: 10.1158/0008-5472.CAN-16-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen K., Bertran-Alamillo J., Molina M.A. Convergent Akt activation drives acquired EGFR inhibitor resistance in lung cancer. Nat. Commun. 2017;8(1):410. doi: 10.1038/s41467-017-00450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.L., Riely G.J., Rizvi N.A. Phase II trial of dasatinib for patients with acquired resistance to treatment with the epidermal growth factor receptor tyrosine kinase inhibitors erlotinib or gefitinib. J. Thorac. Oncol. 2011;6(6):1128–1131. doi: 10.1097/JTO.0b013e3182161508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karachaliou N., Gimenez-Capitan A., Drozdowskyj A. ROR1 as a novel therapeutic target for EGFR-mutant non-small-cell lung cancer patients with the EGFR T790M mutation. Transl. Lung Cancer Res. 2014;3(3):122–130. doi: 10.3978/j.issn.2218-6751.2014.03.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T., Yamamoto H., Lockwood W.W. MET gene amplification or EGFR mutation activate MET in lung cancers untreated with EGFR tyrosine kinase inhibitors. Int. J. Cancer. 2009;124(8):1778–1784. doi: 10.1002/ijc.24150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy C., Shen Q., Strande V. CUB-domain-containing protein 1 overexpression in solid cancers promotes cancer cell growth by activating Src family kinases. Oncogene. 2015;34(44):5593–5598. doi: 10.1038/onc.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linger R.M., Cohen R.A., Cummings C.T. Mer or Axl receptor tyrosine kinase inhibition promotes apoptosis, blocks growth and enhances chemosensitivity of human non-small cell lung cancer. Oncogene. 2013;32(29):3420–3431. doi: 10.1038/onc.2012.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch T.J., Bell D.W., Sordella R. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Mok T.S., Wu Y.L., Thongprasert S. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N. Engl. J. Med. 2009;361(10):947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- Murakami Y., Sonoda K., Abe H. The activation of SRC family kinases and focal adhesion kinase with the loss of the amplified, mutated EGFR gene contributes to the resistance to afatinib, erlotinib and osimertinib in human lung cancer cells. Oncotarget. 2017;8:70736–70751. doi: 10.18632/oncotarget.19982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan R.S., Fedrigo C.A., Brands E. The allosteric AKT inhibitor MK2206 shows a synergistic interaction with chemotherapy and radiotherapy in glioblastoma spheroid cultures. BMC Cancer. 2017;17(1):204. doi: 10.1186/s12885-017-3193-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez J.G., Janne P.A., Lee J.C. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Park K., Tan E.H., O'Byrne K. Afatinib versus gefitinib as first-line treatment of patients with EGFR mutation-positive non-small-cell lung cancer (LUX-Lung 7): a phase 2B, open-label, randomised controlled trial. Lancet Oncol. 2016;17(5):577–589. doi: 10.1016/S1470-2045(16)30033-X. [DOI] [PubMed] [Google Scholar]
- Rikova K., Guo A., Zeng Q. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131(6):1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- Rosell R., Moran T., Queralt C. Screening for epidermal growth factor receptor mutations in lung cancer. N. Engl. J. Med. 2009;361(10):958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- Rosell R., Carcereny E., Gervais R. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13(3):239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- Rosell R., Dafni U., Felip E. Erlotinib and bevacizumab in patients with advanced non-small-cell lung cancer and activating EGFR mutations (BELIEF): an international, multicentre, single-arm, phase 2 trial. Lancet Respir. Med. 2017;5(5):435–444. doi: 10.1016/S2213-2600(17)30129-7. [DOI] [PubMed] [Google Scholar]
- Santo E.E., Ebus M.E., Koster J. Oncogenic activation of FOXR1 by 11q23 intrachromosomal deletion-fusions in neuroblastoma. Oncogene. 2012;31(12):1571–1581. doi: 10.1038/onc.2011.344. [DOI] [PubMed] [Google Scholar]
- Sausgruber N., Coissieux M.M., Britschgi A. Tyrosine phosphatase SHP2 increases cell motility in triple-negative breast cancer through the activation of SRC-family kinases. Oncogene. 2015;34(17):2272–2278. doi: 10.1038/onc.2014.170. [DOI] [PubMed] [Google Scholar]
- Sordella R., Bell D.W., Haber D.A. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305(5687):1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- Soria J.C., Ohe Y., Vansteenkiste J. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N. Engl. J. Med. 2018;378(2):113–125. doi: 10.1056/NEJMoa1713137. [DOI] [PubMed] [Google Scholar]
- Stommel J.M., Kimmelman A.C., Ying H. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318(5848):287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- Sun X., Song Q., He L. Receptor tyrosine kinase phosphorylation pattern-based multidrug combination is an effective approach for personalized cancer treatment. Mol. Cancer Ther. 2016;15(10):2508–2520. doi: 10.1158/1535-7163.MCT-15-0735. [DOI] [PubMed] [Google Scholar]
- Tricker E.M., Xu C., Uddin S. Combined EGFR/MEK inhibition prevents the emergence of resistance in EGFR-mutant lung cancer. Cancer Discov. 2015;5(9):960–971. doi: 10.1158/2159-8290.CD-15-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao M.S., Sakurada A., Cutz J.C. Erlotinib in lung cancer - molecular and clinical predictors of outcome. N. Engl. J. Med. 2005;353(2):133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- Wu Y.L., Cheng Y., Zhou X. Dacomitinib versus gefitinib as first-line treatment for patients with EGFR-mutation-positive non-small-cell lung cancer (ARCHER 1050): a randomised, open-label, phase 3 trial. Lancet Oncol. 2017;18(11):1454–1466. doi: 10.1016/S1470-2045(17)30608-3. [DOI] [PubMed] [Google Scholar]
- Xu M.Z., Chan S.W., Liu A.M. AXL receptor kinase is a mediator of YAP-dependent oncogenic functions in hepatocellular carcinoma. Oncogene. 2011;30(10):1229–1240. doi: 10.1038/onc.2010.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Zhang G., Smith M.A. Tyrosine phosphoproteomics identifies both codrivers and cotargeting strategies for T790M-related EGFR-TKI resistance in non-small cell lung cancer. Clin. Cancer Res. 2014;20(15):4059–4074. doi: 10.1158/1078-0432.CCR-13-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai D., Deng W., Huang Z. Abstract 2132: the novel, rationally-designed, ALK/SRC inhibitor TPX-0005 overcomes multiple acquired resistance mechanisms to current ALK inhibitors. Cancer Res. 2016;76(14 Suppl):2132. [Google Scholar]
- Zhang Z., Lee J.C., Lin L. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat. Genet. 2012;44(8):852–860. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Correlation among the most relevant biomarkers explored in the Cohort 1 of the study. Heat maps of Spearman's rank correlation coefficients.
Kaplan-Meier survival analysis of the selected genes (AXL and CDCP1) as indicated in the R2: Genomics Analysis and Visualization Platform database. A. Overall survival probability according to AXL expression levels. B. Overall survival probability according to CDCP1 expression levels.
Effects of genetic inhibition of STAT3, YAP1 and SFKs in H1975 cells treated with osimertinib. A. H1975 cells were transiently transfected with STAT3, Src, YAP1, YES, LYN or control siRNA. 24 h later cells were treated with serial dilutions of osimertinib. Cell viability was assessed by MTT assay after 72 h of treatment. Plots shown are representative of three independent experiments. B. Effects of negative control siRNA or STAT3, YAP1, SRC, YES and LYN siRNA on STAT3, YAP1, SRC, YES and LYN mRNA expression after 24 h of transfection. The control condition is set at 1.
Effect of TPX-0005 on STAT3 and paxillin phosphorylation. PC9 and H1975 cells were treated with TPX-0005 at indicated concentrations for 24 h. Extracts were analyzed using the indicated antibodies. Similar results were obtained in three independent experiments.
Effect of gefitinib, TPX-0005 and the double combination on the migration of PC9 cells. The cell migration of PC9 cells untreated and treated with gefitinib, TPX-0005 and gefitinib plus TPX-0005 was determined by wound healing assay A cross-shaped wound in a confluent monolayer of PC9 cells was created by scratching with a pipette tip and cells were incubated with or without the indicated drugs. Cell migration at the wound edge to the wound space was captured using an OLYMPUS CKX41 microscope equipped with an NIKON DXM1200C camera, after 12 h of incubation, and the migration distance was calculated using Image J analysis software. A. Representative images of wound healing/cell migration. B. Migration distance of cells treated with the indicated compounds. Data are presented as mean ± standard deviation of three independent experiments. One-way ANOVA test, *p <0.05, **p < 0.01 and ***P ≤ 0.001 versus values at t = 12 h.
Effect of TPX-0005 and osimertinib combination treatment on the body weight of xenograft models. A. Effect of TPX-0005 and osimertinib combination treatment on the body weight of xenograft PC9 tumor. Statistically significant differences on day 21 are shown for osimertinib plus TPX-0005 vs osimertinib alone alone (*p = 0.0309), Statistically significant differences on day 21 are shown osimertinib plus TPX-0005 versus TPX-0005 alone (§p = 0.1741). B. Effect of TPX-0005 and osimertinib combination treatment on the body weight of xenograft H1975 tumor. Statistically significant differences on day 21 are shown for osimertinib plus TPX-0005 vs osimertinib alone (*p = 0.0078), Statistically significant differences on day 21 are shown osimertinib plus TPX-0005 versus TPX-0005 alone (§p = 0.0019).
Supplementary material