Abstract
p204, a murine member of an interferon-inducible p200 family, was reported to recognize intracellular viral and bacterial DNAs, however, its role in the innate immunity in vivo remains unknown due to the lack of p204-deficient animal models. In this study we first generated the p204−/− mice. Unexpectedly, p204 deficiency led to significant defect in extracellular LPS signaling in macrophages, as demonstrated by dramatic reductions of LPS-mediated IFN-β and pro-inflammatory cytokines. The serum levels of IFN-β and pro-inflammatory cytokines were also significantly reduced in p204−/− mice following LPS challenge. In addition, p204−/− mice were resistant to LPS-induced shock. LPS-activated NF-ĸB and IRF-3 pathways were all defective in p204-deficient macrophages. p204 binds to TLR4 through its Pyrin domain, and it is required for the dimerization of TLR4 following LPS-challenge. Collectively, p204 is a critical component of canonical LPS-TLR4 signaling pathway, and these studies also suggest that p204 could be a potential target to prevent and treat inflammatory and infectious diseases.
Keywords: p204, LPS, TLR4, IFN-β, Inflammatory responses, Macrophages
Highlights
-
•
p204 deficiency leads to significant defect in extracellular LPS signaling in macrophages.
-
•
Serum levels of IFN-β and pro-inflammatory cytokines were also significantly reduced in p204-/- mice following LPS challenge.
-
•
p204-/- mice were resistant to LPS-induced shock.
-
•
p204 binds to TLR4 through its Pyrin domain, and it is required for the dimerization of TLR4 following LPS-challenge.
Effective anti-pathogenic responses, including production of type I IFNs and inflammatory response, are critical for host defense. p200 family members, including IFI16 and AIM2, have been reported to function as the sensors of pathogen components. However, investigation of their roles has largely focused on intracellular pathogen components, independent of extracellular pathogen receptors, such as TLRs. Here, we provide unexpected evidences demonstrating that p204, a murine counterpart of human IFI16, is required for extracellular but not intracellular LPS signaling. These results provide not only evidence of functional crosstalk and cooperation between intracellular p204 and extracellular LPS through TLR4 pathways in macrophage-mediated innate immunity, but also new insights into the mechanisms underlying p200 family proteins mediated antiviral and antibacterial infections.
1. Introduction
Detection and elimination of invading pathogens are fundamental mechanisms of host defense. Among immune cells, macrophages are first line of defense cells that play a pivotal role in innate immunity through detecting conserved microbial structures and molecules, such as nucleic acids and cell wall components, called pathogen-associated molecular patterns (PAMPs), which are crucial for pathogen survival and virulence (Vance et al., 2009; Janeway and Medzhitov, 2002; Gay et al., 2014). Macrophages recognize extracellular pathogen components through their conserved pattern recognition receptors (PPRs), especially toll-like receptors (TLRs), and also detect intracellular pathogen components through various intracellular RIG-I-like receptors (RLRs) and sensor proteins (Akira et al., 2001; Iwasaki and Medzhitov, 2004; Gay et al., 2014; Goubau et al., 2014). The recognition of extracellular and intracellular pathogen components initiates signal transduction cascades that lead to the production and release of type I interferon (IFN), various pro-inflammatory cytokines, and chemokines, resulting in the recruitment of other immune cells to the pathogenic lesions for stimulating further adaptive immune responses and the elimination of pathogens (Pandey et al., 2015; Stetson and Medzhitov, 2006b; Stetson and Medzhitov, 2006a).
Traditionally, research concerning the recognition of invading pathogens and the mechanisms by which macrophages initiate innate immunity through binding with pathogen components has largely focused on extracellular pathogen components and their cell surface receptors, including TLRs, scavenger receptors and c-type lectins (Pluddemann et al., 2011). Among these receptors, TLRs are the representative receptors expressed on the surfaces and endosomes of macrophages to recognize a variety of extracellular pathogen components, including lipopeptides, single and double strand nucleic acids, flagellins, and lipopolysaccharide (LPS) (Medzhitov, 2001; Barton and Medzhitov, 2003). TLR1, TLR2 and TLR6 expressed on cell surfaces recognize extracellular di and/or triacylated lipopeptides, and TLR4 and TLR5, also expressed on cell surfaces, recognize extracellular LPS and flagellin, respectively. Through the recognition of extracellular pathogen components by TLRs, macrophages activate and transduce pathogen component-mediated signaling pathways necessary for the innate immunity against pathogen invasion. Interestingly, the recognition of intracellular pathogen components by intracellular sensors has recently been regarded as another critical strategy to detect and eliminate invading pathogens. Intensive efforts have identified a number of intracellular sensors and their specific ligands, and examined anti-pathogenic roles mediated by these intracellular sensors in innate immunity (Wu and Chen, 2014; Takaoka et al., 2007; Kondo et al., 2013; Chiu et al., 2009; Unterholzner et al., 2010; Sun et al., 2013). Cytoplasmic RLRs detect microbial nucleic acids accumulated in the cytosol of infected cells. RIG-I detects short single and double strand RNA, and MDA5 and LGP2 recognize double strand RNA (Takeuchi and Akira, 2010; Pandey et al., 2015). DDX3 also detects viral RNA for host defense (Pandey et al., 2015; Pluddemann et al., 2011). NOD-like receptors, such as NOD1, NOD2, NLRP1, NLRP3, NLRC4 and Naip5 are intracellular sensors to detect intracellular pathogen components (Pluddemann et al., 2011). TLRs localized at intracellular endosomes have also been reported to sense intracellular microbial nucleic acids for host defense. TLR7, TLR8, TLR9 and TLR13 recognize microbial nucleic acids, and TLR11 and TLR12 also recognize pathogen proteins, such as profilin and flagellin (Pandey et al., 2015, Pluddemann et al., 2011).
Emerging studies have actively focused on the roles of p200 protein family as intracellular sensors of pathogen components in macrophage-mediated innate immunity. p200 proteins, also called PYHIN proteins, are interferon-inducible proteins which consist of an amino-terminus pyrin domain (PYD) and one or two carboxyl-terminus hematopoietic interferon-inducible nuclear antigen (HIN)-200 domains (Luan et al., 2008a). Previous studies have reported that p200 proteins act as modulators of many cellular functions, including cell proliferation, differentiation, apoptosis, and senescence (Zhao et al., 2015). In addition to these functions, p200 proteins have been reported as sensors for intracellular pathogen components in innate immunity. Among the human p200 proteins, IFN-inducible protein 16 (IFI16) was reported as an intracellular sensor that directly recognizes cytoplasmic nucleic acids derived from pathogens and subsequently induces the production of IFN-β as well as other pro-inflammatory mediators (Unterholzner et al., 2010; Monroe et al., 2014). Another human p200 protein, absent in melanoma 2 (AIM2), was also reported as a sensor for intracellular nucleic acids to activate inflammasome and caspase-1 signaling pathways, leading to the release of interleukin 1 beta (IL-1β) and cell death (Burckstummer et al., 2009; Roberts et al., 2009; Hornung et al., 2009; Fernandes-Alnemri et al., 2009). p204, regarded as a murine homologue of IFI16, was reported to be an intracellular sensor to sense both viral and bacterial dsDNAs, leading to the production of type I IFNs (Storek et al., 2015; Unterholzner et al., 2010; Conrady et al., 2012). However, due to unavailability of p204 knockout mouse model, the in vivo significance of p204's reported importance for sensing viral and bacterial dsDNAs and whether it is also involved in additional innate immunity pathways in vivo remains unknown.
In this study, we generated p204 knockout mice using a conventional gene-editing approach and p204-deficient macrophages using the CRISPR/Cas9 system. With p204-deficient macrophages and p204−/− mice, we confirmed previous reports that p204 deficiency led to strong inhibition of intracellular double strand (ds)VACV 70mer- and HSV 60mer-mediated IFNβ induction (Unterholzner et al., 2010). Most importantly, p204 was unexpectedly found to also be required for canonical extracellular LPS/TLR4 signaling pathways, but not for intracellular LPS activated signaling pathways. Taken together, these results indicate that in addition to functioning as an intracellular viral dsDNA sensor, p204 is also a critical intracellular mediator essential for extracellular LPS/TLR4 against pathogen infection in macrophage-mediated innate immunity.
2. Materials and Methods
2.1. Generation of p204−/− Mice
We used mouse strain 129 to generate loxP-floxed p204 mice in which exon2 and exon5 of the p204 gene were flanked by loxP sequences. The floxed p204 mice were then crossed with Sox2-Cre mice (which directly express Cre in epiblast at E6.5) to generate p204+/− mice. For the purpose of genetic background consistency, p204+/− mice were used as parental mice to produce mice of p204−/− (KO) and p204+/+ (WT) genotypes (Fig. 1A). p204−/− (KO) mice were backcrossed with C57BL/6 for 10 generations before used for in vitro and in vivo experiments.
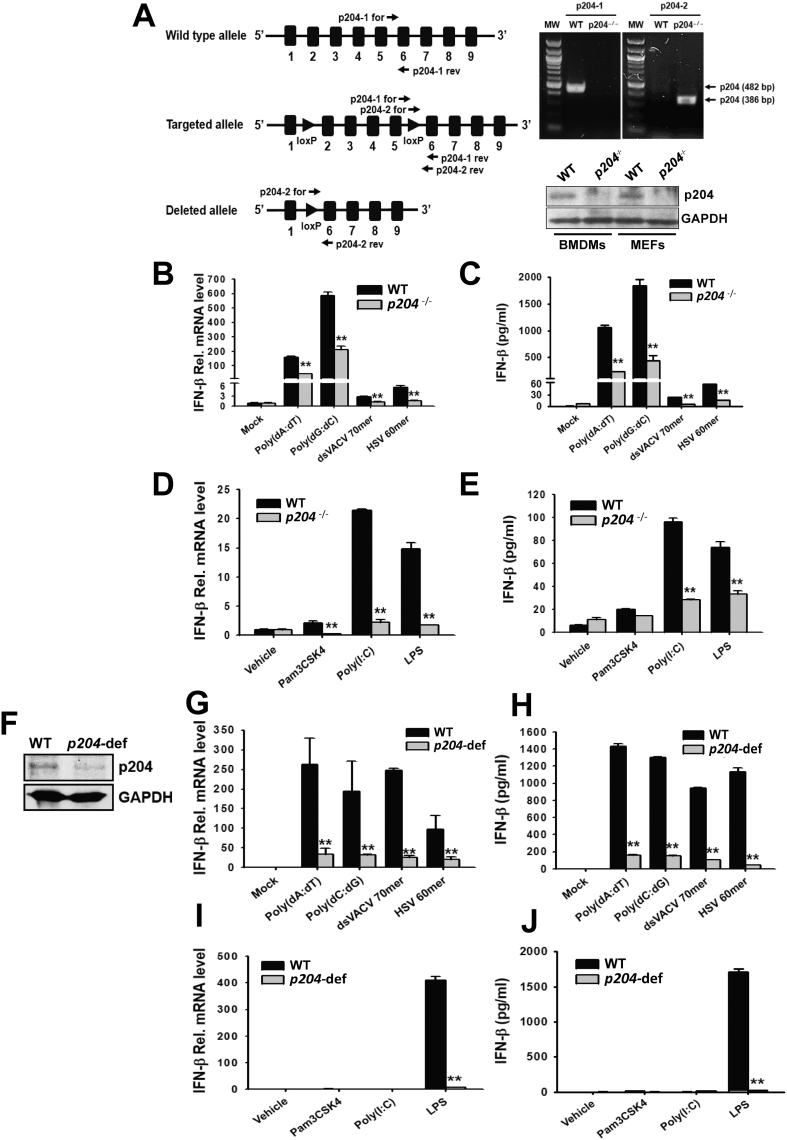
Fig. 1.
p204-deficiency suppresses IFN-β production in macrophages transfected with viral DNA sequences or treated with bacterial components. (A) Gene targeting strategy for the generation of p204−/− mice, and confirmation of p204 deletion by PCR using two different pairs of primers, p204–1 (482 bp) and p204–2 (386 bp) in the BMDMs isolated from p204−/− mice and by Western blot analysis in the indicated tissues of the p204−/− mice. Black boxes and numbers represent exons and exon numbers, respectively. The mRNA expression and the release of IFN-β in WT and p204−/− BMDMs transfected with indicated viral DNA sequences (B–C) or treated with indicated bacterial components (D–E). (F) Western blot analysis of targeted knock-down of p204 expression by CRISPR/Cas9 system in Raw264.7 macrophages. mRNA expression and release of IFN-β in WT and p204-deficient Raw264.7 macrophages transfected with indicated viral DNA sequences (G–H) or treated with indicated bacterial components (I–J). For the analysis of mRNA expression and release of IFN-β, BMDMs and Raw264.7 macrophages were transfected or treated for 6 h and 18 h, respectively. Bar graphs are presented as the mean ± SD of three independent experiments. *p < 0.05 and **p < 0.01.
2.2. Mice
C57BL/6 WT, Casp-11−/− and Tlr4−/− mice were purchased from Jackson Laboratory. All mice used in this study were bred and maintained in a specific pathogen-free facility at New York University Medical Center. All experiments were performed according to the experimental protocols approved by the Institutional Animal Care and Use Committee of New York University.
2.3. Generation of p204-deficient Raw264.7 Macrophages
p204 sgRNA sense (5′-CAG CGA AGT TGT TGC TGA GCC TTC C-3′) and anti-sense oligos (5′-AAA CGG AAG GCT CAG CAA CAA CTT C-3′) were synthesized and p204 sgRNA expression construct (sgp204) was generated by inserting p204 sgRNA duplex into a lentiCRISPRv2 plasmid (Addgene) as described previously (Shalem et al., 2014). Raw264.7 macrophages were transfected with sgp204 constructs using Lipofectamine® 2000 (Invitrogen) according to the manufacturer's instruction, and the transfected cells were selected using 2.0 μg/ml puromycine, Dihydrochloride (Millipore) in Dulbecco's Modified Eagle's medium (DMEM; Invitrogen) containing 10% fetal bovine serum (FBS; Atlanta Biologicals) until all non-transfected cells were dead.
2.4. Isolation of Mouse Bone Marrow-derived Macrophages (BMDMs)
Mouse BMDMs were isolated as described previously with slight modification. Briefly, BMDMs were isolated by flushing the femurs and tibiae of WT, p204−/−, Casp-11−/− and Tlr4−/− C57BL/6 mice (8–10 weeks) and plated in DMEM conditional media (10% FBS and 25% L929 mouse fibroblasts cell culture media in DMEM) providing macrophage-colony-stimulating factor (M-CSF). After 24 h, the suspended cells were re-plated in DMEM conditional media and differentiated for 5–6 days.
2.5. Cell Culture, Treatment and Transfection
BMDMs, Raw264.7 macrophages and HEK293T cells were maintained in DMEM supplemented with 10% FBS and grown at 37 °C in 5% CO2 humidified incubator. BMDMs and Raw264.7 macrophages were transfected with poly(dA:dT) (1 μg/ml), poly(dC:dG) (1 μg/ml), dsVACV70mer (1 μg/ml) or HSV60mer (1 μg/ml) using either Lipofectamine® 2000 according to the manufacturer's instruction or treated with Pam3CSK4 (10 ng/ml; InvivoGen), poly(I:C) (200 ng/ml; InvivoGen) or LPS (Serotype O111:B4; 1 μg/ml; InvivoGen) for 6 h or 18 h. BMDMs were also transfected with various serotypes of LPS (O111:B4, O55:B5, E. coli K12, P. gingivalis, S. Minnesota; InvivoGen and O127:B8; Sigma-Aldrich; 2 μg/ml) for 18 h using FuGENE® HD (Promega) according to the manufacturer's instruction. The cells and culture media were immediately used for RNA isolation and enzyme-linked immunosorbent assay (ELISA) experiments.
2.6. Enzyme-linked Immunosorbent Assay (ELISA)
Cell culture media was harvested from the transfected or treated BMDMs and Raw264.7 macrophages. Sera of the mice (8–10 weeks) were collected from whole blood by centrifugation using BD Microtainer® Amber Tubes with Serum Separator (BD) at 0 h, 1 h, 3 h and 6 h after intraperitoneal injection of LPS (1 mg/kg). The cell culture media and the mouse sera were used for ELISAs to measure the amount of IFN-β (PBL Assay Science), TNF-α(eBioscience), IL-6 (eBioscience), IL-1β(eBioscience) and IL-18 (eBioscience) according to the manufacturers' instructions. All experiments were repeated at least three times for statistical analysis.
2.7. Survival Rate
WT and p204−/− C57BL/6 mice (8–10 weeks) were intraperitoneally injected with lethal dose of LPS (Serotype O111:B4; 30 mg/kg bodyweight; Sigma-Aldrich) and the mice were monitored for signs of lethality twice daily for 4 days (n = 6 per group).
2.8. Preparation of Whole Cell Lysates, Nuclear and Cytoplasmic Proteins of Macrophages and Western Blot Analysis
Whole cell lysates of BMDMs and Raw264.7 macrophages were obtained by incubation with radioimmunoprecipitation assay (RIPA) buffer (50 mM Tric-HCl (pH 8.0), 150 mM NaCl, 1% nonidet p (NP)-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate) containing proteinase inhibitor cocktail (Sigma-Aldrich), phenylmethanesulfonyl fluoride (PMSF; Sigma-Aldrich) and sodium orthovanadate (Sigma-Aldrich) on ice for 30 min followed by centrifugation at 16,000 ×g for 10 min at 4 °C to remove cell debris. The supernatants of cell lysates were transferred to clean Eppendorf tubes and stored at −20 °C until use. Nuclear and cytoplasmic proteins of Raw264.7 macrophages were fractionated using Cytoplasmic and Nuclear Protein Extraction Kit (101 Bio) according to the manufacturer's instruction and stored at −20 °C until use.
For Western blot analysis, whole cell lysates, nuclear or cytoplasmic proteins of the cells were loaded and separated by SDS-polyacrylamide gels electrophoresis and transferred to polyvinylidenedifluoride membranes. After blocking the membranes with 3% bovine serum albumin (Sigma-Aldrich) in 0.1% Tris-buffered saline (TBS)-T (10 mM Tric-HCl (pH 7.5), 150 mM NaCl, 0.1% Tween-20) for 1 h at room temperature, the membranes were incubated with primary antibodies specific for p204 (Santa Cruz), p-TBK1 (Cell Signaling Technology), p-PI3K/p85 (Cell Signaling Technology), p-AKT (Cell Signaling Technology), p-IKKα/β (Cell Signaling Technology), IκBα (Santa Cruz), p- IκBα (Santa Cruz), NF-κB/p65 (Santa Cruz), IRF-3 (Santa Cruz), p-IRF-3 (Cell Signaling Technology), Lamin B (Santa Cruz), GFP (Santa Cruz), FLAG (Sigma-Aldrich) and GAPDH (Cell Signaling Technology) for 1 h at room temperature and washed three times with 0.1% TBS-T. The membranes were then incubated with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature and washed three times with 0.1% TBS-T. The specific bands were visualized using an Enhanced Chemiluminescence system (PerkinElmer).
2.9. Immunoprecipitation
BMDM from WT and p204 null mice were treated with LPS (1 μg/ml) for 4 h, and cells were lysed by RIPA lysis buffer. In another experiment, 293 T cells were transfected with GFP, p204-GFP, as well as p204 mutants tagged with GFP, and 24 h later cells were stimulated (1 μg/ml) for 4 h. Totally, 400 μg protein for each sample was used for immunoprecipitation. 2 μg/ml normal mouse and rabbit antibodies and 20 μl protein A/G agarose-beads were added, and incubated for 1 h at 4 °C to reduce non-specific binding followed by centrifugation at 3000 rpm for 5 min to pellet the beads. The supernatant was transferred to a new tube and 2 μg/ml primary antibodies were added and incubated for 1 h at 4 °C, then 20 μl protein A/G agarose-beads were added and incubated overnight. The beads were washed with RIPA lysis buffer 6–8 times, the samples were run on SDS-PAGE, and targeted proteins were probed with antibody and visualized by western-blot.
2.10. Flow Cytometry
BMDMs from WT and p204−/− mice were stimulated with LPS (100 ng/ml) for 2 h, and the cell surface TLR4-MD2 complex was stained with a specific antibody (MTS510). The percentage of the TLR4/MD-2 monomer was determined by the ratio of the MFI values of MTS510 staining of stimulated cells to those of the un-stimulated cells. The percentage of TLR4 dimerization was calculated as 100%-TLR4 monomer% as previous reported (Zanoni et al., 2016),
2.11. Real Time Polymerase Chain Reaction (Real time PCR)
Total RNAs were isolated from the BMDMs and Raw264.7 macrophages transfected or treated with each reagent using TRIzol® Reagent (Ambion) and first strand cDNAs were synthesized from the isolated total RNAs using reverse transcriptase (Promega) according to the manufacturers' instructions. Quantification and comparison of mRNA expression (IFN-β, TNF-α, IL-6, IL-1 β) were performed by real time PCR with SYBR® Green PCR Master Mix (Applied Biosystems) using a Real Time PCR System (Applied Biosystems). All experiments were repeated at least three times for statistical analysis. The primer sequences used for real time PCR in this study are listed in Table 1.
2.12. Luciferase Reporter Gene Assay
Raw264.7 macrophages were transfected with either NF-kB (Addgene) or IRF-3 luciferase reporter construct (gifted from Dr. Joanna Shisler) along with the construct expressing β-galactosidase using FuGENE® HD (Promega) according to the manufacturer's instruction. 24 h after transfection, the cells were treated with LPS (Serotype O111:B4; 1 μg/ml; InvivoGen) and further incubated for 24 h. The cells were then lysed and luciferase activities in the cell lysates were measured by Luciferase Assay System (Promega) according to the manufacturer's instruction. All experiments were repeated at least three times for statistical analysis.
2.13. Cytotoxicity
BMDMs were transfected with LPS (Serotype O111:B4; 2 μg/ml) using FuGENE® HD (Promega) for 18 h, and the amount of lactate dehydrogenase (LDH) in cell culture media were measured using Pierce® LDH Cytotoxicity Assay Kit (Thermo Fisher Scientific) according to the manufacturer's instruction. All experiments were repeated at least three times for statistical analysis.
2.14. Streptavidin Pull-down Assay
HEK293T cells were transfected with either GFP-p204 or FLAG-Casp-11 (Addgene) expressing construct using Lipofectamine® 2000 according to the manufacturer's instruction for 48 h, and pull-down assay was performed as described previously (Shi et al., 2014) with slight modification. Briefly, the transfected cells were lysed in the lysis buffer (50 mM Tric-HCl (pH 7.6), 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, proteinase inhibitor cocktail, PMSF and sodium orthovanadate) for 30 min on ice followed by centrifugation at 16,000 ×g for 10 min at 4 °C to collect supernatant. Supernatant was incubated with streptavidin agarose resin (Thermo Fisher Scientific) for its pre-clearance for 1 h at 4 °C with constant rotation. Biotin conjugated LPS (Biotin-LPS; InvivoGen) was immobilized onto streptavidin agarose resin, and unbound Biotin-LPS was removed by washing the resin three times with the lysis buffer. Pre-cleared supernatant was added to the Biotin-LPS bound streptavidin agarose resins for 1 h at 4 °C with constant rotation, and the resins were washed three times with the lysis buffer. The precipitates were eluted in 1 X SDS sample buffer followed by Western blot analysis.
2.15. Site-directed Mutagenesis
pEGFP vector containing the p204 or CD3 sequence was used as a template to create serial deletion mutants using a site-directed mutagenesis kit (Stratagene Ipswich, MA). RKR motifs were replaced with an AAA sequence in the p204 full length vector or CD3-GFP vector. All mutant constructs were confirmed by DNA sequencing and their expressions were examined by western-blot.
2.16. Statistics
All the data presented in this study are expressed as mean ± SD. For statistical comparison, all results were analyzed using paired Student's t-test using the Statistical Package for the Social Sciences Software 9SPSS version 15.0; IBM Corporation). For mouse septic shock study, Laplan-Meier survival curves were generated and analyzed for statistical significance. P value < 0.05 was considered to be significantly different between groups for all experiments.
3. Results
3.1. IFN-β Production Was Potently Suppressed in p204-deficient Macrophages in Response to Intracellular Viral DNAs and Extracellular Bacterial Components
To determine whether p204 is responsible for the production of type I IFN, p204−/− mice were generated by deleting exon 2 to exon 5 of p204 gene (Fig. 1A). Deletion of p204 gene was confirmed by PCR using two different pairs of primers, p204-1, which produces 482 base pairs only in wildtype (WT), and p204-2, which produces 386 base pairs only in p204−/− mice. The loss of p204 protein expression was further confirmed in bone marrow derived macrophages (BMDMs) and mouse embryonic fibroblasts by Western blot analysis (Fig. 1A). IFN-β production was measured in BMDMs isolated from p204−/− mice after transfection with various viral DNA sequences. IFN-β mRNA level and release were significantly suppressed in the p204−/− BMDMs transfected with viral DNA sequences compared to those of WT BMDMs (Fig. 1B, C). The reduced production of IFN-β is not due to the reduced number of BMDMs, as differentiations of macrophages and dendritic cells were not affected by p204 gene deletion (Fig. S1). IFN-β production was also measured in the BMDMs isolated from p204−/− mice treated with various bacterial components, such as Pam3CSK4, poly(I:C), and LPS. Unexpectedly, IFN-β mRNA expression level and release were also dramatically suppressed in the p204−/− BMDMs treated with poly(I:C) and LPS, but not Pam3CSK4 compared to those of WT BMDMs (Fig. 1D, E). In addition, p204−/− BMDMs lost responses to cytosolic DNA sensor, cGAMP, and PAM3Cys, the ligand of TLR2, to produce TNFα and IL-6 (Fig. S2A–D). Interestingly, p204−/− BMDMs also shown defective response to IFN- β to release pro-inflammatory cytokines, TNFα and IL-6 (Fig. S2E, F), suggesting that p204 may be involved in the regulation of the IFN-β autocrine loop.
To confirm these results in Raw264.7 macrophages, p204-deficient Raw264.7 macrophages were generated by using CRISPR/Cas9 system (Fig. 1F), and IFN-β production was measured in the p204-deficient Raw264.7 macrophages. Similar to the results from BMDMs, mRNA expression and release of IFN-β was significantly suppressed in the p204-deficient Raw264.7 macrophages transfected with viral DNA sequences (Fig. 1G, H) as well as treated with LPS (Fig. 1I, J) compared to those of WT Raw264.7 macrophages. However, unlike our observations in BMDMs, poly(I:C) did not induce the production of IFN-β in WT Raw264.7 macrophages, and no significant difference in IFN-β production was observed between WT and p204-deficient Raw264.7 macrophages transfected with poly(I:C) (Fig. 1I, J). These results demonstrate that p204 is responsible for the production of IFN-β in response to viral and bacterial challenges in macrophages.
3.2. Production of Pro-inflammatory Cytokines Was Strongly Suppressed in p204-deficient Macrophages Challenged by LPS
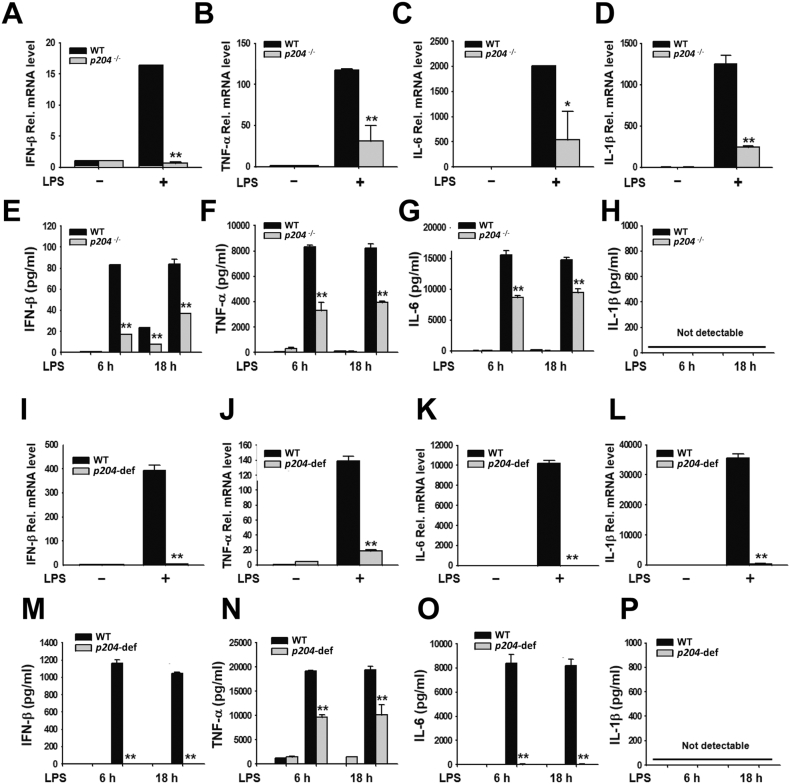
To examine whether p204 is responsible for inflammatory response, the production of pro-inflammatory cytokines was measured in p204-deficient macrophages. BMDMs were isolated and treated with LPS, and the production of IFN-β, TNF-α, IL-6 and IL-1β was determined. mRNA expression (Fig. 2A–C) and the secretion of IFN-β, TNF-α and IL-6 (Fig. 2E–G) were suppressed in the p204−/− BMDMs treated with LPS compared to those of WT BMDMs. Interestingly, mRNA expression of IL-1β was highly induced in LPS-treated WT BMDMs and significantly suppressed in LPS-treated p204−/− BMDMs compared to that of WT BMDMs, while its secretion in the cell culture media was not detectable in WT or p204−/− BMDMs treated with LPS (Fig. 2D, H). To confirm these results in Raw264.7 macrophages, p204-deficient Raw264.7 macrophages were treated with LPS, and the production of IFN-β, TNF-α, IL-6 and IL-1β was determined. In accordance with BMDM results, mRNA expression (Fig. 2I–K) and protein secretion in the cell culture media (Fig. 2M–O) of IFN-β, TNF-α and IL-6 were potently suppressed in the p204-deficient Raw264.7 macrophages treated with LPS as compared to WT Raw264.7 macrophages treated with LPS. mRNA expression and secretion of IL-1β in the Raw264.7 macrophages showed similar pattern to those of BMDMs. mRNA expression of IL-1β was suppressed in LPS-treated p204-deficient Raw264.7 macrophages compared to that of LPS-treated WT Raw264.7 macrophages, and its secretion in the cell culture media was not detectable in either WT or p204-deficient Raw264.7 macrophages treated with LPS (Fig. 2L, P). These results demonstrate that p204 is required for the production of not only type I IFN, IFN-β, but also pro-inflammatory cytokines, including TNF-α, IL-6 and IL-1β in response to LPS in macrophages in vitro.
Fig. 2.
Production of pro-inflammatory cytokines was suppressed in p204-deficient macrophages treated with LPS in vitro. mRNA expression (A–D) and release (E–H) of IFN-β, TNF-α, IL6 and IL-1β in WT and p204−/− BMDMs treated with LPS (1 μg/ml). mRNA expression (I–L) and release (M–P) of IFN-β, TNF-α, IL6 and IL-1β in WT and p204-deficient Raw264.7 macrophages treated with LPS (1 μg/ml). BMDMs and Raw264.7 macrophages were treated with LPS for 6 h and the indicated time for the analysis of mRNA expression and release of cytokines, respectively. Bar graphs are presented as the mean ± SD of three independent experiments. *p < 0.05 and **p < 0.01.
3.3. p204 Deficiency Suppressed the Production of Pro-inflammatory Cytokines in LPS-challenged Mice and Enhanced Mouse Survival Under LPS Septic Shock
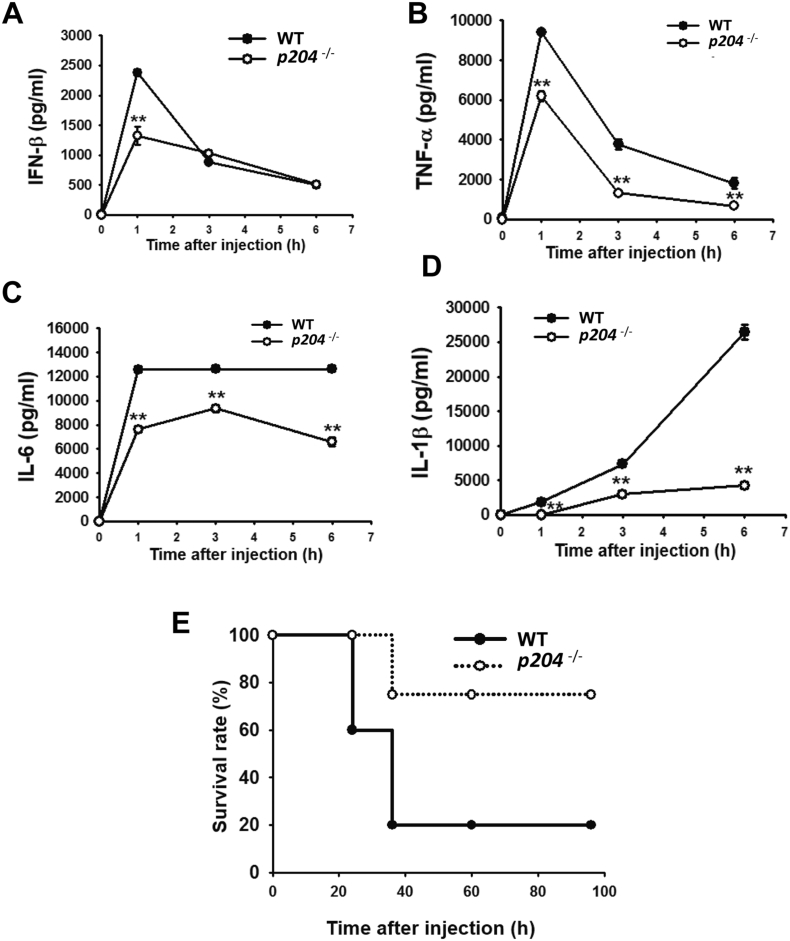
The production of pro-inflammatory cytokines was also examined in the sera of p204−/− mice. WT and p204−/− mice were intraperitoneally injected with LPS, and the serum levels of IFN-β, TNF-α, IL-6 and IL-1β were compared at different time points. The serum levels of all cytokines were lower in p204−/− mice than WT mice with LPS injection (Fig. 3A–D). The serum level of IFN-β was lower in p204−/− mice than WT mice at 1 h after LPS injection, but its levels were comparable between WT and p204−/− mice at 3 h and 6 h after LPS injection (Fig. 3A). Unlike IFN-β, the serum levels of TNF-α, IL-6 and IL-1β were continuously lower in p204−/− mice than WT mice from 1 h up to 6 h after LPS injection (Fig. 3B–D). Interestingly, the serum levels of IFN-β and TNF-α were highest at 1 h after LPS injection and gradually decreased at 3 h and 6 h (Fig. 3A–B). The serum level of IL-6 was also highest at 1 h after LPS injection, but the level was maintained up to 6 h in WT mice, while in p204−/− mice it was slightly increased at 3 h and decreased again at 6 h (Fig. 3C). The serum level of IL-1β was gradually increased over 1 h to 6 h after LPS injection (Fig. 3D). Next, the significance of in vitro and in vivo production of pro-inflammatory cytokines was examined with a mouse model of acute septic shock. WT and p204−/− mice were injected with a lethal dose of LPS (30 mg/kg), and their survival rates were measured. LPS injection caused severe morbidity in 80% of WT mice within 40 h, whereas onset of morbidity was significantly delayed in p204−/− mice and 75% of the p204−/− mice survived up to 100 h under lethal dose of LPS challenge (Fig. 3E). These results demonstrate that p204 is critical for the production of pro-inflammatory cytokines and host defense in vivo.
Fig. 3.
Serum levels of pro-inflammatory cytokines were suppressed and survival rate was increased in p204−/− mice injected with LPS in vivo. (A–D) Serum levels of IFN-β, TNF-α, IL-6 and IL-1β in WT and p204−/− mice injected intraperitoneally with LPS (1 mg/kg bodyweight) for the indicated time. Plots are presented as the mean ± SD of three independent experiments. (E) Kaplan-Meier survival plot for WT and p204−/− mice injected intraperitoneally with a lethal dose of LPS (30 mg/kg bodyweight) for the indicated time (n = 6). *p < 0.05 and **p < 0.01.
3.4. p204 Did Not Recognize Intracellular LPS and Was Dispensable for Inflammasome Activation
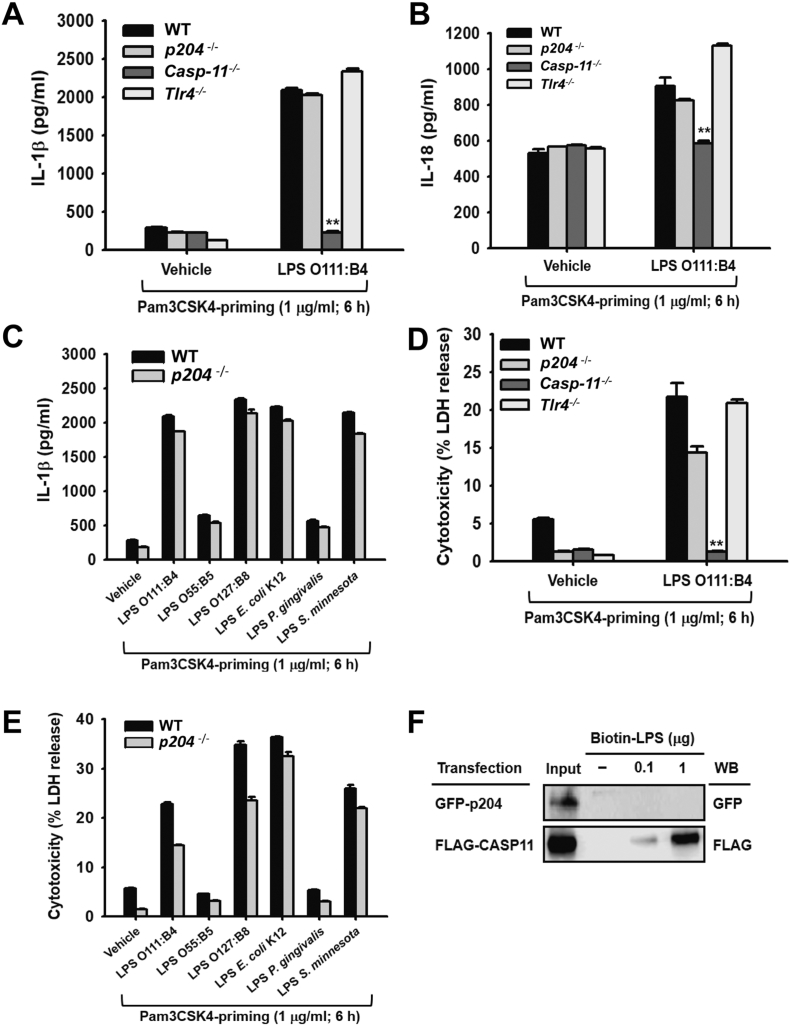
We next examined the possibility that p204 is a sensor/receptor for intracellular LPS in macrophages. BMDMs primed with Pam3CSK4 in advance were transfected with LPS, and the secretion of inflammasome-activated inflammatory cytokines, such as IL-1β and IL-18, and BMDM pyroptosis were determined. As expected, the secretion of IL-1β was highly induced in WT and Tlr4−/− BMDMs and was not induced in Casp-11−/− BMDMs with LPS transfection, while its secretion level in LPS-transfected p204−/− BMDMs was comparable with those of WT and Tlr4−/− BMDMs (Fig. 4A). The secretion of IL-18, another representative cytokine released by intracellular LPS-mediated inflammasome activation, was also comparable between LPS-transfected WT and p204−/− BMDMs, while its secretion was not induced in Casp-11−/− BMDMs (Fig. 4B). IL-1β secretion was measured in the p204−/− BMDMs transfected with various serotypes of LPS to test the possibility that different serotypes of LPS elicit suppressed levels of IL-1β release in LPS-transfected p204−/− BMDMs compared to WT BMDMs, and IL-1β secretion was comparable between LPS-transfected WT and p204−/− BMDMs regardless of LPS serotype (Fig. 4C). Moreover, LPS transfection significantly induced pyroptosis of WT and Tlr4−/− BMDMs and did not induce the pyroptosis of Casp-11−/− BMDMs, while the pyroptosis of p204−/− BMDMs was not dramatically reduced compared to that of WT BMDMs (Fig. 4D). A comparable level of pyroptosis was also observed between LPS-transfected WT and p204−/− BMDMs regardless of LPS serotypes (Fig. 4E). In addition, the binding of p204 with LPS was examined by streptavidin pull-down assay. LPS bound to Casp-11 in a dose-dependent manner, but did not bind with p204 (Fig. 4F). These results demonstrate that p204 does not recognize intracellular LPS, induce the production of IL-1β and IL-18, nor promote BMDM pyroptosis mediated by inflammasome activation in macrophages.
Fig. 4.
p204 does not recognize intracellular LPS and is dispensable for the activation of inflammasome signaling pathways in macrophages. (A) IL-1β and (B) IL-18 released from the BMDMs isolated from WT, p204−/−, CASP-11−/− and Tlr4−/− mice transfected with either vehicle or LPS (Serotype O111:B4, 2 μg/ml) for 18 h after Pam3CSK4 (1 μg/ml) priming for 6 h. (C) IL-1β released from the BMDMs isolated from WT and p204−/− mice transfected with either vehicle or indicated LPSs (2 μg/ml) for 18 h after Pam3CSK4 (1 μg/ml) priming for 6 h. (D) LDH levels in the cell culture media of the BMDMs isolated from WT, p204−/−, CASP-11−/− and Tlr4−/− mice transfected with either vehicle or LPS (Serotype O111:B4, 2 μg/ml) for 18 h after Pam3CSK4 (1 μg/ml) priming for 6 h. (E) LDH levels in the cell culture media of the BMDMs isolated from WT and p204−/− mice transfected with either vehicle or indicated LPSs (2 μg/ml) for 18 h after Pam3CSK4 (1 μg/ml) priming for 6 h. (F) Western blot analysis of GFP and FLAG of the whole cell lysates (input) and streptavidin pull-downs of either GFP-p204 or FLAG-CASP-11-transfected HEK293T cell lysates incubated with the indicated amounts of Biotin-LPS. Bar graphs are presented as the mean ± SD of three independent experiments. *p < 0.05 and **p < 0.01.
3.5. LPS-activated NF-κB and IRF-3 Signaling Pathways Were Suppressed in p204-deficient Macrophages
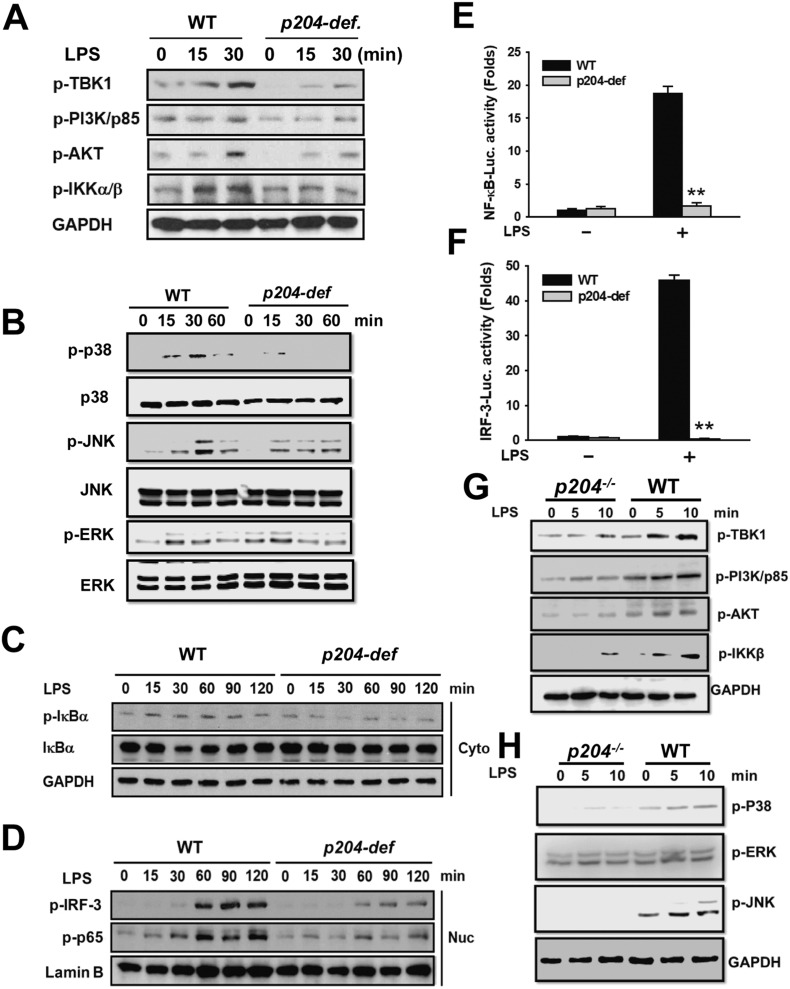
To investigate the molecular mechanisms by which p204-deficiency suppresses extracellular, but not intracellular, LPS-induced production of IFN-β and pro-inflammatory cytokines in macrophages, the activities of intracellular signaling molecules of NF-κB and IRF-3 pathways were examined in p204-deficient Raw264.7 macrophages with extracellular LPS challenge. The phosphorylation of intracellular signaling molecules of NF-ĸB pathway, including phosphoinositide 3-kinase (PI3K)/p85, AKT, inhibitor of NF-κB (IκBα) kinase alpha/beta (IKKα/β) and IκBα, and TRAF family member-associated NF-κB activator (TNAK)-binding kinase 1 (TBK1), a key intracellular signaling molecule of IRF-3 pathway, as well as MARK pathways were activated by extracellular LPS, as expected, in WT Raw264.7 macrophages, while their activations, with the exception of ERK, were reduced in p204-deficient Raw264.7 macrophages treated with LPS. Note that ERK activation was not reduced in p204-deficient cells (Fig. 5A, B). Nuclear translocation of transcription factors NF-κB/p65 and p-IRF-3 also showed time-dependent increase in LPS-treated WT Raw264.7 macrophages; however, their nuclear translocation was clearly inhibited in p204-deficient Raw264.7 macrophages treated with LPS (Fig. 5C, D). Next, transcriptional activities of NF-κB/p65 and p-IRF-3 were examined by luciferase reporter gene assay. Consistent with nuclear translocation results, transcriptional activities of both NF-κB/p65 and p-IRF-3 were activated in LPS-treated WT Raw264.7 macrophages, while their transcriptional activities were dramatically decreased in p204-deficient Raw264.7 macrophages treated with LPS (Fig. 5E, F). In addition, reduced activations of PI3K-AKT pathways and downstream NF-κB activation, as well as MAPK pathways p38 and JNK, were also observed in p204−/− BMDMs (Fig. 5G, H). These results demonstrate that p204 is needed for canonical LPS-activated NF-κB and IRF-3 signaling pathways for the production of pro-inflammatory cytokines and IFN in macrophages.
Fig. 5.
NF-κB and IRF-3 signaling pathways are suppressed in p204-deficient macrophages treated with LPS. WT and p204-deficient Raw264.7 macrophages were treated with LPS (1 μg/ml) for the indicated time. (A) Phosphorylated forms of TBK1, PI3K/p85, AKT, and IKKα/β were determined by Western blot analysis in the whole lysates of the cells. (B) Activation of MAPK pathway in WT and p204-deficient Raw264.7 macrophages was determined by Western blotting. (C)Phosphorylated and total form of IκBα was determined by Western blot analysis in the cytoplasmic fraction (Cyto) of the cells. (D) Phosphorylated IRF-3 and NF-κB/p65 were determined by Western blot analysis in the nuclear fraction (Nuc) of the cells. GAPDH and lamin B were used for the internal control for the cytoplasmic and nuclear fraction, respectively. Luciferase reporter gene assay of (E) NF-κB promoter and (F) IRF-3 promoter activity in WT and p204-deficient Raw264.7 macrophages treated with LPS (1 μg/ml) for 24 h. (G) WT and p204−/− BMDMs were treated with LPS (1 μg/ml) for the indicated time, and phosphorylation of TBK1, PI3K/p85, AKT, and IKKα/β was determined by Western blotting. (H) Phosphorylation of MAPK pathways in WT and p204−/− BMDMs following LPS treatment at indicated time points was determined by Western blotting. Bar graphs are presented as the mean ± SD of three independent experiments. *p < 0.05 and **p < 0.01.
3.6. p204 binds to TLR4 and it is Required for TLR4 Dimerization
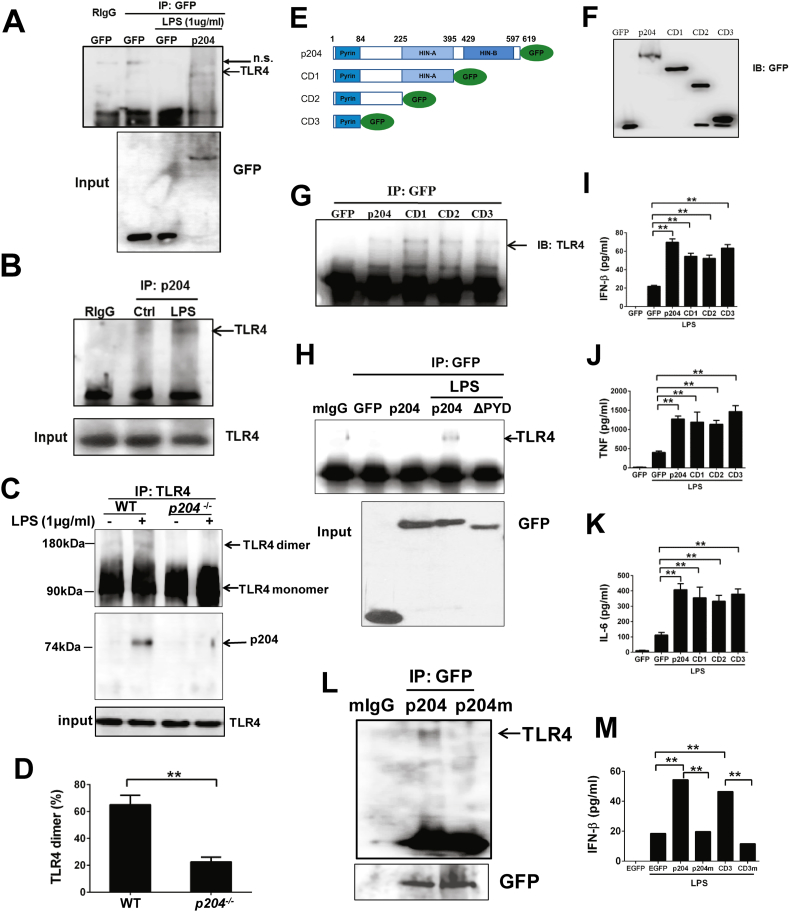
The findings that extracellular, but not intracellular, LPS signaling is defective in p204 deficient cells (Fig. 1, Fig. 2, Fig. 3, Fig. 4), and that the LPS/TLR4 signaling pathway, evidenced by loss of PI3K and TBK1 activation, is blocked in p204 null cells (Fig. 5A), led us to hypothesize that p204 may function as an early component of the TLR4 pathway. We thus tested whether p204 could associate with TLR4. First we performed a co-immunoprecipitation assay using 293 T cells co-transfected with a p204-GFP expression plasmid with a combination of TLR4, MD2 and CD14 expression constructs. The results demonstrated that p204 bound to TLR4 following LPS stimulation (Fig. 6A). LPS-dependent association between endogenous p204 and TLR4 was further confirmed by co-immunoprecipitation with BMDMs from WT mice (Fig. 6B).
Fig. 6.
p204 binds to TLR4 and is required for LPS-triggered TLR4 dimerization. (A) p204 binds to TLR4 in overexpressed 293T cells. GFP control vector and p204-GFP expression plasmid were co-transfected with a combination of TLR4, MD2 and CD14 expressing constructs, followed by LPS challenge. The cell lysates were pulled down by GFP antibody, and probed with TLR4 antibody (upper panel) and the inputs were probed with GFP antibody (lower panel). (B) Endogenous p204 binds to TLR4 followed LPS stimulation. Raw cells were treated with LPS (1 μg/ml) for 2 h, and the cell lysates were used for immunoprecipitation with p204 antibody. The interaction between p204 and TLR4 was detected with TLR4 antibody. (C) p204 is required for TLR4 dimerization. BMDM isolated from WT and p204 KO mice were treated with LPS (1 μg/ml) for 2 h, then cell lysates were prepared. The TLR4 antibody was used for immunoprecipitation, and the samples were run on non-reducing gel to visualize the dimerization of TLR (upper panel), and p204 binds to TLR4 in WT, but not in p204−/− BMDMs, following LPS treatment (middle panel), and the input of TLR4 was comparable (lower panel). (D) Dimerization of membrane TLR4 is defective in p204−/− BMDMs. BMDMs from WT and p204−/− were stimulated with LPS (100 ng/ml) for 2 h. The TLR4-MD2 complex was stained with the specific antibody (MTS510). Percentage of dimerization of TLR4 was calculated based on previous report (Zanoni et al., 2016). (E) Structure of p204 serial deletion mutants. To determine the binding domain that mediates the interaction between p204 and TLR4, we generated serial p204 deletions from its C-terminus, and all the constructs were fused with a GFP tag. (F) The expression of p204 constructs in (E) was examined by western-blot. (G) Pyrin domain alone is sufficient for binding to TLR4. p204 serial constructs were co-transfected with TLR4-expression plasmid into 293 T cells, followed by LPS stimulation. The IP experiment was performed by using antibody against GFP, and samples were probed with TLR4 antibody. (H) Pyrin domain is required to mediate the interaction between p204 and TLR4. The expression constructs of GFP, p204-GFP, or p204-GFP lacking Pyrin domain (ΔPYD) were transfected into 293 T cells, and the cells were stimulated with LPS. The immunoprecipitation was performed with anti-GFP antibody, and the immunoprecipitated complexes were detected with anti-TLR4 antibody. (I–K) Pyrin domain also can restore the response to LPS in p204-deficient RAW cells. p204-deficient RAW cells were transiently transfected with GFP control vector, p204-GFP, and its serial deletion mutants. After 24 h after transfection, LPS (1 μg/ml) were added to the cell culture medium for additional 24 h. The levels of IFN-β (I), TNF-α (J), and IL-6 (K) were measured by ELISA. (L) RKR motif was replaced with AAA by site-directed mutagenesis in p204-GFP expressing plasmids. The p204-GFP and p204m-GFP plasmids were co-transfected with TLR4 expression plasmid into 293 T cells. After LPS treatment, The immunoprecipitation was performed with anti-GFP antibody, and the immunoprecipitated complexes were detected with anti-TLR4 antibody. (M) RKR motif is critical to restore LPS-triggered IFN-β production in p204-deficient RAW cells. RKR motif was replaced with AAA motif using site-directed mutagenesis in CD3-GFP expressing plasmid. GFP, p204-GFP, CD3-GFP, and CD3m-GFP were transiently transfected into p204-deficient RAW cells and stimulated with LPS for 24 h. The IFN-β levels were measured by ELISA.
As the dimerization of TLR4 is important for the activation of its downstream signaling pathway, we next tested whether p204 affected TLR dimerization. Briefly, BMDM from WT and p204 null mice were treated with LPS and immunoprecipitated with TLR4 antibody, and the TLR dimerization was analyzed in a non-reducing gel. As shown in Fig. 6C, TLR4 dimer is detectable in WT BMDMs, and the amount of dimer is significantly enhanced following LPS treatment. However, in p204-deficient BMDMs, TLR4 is exclusively in monomer form, as the TLR4 dimer was undetectable, even after LPS stimulation (Fig. 6C). These results indicated that p204 is required for LPS-trigged TLR4 dimerization. This result was further confirmed by staining with TLR4/MD2 specific antibody MTS510 (Zanoni et al., 2016). Around 70% TLR4 dimerization was detected following LPS stimulation in WT BMDMs, whereas the percentage of TLR4 dimerization was markedly reduced in p204−/− BMDMs (Fig. 6D).
p204 contains a N-terminal Pyrin domain, and HIN200 domains (HinA and HinB) in the C-terminal. To further dissect the binding domain that mediates the interaction between p204 and TLR4, we created serial p204 C-terminal deletion mutations and expressed mutations with a GFP tag (Fig. 6E, F). All p204 constructs were co-transfected with a TLR4 expressing plasmid into 293 T cells. Immunoprecipitation was performed using GFP antibody and TLR4 antibody probe. The results revealed that p204 CD3 alone was sufficient to bind to TLR4 (Fig. 6G), indicating that the N-terminal Pyrin domain of p204 was responsible for its interaction with TLR4. In addition, deletion of Pyrin domain abolished the interaction between p204 and TLR4 (Fig. 6H). Next, we examined whether these p204 mutants were able to rescue the defective response to extracellular LPS seen in p204 deficient cells. After transient transfection of these p204 mutants into p204 deficient Raw cells, we found that transfection of full-length p204 could restore the p204 deficient Raw cells' response to LPS treatment in terms of pro-inflammatory cytokine expressions, including IFN-β, TNF-α, and IL-6 (<10% transfection rate) (Fig. 6I–K). Intriguingly, the Pyrin domain alone could also restore the response to LPS in p204 deficient Raw cells (Fig. 6I–K). These data further indicate the Pyrin domain of p204 is sufficient for binding to TLR4 and for the activation of TLR4 signaling pathway following LPS treatment.
Since the Pyrin domain is common to the majority of murine p200 family members, we next determined whether the Pyrin domain of other members in this family could restore the defective response to LPS in p204-deficient cells. As shown in Fig. S3C, Pyrin domains of p206 and AIM2 failed to rescue the defective responses to LPS in p204-deficient cells. These results led us to examine whether the Pyrin domain of p204 contains a unique motif that may be important for its interaction with TLR4. By comparing the sequences of Pyrin domain from murine p200 family members, we found three consecutive positively charged amino acids RKR motif is present in p204 and its closest homologs, p207 and p205, but absent in the rest of murine p200 family members (Fig. S3 A, B). Both p207 and p205 were undetectable in wildtype and p204 null macrophages in a gene-array screening (data not shown). Interestingly, a motif of three consecutive, negatively charged amino acids (EEE) was present in the intracellular domain of murine TLR4 (Kim et al., 2007). To test whether the RKR motif is important for the interaction of p204 with TLR4, we mutated RKR motif to AAA in both p204 full-length and the p204 CD3 fragment by site-directed mutagenesis, and confirmed the mutation by DNA sequencing. After co-transfection with TLR4 into 293 T cells, we found that RKR-AAA mutation in p204 (p204m) abolished its binding activity to TLR4 (Fig. 6L). In addition, similar RKR-AAA mutation in CD3 (CD3m) blunted the ability of the CD3 fragment to restore LPS-triggered IFN-β production in p204−/− Raw cells (Fig. 6M). Taken together, these results demonstrate that p204 is required for TLR4 dimerization through its Pyrin domain in which the RKR motif is essential for p204/TLR4 association.
4. Discussion
Several human p200 protein family members, such as IFI16 and AIM2, have been reported to recognize intracellular pathogen components, leading to the production of type I IFNs in cooperation with other intracellular sensors and inflammasomes in macrophages for host defense (Unterholzner et al., 2010; Jakobsen et al., 2013; Ansari et al., 2015; Dell'Oste et al., 2015; Diner et al., 2015; Storek et al., 2015; Watson et al., 2015; Herzner et al., 2015; Orzalli et al., 2015; Cai et al., 2014; Hansen et al., 2014; Meunier et al., 2015; Karki et al., 2015). p204 is a murine p200 protein family member that regulates cell proliferation, differentiation and apoptosis (Zhao et al., 2015, Lengyel and Liu, 2010, Luan et al., 2008a, Ding and Lengyel, 2008, Ding et al., 2006a, Ding et al., 2006b, Liu et al., 2000, Liu et al., 2005, Liu et al., 2002, Liu et al., 1999, Luan et al., 2007, Luan et al., 2008b, De Andrea et al., 2002b, De Andrea et al., 2002a). Recently, p204 was also reported as a sensor against bacterial infection that produces type I IFNs in cooperation with cGAS (Storek et al., 2015), however, how and what pathogen components stimulate p204-mediated innate immunity in macrophages remains unknown. In this study, we demonstrated that p204 is a critical intracellular mediator for induction of IFN-β production and inflammatory responses in macrophages in response to extracellular LPS, one of the most dominant and pathogenic components of gram negative bacteria through TLR4 signaling pathways. More importantly, we provide evidence demonstrating the association of p204/TLR4 and the importance of p204 in LPS-induced TLR4 dimerization and subsequent activation of its downstream signaling pathways.
We have generated the first reported p204−/− mice for our studies allowing us to extend findings from previous studies that have used p204 siRNA, which only partially reduces p204 expression (Unterholzner et al., 2010; Storek et al., 2015). The size and body weight of p204−/− mice were comparable with those of WT mice at same age, and no defect of appearance, behavior and reproduction was observed (data not shown). p204 is an ortholog of human IFI16, also reported as a sensor for some viral DNA sequences, such as dsVACV 70mer and HSV 60mer (Unterholzner et al., 2010) and bacterial infection (Hornung et al., 2009). Therefore, we examined the possibility of whether p204 is a murine counterpart of IFI16 in function, and IFN-β production in p204-deficient macrophages transfected with various viral DNA sequences was examined. Similar to IFI16 results, IFN-β production was significantly suppressed in p204-deficient macrophages transfected with not only previously reported viral DNA sequences, dsVCAV 70mer and HSV 60mer (Unterholzner et al., 2010), but also newly tested viral DNA sequences, poly(dA:dT) and poly(dC:dG). These results indicate that p204 is a sensor of intracellular viral DNA as reported previously (Unterholzner et al., 2010).
While investigating p204 as a sensor of intracellular viral DNAs in p204 null mice, we, unexpectedly, found that BMDMs from p204 null mice lost response to LPS treatment, originally implemented as a negative control for viral DNA. The expressions of IFN-β and inflammatory cytokines, including IL-1β, were significantly reduced in p204 null BMDMs (Fig. 2A–G). Similar data were also obtained with p204-deficicent RAW cells generated with CRISPR-Cas9 technology (Fig. 2I–O). These in vitro data clearly showed that p204 is a critical intracellular protein not only acting as a viral DNA sensor, but is also required to trigger innate immune response against bacterial infection, specifically Gram-negative bacteria.
It has been widely reported that LPS can cause severe systemic inflammation that mimics septic shock. To confirm the critical function of p204 in LPS-triggered response in vivo, we I.P. injected LPS in both WT and p204 null mice. As expected, serum levels of all pro-inflammatory cytokines were lower in p204−/− mice than WT mice with LPS challenge. However, production profiles of pro-inflammatory cytokines were different across time points, demonstrating that the production and metabolism of each pro-inflammatory cytokine is differentially regulated by p204 (Fig. 3). We further evaluated the survival rate of mice injected with a lethal dose of LPS. Many studies have reported that production of type I IFNs and inflammation in the context of bacterial infection strongly correlates with decreased host survival (Auerbuch et al., 2004). Indeed, p204−/− mice were much more resistant to the lethal dose of LPS injection and showed a significantly higher survival rate than WT mice, strongly suggesting that p204 plays a pivotal role for host defense against bacterial infection through producing type I IFNs and activating inflammatory responses in macrophages.
It is well-established that LPS induces the production of pro-inflammatory cytokines and various inflammatory mediators in macrophages through activating NF-κB signaling pathway which is regulated by a number of upstream signaling molecules, including spleen tyrosine kinase (SYK), SRC, PI3K/p85, AKT, IKKα/β and IκBα. In contrast, LPS induces the production of type I IFNs in macrophages through activating the IRF-3 signaling pathway, which is mainly regulated by the key upstream signaling molecule TBK1. Therefore, we evaluated the activities of these signaling molecules in p204-deficient macrophages with LPS challenge. In accordance with the reduced production of pro-inflammatory cytokines and IFN-β in p204-deficient macrophages and mice, the activities of signaling molecules involved in both NF-κB and IRF-3 signaling pathways were significantly decreased, and nuclear translocation as well as the transcriptional activities of both transcription factors NF-κB/p65 and phosphor-IRF-3 were also dramatically suppressed in LPS-stimulated p204-deficient macrophages. These findings demonstrate that p204 is a critical modulator of both NF-κB and IRF-3 signaling pathways in response to LPS, leading to the production of pro-inflammatory cytokines and type I IFNs in macrophages (Please see proposed working model in Fig. 7). Interestingly, the activity of PI3K/p85, which is one of the early upstream signaling molecules in NF-κB pathway, was significantly suppressed in p204-deficient macrophages, suggesting that p204 is an upstream regulator of signaling pathways and that its effect is efficiently amplified downstream (Fig. 4, Fig. 7). Intriguingly, it was reported that the extracellular IFI16 propagated inflammation in endothelial cells via p38 MAPK and NFĸB p65 activation (Bawadekar et al., 2015). Whether p204 is also secreted and if it is, whether extracellular p204 activates MAPK and NF-κB and regulates inflammation, need further investigation.
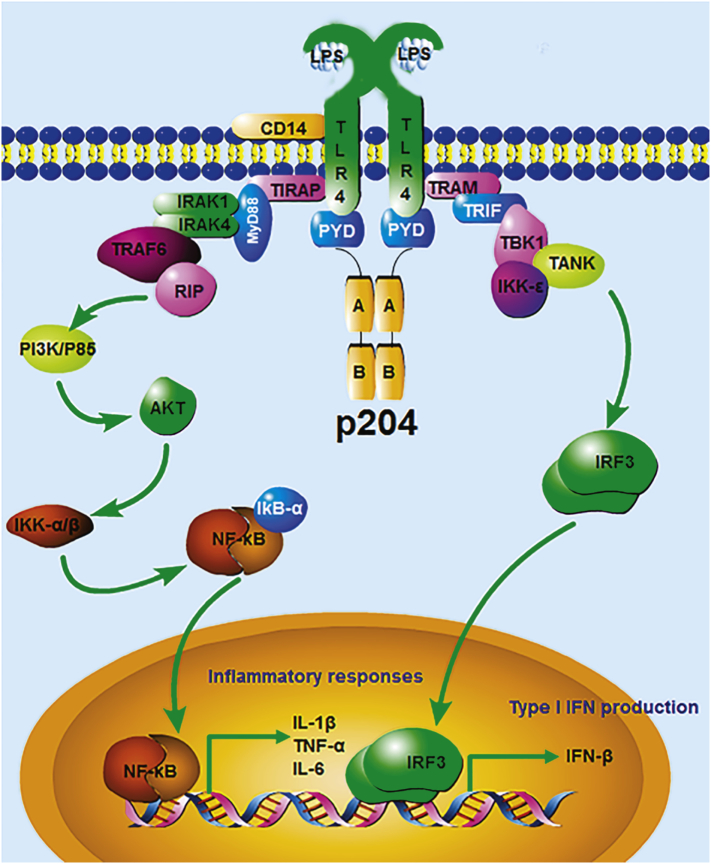
Fig. 7.
Illustration of p204 function in LPS/TLR4 signaling pathway. p204 is recruited to TLR4 receptor complex following LPS stimulation. p204, known to form a homodimer through its C-terminal HIN-200 domain, induces the dimerization of TLR4 through the binding of its N-terminal Pyrin domain to TLR4, followed by the activation of downstream IRF-3 and NF-ĸB signaling pathways and the release of IFN-β and pro-inflammatory cytokines, respectively. “PYD”: Pyrin domain; “A” and “B”: p204 C-terminal HIN-A and HIN-B domain respectively.
Intracellular p204 binds to TLR4, and it is required for TLR4 dimerization after LPS challenge (Fig. 6). In addition, the N-terminal Pyrin domain of p204 mediates binding and dimerization of TLR4. Furthermore, the RKR motif in p204's Pyrin domain is essential for its binding to TLR4 and activation. TLR4 dimerization is driven by the extracellular binding of an MD2/LPS complex that subsequently permits re-orientation of the intracellular Toll–interleukin receptor resistance (TIR) domains of two TLR4 receptors and the assembly of the “MyDDosome” complex (Ferrao et al., 2012; Ferrao and Wu, 2012; Lin et al., 2010). In addition, the “MyDDosome” has been shown to differ substantially in mouse and human cells through selective utilization of the components in the complex (Perkins and Vogel, 2016). The mechanisms by which p204 enhances TLR4 dimerization, such as whether and how p204 affects MyDDosome formation, whether p204 affects the translocation or degradation of TLR4, and whether the effect is specific to mice or whether it also applies to other species, warrant further investigations. Intriguingly, p204 also interacts with TLR3 (Fig. S4), and p204 deficiency also impairs poly(I:C)-induced IFN-β production (Fig. 1), suggesting that p204 may associate with several members in TLR family, which also warrants further studies.
Most of the previous studies regarding LPS have focused predominately on extracellular LPS because TLR4 was identified as a LPS-specific cell surface receptor and binding of extracellular LPS to TLR4 activates a variety of signaling pathways in innate immune cells, including macrophages. However, macrophages capture LPS on gram negative bacteria and phagocytose the bacteria into the cytoplasm for digestion and initiate innate immune responses. Therefore, LPS could be released in the cytoplasm and it is highly possible that cytoplasmic sensors could recognize intracellular LPS and induce immune responses in macrophages. Indeed, several groups have recently tested this possibility and reported that intracellular LPS is directly recognized by one of the inflammatory caspases, caspase-11 (CASP-11), followed by activation of non-canonical inflammasome pathways in macrophages, subsequently leading to the induction of pyroptosis and the release of IL-1β and IL-18, which are representative inflammasome-activated inflammatory cytokines (Kayagaki et al., 2011; Kayagaki et al., 2013; Hagar et al., 2013; Shi et al., 2014; Kayagaki et al., 2015; Shi et al., 2015). Inspired by these studies, we tested the possibility whether p204 directly recognizes intracellular LPS to induce pyroptosis and the release of IL-1β and IL-18 in macrophages. However, unlike CASP-11, p204 did not directly recognize intracellular LPS, moreover, it did not induce pyroptosis of macrophages and the release of IL-1β and IL-18 regardless of LPS serotypes in macrophages, suggesting that p204, which is essential for extracellular LPS/TLR4 signaling, is not an intracellular receptor for intracellular LPS and is dispensable for intracellular LPS-mediated inflammasome activation in macrophages.
In summary, we present in vivo evidences from the first p204 knockout line that confirms p204 as an intracellular viral DNA sensor. Most significantly, we also present evidence demonstrating that p204 is a critical intracellular mediator of IFN-β and inflammatory response in response to extracellular, but not intracellular, LPS, the most significant pathogenic component of gram negative bacteria. p204 binds to TLR4 through its Pyrin domain, and it is required for TLR4 dimerization following LPS challenge. These results not only present p204 as a previously-unknown component of canonical LPS signaling, but may also provide new insights into the complicated host defense during gram negative bacterial infection.
Conflict of Interest
We herein declare that we have no conflict of interest.
Author's Contribution
Y. Yi, J. Jian, E. Gonzalez-Gugel, Y. Shi designed and performed experiments, collected and analyzed data, and wrote the paper. Q. Tian, W. Fu, A. Hettinghouse, W. Song, R. Liu, M. He, H. Qi, J. Yang, and X. Du performed experiments, collected and analyzed data. G. Xiao analyzed data and edited the manuscript. L. Chen and C.J. Liu designed and supervised this study, analyzed data, and edited the manuscript. All authors contributed discussions and interpretations.
Acknowledgements
We thank Dr. Joanna Shisler (University of Illinois at Urbana Champaign) for providing IRF-3 luciferase expressing construct. This work was supported partly by NIH research grants R01AR062207, R01AR061484, R01NS103931, and a DOD research grant W81XWH-16-1-0482. It's also supported by the Special Funds for Major State Basic Research Program of China (973 program) (No. 2014CB942904), National Natural Science Foundation of China (No. 81530071, No. 81630066), Natural Science Foundation of Shandong Province (No. ZR2016CM13). Funders had no role in study design, data collection, data analysis, interpretation, writing of the report.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.02.012.
Contributor Information
Lin Chen, Email: linchen70@163.com.
Chuan-ju Liu, Email: chuanju.liu@nyumc.org.
Appendix A. Supplementary Data
Supplementary material
References
- Akira S., Takeda K., Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat. Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- Ansari M.A., Dutta S., Veettil M.V., Dutta D., Iqbal J., Kumar B., Roy A., Chikoti L., Singh V.V., Chandran B. Herpesvirus genome recognition induced acetylation of nuclear IFI16 is essential for its cytoplasmic translocation, inflammasome and IFN-beta responses. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbuch V., Brockstedt D.G., Meyer-Morse N., O'riordan M., Portnoy D.A. Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. J. Exp. Med. 2004;200:527–533. doi: 10.1084/jem.20040976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton G.M., Medzhitov R. Toll-like receptor signaling pathways. Science. 2003;300:1524–1525. doi: 10.1126/science.1085536. [DOI] [PubMed] [Google Scholar]
- Bawadekar M., De Andrea M., Lo Cigno I., Baldanzi G., Caneparo V., Graziani A., Landolfo S., Gariglio M. The extracellular IFI16 protein propagates inflammation in endothelial cells via p38 MAPK and NF-kappaB p65 activation. J. Interf. Cytokine Res. 2015;35:441–453. doi: 10.1089/jir.2014.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckstummer T., Baumann C., Bluml S., Dixit E., Durnberger G., Jahn H., Planyavsky M., Bilban M., Colinge J., Bennett K.L., Superti-Furga G. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat. Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- Cai X., Chiu Y.H., Chen Z.J. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol. Cell. 2014;54:289–296. doi: 10.1016/j.molcel.2014.03.040. [DOI] [PubMed] [Google Scholar]
- Chiu Y.H., Macmillan J.B., Chen Z.J. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrady C.D., Zheng M., Fitzgerald K.A., Liu C., Carr D.J. Resistance to HSV-1 infection in the epithelium resides with the novel innate sensor, IFI-16. Mucosal Immunol. 2012;5:173–183. doi: 10.1038/mi.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Andrea M., Ravotto M., Noris E., Ying G.G., Gioia D., Azzimonti B., Gariglio M., Landolfo S. The interferon-inducible gene, Ifi204, acquires malignant transformation capability upon mutation at the Rb-binding sites. FEBS Lett. 2002;515:51–57. doi: 10.1016/s0014-5793(02)02431-6. [DOI] [PubMed] [Google Scholar]
- De Andrea M., Zannetti C., Noris E., Gariglio M., Azzimonti B., Landolfo S. The mouse interferon-inducible gene Ifi204 product interacts with the Tpr protein, a component of the nuclear pore complex. J. Interf. Cytokine Res. 2002;22:1113–1121. doi: 10.1089/10799900260442539. [DOI] [PubMed] [Google Scholar]
- Dell'Oste V., Gatti D., Giorgio A.G., Gariglio M., Landolfo S., De Andrea M. The interferon-inducible DNA-sensor protein IFI16: a key player in the antiviral response. New Microbiol. 2015;38:5–20. [PubMed] [Google Scholar]
- Diner B.A., Lum K.K., Javitt A., Cristea I.M. Interactions of the antiviral factor interferon gamma-inducible protein 16 (IFI16) mediate immune signaling and herpes simplex virus-1 immunosuppression. Mol. Cell. Proteomics. 2015;14:2341–2356. doi: 10.1074/mcp.M114.047068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B., Lengyel P. p204 protein is a novel modulator of Ras activity. J. Biol. Chem. 2008;283:5831–5848. doi: 10.1074/jbc.M709680200. [DOI] [PubMed] [Google Scholar]
- Ding B., Liu C.J., Huang Y., Hickey R.P., Yu J., Kong W., Lengyel P. p204 is required for the differentiation of P19 murine embryonal carcinoma cells to beating Cardiac myocytes: its expression is activated by the cardiac GATA4, NKX2.5, and TBX5 proteins. J. Biol. Chem. 2006;281:14882–14892. doi: 10.1074/jbc.M511747200. [DOI] [PubMed] [Google Scholar]
- Ding B., Liu C.J., Huang Y., Yu J., Kong W., Lengyel P. p204 protein overcomes the inhibition of the differentiation of P19 murine embryonal carcinoma cells to beating cardiac myocytes by Id proteins. J. Biol. Chem. 2006;281:14893–14906. doi: 10.1074/jbc.M511748200. [DOI] [PubMed] [Google Scholar]
- Fernandes-Alnemri T., Yu J.W., Datta P., Wu J., Alnemri E.S. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrao R., Wu H. Helical assembly in the death domain (DD) superfamily. Curr. Opin. Struct. Biol. 2012;22:241–247. doi: 10.1016/j.sbi.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrao R., Li J., Bergamin E., Wu H. Structural insights into the assembly of large oligomeric signalosomes in the Toll-like receptor-interleukin-1 receptor superfamily. Sci. Signal. 2012;5 doi: 10.1126/scisignal.2003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay N.J., Symmons M.F., Gangloff M., Bryant C.E. Assembly and localization of Toll-like receptor signalling complexes. Nat. Rev. Immunol. 2014;14:546–558. doi: 10.1038/nri3713. [DOI] [PubMed] [Google Scholar]
- Goubau D., Schlee M., Deddouche S., Pruijssers A.J., Zillinger T., Goldeck M., Schuberth C., van der Veen A.G., Fujimura T., Rehwinkel J., Iskarpatyoti J.A., Barchet W., Ludwig J., Dermody T.S., Hartmann G., Reis E Sousa C. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature. 2014;514:372–375. doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagar J.A., Powell D.A., Aachoui Y., Ernst R.K., Miao E.A. Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science. 2013;341:1250–1253. doi: 10.1126/science.1240988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen K., Prabakaran T., Laustsen A., Jorgensen S.E., Rahbaek S.H., Jensen S.B., Nielsen R., Leber J.H., Decker T., Horan K.A., Jakobsen M.R., Paludan S.R. Listeria monocytogenes induces IFNbeta expression through an IFI16-, cGAS- and STING-dependent pathway. EMBO J. 2014;33:1654–1666. doi: 10.15252/embj.201488029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzner A.M., Hagmann C.A., Goldeck M., Wolter S., Kubler K., Wittmann S., Gramberg T., Andreeva L., Hopfner K.P., Mertens C., Zillinger T., Jin T., Xiao T.S., Bartok E., Coch C., Ackermann D., Hornung V., Ludwig J., Barchet W., Hartmann G., Schlee M. Sequence-specific activation of the DNA sensor cGAS by Y-form DNA structures as found in primary HIV-1 cDNA. Nat. Immunol. 2015;16:1025–1033. doi: 10.1038/ni.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V., Ablasser A., Charrel-Dennis M., Bauernfeind F., Horvath G., Caffrey D.R., Latz E., Fitzgerald K.A. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A., Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 2004;5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- Jakobsen M.R., Bak R.O., Andersen A., Berg R.K., Jensen S.B., Tengchuan J., Laustsen A., Hansen K., Ostergaard L., Fitzgerald K.A., Xiao T.S., Mikkelsen J.G., Mogensen T.H., Paludan S.R. IFI16 senses DNA forms of the lentiviral replication cycle and controls HIV-1 replication. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E4571–80. doi: 10.1073/pnas.1311669110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway C.A., Jr., Medzhitov R. Innate immune recognition. Annu. Rev. Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- Karki R., Man S.M., Malireddi R.K., Gurung P., Vogel P., Lamkanfi M., Kanneganti T.D. Concerted activation of the AIM2 and NLRP3 inflammasomes orchestrates host protection against Aspergillus infection. Cell Host Microbe. 2015;17:357–368. doi: 10.1016/j.chom.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki N., Warming S., Lamkanfi M., Vande Walle L., Louie S., Dong J., Newton K., Qu Y., Liu J., Heldens S., Zhang J., Lee W.P., Roose-Girma M., Dixit V.M. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- Kayagaki N., Wong M.T., Stowe I.B., Ramani S.R., Gonzalez L.C., Akashi-Takamura S., Miyake K., Zhang J., Lee W.P., Muszynski A., Forsberg L.S., Carlson R.W., Dixit V.M. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science. 2013;341:1246–1249. doi: 10.1126/science.1240248. [DOI] [PubMed] [Google Scholar]
- Kayagaki N., Stowe I.B., Lee B.L., O'rourke K., Anderson K., Warming S., Cuellar T., Haley B., Roose-Girma M., Phung Q.T., Liu P.S., Lill J.R., Li H., Wu J., Kummerfeld S., Zhang J., Lee W.P., Snipas S.J., Salvesen G.S., Morris L.X., Fitzgerald L., Zhang Y., Bertram E.M., Goodnow C.C., Dixit V.M. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526:666–671. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- Kim H.M., Park B.S., Kim J.I., Kim S.E., Lee J., Oh S.C., Enkhbayar P., Matsushima N., Lee H., Yoo O.J., Lee J.O. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell. 2007;130:906–917. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Kondo T., Kobayashi J., Saitoh T., Maruyama K., Ishii K.J., Barber G.N., Komatsu K., Akira S., Kawai T. DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proc. Natl. Acad. Sci. U. S. A. 2013;110:2969–2974. doi: 10.1073/pnas.1222694110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengyel P., Liu C.J. The p200 family protein p204 as a modulator of cell proliferation and differentiation: a brief survey. Cell. Mol. Life Sci. 2010;67:335–340. doi: 10.1007/s00018-009-0195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.C., Lo Y.C., Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.J., Wang H., Lengyel P. The interferon-inducible nucleolar p204 protein binds the ribosomal RNA- specific UBF1 transcription factor and inhibits ribosomal RNA transcription. EMBO J. 1999;18:2845–2854. doi: 10.1093/emboj/18.10.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Wang H., Zhao Z., Yu S., Lu Y.B., Meyer J., Chatterjee G., Deschamps S., Roe B.A., Lengyel P. MyoD-dependent induction during myoblast differentiation of p204, a protein also inducible by interferon. Mol. Cell. Biol. 2000;20:7024–7036. doi: 10.1128/mcb.20.18.7024-7036.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.J., Ding B., Wang H., Lengyel P. The MyoD-inducible p204 protein overcomes the inhibition of myoblast differentiation by id proteins. Mol. Cell. Biol. 2002;22:2893–2905. doi: 10.1128/MCB.22.9.2893-2905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.J., Chang E., Yu J., Carlson C.S., Prazak L., Yu X.P., Ding B., Lengyel P., Di Cesare P.E. The interferon-inducible p204 protein acts as a transcriptional coactivator of Cbfa1 and enhances osteoblast differentiation. J. Biol. Chem. 2005;280:2788–2796. doi: 10.1074/jbc.M412604200. [DOI] [PubMed] [Google Scholar]
- Luan Y., Yu X.P., Xu K., Ding B., Yu J., Huang Y., Yang N., Lengyel P., Di Cesare P.E., Liu C.J. The retinoblastoma protein is an essential mediator of osteogenesis that links the p204 protein to the Cbfa1 transcription factor thereby increasing its activity. J. Biol. Chem. 2007;282:16860–16870. doi: 10.1074/jbc.M610943200. [DOI] [PubMed] [Google Scholar]
- Luan Y., Lengyel P., Liu C.J. p204, a p200 family protein, as a multifunctional regulator of cell proliferation and differentiation. Cytokine Growth Factor Rev. 2008;19:357–369. doi: 10.1016/j.cytogfr.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan Y., Yu X.P., Yang N., Frenkel S., Chen L., Liu C.J. p204 protein overcomes the inhibition of Cbfa1-mediated osteogenic differentiation by id helix-loop-helix proteins. Mol. Biol. Cell. 2008;19:2113–2126. doi: 10.1091/mbc.E07-10-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- Meunier E., Wallet P., Dreier R.F., Costanzo S., Anton L., Ruhl S., Dussurgey S., Dick M.S., Kistner A., Rigard M., Degrandi D., Pfeffer K., Yamamoto M., Henry T., Broz P. Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat. Immunol. 2015;16:476–484. doi: 10.1038/ni.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe K.M., Yang Z., Johnson J.R., Geng X., Doitsh G., Krogan N.J., Greene W.C. IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science. 2014;343:428–432. doi: 10.1126/science.1243640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzalli M.H., Broekema N.M., Diner B.A., Hancks D.C., Elde N.C., Cristea I.M., Knipe D.M. cGAS-mediated stabilization of IFI16 promotes innate signaling during herpes simplex virus infection. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E1773–81. doi: 10.1073/pnas.1424637112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S., Kawai T., Akira S. Microbial sensing by Toll-like receptors and intracellular nucleic acid sensors. Cold Spring Harb. Perspect. Biol. 2015;7:a016246. doi: 10.1101/cshperspect.a016246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D.J., Vogel S.N. Inflammation: species-specific TLR signalling — insight into human disease. Nat. Rev. Rheumatol. 2016;12:198–200. doi: 10.1038/nrrheum.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluddemann A., Mukhopadhyay S., Gordon S. Innate immunity to intracellular pathogens: macrophage receptors and responses to microbial entry. Immunol. Rev. 2011;240:11–24. doi: 10.1111/j.1600-065X.2010.00989.x. [DOI] [PubMed] [Google Scholar]
- Roberts T.L., Idris A., Dunn J.A., Kelly G.M., Burnton C.M., Hodgson S., Hardy L.L., Garceau V., Sweet M.J., Ross I.L., Hume D.A., Stacey K.J. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelsen T.S., Heckl D., Ebert B.L., Root D.E., Doench J.G., Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Zhao Y., Wang Y., Gao W., Ding J., Li P., Hu L., Shao F. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014;514:187–192. doi: 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- Shi J., Zhao Y., Wang K., Shi X., Wang Y., Huang H., Zhuang Y., Cai T., Wang F., Shao F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- Stetson D.B., Medzhitov R. Antiviral defense: interferons and beyond. J. Exp. Med. 2006;203:1837–1841. doi: 10.1084/jem.20061377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson D.B., Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Storek K.M., Gertsvolf N.A., Ohlson M.B., Monack D.M. cGAS and Ifi204 cooperate to produce type I IFNs in response to Francisella infection. J. Immunol. 2015;194:3236–3245. doi: 10.4049/jimmunol.1402764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Wu J., Du F., Chen X., Chen Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka A., Wang Z., Choi M.K., Yanai H., Negishi H., Ban T., Lu Y., Miyagishi M., Kodama T., Honda K., Ohba Y., Taniguchi T. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Takeuchi O., Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Unterholzner L., Keating S.E., Baran M., Horan K.A., Jensen S.B., Sharma S., Sirois C.M., Jin T., Latz E., Xiao T.S., Fitzgerald K.A., Paludan S.R., Bowie A.G. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance R.E., Isberg R.R., Portnoy D.A. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson R.O., Bell S.L., Macduff D.A., Kimmey J.M., Diner E.J., Olivas J., Vance R.E., Stallings C.L., Virgin H.W., Cox J.S. The cytosolic sensor cGAS detects mycobacterium tuberculosis DNA to induce type I interferons and activate autophagy. Cell Host Microbe. 2015;17:811–819. doi: 10.1016/j.chom.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Chen Z.J. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 2014;32:461–488. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- Zanoni I., Tan Y., DI Gioia M., Broggi A., Ruan J., Shi J., Donado C.A., Shao F., Wu H., Springstead J.R., Kagan J.C. An endogenous caspase-11 ligand elicits interleukin-1 release from living dendritic cells. Science. 2016;352:1232–1236. doi: 10.1126/science.aaf3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Gonzalezgugel E., Cheng L., Richbourgh B., Nie L., Liu C. The roles of interferon-inducible p200 family members IFI16 and p204 in innate immune responses, cell differentiation and proliferation. Genes Dis. 2015;2:46–56. doi: 10.1016/j.gendis.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material