Abstract
Background
The study was carried out to assess the genetic variability present in ashwagandha and to examine the nature of associations of various traits to the root yield of the plant.
Methods
Fifty-three diverse genetic stocks of ashwagandha (Withania somnifera) were evaluated for 14 quantitative characteristics. Analysis of variance, correlation, and path coefficient analysis were performed using the mean data of 2 years.
Results
Analysis of variance revealed that the genotypes differed significantly for all characteristics studied. High heritability in conjunction with high genetic advance was observed for fresh root weight, 12 deoxywithastramonolide in roots, and plant height, which indicated that selection could be effective for these traits. Dry root weight has a tight linkage with plant height and fresh root weight. Further, in path coefficient analysis, fresh root weight, total alkaloid (%) in leaves, and 12 deoxywithastramonolide (%) in roots had the highest positive direct effect on dry root weight.
Conclusion
Therefore, these characteristics can be exploited to improve dry root weight in ashwagandha genotypes and there is also scope for the selection of promising and specific chemotypes (based on the alkaloid content) from the present germplasm.
Keywords: ashwagandha, direct effect, genetic advance, heritability, path coefficient
1. Introduction
Ashwagandha (Withania somnifera (L.) Dunal) (2n = 48) is also known as gooseberry or winter cherry and belongs to the Solanaceae family. It originates in North-Western and Central India as well as Mediterranean region of North Africa. Dry and subtropical climatic regions are best suited for growth of this crop. In India ashwagandha is cultivated in an area of around 10,780 ha, with a total dry root production of 8,429 tones. The annual demand for this herb increased from 7,028 tones (2001–2002) to 9,127 tones (2004–2005). This 29.8% increase in the demand for W. somnifera has increased the area used for its cultivation, which has led to higher production of good-quality plants [1]; Madhya Pradesh alone cultivates an area of over 4,000 ha in the drier parts, especially in Manasa, Neemuch, and Jawad Tehsils in the Mandsaur District [2]. It is also cultivated in the states of Rajasthan, Gujarat, Maharashtra, Punjab, and Uttar Pradesh in India.
India is also an exporter of Indian ginseng roots to other countries. India exported W. somnifera worth 8.17 crores (USD 1,202,740) with a total quantity of about 132.72 tons during the period from January 2014 to November 2016. The United States is the largest buyer of Withania from India accounting for exports worth 5.2 crores (USD 779,111), followed by Australia and China who imported Withania worth 57.7 lakhs (USD 84,946) and 54.6 lakhs (USD 80,407), respectively [3].
This herb is commonly known as rasayana in Ayurvedic practice, which means it functions as a tonic for vitality and longevity [4]. The plant is also known to trigger the immune system cells, lymphocytes, and phagocytes, which also help to control the effects of stress and promote wellness [5], [6], [7]. The plant is also used as a liver tonic, anti-inflammatory agent, astringent, and to treat bronchitis, asthma, emaciation, insomnia, dementia, neurological disorders, inflammation, and Parkinson's disease.
The roots of ashwagandha have properties similar to ginseng roots, used for the maintenance and restoration of health; hence it is also known as Indian ginseng [8]. The roots are considered to be the most important part of the whole plant, as they possess a wide number of therapeutic agents. Reports show that the total alkaloidal content in the Indian roots vary between 0.13% and 0.31%, although a much higher content (up to 4.3%) have been recorded elsewhere [9], [10]. The drugs obtained from its roots are used in the treatment of rheumatic pain, inflammation of joints [11], nervous disorders, female disorders, hiccups, coughs and colds, ulcers, leprosy, as a sedative, etc. The main chemical constituents are alkaloids and steroidal lactones. Its leaves contain withanolides, of which withaferin A and withanolide D are the most important. The medicinal properties of the plant are due to the presence of withanolides [12].
The plant (Indian chemotype) is reported to contain 12 withanolides, five unidentified alkaloids, many free amino acids, chlorogenic acid, glycosides, glucose, condensed tannins, and flavonoids in the leaves [13]. Ayurvedic and Unani systems use the leaves for treatment of tumors and tubercular glands [14] as they possess antibacterial, -fungal, and -tumor properties [15]. Ashwagandha has also found calm the mind, relieve weakness and nervous exhaustion, build sexual energy, and promote healthy sleep.
In spite of its high nutritive value, there is a lack of wider genetic variability in ashwagandha. The success of any plant-breeding program depends mainly on the genetic variability available in the population. Availability of a wide variability provides the breeder with a greater chance of selecting desired material. Besides knowledge of the variability, a detailed knowledge of the association of characteristics with yield is also necessary. Using correlation analysis we can find the linkage relationship among the traits, which gives the opportunity of mutual improvement of two desirable traits by common selection program. Path coefficient analysis helps to evaluate the relative contribution of each trait, both direct and indirect, to the yield. The information on genetic diversity in W. somnifera is very scarce and scanty [16]. Therefore, an investigation was undertaken to the variability present in ashwagandha and to examine the magnitude of the genetic associations of various traits to the root yield of the plant.
2. Materials and methods
The experimental material consisted of 53 genotypes of W. somnifera (ashwagandha) collected from different geographical regions of India (Table 1). These genotypes were grown for 2 consecutive years (monsoon season of 2012–2013 and 2013–2014) at the research farm of the Central Institute of Medicinal and Aromatic Plants, Lucknow, India, located within longitude 80°59́ E and latitude 26°55́ E, in the subtropical climatic region. In the year 2012–2013 the seeds were sown on August 20, 2012 and transplanted to the field on October 5, 2012, and in 2013–2014 the seeds were sown on August 27, 2013 and transplanted on October 17, 2013. A randomized block design with three replications was followed. Each genotype was grown in 3 m long rows with a row-to-row and plant-to-plant distance of 50 cm × 20cm. The soil of the experimental plot was sandy loam with good organic matter and drainage, with a soil pH of 7.6, mean rainfall 972 mm, and mean temperature of 18–31°C. Recommended cultural practices were followed to raise the crop [17]. The roots were dug 200 d after transplantation, washed with plain water, and dried in the oven. After drying the roots until their moisture content was 7–8% of the original level, they were stored in air-tight packs for further chemical analysis. Five competitive plants from each replication were randomly selected to record the observations on plant height (cm), fresh root weight (g), dry root weight (g), and 1,000-seed weight (g). The percentages of total alkaloid, withanolide, withaferin A, withanolide A, and 12 deoxywithastramonolide were also estimated in the properly dried leaves and root samples of all the 53 genotypes.
Table 1.
Origin of genotypes of Withania somnifera L. used in the study
| S. No. | Acc No. | Place of collection | S. No. | Acc. No. | Place of collection |
|---|---|---|---|---|---|
| 1 | CWS-1 | CIMAP | 28 | CWS-31 | CIMAP |
| 2 | CWS-2 | CIMAP | 29 | CWS-33 | CIMAP |
| 3 | CWS-3 | Malda | 30 | CWS-34 | CIMAP |
| 4 | CWS-4 | Krishinagar | 31 | CWS-35 | CIMAP |
| 5 | CWS-5 | Bhagalpur | 32 | CWS-36 | CIMAP |
| 6 | CWS-6 | Udaipur | 33 | CWS-37 | Malda |
| 7 | CWS-7 | Patna | 34 | CWS-51 | Malda |
| 8 | CWS-9 | Udaipur | 35 | CWS-52 | Malda |
| 9 | CWS-10 | Udaipur | 36 | CWS-54 | Malda |
| 10 | CWS-11 | Udaipur | 37 | CWS-55 | Malda |
| 11 | CWS-12 | Bagidora | 38 | CWS-56 | Malda |
| 12 | CWS-14 | Fatehpur | 39 | CWS-57 | Malda |
| 13 | CWS-15 | Bharatpur | 40 | CWS-58 | Malda |
| 14 | CWS-16 | Agra | 41 | CWS-60 | Krishinagar |
| 15 | CWS-17 | CIMAP | 42 | CWS-61 | Krishinagar |
| 16 | CWS-18 | CIMAP | 43 | CWS-62 | Krishinagar |
| 17 | CWS-19 | CIMAP | 44 | CWS-64 | Krishinagar |
| 18 | CWS-20 | CIMAP | 45 | CWS-65 | Krishinagar |
| 19 | CWS-21 | CIMAP | 46 | CWS-66 | Krishinagar |
| 20 | CWS-23 | CIMAP | 47 | CWS-67 | Krishinagar |
| 21 | CWS-24 | CIMAP | 48 | CWS-68 | Krishinagar |
| 22 | CWS-25 | CIMAP | 49 | CWS-70 | Hazipur |
| 23 | CWS-26 | CIMAP | 50 | CWS-74 | Udaipur |
| 24 | CWS-27 | CIMAP | 51 | CWS-76 | Udaipur |
| 25 | CWS-28 | CIMAP | 52 | CWS-82 | Fatehpur |
| 26 | CWS-29 | CIMAP | 53 | CWS-83 | Fatehpur |
| 27 | CWS-30 | CIMAP |
Acc. No., accession number; CIMAP, Central Institute of Medicinal and Aromatic Plants; S. No., sample number
2.1. Statistical analysis
The variance analysis (ANOVA) was carried out using Statistical Software version 4.0, available in the Division of Genetics and Plant Breeding of the Council of Scientific and Industrial Research, Central Institute of Medicinal and Aromatic Plants, as prescribed by Singh and Chaudhary [18]. The 2 years pooled mean data of all the plant characteristics were used to perform the correlation and path coefficient analysis. The genetic associations and path analysis were determined using the method described by Dewey and Lu [19]. Significance of correlation was tested following the method of Fisher and Yates [20].
2.2. Extraction of total alkaloids and withanolides
For the extraction of total alkaloid and withanolide content, the roots and leaves were dried in a hot air oven at 40–45°C. Then the dried material was crushed and stored at room temperature. Leaf and root powder (5 g each) were extracted separately with methanol (50 mL × 3), filtered and evaporated using the Rotary evaporator (vacuum 337 mbar for at 40°C). The obtained crude mass was then defatted with n-hexane (10 mL × 3) and then with 1% H2SO4 (50 mL). The H2SO4 solution was extracted with diethyl ether (30 mL × 3) in a separating funnel. The two layers thus obtained were separated. The upper layer which was ether soluble was dried over anhydrous Na2SO4, filtered, evaporated, and the crude withanolide content was estimated. The lower layer which was acid soluble was basified with ammonia and then extracted with CHCl3 (30 mL × 3) to separate two distinct layers (upper insoluble CHCl3 layer and lower soluble layer). Using anhydrous Na2SO4, the soluble layer was dried, filtered, and evaporated on the rotary evaporator (vacuum 474 mbar for at 40°C). This was estimated as the total alkaloid content and the upper layer was discarded [21].
2.3. Extraction of withaferin A, withanolide A, and 12 deoxywithastramonolide
For the extraction of withaferin A, withanolide A, and 12 deoxywithastramonolide from the roots and leaves, reverse HPLC analysis was carried out at the analytical chemistry division of the institute. LC-10A gradient HPLC instrument with two LC-10AD pumps controlled by a CBM-10 interface module, a model 7725 I manual injector valve, a 20 μL sample loop, and a multidimensional UV-visible detector, SPD-10 was used for analysis. For peak purity tests of the compound, a SPDM10AVP photodiode array detector was used. The solvents were filtered using a filtering system (Millipore, Bedford, MA, USA). As per HPLC method [22], the different accessions of Withania were analyzed with slight modifications [23] using a Waters Spherisorb C18 analytical column (4.6 mm × 250 mm, 10 μm ODS); mobile-phase acetonitrile; 0.1% tri-flouro acetic acid in water (40:60; flow rate 1.0 mL/min; column temperature 26°C, detector wavelength 220 nm.
3. Results and discussion
Analysis of variance is a statistical procedure which separates the total variation into different components. The result from analysis of variance for different characteristics is presented in Table 2. The mean sum of squares due to genotypes showed a significant difference for all characteristics in this study at 1% and 5% levels of significance. This indicates the presence of ample genetic variability among the ashwagandha genotypes and that the genotypes are genetically distinct. The total variation present in a population arises as a result of genotypic and environmental effects. Coefficient of variation is a true relative measure of variance among different traits. The estimates of genotypic coefficient of variation (GCV), phenotypic coefficient of variation (PCV), heritability, and genetic advance for all 14 characteristics studied are provided in Table 3. It was observed that the relative magnitude of PCV was marginally higher compared to corresponding GCV for all characteristics. This indicates that to some extent the characteristics were a result of the environment. A wide range of PCV was observed for all characteristics ranging from 3.09 to 317.98. Similarly, the GCV ranged from 2.83 to 314.46. Joshi et al [24] also reported high values of GCV and PCV for withanolide A content in ashwagandha. A relatively high difference between GCV and PCV for some characteristics suggested a greater influence of the environment in the expression of these traits, whereas low difference indicated less environmental influence on expression of those characteristics. The proportion of genotypic variability which is transmitted from parents to progeny is reflected by heritability. The heritability estimates was found to be high (> 60%) for all characteristics, except the total alkaloid content in roots (25.63). Such a high level of heritability may be due to the major role of additive gene action in the expression of these characteristics. The highest heritability was recorded for fresh root weight (99.95), followed by plant height (99.71), 12 deoxywithastramonolide in roots (99.62), and withanolide (%) in leaves (99.30). Additive gene action plays a major role in the expression of traits showing high heritability and these can be improved through individual plant selection. Heritability alone does not give any clear picture about the nature of inheritance of a trait. Genetic advance represents the improvement in the mean of selected families over the base population. Heritability in conjunction with genetic advance over mean can suggest the exact nature of inheritance of a trait. Johnson et al [25] suggested that consideration of heritability and genetic advance together would be more useful in predicting the effect of selection on phenotypic expression of traits. In the present study, high genetic advance coupled with high heritability was observed for the characteristics of fresh root weight, 12 deoxywithastramonolide, and plant height. This suggested the major role of additive gene action with low environmental influence and could be effective in phenotypic selection.
Table 2.
Analysis of variance for 14 characteristics of Withania somnifera
| S. No. | Characteristics | Mean sum of squares |
||
|---|---|---|---|---|
| Replication (d.f. = 2) | Treatment (d.f. = 52) | Error (d.f. = 104) | ||
| 1 | Plant height (cm) | 7.13 | 1,713.05** | 1.67 |
| 2 | Fresh root weight (g) | 7.75 | 12,656.34** | 2.04 |
| 3 | Dry root weight (g) | 1.30 | 2,573.20** | 3.25 |
| 4 | 1,000 seed weight (g) | 0.0023 | 0.008** | 0.0005 |
| 5 | Total alkaloid % (L) | 0.002 | 0.072** | 0.063 |
| 6 | Withanolide % (L) | 0.002 | 0.085** | 0.001 |
| 7 | Withaferin A % (L) | 0.004 | 0.662** | 0.002 |
| 8 | Withanolide A % (L) | 0.0001 | 0.576** | 0.0002 |
| 9 | 12 deoxywithastramonolide (L) | 0.0001 | 0.032** | 0.0002 |
| 10 | Total alkaloid % (R) | 0.0017 | 0.070** | 0.0007 |
| 11 | Withanolide % (R) | 0.245 | 0.035** | 0.017 |
| 12 | Withaferin A % (R) | 0.00 | 0.0002** | 0.00 |
| 13 | Withanolide A % (R) | 0.0001 | 0.010** | 0.0002 |
| 14 | 12 deoxywithastramonolide (R) | 0.0005 | 0.013** | 0.0001 |
* Significant at 1% probability level
** Significant at 5% probability level
d.f., degrees of freedom; L, leaves; R, root
Table 3.
Estimates of genetic parameters for 14 characters of Withania somnifera based on pooled mean data of 2 years
| S. No. | Characters | Coefficient of variation |
Heritability (%) | Genetic advance | |
|---|---|---|---|---|---|
| GCV | PCV | ||||
| 1 | Plant height | 34.50 | 34.56 | 99.71 | 49.06 |
| 2 | Fresh root weight | 57.78 | 57.80 | 99.95 | 133.73 |
| 3 | Dry root weight | 2.83 | 3.09 | 83.40 | 0.084 |
| 4 | 1,000 seed weight | 38.56 | 39.06 | 97.50 | 0.309 |
| 5 | Total alkaloid % (L) | 35.59 | 36.27 | 96.25 | 0.332 |
| 6 | Withanolide % (L) | 113.243 | 113.643 | 99.30 | 0.965 |
| 7 | Withaferin A % (L) | 137.93 | 137.98 | 99.93 | 0.90 |
| 8 | Withanolide A % (L) | 314.46 | 317.98 | 97.80 | 0.206 |
| 9 | 12 deoxywithastramonolide (L) | 42.05 | 42.73 | 96.83 | 0.301 |
| 10 | Total alkaloid % (R) | 19.85 | 39.21 | 25.63 | 0.040 |
| 11 | Withanolide % (R) | 123.96 | 159.91 | 60.12 | 0.009 |
| 12 | Withaferin A % (R) | 83.29 | 85.62 | 94.63 | 0.110 |
| 13 | Withanolide A % (R) | 251.96 | 255.37 | 97.35 | 0.131 |
| 14 | 12 deoxywithastramonolide (R) |
63.70 | 63.82 | 99.62 | 60.06 |
GCV, genotypic coefficient of variation; L, leaves; PCV, phenotypic coefficient of variation; R, root
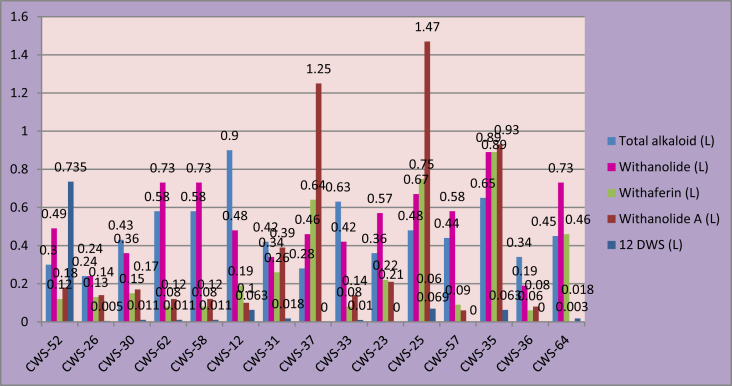
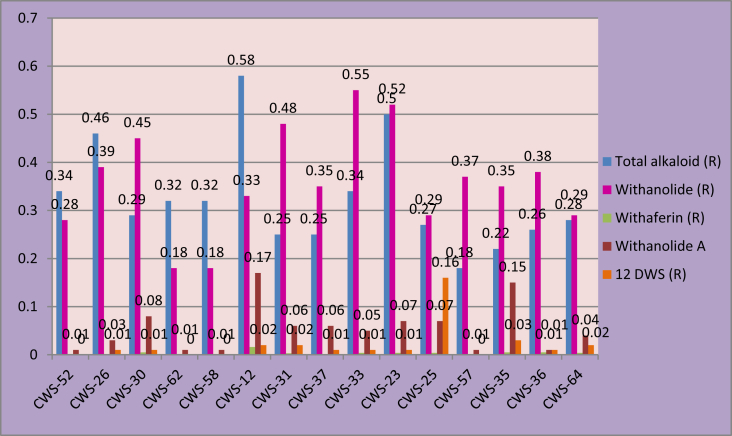
The 15 best genotypes were identified on the basis of the dry root yield and the percentages of total alkaloid, withanolide, withaferin A, withanolide A, and 12 deoxywithastramonolide content in leaves and roots were compared (Fig. 1, Fig. 2). From the graph it is clear that the genotypes CWS 12(0.9) and CWS 35(0.65) are rich in total alkaloid content in leaves. Whereas, genotypes CWS 35 (0.89), CWS 62 (0.73), and CWS 58(0.73) can be selected for withanolide content in leaves; genotypes CWS 35(0.89) and CWS 25(0.67) for withaferin A content in leaves; genotypes CWS 25(1.47) and CWS 37(1.25) for withanolide A content in leaves; and genotype CWS 52(0.74) for 12 deoxywithastramonolide in leaves. Overall, genotype CWS 35 appears most promising for leaf compounds. In the roots, genotypes CWS 12(0.58) and CWS 23(0.5) can be selected for total alkaloid content and genotypes CWS 33(0.55) and CWS 23 (0.52) for withanolide content. From the study of both figures it appears that the genotypes CWS 12 and CWS 35 are the ideal genotypes considering the content in leaves and roots. However, the other genotypes, having a particular compound specifically in a higher amount, could be used as donor parents in further genetic improvement programs, e.g., CWS 25 and CWS 37 for withanolide A content in leaves and CWS 23, CWS 31, and CWS 33 for withanolide content in roots.
Fig. 1.
Graph representing the content of total alkaloid, withanolide, withaferin A, withanolide A, and 12 deoxywithastramonolide in leaves of the 15 best genotypes of Withania somnifera selected based on dry root yield.
Fig. 2.
Graph representing the content of withanolide, withaferin A, withanolide A, and 12 deoxywithastramonolide in roots of the 15 best genotypes of Withania somnifera selected based on dry root yield.
While selecting a suitable plant type, correlation studies provide reliable information about the nature and direction of the selection, especially when the breeder needs to combine high yield potential with desirable traits. All possible correlations and intercorrelations have been determined and are presented in Table 4. In the present study, the dry root weight exhibited high and strong positive and significant association with fresh root weight (0.82) and plant height (0.42). Das et al [26] also reported positive and significant correlation of dry root weight with plant height. Negative and significant association of dry root weight was observed with withaferin A (%) in leaves (−0.36) and in roots (−0.33). The characteristic of withaferin A (%) in leaves showed a high and strong positive significant association with withaferin A (%) in roots (0.72). A positive association of plant height with fresh root weight (0.50) and 1,000 seed weight (0.47) was observed. Total alkaloid (%) in roots was associated with withanolide (%) in roots (0.39); 1,000 seed weight with withanolide A (%) in leaves (0.38); 12 deoxywithastramonolide in roots with withaferin A (%) in roots (0.31) and withanolide A (%) in roots (0.34). Weak but positive association was recorded for withanolide A (%) in leaves with withanolide A (%) in roots (0.31) and withaferin A (%) in leaves with withanolide A (%) in leaves (0.29). Thus, the correlation between dry root weight and other characteristics and the intercorrelation between all characteristics were taken into account.
Table 4.
Estimates of genotypic correlation coefficients between dry root weight and component characteristics of Withania somnifera
| Characteristics | PH | FRW | 1,000 Seed weight | Total alkaloid (L) | Wid (L) | Wi-A (L) | Wid-A (L) | 12 DWS (L) | Total alk (R) | Wid (R) | Wi-A (R) | Wid-A (R) | 12 DWS (R) | DRW |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PH | — | 0.50** | 0.47** | 0.08 | 0.24 | −0.35** | 0.10 | 0.10 | −0.43** | −0.02 | −0.39** | −0.08 | −0.18 | 0.42** |
| FRW | — | 0.24 | 0.11 | 0.06 | −0.34* | −0.16 | 0.22 | −0.14 | −0.12 | −0.29* | −0.08 | −0.17 | 0.82** | |
| 1,000 seed weight | — | −0.12 | 0.22 | −0.08 | 0.38** | −0.006 | −0.49** | −0.44** | −0.32* | 0.12 | −0.10 | 0.13 | ||
| Total alk (L) | — | 0.32* | 0.05 | −0.16 | −0.05 | 0.15 | 0.17 | 0.17 | 0.15 | 0.02 | 0.11 | |||
| Wid (L) | — | 0.08 | 0.17 | 0.03 | −0.33* | −0.39** | −0.17 | −0.11 | −0.12 | 0.06 | ||||
| Wi-A % (L) | — | 0.29* | 0.009 | −0.10 | −0.26 | 0.72** | −0.05 | 0.07 | −0.36** | |||||
| Wid-A % (L) | — | 0.11 | −0.22 | −0.33* | −0.03 | 0.31* | 0.11 | −0.11 | ||||||
| 12 DWR (L) | — | −0.17 | −0.25 | 0.001 | −0.006 | −0.01 | 0.26 | |||||||
| Total alk (R) | — | 0.39** | 0.17 | 0.23 | −0.04 | −0.12 | ||||||||
| Wid (R) | — | 0.13 | 0.20 | 0.31* | −0.22 | |||||||||
| Wi-A % (R) | — | 0.26 | 0.34* | −0.33* | ||||||||||
| Wid-A % (R) | — | 0.18 | −0.12 | |||||||||||
| 12 DWR (R) | — | −0.11 | ||||||||||||
| DRW | — |
* Significant at 1% probability level
** Significant at 5% probability level
alk, alkaloid; 12 DWS, 12 deoxywithastramonolide; FRW, fresh root weight; L, leaves; PH, plant height; R, roots; Wi-A, withaferin A; Wid, withanolide; Wid-A, withanolide A
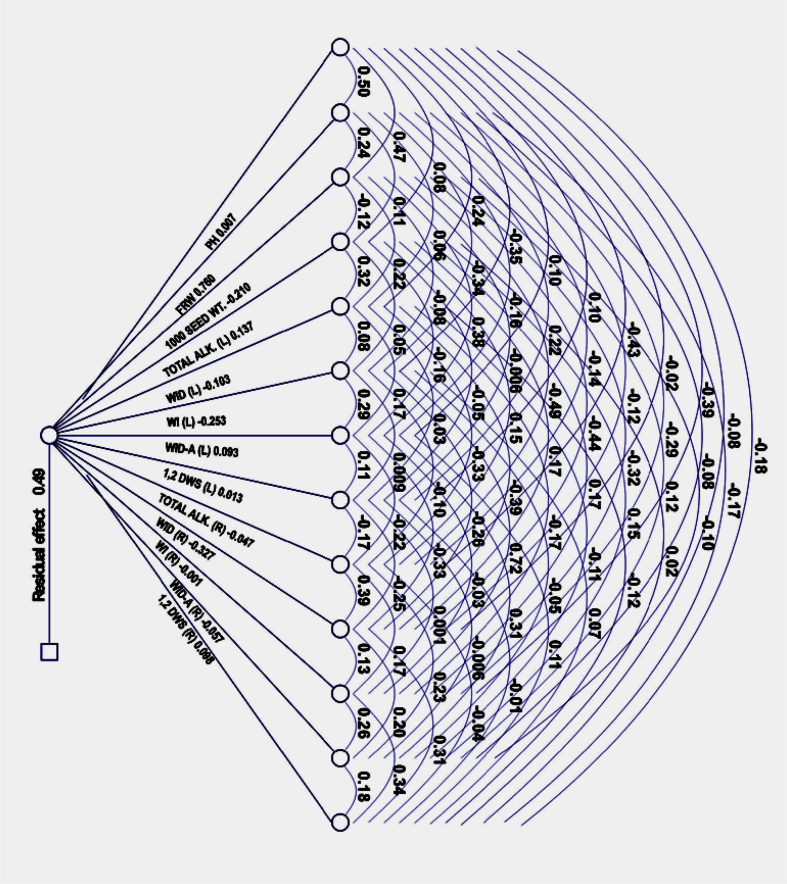
Path coefficient analysis was performed to assess the magnitude of the contributions of various characteristics to dry root weight in the form of cause and effects. The dry root yield was taken as a dependent trait and the other traits were taken as independent. The phenotypic correlation coefficients of dry root yield with component traits were further partitioned into direct and indirect effects. The results of the analysis of direct and indirect contribution of various characteristics on dry root weight are presented in Table 5. The traits of plant height (0.424) and fresh root weight (0.823) showed a positive and significant genetic association with dry root yield. Fig. 3 represents the genotypic path diagram for dry root yield with different characteristics. The fresh root weight (0.76) was found to be the main contributor to the dry root weight. The other characteristics which had a positive direct effect on dry root weight were total alkaloid (%) in leaves (0.137), 12 deoxywithastramonolide in roots (0.098), and withanolide A (%) in leaves (0.093). Plant height and 12 deoxywithastramonolide in leaves also had positive direct effect; however, their value was low. The highest negative direct effect on dry root weight was shown by withanolide (%) in roots (−0.327), followed by withaferin A (%) in leaves (−0.253). The characteristic of plant height exhibited the highest indirect effect on dry root weight via fresh root weight (0.382). The 12 deoxywithastramonolide content in leaves contributed to dry root weight through fresh root weight (0.170). Thus, fresh root weight proved to be the highest direct and indirect contributor of dry root yield in W. somnifera. Residual effect (0.49) suggested that there are still some more traits to be included in any further study. Thus, the present study revealed that fresh root weight and plant height should be considered for direct selection in W. somnifera, as these traits must bring an improvement in the dry root yield of the plant (see Fig. 4).
Table 5.
Estimates of direct and indirect effects of different yield contributing traits on dry root weight in Withania somnifera
| Characteristics | PH | FRW | 1,000 Seed weight | Total alk (L) | Wid (L) | Wi-A (L) | Wid-A (L) | 12 DWR (L) | Total alk. (R) | Wid (R) | Wi-A % (R) | Wid- A % (R) | 12 DWR (R) | Correlation (rg) DRW |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PH | 0.007 | 0.382 | −0.098 | 0.011 | −0.025 | 0.088 | 0.009 | 0.001 | 0.020 | 0.055 | 0.0006 | 0.005 | −0.017 | 0.424** |
| FRW | −0.003 | 0.760 | −0.049 | 0.015 | −0.006 | 0.086 | −0.015 | 0.003 | 0.006 | 0.038 | 0.0004 | 0.004 | 0.017 | 0.823** |
| 1,000 Seed weight | −0.003 | 0.180 | −0.210 | −0.016 | −0.023 | 0.022 | 0.036 | −0.00008 | 0.023 | 0.144 | 0.0005 | −0.007 | −0.010 | 0.137 |
| Total alk (L) | −0.0005 | 0.085 | 0.025 | 0.137 | −0.034 | −0.015 | −0.015 | −0.0007 | −0.008 | −0.057 | −0.0002 | −0.009 | 0.003 | 0.112 |
| Wid (L) | −0.002 | 0.042 | −0.047 | 0.045 | −0.103 | −0.022 | 0.015 | 0.0005 | 0.016 | 0.130 | 0.0003 | 0.006 | −0.012 | 0.068 |
| Wi-A % (L) | 0.0002 | −0.258 | 0.018 | 0.008 | −0.009 | −0.253 | 0.027 | 0.0001 | 0.005 | 0.086 | −0.001 | 0.003 | 0.007 | −0.364** |
| Wid-A % (L) | −0.0007 | −0.125 | −0.082 | −0.023 | −0.017 | −0.075 | 0.093 | 0.001 | 0.011 | 0.110 | 0.00005 | −0.018 | 0.011 | −0.115 |
| 12 DWR (L) | −0.0007 | 0.170 | 0.001 | −0.007 | −0.004 | −0.002 | 0.010 | 0.013 | 0.0008 | 0.084 | −0.000003 | 0.0003 | −0.002 | 0.263 |
| Total alk (R) | 0.003 | −0.096 | 0.104 | 0.022 | 0.035 | 0.028 | −0.021 | −0.0002 | −0.047 | −0.129 | −0.0003 | −0.013 | −0.005 | −0.121 |
| Wid (R) | 0.001 | −0.089 | 0.093 | 0.024 | 0.041 | 0.067 | −0.031 | −0.003 | −0.019 | −0.327 | −0.0002 | −0.011 | 0.031 | −0.224 |
| Wi-A % (R) | 0.003 | −0.225 | 0.068 | 0.024 | 0.018 | −0.183 | −0.003 | 0.00002 | −0.008 | −0.044 | −0.001 | −0.015 | 0.034 | −0.336 |
| Wid-A % (R) | 0.0006 | −0.059 | −0.026 | 0.021 | 0.012 | 0.013 | 0.029 | −0.00007 | −0.011 | −0.066 | −0.0004 | −0.057 | 0.019 | −0.126 |
| 12 DWR (R) | 0.001 | −0.129 | 0.021 | 0.004 | 0.013 | −0.019 | 0.010 | −0.0002 | 0.002 | −0.103 | −0.0005 | −0.011 | 0.098 | −0.111 |
Residual effect = 0.49
alk, alkaloid; 12 DWS, 12 deoxywithastramonolide; FRW, fresh root weight; L, leaves; PH, plant height; R, roots; Wi-A, withaferin A; Wid, withanolide; Wid-A, withanolide A
Fig. 3.
Path diagram showing the values of direct and indirect contribution of yield and yield-related traits on dry root yield in Indian ginseng (Withania somnifera). ALK, alkaloid; 12 DWS, 12 deoxywithastramonolide; FRW, fresh root weight; L, leaves; PH, plant height; R, roots; WI, withaferin; WID, withanolide; WID-A, withanolide A.
Fig. 4.
Field view of different collections of Indian ginseng or ashwagandha (Withania somnifera), collected from different locations of India and grown in the field for their conservation in the field gene bank of the Central Institute of Medicinal and Aromatic Plants, Lucknow, India.
4. Conclusions
In the present study an attempt was made to assess genetic variation and the association between morphological and chemical characteristics and dry root weight, and to estimate the direct and indirect effect of these traits on the root weight. The analysis of variance showed significant differences between accessions for all characteristics studied. High genetic advance in conjunction with high heritability was observed in the characteristics of fresh root weight, 12 deoxywithastramonolide, and plant height. Dry root weight had a strong positive association with fresh root weight and plant height. Path analysis revealed that fresh root weight was the direct contributor towards dry root weight. Therefore, the characteristics of fresh root weight and plant height can be exploited for selection programs to improve the dry root yield in ashwagandha. This study will be helpful in the selection and development of specific chemotypes of ashwagandha based on the content of different alkaloids estimated in the roots and leaves, respectively.
Acknowledgments
The authors thank the National Gene Bank for Medicinal and Aromatic Plants at CSIR-CIMAP, Lucknow for providing the germplasm for the present study. The authors are also grateful to the Director of CSIR–CIMAP, Lucknow for allowing us to use the necessary facilities and infrastructure to carry out the present research work.
Conflicts of interest
All authors have no conflicts of interest to declare.
References
- 1.Tripathi A.K., Shukla Y.N., Kumar S. Ashwagandha (Withania somnifera): a status report. J Med Aromatic Plant Sci. 1996;18:46–52. [Google Scholar]
- 2.Sharma R. 1st ed. Daya Publishing House; New Delhi: 2013. Agro-Techniques of Medicinal Plants; pp. 31–33. [Google Scholar]
- 3.Anonymous . 2016. Export analysis and trends of Withania roots.https://www.zauba.com/exportanalysis-withania+roots-report.html Available from: [Accessed 30 December 2016] [Google Scholar]
- 4.Singh G., Sharma P.K., Dudhe R., Singh S. Biological activities of Withania somnifera. Scholars Res Libr. 2010;1:56–63. [Google Scholar]
- 5.Wagner H., Norr H., Winterhoff H. Plant adaptogens. Phytomedicine. 1994;1:63–76. doi: 10.1016/S0944-7113(11)80025-5. [DOI] [PubMed] [Google Scholar]
- 6.Singh B., Saxena A.K., Chandan B.K., Gupta D.K., Bhutani K.K., Anand K.K. Adaptogenic activity of a novel, withanolide-free aqueous fraction from the root of Withania somnifera. Phytother Res. 2001;15:311–318. doi: 10.1002/ptr.858. [DOI] [PubMed] [Google Scholar]
- 7.Singh B., Chandan B.K., Gupta D.K. Adaptogenic activity of a novel withanolide-free aqueous fraction from the roots of Withania somnifera Dun. (Part II) Phytother Res. 2003:531–536. doi: 10.1002/ptr.1189. [DOI] [PubMed] [Google Scholar]
- 8.Singh S., Kumar S. Central Institute of Medicinal and Aromatic Plants; Lucknow, India: 1998. Withania somnifera. The Indian Ginseng ashwagandha. [Google Scholar]
- 9.Anonymous . Publications and Information Directorate, Council of Scientific and Industrial Research (CSIR); New Delhi: 1982. The Wealth of India. Vol. X (Sp-W) pp. 580–585. [Google Scholar]
- 10.Anonymous . 1st ed. Central Council for Research in Unani Medicine (CCRUM); New Delhi: 2007. Standardisation of Single Drugs of Unani Medicine. Part III; pp. 9–14. [Google Scholar]
- 11.Al-Hindwani M.K., Al-Khafaji S., Abdul-Nabi M. Anti-granuloma activity of Iraqi Withania somnifera. J Ethanopharmacol. 1992;37:113. doi: 10.1016/0378-8741(92)90069-4. [DOI] [PubMed] [Google Scholar]
- 12.Davis L, Girija Kumar, Kuttan G. Immunomodulatory activity of Withania somnifera. [DOI] [PubMed]
- 13.Khare C.P. Springer; New Delhi: 2007. Indian Medicinal Plants–An Illustrated Dictionary; pp. 717–718. [Google Scholar]
- 14.Chopra R.N. Academic Publishers India; New Delhi: 1994. Glossary of Indian Medicinal Plants. [Google Scholar]
- 15.Devi P.U., Sharada A.C., Solomon F.E. Antitumor and radiosensitizing effects of Withania somnifera (Ashwagandha) on a transplantable mouse tumor, Sarcoma-180. Indian J Exp Biol. 1993;31:607–611. [PubMed] [Google Scholar]
- 16.Gupta A.K., Verma S.R., Gupta M.M., Saikia D., Verma R.K., Jhang T. Genetic diversity in germplasms collections of Withania somnifera for root and leaf alkaloids. J Trop Med Plants. 2011;12:59–69. [Google Scholar]
- 17.Patra D.D., Singh K., Mishra H.O., Gupta A.K., Singh J., Singh S.C., Khanuja S.P.S. Agrotechnologies of Ashwagandha (Withania somnifera) J Med Aromatic Plant Sci. 2004;26:332–335. [Google Scholar]
- 18.Singh R.K., Chaudhary B.D. Kalyani; New Delhi: 1979. Biometrical methods in quantitative genetic analysis; p. 120. [Google Scholar]
- 19.Dewey D.R., Lu K.H. A correlation and path coefficient analysis of components of wheat grass and seed production. Agron J. 1959;51:515–518. [Google Scholar]
- 20.Fisher R.A., Yates F. Oliver and Boyd; Edinburgh: 1938. Statistical tables for biological agricultural and medical research. [Google Scholar]
- 21.Khajuria R.K., Suri K.A., Gupta R.K., Satti N.K., Amina M., Suri O.P., Qazi G.N. Separation, identification and quantification of selected withanolides in plant extracts of Withania somnifera by HPLC-UV (DAD)-positive ion electrospray ionization-mass spectrometry. J Sep Sci. 2004;27:541–546. doi: 10.1002/jssc.200301690. [DOI] [PubMed] [Google Scholar]
- 22.Scartezzini P., Antognoni F., Conte L., Maxia A., Troia A., Poli F. Genetic and phytochemical difference between some Indian and Italian plants of Withania somnifera (L.) Dunal. Nat Prod Res. 2007;21:923–932. doi: 10.1080/14786410701500169. [DOI] [PubMed] [Google Scholar]
- 23.Srivastava P., Tiwari N., Yadav A.K., Shanker K., Verma R.K., Gupta M.M., Gupta A.K., Khanuja S.P.S. Simultaneous quantification of withanolides in Withania somnifera by a validated high-performance thin layer chromatographic method. J AOAC Int. 2008;91:1154–1161. [PubMed] [Google Scholar]
- 24.Joshi N.R., Patel M.A., Prajapati K.N., Patel A.D. Genetic variability, correlation and path analysis in Ashwagandha (L.) Dunal) Electron J Plant Breeding. 2014;5:875–880. [Google Scholar]
- 25.Johnson H.W., Robinson H.F., Comstock R.E. Estimates of variance and environmental variability in soybean. Agron J. 1955;47:314–318. [Google Scholar]
- 26.Das A., Datta A.K., Ghose S., Bhattacharya A. Genetic analysis in Poshita and Jawahar 22 varieties of Withania somnifera (L.) Dunal. Plant Arch. 2011;11:59–62. [Google Scholar]