Abstract
Background
Ginseng is a well-known traditional Chinese medicine that has been widely used in a range of therapeutic and healthcare applications in East Asian countries. Microbial transformation is regarded as an effective and useful technology in modification of nature products for finding new chemical derivatives with potent bioactivities. In this study, three minor derivatives of ginsenoside compound K were isolated and the inducing effects in the Wingless-type MMTV integration site (Wnt) signaling pathway were also investigated.
Methods
New compounds were purified from scale-up fermentation of ginsenoside Rb1 by Paecilomyces bainier sp. 229 through repeated silica gel column chromatography and high pressure liquid chromatography. Their structures were determined based on spectral data and X-ray diffraction. The inductive activities of these compounds on the Wnt signaling pathway were conducted on MC3T3-E1 cells by quantitative real-time polymerase chain reaction analysis.
Results
The structures of a known 3-keto derivative and two new dehydrogenated metabolites were elucidated. The crystal structure of the 3-keto derivative was reported for the first time and its conformation was compared with that of ginsenoside compound K. The inductive effects of these compounds on osteogenic differentiation by activating the Wnt/β-catenin signaling pathway were explained for the first time.
Conclusion
This study may provide a new insight into the metabolic pathway of ginsenoside by microbial transformation. In addition, the results might provide a reasonable explanation for the activity of ginseng in treating osteoporosis and supply good monomer ginsenoside resources for nutraceutical or pharmaceutical development.
Keywords: biotransformation, ginsenoside, osteogenic differentiation, Wnt/β-catenin signal
Highlights
-
•
Two new structures of ginsenosides were identified.
-
•
A new crystal structure of ginsenoside was reported.
-
•
The effects on inducing osteogenic differentiation by ginsenosides were identified.
-
•
The mechanism of new ginsenosides on Wnt signaling pathway was investigated.
1. Introduction
Ginseng (the root of Panax ginseng Meyer, Araliaceae) is a well-known traditional Chinese medicine used in a range of therapeutic and healthcare applications in East Asian countries. The major pharmacologically active components of ginseng are dammarane type triterpene saponins, commonly known as ginsenosides, which are mainly classified as protopanaxadiol- and protopanaxatriol-type saponins [1], [2], [3]. In general, natural ginsenosides are poorly absorbed after oral administration, and metabolism of ginsenosides in the intestinal tract is critical for the clinical efficacy of ginseng products [4]. Many studies have been conducted in attempts to elucidate the fate of ginsenosides through the gastrointestinal tract, and now it is known that intact ginsenosides are largely degraded by either acids or human intestinal bacteria before reaching the circulation system [5], [6]. Thus, research focus has shifted from intact ginsenosides to secondary ginsenosides [7], [8].
20-O-β-D-glucopyranosyl-20(S)-protopanaxadiol, or ginsenoside compound K (C-K, Compound 1), is one of the major metabolites detected in blood after oral administration of ginsenoside Rb1, Rb2, or Rc, and it is also speculated to be the major form of protopanaxadiol saponins absorbed from the intestine [4], [9]. We have previously shown that biotransformation of ginsenoside Rb1 by fungus Paecilomyces bainier sp. 229 could effectively produce C-K, and the crystal structure was determined for the first time [10]. The hydrolytic pathway of Rb1→Rd→F2→C-K was confirmed by HPLC, which was the same with intestinal metabolism [11]. Moreover, a new bioactivity of C-K in rheumatoid arthritis (RA) has been found and it is now in Phase I clinical trials in China as an antiinflammatory and bone protection candidate for the treatment of RA [12].
Microbial transformation is effective in modifying natural products to obtain new chemical derivatives with potent bioactivities and physical-chemical characteristics [13], [14]. In this study, we analyzed the metabolites in scale-up microbial transformation of ginsenoside Rb1 by Paecilomyces bainier sp. 229 and then identified three specific dehydrogenated metabolites (Compound 2, 3, and 4).
Moreover, we also investigated the pharmacological mechanism of this skeleton structure in bone protection by studying its activities on some key osteoblast-specific transcription factors at the ribonucleic acid (RNA) level by quantitative real-time polymerase chain reaction (qRT-PCR), including alkaline phosphatase (ALP), type I collagen (Collα1), osteoprotegerin/receptor activator of nuclear factor-B ligand ratio (OPG/RANKL) and runt-related transcription factor 2 (Runx2) in MC3T3-E1 cells. In order to clarify the mechanism of osteoblastogenesis and bone metabolism, we investigated the effects of these metabolites on canonical Wingless-type MMTV integration site (Wnt) signaling of Wnt10b, Wnt11, low density lipoprotein receptor-related protein 5 (Lrp5), and β-catenin. Our study may provide an insight into the molecular mechanism for the potential treatment of osteoarthritis and osteoporosis by these special ginsenosides and the bone protection effect of ginsenosides in RA-induced bone damage.
2. Materials and methods
2.1. Materials
Ginsenoside Rb1 was purchased from Kunming Shanghima Biotech Co. Ltd., (Kunming, Yunnan, China). Alpha modification of Eagle's minimum essential medium (α-MEM) and fetal bovine serum were purchased from Gibco (Grand Island, NY, USA). β-Glycerophosphate, L-ascorbic acid, and methylthiazolyldiphenyl-tetrazolium bromide (MTT) were purchased from Sigma-Aldrich (St. Louis, MO, USA). All solvents were of analytical grade (Sinopharm Chemical Reagent, Shanghai, China).
Column chromatography was performed with Diaion HP-20 resin (Mitsubishi Chemical, Tokyo, Japan) and silica gel (200–300 mesh; Qingdao Haiyang Chemical Co., Ltd., Qingdao, China). Analytical HPLC was performed on an Agilent 1100 HPLC system equipped with a diode-array detector and a quaternary pump system (Agilent Technologies Singapore Pte. Ltd., Singapore) using a Waters Symmetry C18 analytical column (4.6 mm × 150 mm, 5 μm) and a C18 guard column (4.6 mm × 12.5 mm, 5 μm) (Waters Corporation, Milford, MA, USA). Preparative HPLC was performed on an Agilent 1100 preparative instrument with a Kromasil C18 column (250 mm × 20 mm, 5 μm) (AkzoNobel Pulp and Performance Chemicals, Bohus, Sweden) using the mobile phase at 5.0 mL/min. For both analytical and preparative HPLC, the detection wavelength was 203 nm and the column temperature was 40°C.
1H and 13C NMR spectra were recorded on a Bruker Advance-400 spectrometer at 400 MHz and 100 MHz in dimethyl sulfoxide (DMSO)-d6, and chemical shifts were expressed in δ (ppm) relative to tetramethylsilane. High-resolution electronspray ionization mass spectra (HR-ESI-MS) were recorded on a Bruker microtof-QII instrument.
A well-shaped prismatic single crystal of Compound 2 was used for data collection on a Bruker Smart Apex CCD diffractometer with a graphite monochromator. The final unit-cell parameters were based on all reflections. Data collection and cell refinement were performed with the SMART program, and data reduction was carried out with the SAINT software (Bruker AXS Inc., Madison, WI, USA). The structure was solved by direct methods with SHELXS-97 [15], and the models were refined by full-matrix least-squares procedures on F2 using SHELXL-97 [16].
2.2. Microorganism
Paecilomyces bainier sp. 229 was isolated as reported previously [10]. The strain was maintained on a potato dextrose agar slant by periodical transfers and incubated at 28°C for 5 days prior to storage at 4°C until required.
2.3. Biotransformation
Agar slants were inoculated with spores, incubated at 28°C for 5 d, and then used for seed culture inoculation. The seed culture was grown in 500 mL Erlenmeyer flasks containing 60 mL of liquid medium, and then incubated at 28°C on a rotary shaker (220 rpm) for 2 d. The seed medium was composed of soybean powder 30 g/L, glucose 30 g/L, (NH4)2SO4 1 g/L, MgSO4·7H2O 2 g/L, and CaCl2 1 g/L.
Scale-up fermentation was carried out in a 14 L fermentor (BioFlo 110, New Brunswick Scientific Inc. Co., Edison, NJ, USA) containing 6 L (working volume) of fermentation medium (yeast powder 30 g/L, sucrose 30 g/L, bran 5 g/L, (NH4)2SO4 1 g/L, MgSO4·7H2O 2 g/L, and CaCl2 1 g/L). The fermentor was an agitated bioreactor equipped with two turbine impellers and devices to monitor pH, stir speed, and dissolved oxygen on line. Fermentation was performed with 10% (v/v) inoculation of seed culture. All media were sterilized at 121°C for 20 min, and the initial pH was 5.0. The broth was cultured at 28°C at a stirring rate of 600 rpm. After 30 h, 30 g of substrate of ginsenoside Rb1 dissolved in 500 mL of water was added and pH was controlled at 5.0 with 1 N H3PO4. Transformation was terminated 60 h later.
2.4. Isolation of metabolites
After the scale up fermentation, culture broth was collected and the same volume of ethanol was added to extract for 24 h at room temperature. The extract was collected by vacuum filtration and the filtrate was subjected to HP-20 column chromatography for binding ginsenosides and removing pigments. Ginsenosides were eluted with ethanol-water mixtures of increasing polarity, and 70% ethanol chromatographic fractions containing C-K and three new metabolites were collected and then evaporated in vacuum to give a residue (10.3 g). This residue was subjected to column chromatography on a silica gel column with CHCl3-MeOH-EtOAc-H2O = 2:1:4:1 (lower phase) as solvent, which yielded Fraction A (7.6 g) and Fraction B (2.3 g). Fraction B was purified by preparative HPLC with ACN:H2O = 51:49 to yield Metabolite 2 (211 mg). Fraction A was purified by crystal in MeOH:H2O = 60:40 at room temperature, and then the crystal (5.1 g) was subjected to further chromatography on preparative HPLC with ACN:H2O = 51:49 to yield Metabolite 3 (46.3 mg) and Metabolite 4 (72.1 mg).
2.5. Biological activity evaluation
2.5.1. Cell culture
MC3T3-E1 cells obtained from the Type Culture Collection of Chinese Academy of Sciences (Shanghai, China) were maintained in α-MEM supplemented with antibiotics (100 units/mL penicillin A and 100 μg/mL streptomycin) and 10% fetal bovine serum in an atmosphere of 5% CO2 at 37°C. The medium was changed every 3 d. Cells were induced to differentiate in maintenance medium with 10mM β-glycerophosphate and 50μM L-ascorbic acid. Exponentially growing cells were plated at a final concentration of 4 × 104 cells/well in 24-well plates for cell proliferation assay, ALP activity assay, and qRT-PCR analysis. To examine their effects on osteoblast differentiation, the cells were cultured with the differentiation medium in the presence of various concentrations (1–16μM) of tested compounds.
2.5.2. Cell viability
Cell viability was determined by MTT. MC3T3-E1 cells were treated with various concentrations (1–16μM) of tested compounds for 4 d, and the MTT solution (0.5 mg/mL) was added. After 4 h of incubation at 37°C for MTT-formazan formation, the supernatant was removed and 600 μL of DMSO was added into each well. Absorbance at 490 nm was determined spectrophotometrically by using a microplate reader (Epoch, BioTek Instruments, Inc., Winooski, VT, USA).
2.5.3. Quantitative assay of ALP activity
The ALP activity and the total protein concentration were measured to assess the differentiation of preosteoblasts. Four days after induction of osteoblast differentiation, cells were washed three times with phosphate-buffered saline and lysed with 100 μL of lysis buffer containing 1.0% Triton X-100 and 0.1% sodium dodecyl sulfate (SDS). The lysate was centrifuged at 10,000g for 5 min at 4°C, the supernatant was collected for the measurement of the ALP activity using an ALP activity assay kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China), and the total protein concentration was determined using a commercial bicinchoninic acid protein assay kit (Beyotime, Shanghai, China). ALP activity was indicated by nmol p-NP/μg total protein/h to present the ALP activity.
2.5.4. qRT-PCR analysis of osteoblastogenesis and canonical Wnt signaling
On the 4th d, the total RNA was extracted from MC3T3-E1 cells using Trizol reagent (Invitrogen, Carlsbad, CA, USA). Complementary deoxyribonucleic acid (cDNA) was converted with a PrimeScript RT reagent kit with gDNA eraser (Takara Biotechnology Co., Ltd., Dalian, China). qRT-PCR amplification of the cDNA products (2 μL) was performed with SYBR premix (PrimeScript RT Master Mix, Takara, China) and specific primers (Sangon Biotech Co., Ltd., Shanghai, China) used for detecting mRNA transcripts are shown in Table 1. PCR was performed using a StepOnePlus PCR system (Applied Biosystems, Foster City, CA, USA) and the transcripts were normalized to glyceraldehyde 3-phosphate dehydrogenase transcript levels.
Table 1.
Primer sequences used for quantitative real-time polymerase chain reaction (qRT-PCR) analysis
| Gene | Sequence |
|
|---|---|---|
| Forward(5′-3′) | Reverse(3′-5′) | |
| ALP | GAGATGGTATGGGCGTCTC | GTTGGTGTTGTACGTCTTGGA |
| Collα1 | GACATGTTCAGCTTTGTGGACCTC | GGGACCCTTAGGCCATTGTGTA |
| OPG | AGCTGCTGAAGCTGTGGAA | GGTTCGAGTGGCCGAGAT |
| RANKL | CGCTCTGTTCCTGTACTTTCGAGCG | TCGTGCTCCCTCCTTTCATCAGGTT |
| Runx2 | GCACAAACATGGCCAGATTCA | AAGCCATGGTGCCCGTTAG |
| Wnt10b | GGCTGTAACCACGACATGGAC | ACGTTCCATGGCATTTGCAC |
| Lrp5 | CACCATTGATTATGCCGACCAG | TGAGTCAGGCCAAACGGGTAG |
| β-catenin | ATTTGATGGAGTTGGACATGG | TGTTCTTGAGTGAAGGACTGA |
| GAPDH | CCA CAC CTT CTACAA TGA GC | TGA GGT AGT CAG TCAGGT C |
ALP, alkaline phosphatase; Collα1, type I collagen; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; Lrp5, low density lipoprotein receptor related protein 5; OPG, osteoprotegerin; RANKL, receptor activator of nuclear factor-B ligand; Runx2, runt-related transcription factor 2; Wnt, wingless-type MMTV integration site
3. Results
3.1. Structure identification
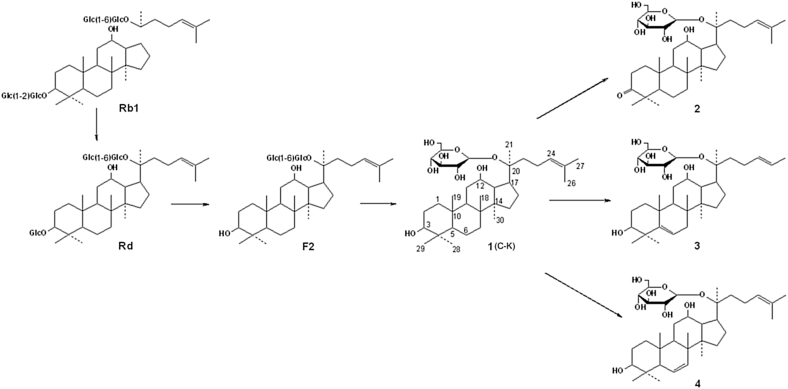
In our previous studies, the amounts of main metabolites in the fermentation broth during the biotransformation of ginsenoside Rb1 by Paecilomyces bainier sp. 229 were detected by a gradient HPLC method, and the biotransformation pathway was identified as Rb1→Rd→F2→C-K [10]. However, several minor products were also observed in HPLC analysis, which could be ginsenoside metabolites (Fig. S1). Thus, preparative scale biotransformation was carried out, and four metabolites were obtained including C-K, Compounds 2, 3, and 4. Compounds 3 and 4 were new compounds, while Compound 2 was reported previously [17].
Compound 2: colorless crystal solid; 1H NMR (DMSO-d6, 400 MHz) δ0.85 (3H, s, H-30), 0.87 (3H, s, H-19), 0.94 (3H, s, H-18), 0.95 (3H, s, H-29), 1.00 (3H, s, H-28), 1.25 (3H, s, H-21), 1.57 (3H, s, H-26), 1.64 (3H, s, H-27), 5.08 (1H, t, H-24); 13C NMR (DMSO-d6, 100 MHz) spectroscopic data see Table 2. HR-ESI-MS m/z 643.4180 [M + Na]+ (scaled for C36H60NaO8, 643.4180). A distinctive signal of H-3 at about 2.98 ppm was absent in 1H NMR spectra of Compound 2 compared with that of C-K. The 13C NMR and distortionless enhancement by polarization transfer (DEPT) spectra showed a new carbonyl carbon at 216.85 ppm, and a distinctive signal of oxygen-bearing carbon (δC 77.22) disappeared, indicating that it was a dehydrogenative product. The carbonyl group was located at C-3 on the basis of heteronuclear multiple bond correlation correlations between H-29 (δH 0.95), H-28 (δH 1.00), H-1 (δH 1.82), and H-2 (δH 2.39, δH 2.45). Accordingly, the signals of C-2 shifted downfield from 27.64 to 34.10, and that of C-4 shifted downfield from 39.03 to 47.06, respectively, due to the inductive effect of the C-3 carbonyl group. Thus, Compound 2 was identified as 3-one-20-O-β-D-glucopyranosyl-20(S)-protopanaxadiol.
Table 2.
13C nuclear magnetic resonance (NMR) chemical shifts of Compounds 1, 2, 3, and 4 in dimethyl sulfoxide (DMSO)-d6
| Carbon position | 1(C-K) | 2 | 3 | 4 |
|---|---|---|---|---|
| 1 | 38.98 | 39.45 | 39.43 | 36.63 |
| 2 | 27.64 | 34.10 | 27.60 | 27.71 |
| 3 | 77.22 | 216.85 | 76.60 | 77.21 |
| 4 | 39.03 | 47.06 | 42.39 | 38.53 |
| 5 | 55.81 | 54.68 | 147.37 | 55.28 |
| 6 | 18.40 | 19.63 | 119.09 | 125.07 |
| 7 | 34.82 | 34.10 | 34.44 | 135.79 |
| 8 | 39.69 | 39.45 | 36.78 | 43.77 |
| 9 | 50.90 | 48.82 | 51.04 | 50.57 |
| 10 | 37.00 | 36.64 | 37.24 | 35.84 |
| 11 | 30.53 | 30.98 | 32.20 | 29.06 |
| 12 | 69.41 | 69.34 | 69.25 | 70.17 |
| 13 | 48.91 | 49.08 | 46.55 | 47.69 |
| 14 | 51.17 | 51.20 | 50.73 | 50.43 |
| 15 | 30.53 | 30.55 | 31.08 | 30.09 |
| 16 | 26.18 | 26.17 | 26.25 | 25.87 |
| 17 | 49.69 | 50.86 | 48.92 | 49.00 |
| 18 | 16.01 | 15.66 | 16.89 | 17.11 |
| 19 | 28.60 | 15.99 | 20.26 | 17.81 |
| 20 | 82.49 | 82.51 | 82.53 | 82.44 |
| 21 | 21.99 | 22.00 | 22.12 | 21.86 |
| 22 | 35.77 | 35.79 | 35.95 | 35.43 |
| 23 | 22.64 | 22.64 | 22.71 | 22.57 |
| 24 | 125.78 | 125.78 | 125.74 | 125.79 |
| 25 | 130.62 | 130.62 | 130.63 | 130.64 |
| 26 | 18.05 | 18.04 | 18.04 | 18.02 |
| 27 | 25.98 | 25.95 | 25.94 | 25.94 |
| 28 | 16.30 | 26.90 | 28.70 | 28.27 |
| 29 | 16.28 | 21.20 | 23.72 | 16.60 |
| 30 | 17.41 | 17.34 | 18.00 | 19.32 |
| 20-iner-glc | ||||
| 1′ | 97.02 | 97.03 | 97.08 | 97.00 |
| 2′ | 74.29 | 74.32 | 74.28 | 74.36 |
| 3′ | 77.72 | 77.74 | 77.63 | 77.80 |
| 4′ | 70.56 | 70.62 | 70.55 | 70.61 |
| 5′ | 77.09 | 77.08 | 77.06 | 77.06 |
| 6′ | 61.60 | 61.66 | 61.59 | 61.66 |
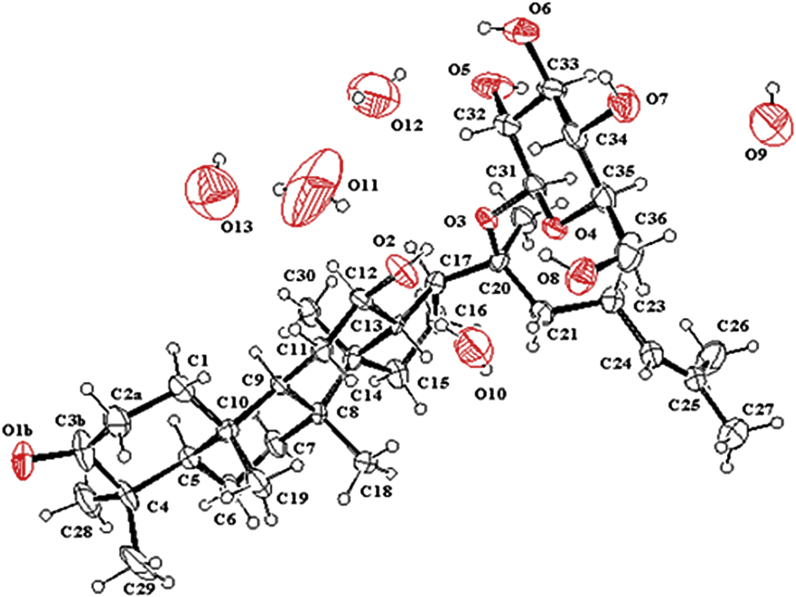
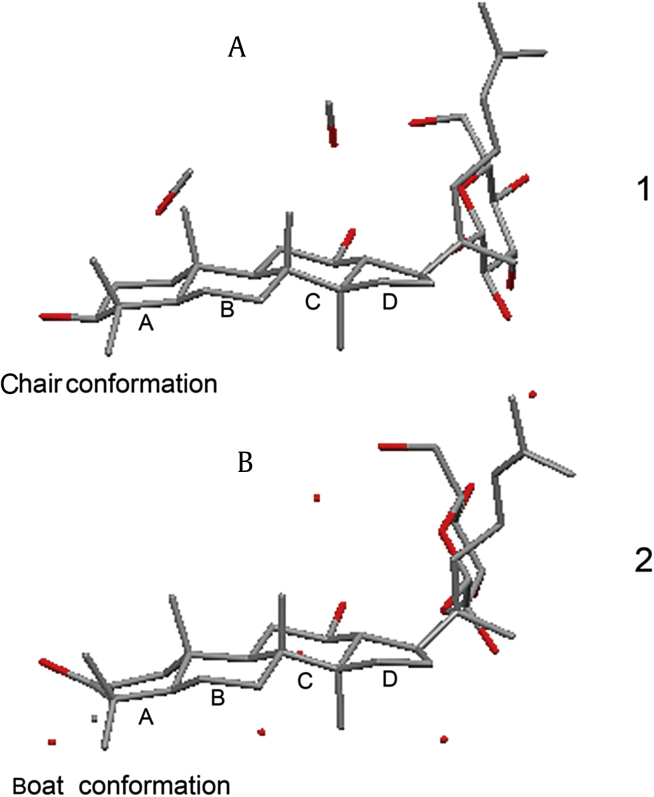
Recrystallization from MeOH-H2O = 60:40 afforded colorless needles of Compound 2, and the crystal structure was solved by means of X-ray diffraction data collected with MoKa radiation. Crystal data, data-collection procedures, structure determination methods, and refinement results are shown in Table 3. (CCDC 1502281 contains the supplementary crystallographic data for this paper. These data can be obtained free of charge via http://www.ccdc.cam.ac.uk/conts/retrieving.html). Compound 2 crystallized in the orthorhombic space group P 21 21 2, and each unit cell contained one Compound 2 molecule and five H2O molecules (O9, O10, O11, O12, and O13) (Fig. 1). The structure consisted of a glucopyranosyl group and a dammarane type triterpene aglycone composed of one five-membered ring D and three six-membered rings A, B, and C. The A/B, B/C, and C/D ring junctions all approach trans characteristics as C-K (Table S1), and the relative configuration of C20 was also S. A carbonyl carbon at C-3 was observed on ring A with a bond length of 1.266 Å, which was shorter than that of the O-C bond of C-K at C-3 (1.443 Å) [18]. The conformation of ring A changes from the chair form to an ideal boat form due to the rigidity and stretching effect of carbonyl bound at C-3 (Fig. 2).
Table 3.
Crystal data and structure refinement of Compound 2
| Formula | C36 H67.50 O11.75 |
|---|---|
| Formula weight | 688.40 |
| Wavelength (Å) | 0.71073 |
| Temperature (K) | 293 (2) |
| Crystal system | Orthorhombic |
| Space group | P 21 21 2 |
| Unit cell dimensions (Å) | a = 32.529(16)Å, b = 10.549(5) Å, c = 11.454(6) Å |
| β = 96.862 (9) ° | |
| V (Å3) | 3,930(3) |
| Z | 4 |
| Calc. density (Mg/m3) | 1.163 |
| Absorption coefficient (mm−1) | 0.085 |
| F(000) | 1,510 |
| Crystal size (mm) | 0.15 × 0.10 × 0.08 |
| θ range for data collection (°) | 1.78–25.10° |
| Range of h, k, & l | −38/38, −12/10, −10/113 |
| Reflection collected | 16,685 |
| Independent reflections | 7,002 [Rint = 0.1178] |
| Completeness to θ = 27.00° | 99.8% |
| Refinement method | Full-matrix least-squares on F2 |
| Data/restraints/parameters | 7,002/29/493 |
| Goodness-of-fit on F2 | 1.047 |
| Final R indices [I > 2σ (I )] | R1 = 0.0963; ωR2 = 0.1277 |
| R indices (all data) | R1 = 0.1888; ωR2 = 0.1507 |
| Δρmax, Δρmin (e Å−3) | 0.255, −0.311 |
Fig. 1.
X-Ray crystal structure of one molecule of Compound 2 with five H2O solvent molecules (O9-O13). Ellipsoids are drawn at the 50% probability level.
Fig. 2.
The conformation and junction comparisons of four rings (A, B, C, and D) in dammarane triterpene structure of Compound 1 (A) and 2 (B). All of the ring junctions (A/B, B/C, and C/D) approach trans characteristics in these two compounds. The conformation of ring A is chair form in Compound 1 and boat form in Compound 2.
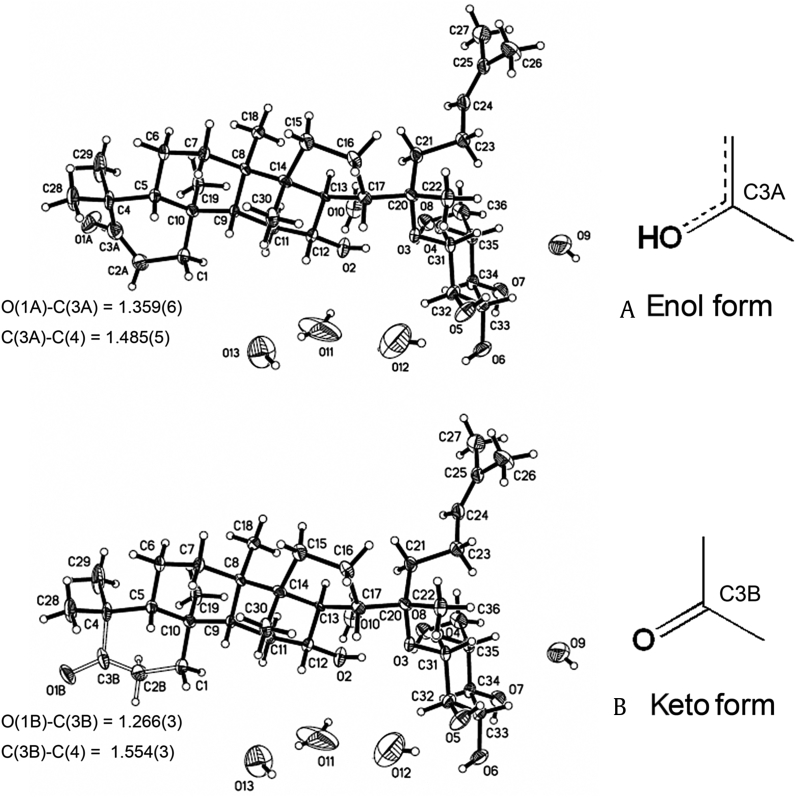
Keto-enol tautomerism was found in the crystal of Compound 2 with different O-C lengths at C-3 (1.266 Å and 1.359 Å for keto and enol tautomerism, respectively) (Fig. 3). They were disorderly. The keto form accounted for 70%, while the enol form accounted for only 30%. This was because the enol form had relatively high lattice energy with a C3-C4 bond length of C(3A)-C(4) = 1.485 (5), and could be converted to the more stable keto form with a longer bond length of C(3B)-C(4) = 1.554 (3) (Table S2).
Fig. 3.
Two molecular forms of Compound 2 in the crystal. (A) enol form (30%, with bond lengths of O(1A)-C(3A) = 1.359(6) and C(3A)-C(4) = 1.485(5) Å); (B) keto form (70%, with bond lengths of O(1B)-C(3B) = 1.266(3) and C(3B)-C(4) = 1.554(3) Å).
Compound 3: white amorphous powder; 1H NMR (DMSO-d6, 400 MHz) δ0.80 (3H, s, H-30), 0.90 (3H, s, H-18), 0.98 (3H, s, H-29), 1.05 (3H, s, H-19), 1.06 (3H, s, H-28), 1.25 (3H, s, H-21), 1.56 (3H, s, H-26), 1.64 (3H, s, H-27), 5.07 (1H, t, H-24), 5.51 (1H, s, H-6); 13C NMR (DMSO-d6, 100 MHz) spectroscopic data see Table 1. ESI-MS m/z = 643.37 [M+Na]+, m/z = 665.39 [M+HCOO]−. Its molecular formula was determined to be C36H60O8 by HR-ESI-MS ([M + Na]+, m/z 643.4037) with two hydrogen atoms less than Compound 1. Compared with Compound 1, the 1H NMR and 13C NMR spectra of Compound 3 showed the presence of a new olefinic proton signal at δH 5.51 (1H, s), a new olefinic carbon at δC 119.09, and a new quaternary carbon signal at δC 147.37, but the absence of a distinctive signal of C-5 at about 55.28 ppm, indicating the presence of a new double bond beside C24-C25 in Compound 3. In the heteronuclear multiple quantum coherence (HMQC) spectrum, the new olefinic proton signal at δH 5.51 was related to the carbon at δC 119.09 and showed correlations with H-7 (δH 1.56, δH 2.23) in a 1H-1H COSY [(homonuclear chemical shift) COrrelation SpectroscopY] experiment, suggesting that the double bond occurred at C5-C6. Thus, Metabolite 3 was assigned as 5,6-ene-20-O-β-D-glucopyranosyl-20(S)-protopanaxadiol.
Compound 4: white amorphous powder; 1H NMR (DMSO-d6, 400 MHz) δ 0.68 (3H, s, H-29), 0.79 (3H, s, H-30), 0.82 (3H, s, H-19), 0.926 (3H, s, H-28),1.01 (3H, s, H-18), 1.25 (3H, s, H-21), 1.56 (3H, s, H-26), 1.64 (3H, s, H-27), 5.08 (1H, t, H-24), 5.44 (1H, dd, H-6), 5.58 (1H, d, H-6); 13C NMR (DMSO-d6, 100 MHz) spectroscopic data see Table 1. ESI-MS m/z = 643.40 [M+Na]+, m/z = 1,263.33 [2M+Na]+, m/z = 665.71 [M+HCOO]−, m/z = 1,239.63 [2M-H]−, m/z = 1,285.20 [2M+HCOO]−. Its molecular formula was determined to be C36H60O8 by HR-ESI-MS ([M + Na]+, m/z 643.4071). Compared with Compound 1, the 1H NMR and 13C NMR spectra of Compound 4 exhibited two new olefinic proton signals at δH 5.58 (1H, d) and δH 5.54 (1H, dd) and two new olefinic carbons at δC 125.07 and 135.79, indicating the presence of a new carbon-carbon double bond without quaternary carbon in Compound 4. δH 1.56 correlating with C5 (δC 55.28) in the HMQC spectrum showed correlations with the new olefinic proton signal at δH 5.58 in 1H-1H COSY spectra, indicating that this new double bond was located at C6-C7. Consequently, Compound 4 was determined as 6,7-ene-20-O-β-D-glucopyranosyl-20(S)-protopanaxadiol.
3.2. The effects on osteoblastic differentiation
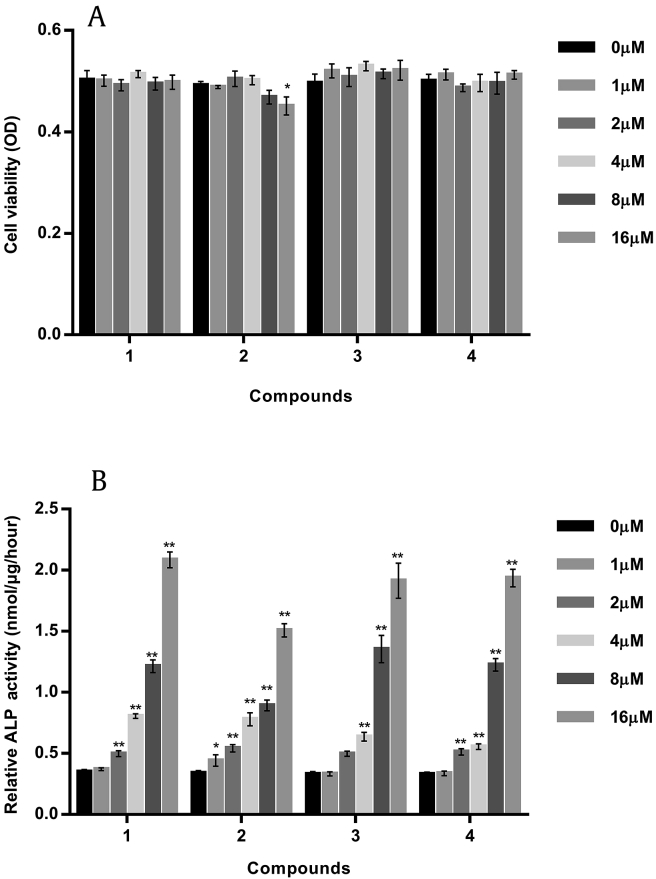
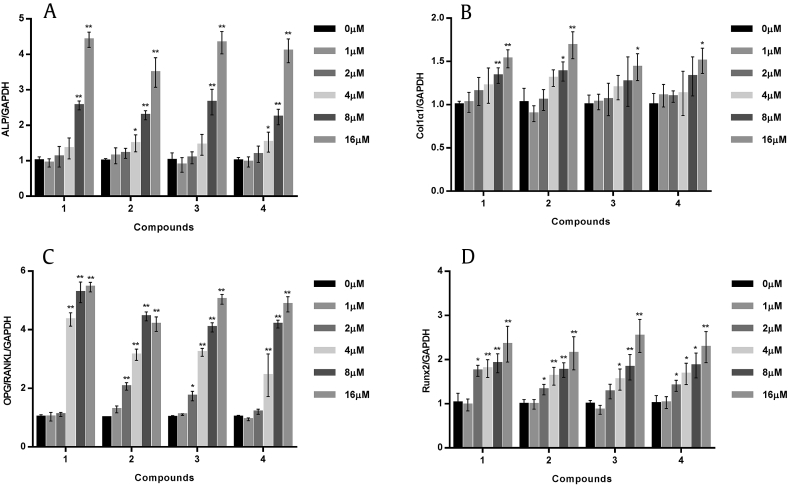
Fig. 4A shows that cell proliferation was not significantly enhanced or inhibited in all compounds except Compound 2 on the 4th d, indicating no significant cytotoxic effect of Compounds 1, 3, and 4 on MC3T3-E1 viability at concentrations of 1–16μM. Compound 2 showed cytotoxic effects at 8μM and significant inhibitory effects on cell proliferation at 16μM (p < 0.05). ALP is an early phenotypic marker and an essential enzyme for osteoblast differentiation [19]. All the tested compounds significantly increased ALP activity in a dose-dependent manner (Fig. 4B). The maximal effect was observed at 16μM of C-K, which led to an increase of ALP activity by six times as compared with the control. However, the induction effect of Compound 2 on ALP was much lower than other analogs due to its stronger cytotoxicity.
Fig. 4.
Effects of tested compounds on cell viability (A) and alkaline phosphatase (ALP) activity (B) in MC3T3-E1 cells. The results were expressed as mean ± standard deviation (SD) (% control) of three independent experiments. * p < 0.05 and ** p < 0.01 (0μM as control).
3.3. The mRNA expression of osteogenic differentiation related marker genes
Preosteoblastic cells produced extracellular matrix proteins, such as ALP, collagen, and Runx2, which induced deposition and mineralization of osteoblasts [20]. The mRNA expression of osteoblast differentiation markers, such as ALP, Collα1, and Runx2, was significantly increased by these analogies in a dose-dependent manner. C-K resulted in a three times increase of ALP mRNA expression compared with the control (Fig. 5A). Compound 2 had slightly weaker activities on ALP gene expression, but it showed the highest effect on Collα1 mRNA with an increase of about 70% at 16μm (Fig. 5B). Compound 3 was the best one on Runx2 expression with 2.5 fold promotion (Fig. 5D). The OPG/RANKL system is another key factor in the bone remodeling process (Fig. 5C). The OPG-RANKL interaction played a dominant role in osteoclastogenesis and it was a key link between bone formation and resorption [21], [22]. Ginsenoside metabolites also showed appreciable increases in this aspect and the best activity was found in C-K treatment with more than five fold increase at 16μM. Thus, C-K and derivatives had special activity on promoting bone formation by increasing markers of osteoblast differentiation.
Fig. 5.
Effects of tested compounds on osteoblast differentiation in MC3T3-E1 cells. The mRNA expression of osteoblast differentiation factors, such as alkaline phosphatase (ALP) (A), Collα1 (B), osteoprotegerin/receptor activator of nuclear factor-B ligand ratio (OPG/RANKL) (C), and runt-related transcription factor 2 (Runx2) (D) were detected by quantitative real-time polymerase chain reaction (qRT-PCR). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as internal control. The results are expressed as the mean ± standard deviation (SD) (% control) of three independent experiments. * p < 0.05 and ** p < 0.01 (0μM as control).
3.4. Modulation of canonical Wnt signaling
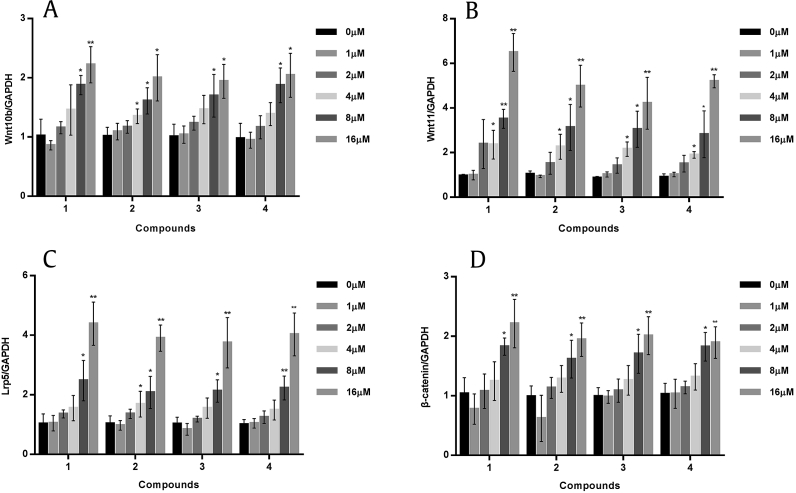
C-K and these new derivatives significantly upregulated the mRNA expression of genes in the Wnt/β-catenin signaling pathway related regulators, including Wnt10b, Wnt11, Lrp5, and β-catenin (Fig. 6). Our results showed that these analogies selectively activated two most critical bone metabolism related Wnt genes, Wnt10b and Wnt11 (Figs. 6A and 6B) [23]. The highest increase was obtained by C-K at 16μM, with an increase of 2.2 times and 6.5 times, respectively. Lrp 5, one of the main transmembrane receptor proteins [24], was also significantly increased by about four times by all compounds (Fig. 6C). β-catenin, a transcriptional cofactor located downstream of the Wnt/β-catenin pathway, was doubled by all the compounds, while the highest one was obtained by C-K (Fig. 6D). Furthermore, western blotting and ALP staining assay also showed canonical Wnt signaling to affect osteoblast differentiation in the presence of ginsenoside C-K (Figs. S2 and S3). From the results presented above, we firmly believed that these minor ginsenosides can induce osteogenic differentiation of MC3T3-E1 cells via partially or maybe dependently controlling the canonical Wnt signaling pathway [23], [25].
Fig. 6.
Effects of tested compounds on wingless-type integration site (Wnt)/β-catenin signaling pathway in MC3T3-E1 cells. The mRNA expression of Wnt/β-catenin signal related markers, such as Wnt10b (A), Wnt11 (B), Lrp5 (C), and β-catenin (D) were detected by real-time polymerase chain reaction (RT-PCR). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as internal control. The results are expressed as the mean ± standard deviation (SD) (% control) of three independent experiments. * p < 0.05 and ** p < 0.01 (0μM as control).
4. Discussion
Numerous fungi and microorganisms metabolize saponins in a pathway similar to that of the intestinal system, and thus they can serve as microbial models to identify and prepare bioactive compounds that are absorbable by mammalian body. The metabolism of ginsenoside Rb1 by intestinal bacteria and fungi has been extensively studied and the main deglycosylated metabolic pathway was identified [7]. Moreover, few modifications of dammarane skeleton by fungus, such as selective C-3/C-12 carbonylation and sidechain oxidation/reduction, were also conducted to find new chemical derivatives with potent characteristics [26], [27]. The purposes of this study were to isolate and identify new dammarane skeleton modified compounds in scale-up fermentation of ginsenoside Rb1 with Paecilomyces bainier sp. 229, and to explore new metabolic pathways of ginsenoside Rb1 by fungus. Three minor dehydrogenated metabolites were obtained, including a known C-3 carbonylation Compound 2 and two new dammarane skeleton dehydrogenation compounds, Compounds 3 and 4. To our knowledge, the structures of Metabolites 3 and 4 were first reported in biotransformation of ginsenosides. This was also the first study about the structural modification on the rings of ginsenoside skeleton, which enriched the ginsenoside metabolic pathway extensively (Fig. 7). These results show the advantages of microbial transformations in structure and conformation modification of ginsenosides, which is difficult to realize by chemical synthetic methods [7], [8], [9], [10], [26], [27], [28].
Fig. 7.
The proposed biotransformation pathway of ginsenoside Rb1 by Paecilomyces bainier sp. 229. Major pathway: Rb1→Rd→F2→C-K (previous work, [10], [11]); minor pathway: C-K→Compounds 2, 3, and 4, this work).
The crystal structure of Compound 2 was also identified for the first time in this work and its configuration and conformation were studied. The relative configuration of C-20 of Compound 2 is S and the junctions of rings are all trans characteristic, which is the same as that of the other two metabolites, ginsenoside Mx and C-K [18], [29]. However, the conformation of ring A is boat, which is different for the other two. This conformational distortion is due to the carbonyl effects at C-3, which also induces the keto-enol tautomerism. These crystallographic data of three ginsenoside metabolites would be useful for studying spatial structures and structure-activity relationships of ginsenosides.
The Wnt signaling shows multiple effects on cell proliferation and differentiation, and alterations of the β-catenin-dependent pathway lead to various diseases [30]. The Wnt/β-catenin signaling pathway also cooperatively controls bone formation and osteoblast differentiation, which stimulates the generation of osteoblasts and thus leads to bone formation by promoting the expression of key osteoblast-specific transcription factors, such as Runx2 and osterix (Osx), accompanied by the increased production of bone matrix proteins, such as ALP, Collα1, and OPG/RANKL ratio [31], [32], [33].
Ginsenosides, as the main active components of ginseng, have various physiological and pharmacological properties [1], [2], [34]. However, studies on the Wnt/β-catenin signal remain in their infancy. Ginsenoside Rg3 could inhibit the growth of colon cancer cells by downregulating β-catenin/Tcf activity, and ginsenoside F2 improved hair growth by increasing β-catenin/Lef-1 and decreasing DKK-1 expression [35], [36], [37]. In this study, C-K and metabolic analogies were found to be able to promote obsteoblastogenesis through the Wnt/β-catenin signal, which would be a new supplement for the studies of ginsenoside in the Wnt pathway.
Ginseng could prevent osteoporosis by reducing bone loss induced by ovariectomy or glucocorticoid [38]. However, the mechanism is still unclear. We found that ginsenoside C-K and its dehydrogenated analogs had a special effect on Wnt10b and Wnt11, and upregulated downstream targets, including β-catenin, ALP, collagen, and Runx2, a pivotal pathway for modulating osteoblast differentiation [39]. Thus, our study may provide an insight into the activity of ginseng in treating osteoporosis and supplies a good monomer ginsenoside resource for drug or functional food development.
5. Conclusion
In this study, three minor derivatives of ginsenoside compound K were isolated from the preparative scale fermentation of ginsenoside Rb1 by Paecilomyces bainier sp. 229, including a known 3-keto derivative and two new dehydrogenated metabolites. The crystal structure of C-3 carbonylzation metabolite was reported for the first time and the conformation difference with C-K was also identified. The effective activities of these compounds on conical Wnt/β-catenin signal and osteogenesis-related downstream targets were first investigated, which indicated food or pharmaceutical use in osteoporosis treatment.
Conflicts of interest
The authors declare that they do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted.
Acknowledgments
The authors acknowledge the financial support from The National Nature Fund of China (Number 81503119) and The Shanghai Scientific Fund Committee (Number 14DZ1930304).
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jgr.2017.03.004.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Park J.D., Rhee D.K., Lee Y.H. Biological activities and chemistry of saponins from Panax ginseng C.A. Meyer. Phytochem Rev. 2005;4:159–175. [Google Scholar]
- 2.Jia L., Zhao Y.Q., Liang X.J. Current evaluation of the millennium phytomedicine-ginseng (ii): Collected chemical entities, modern pharmacology, and clinical applications emanated from traditional Chinese medicine. Cur Med Chem. 2009;16:2924–2942. doi: 10.2174/092986709788803204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shin B.K., Kwon S.W., Park J.H. Chemical diversity of ginseng saponins from Panax ginseng. J Gin Res. 2015;39:287–298. doi: 10.1016/j.jgr.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karikura M., Miyase T., Tanizawa H., Taniyama T., Takino Y. Studies on absorption, distribution, excretion and metabolism of ginseng saponins. VII. Comparison of the decomposition modes of ginsenoside-Rb1 and -Rb2 in the digestive tract of rats. Chem Pharm Bull. 1991;39:2357–2361. doi: 10.1248/cpb.39.2357. [DOI] [PubMed] [Google Scholar]
- 5.Takino T. Studies on the pharmacodynamics of ginsenoside Rg1, Rb1, and Rb2 in rats. Yakugaku Zasshi. 1994;114:550–564. [PubMed] [Google Scholar]
- 6.Hasegawa H. Proof of the mysterious efficacy of ginseng. Basic and clinical trials: Metabolic activation of ginsenoside: Deglycosylation by intestinal bacteria and esterification with fatty acid. J Pharmacol Sci. 2004;95:153–157. doi: 10.1254/jphs.fmj04001x4. [DOI] [PubMed] [Google Scholar]
- 7.Park C.S., Yoo M.H., Noh K.H., Oh D.K. Biotransformation of ginsenosides by hydrolyzing the sugar moieties of ginsenosides using microbial glycosidases. Appl Microbiol Biotechnol. 2010;87:9–19. doi: 10.1007/s00253-010-2567-6. [DOI] [PubMed] [Google Scholar]
- 8.Cui L., Wu S.Q., Zhao C.A., Yin C.R. Microbial conversion of major ginsenosides in ginseng total saponins by Platycodon grandiflorum endophytes. J Gin Res. 2016;40:366–374. doi: 10.1016/j.jgr.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bae E.A., Choo M.K., Park E.K., Park S.Y., Shin H.Y., Kim D.H. Metabolism of ginsenoside Rc by intestinal bacteria and its related antiallergic activity. Chem Pharm Bull. 2002;25:743–747. doi: 10.1248/bpb.25.743. [DOI] [PubMed] [Google Scholar]
- 10.Zhou W., Yan Q., Li J.Y., Zhang X.C., Zhou P. Biotransformation of Panax notoginseng saponins into ginsenoside compound K production by Paecilomyces bainier sp.229. J Appl Microbiol. 2008;104:699–706. doi: 10.1111/j.1365-2672.2007.03586.x. [DOI] [PubMed] [Google Scholar]
- 11.Zhou W., Li J.Y., Li X.W., Yan Q., Zhou P. Development and validation of a reversed-phase HPLC method for quantitative determination of ginsenosides Rb1, Rd, F2, and compound K during the process of biotransformation of ginsenoside Rb1. J Sep Sci. 2008;31:921–925. doi: 10.1002/jssc.200700406. [DOI] [PubMed] [Google Scholar]
- 12.Liu QH, Zhou P, Bai H, Zhou W, Li JJ, Feng MQ, Hua ML, Xu JY. New use of ginsenoside compound-K for the prevention of rheumatoid arthritis. PCT/CN2007/002354.
- 13.Kuban M., Ögen G., Kha I.A., Bedi E. Microbial transformation of cycloastragenol. Phytochemistry. 2013;88:99–104. doi: 10.1016/j.phytochem.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 14.Bhattia H.N., Khan S.S., Khan A., Rani M., Ahmad V.U., Choudhary M.I. Biotransformation of monoterpenoids and their antimicrobial activities. Phytomedicine. 2014;21:1597–1626. doi: 10.1016/j.phymed.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 15.Sheldrick G.M. University of Götingen; Götingen, Germany: 1997. SHELXS-97, Program for the solution of crystal structure. [Google Scholar]
- 16.Sheldrick G.M. University of Götingen; Götingen, Germany: 1997. SHELXL-97, Program for the refinement of crystal structure. [Google Scholar]
- 17.Chen G.T., Yang M., Song Y., Lu Z.Q., Zhang J.Q., Huang H.L., Wu L.J., Guo D.A. Microbial transformation of ginsenoside Rb1 by Acremonium strictum. Appl Microbiol Biotechnol. 2008;77:1345–1350. doi: 10.1007/s00253-007-1258-4. [DOI] [PubMed] [Google Scholar]
- 18.Zhou W., Feng M.Q., Li X.W., Yan Q., Zhou C.Q., Li J.Y., Zhou P. X-ray structure investigation of 20(S)-O-β-D-glucopyranosyl-protopanaxadiol and antitumor effect on Lewis lung carcinoma in vivo. Chem Biodivers. 2009;6:380–388. doi: 10.1002/cbdv.200700444. [DOI] [PubMed] [Google Scholar]
- 19.Serigano K., Sakai D., Hiyama A., Tamura F., Tanaka M., Mochida J. Effect of cell number on mesenchymal stem cell transplantation in a canine disc degeneration model. J Orthop Res. 2010;28:1267–1275. doi: 10.1002/jor.21147. [DOI] [PubMed] [Google Scholar]
- 20.Lee H.S., Jung E.Y., Bae S.H., Kwon K.H., Kim J.M., Suh H.J. Stimulation of osteoblastic differentiation and mineralization in MC3T3-E1 cells by yeast hydrolysate. Phytother Res. 2011;25:716–723. doi: 10.1002/ptr.3328. [DOI] [PubMed] [Google Scholar]
- 21.Thomas G.P., Baker S.U., Eisman J.A., Gardiner E.M. Changing RANKL/OPG mRNA expression in differentiating murine primary osteoblasts. J Endocrinol. 2001;170:451–460. doi: 10.1677/joe.0.1700451. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka H., Mine T., Ogasa H., Taguchi T., Liang C.T. Expression of RANKL/OPG during bone remodeling in vivo. Biochem Biophys Res Commun. 2011;411:690–694. doi: 10.1016/j.bbrc.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 23.Bennett C.N., Ouyang H., Ma Y.L., Zeng Q., Gerin I., Sousa K.M., Lane T.F., Krishnan V., Hankenson K.D., MacDougald O.A. Wnt10b increases postnatal bone formation by enhancing osteoblast differentiation. J Bone Miner Res. 2007;22:1924–1932. doi: 10.1359/jbmr.070810. [DOI] [PubMed] [Google Scholar]
- 24.Gordon M.D., Nusse R. Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. J Biol Chem. 2006;281:22429–22433. doi: 10.1074/jbc.R600015200. [DOI] [PubMed] [Google Scholar]
- 25.Krishnan V., Bryant H.U., Macdougald O.A. Regulation of bone mass by Wnt signaling. J Clin Invest. 2006;116:1202–1209. doi: 10.1172/JCI28551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H.F., Ye M., Guo H.Z., Tian Y., Zhang J., Zhou J.P., Hu Y.C., Guo D. Biotransformation of 20(S)-protopanaxadiol by Mucor spinosus. Phytochemistry. 2009;70:1416–1420. doi: 10.1016/j.phytochem.2009.07.041. [DOI] [PubMed] [Google Scholar]
- 27.Chen G.T., Yang X., Li J.L., Ge H.J., Songa Y., Ren J. Biotransformation of 20(S)-protopanaxadiol by Aspergillus niger AS 3.1858. Fitoterapia. 2013;91:256–260. doi: 10.1016/j.fitote.2013.09.019. [DOI] [PubMed] [Google Scholar]
- 28.Wang J.R., Yau L.F., Zhang R., Xia Y., Ma J., Ho H.M., Hu P., Hu M., Liu L., Jiang Z.H. Transformation of ginsenosides from notoginseng by artificial gastric juice can increase cytotoxicity toward cancer cells. J Agric Food Chem. 2014;62:2558–2573. doi: 10.1021/jf405482s. [DOI] [PubMed] [Google Scholar]
- 29.Li X.W., Zhou W., Yan Q., Zhou P. 20-O-D-xylopyranosyl(1,6)-β-D-glucopyranosyl-20(S)-protopanaxdiol methanol solvate. Acta Crystalgr Sect E. Struct Rep Online. 2008;E64:o165. doi: 10.1107/S1600536807063118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moon R.T., Kohn A.D., De Ferrari G.V., Kaykas A. WNT and beta-catenin signalling: diseases and therapies. Nat Rev Genet. 2004;5:691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- 31.Lacey D.L., Timms E., Tan H.L., Kelley M.J., Dunstan C.R., Burgess T., Elliott R., Colombero A., Elliott G., Scully S. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 1998;93:165–176. doi: 10.1016/s0092-8674(00)81569-x. [DOI] [PubMed] [Google Scholar]
- 32.Almeida M., Han L., Bellido T., Manolagas S.C., Kousteni S. Wnt proteins prevent apoptosis of both uncommitted osteoblast progenitors and differentiated osteoblasts by beta-catenin-dependent and -independent signaling cascades involving Src/ERK and phosphatidylinositol 3-kinase/AKT. J Biol Chem. 2005;280:41342–41351. doi: 10.1074/jbc.M502168200. [DOI] [PubMed] [Google Scholar]
- 33.Glass D.A., Bialek P., Ahn J.D., Starbuck M., Patel M.S., Clevers H., Taketo M.M., Long F., McMahon A.P., Lang R.A. Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. Dev Cell. 2005;8:751–764. doi: 10.1016/j.devcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 34.Lim S.I., Cho C.W., Choi U.K., Kim Y.C. Antioxidant activity and ginsenoside pattern of fermented white ginseng. J Gin Res. 2010;34:168–174. [Google Scholar]
- 35.He B.C., Gao J.L., Luo X., Luo J., Shen J., Wang L., Zhou Q., Wang Y.T., Luu H.H., Haydon R.C. Ginsenoside Rg3 inhibits colorectal tumor growth through the down-regulation of Wnt/β-catenin signaling. Int J Oncol. 2011;38:437–445. doi: 10.3892/ijo.2010.858. [DOI] [PubMed] [Google Scholar]
- 36.Shin H.S., Park S.Y., Hwang E.S., Lee D.G., Song H.G., Mavlonov G.T., Yi T.H. The inductive effect of ginsenoside F2 on hair growth by altering the WNT signal pathway in telogen mouse skin. Eur J Pharmacol. 2014;740:82–89. doi: 10.1016/j.ejphar.2014.02.024. [DOI] [PubMed] [Google Scholar]
- 37.Bi X.L., Xia X.C., Mou T., Jiang B.W., Fan D.D., Wang P., Liu Y.F., Hou Y., Zhao Y.Q. Anti-tumor activity of three ginsenoside derivatives in lung cancer is associated with Wnt/β-catenin signaling inhibition. Eur J Pharmacol. 2014;742:145–152. doi: 10.1016/j.ejphar.2014.08.032. [DOI] [PubMed] [Google Scholar]
- 38.Cui L., Wu T., Li Q.N., Lin L.S., Liang N.C. Preventive effects of ginsenosides on osteopenia of rats induced by ovariectomy. Acta Pharmacol Sin. 2001;22:428–434. [PubMed] [Google Scholar]
- 39.Baron R., Kneissel M. WNT signaling in bone homeostasis and disease: from human mutations to treatments. Nat Med. 2013;19:179–192. doi: 10.1038/nm.3074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.