ABSTRACT
Bartonella spp. are bacteria of worldwide distribution that cause asymptomatic to fatal infections in animals and humans. The most common zoonotic species is Bartonella henselae, for which cats are the major natural reservoir host. To better understand Bartonella sp. diagnostic limitations, we determined the frequency of bloodstream infection in 112 cats by comparing and combining the results of multiple conventional and nested PCRs from blood and liquid culture samples. Using liquid culture conventional PCR, Bartonella sp. DNA was amplified from 27.7% of samples (31/112) compared to 90.2% of samples (101/112) by combining nested PCR from blood and liquid culture, indicating that PCR testing of more than one type of sample provides better sensitivity than a standalone PCR and that bloodstream infection is very frequent among cats in southeastern Brazil. This study reinforces the need for multistep testing for Bartonella sp. infection to prevent false-negative diagnostic results, even in reservoir hosts such as cats that typically maintain higher bacteremia levels.
KEYWORDS: bacteremia, Bartonella, cats, diagnosis, PCR
INTRODUCTION
Bartonella spp. are a group of reemerging bacteria that are distributed worldwide (1). These bacteria cause diverse disease manifestations in humans, ranging from asymptomatic bacteremia to chronic debilitation and death (2). Despite their fastidious behavior, we use the term bacteremia only for samples with Bartonella colony isolation on solid agar and bloodstream infection for any positive Bartonella DNA sample.
Bartonella henselae is the most relevant zoonotic species (3). Cats are considered the major reservoir of this species, and their fleas are considered the principal vector for transmission among cats and potentially humans (4). Previous studies from Brazil found a bloodstream infection prevalence in cats varying between 1.6% and 97.0% (5, 6), a rate much higher than that in asymptomatic human blood donors (2). Subsequently, cat contact was reported to be a risk factor associated with Bartonella sp. infection in asymptomatic Brazilian blood donors (7). We also found that the predictive value of serology for confirming Bartonella infection in asymptomatic blood donors was low. Only three of 16 Bartonella-infected donors were B. henselae or Bartonella quintana seroreactive. Therefore, antibody status should not be used as the sole diagnostic method to determine Bartonella infection status (2).
The fastidious growth characteristics and requirements for special culture conditions, combined with low bacteremia, make the laboratory isolation of Bartonella spp. from opportunistic hosts, such as humans, diagnostically challenging (8–10).
Many reports describe these challenges and show the utility of enrichment blood culture and subculture in enhancing the diagnosis of Bartonella sp. bloodstream infections (11–13).
We performed a study to gain a better understanding of diagnostic sensitivity when using the different techniques for the documentation of Bartonella bloodstream infection in cats.
Cats were selected because they are a natural reservoir host for several Bartonella spp. (14, 15) and are frequently and chronically B. henselae bacteremic (16) and because of their close proximity with human at-risk populations, including children, the elderly, and immunocompromised individuals (7, 17). Therefore, the specific objective of this study was to compare the frequencies of Bartonella bloodstream infection in cats from Campinas, Sao Paulo, Brazil, using a conventional and a nested PCR assay to test DNA extracted from cat blood, DNA extracted from liquid blood culture medium, and DNA extracted from agar subculture isolates.
MATERIALS AND METHODS
Study design.
This project was submitted to and approved by the Institutional Animal Care and Use Committee of the University of Campinas (UNICAMP) under protocol number 2284-1. Between May and September 2009, a convenience sampling of 112 cats from Campinas, Sao Paulo, Brazil (22°54′20″S, 47°03′38″W) was enrolled. Approximately 3 ml of blood was aseptically obtained from each cat by a jugular venipuncture when the cat was presented for ovariohysterectomy or orchiectomy at a local veterinary clinic. Each whole-blood sample was collected into an EDTA tube and stored frozen at −20°C for 9 to 13 months prior to analysis. Sex, estimated age, and origin (stray or pet) data were collected. All experiments were performed at the Laboratory of Applied Research in Dermatology and Bartonella Infection (LARDBI), UNICAMP, Brazil.
Blood culture.
Blood culture using liquid medium was performed as previously described by Duncan et al. (18) and Maggi et al. (19) with modifications. After blood was thawed, an aliquot of 500 μl was inoculated into filter cap cell culture flasks with 2 ml of liquid Bartonella alphaproteobacterium growth medium (BAPGM). A negative-control flask (liquid culture without inoculation) was prepared simultaneously with the cat blood cultures. All flasks were incubated at 37°C in 5% CO2 in a water-saturated atmosphere and maintained with a constant shaking motion for 10 days. After this incubation, a 500-μl aliquot was seeded over solid medium slant tubes prepared as follows: 6 g of Bordet-Gengou agar base in 117 ml of distilled water and 1.167 ml of glycerol and, after sterilization and cooling until 50°C, supplementation with ∼30% (50 ml) sheep blood (Bartonella negative). Sheep blood was confirmed Bartonella sp. negative by PCR and culture, as suggested in previous studies (20, 21). Slant tube cultures were incubated at 37°C in 5% CO2 in a water-saturated atmosphere and examined weekly for up to 45 days. If growth was detected, colonies were Gram stained, and those isolates with consistent Bartonella sp. morphology were suspended and frozen in brain heart infusion (BHI) for future identification by PCR amplification and DNA sequencing. All Bartonella sp. culture methods were carried out in a class 2 biosafety cabinet in order to prevent the possibility of specimen contamination and to protect laboratory personnel.
DNA extraction and quality control.
DNA was extracted from 200 μl of whole blood, 1 ml of BAPGM liquid culture, and colonies using a QIAamp DNA minikit (Qiagen Inc., USA), according to the manufacturer's instructions. We added one positive control (B. henselae) and one negative control (flask only with reagents) per 28 test subject samples. We also performed a PCR specific for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) to test all samples for quality of extracted genomic DNA and for the absence of amplification inhibitors in samples extracted from blood and liquid culture.
Molecular techniques were performed in five separate rooms to avoid DNA contamination as previously described (2).
DNA amplification.
Conventional PCR was performed using DNA extracted from liquid and solid cultures, using the primers 314A-s/314B-s and 357-as targeting a hypervariable region of the Bartonella genus 16S-23S rRNA internal transcribed spacer (ITS) region, as previously described (2). Depending on the Bartonella sp., the length of the expected ITS amplicon was between 157 and 271 bp. Because of the presence of nonspecific bands under LARDBI conditions, the ITS PCR was not utilized to test DNA samples extracted from cat blood samples. A known concentration of B. henselae DNA was serially diluted 10-fold from 109 to 1 genome equivalent (GE) per microliter to determine the sensitivity of the PCR assay. The sensitivity of this PCR assay was established at a minimum of 50 GE of B. henselae per reaction tube. Thus, all blood, liquid culture, and isolate DNA extraction samples were tested by a nested PCR that amplifies the ftsZ gene and is B. henselae specific (22). The detection limit for this reaction was 10 GE of B. henselae per reaction tube, and the expected amplicon length was 218 bp.
The initial concentration of B. henselae GE was estimated as follows: the number of base pairs from the complete B. henselae genome was obtained from GenBank. The molecular weight of the entire genome was calculated using an online mathematical tool (http://www.changbioscience.com/genetics/mw.html). Using the type strain of B. henselae (Houston-1, ATCC 49882), DNA was extracted and quantified by spectrophotometry (NanoDrop). From these two pieces of data, it was then possible to calculate the number of copies per microliter present in the DNA extracted from B. henselae Houston-1. Tenfold serial dilutions were performed and then were tested with 10 independent PCRs. The lowest copy number amplified in all 10 reactions was used as the limit of detection. The limit of detection for the conventional ITS PCR used in this study was more than 25 GE per μl compared to 4 GE per μl for the fstZ nested PCR.
All PCR products were analyzed by horizontal electrophoresis in a 2% agarose gel stained with ethidium bromide. Amplicons generated from conventional ITS PCR were sequenced for bacterial species identification.
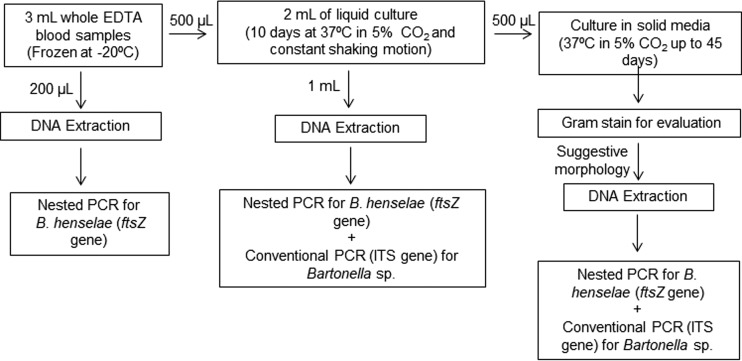
In summary, the approach and methodologies used to document Bartonella sp. bacteremia from each cat blood sample were as follows (Fig. 1).
FIG 1.
Flow chart of culture- and PCR-based procedures performed to determine Bartonella sp. bloodstream infection in cats from Campinas, Sao Paulo, Brazil.
Data analysis.
All cats with at least one PCR amplification from any sample source were considered positive for Bartonella bloodstream infection, which was used as the gold standard for further analyses. The associations between the positive results from the three methods, coupled with approximate cat age (> or <1 year), origin of the animal (stray or pet), and sex (male or female), were also analyzed. Potential associations were first compared in a univariate analysis using Fisher's exact test or the Fisher-Freeman-Halton test. All risk factors significant at the P < 0.25 level were entered into a stepwise logistic regression model, and variables significant to P < 0.05 were retained. Univariate odds ratios (OR), adjusted odds ratios (aOR), and 95% confidence intervals (CI) were calculated. PCR-positive bloodstream infection and the prevalence of Bartonella sp. bacteremia were described as absolute frequencies and percentages, with 95% confidence intervals computed using the score method. Sensitivity and specificity were also determined for each PCR assay, with 95% confidence intervals. A chi-square automatic interaction detection (CHAID) method was used to determine the assay most significantly associated with Bartonella bloodstream infection. Significant values were considered <0.05 and adjusted using the Bonferroni method where appropriate analyses were performed with JMP Pro 13 for Windows (SAS Institute Inc., Cary, NC).
RESULTS
Of 112 cats, 28 (25.0%) were classified as pets and 84 (75.0%) were classified as strays. Seventy-one cats (63.4%) were under 1 year of age, and 41 (36.6%) were more than 1 year of age. Eighty-three (74.1%) were female, and 29 (25.9%) were male.
The results of each PCR assay for each type of sample (blood and liquid culture) are provided in Table 1. All samples were tested for the constitutive gene GAPDH and were PCR positive. All negative controls from each stage in the PCR process (DNA extraction, culture, and master mix) remained negative throughout the study.
TABLE 1.
Bartonella multistep microbiological and molecular method results for pet and stray cats in Brazil
| Group (n) | No. of Bartonella-positive PCR samples by method (% of total per group) |
No. (%) with positive result in at least one sample source and one PCR | ||
|---|---|---|---|---|
| Blood, nested (ftsZ) | Liquid culture |
|||
| Nested (ftsZ) | Conventional (ITS) | |||
| Pet (n = 28) | 15 (53.6) | 18 (64.0) | 11 (39.3) | 21 (75.0) |
| Stray (n = 84) | 71 (84.5) | 33 (39.0) | 20 (23.8) | 80 (95.0) |
| Total (n = 112) | 86 (76.8) | 51 (45.5) | 31 (27.7) | 101 (90.2) |
Using ftsZ nested PCR, B. henselae DNA was amplified from 86/112 (76.8%) cat blood specimens and 51/112 (45.5%) liquid culture samples. Amplicons of sufficient quality for DNA sequencing from 11 blood and 19 liquid culture samples had 100% similarity with the B. henselae complete genome (GenBank accession number HG969191). By combining results from both PCR assays and all sample sources, bloodstream Bartonella sp. was detected in 90.2% (101/112) of cats. The ftsZ species-specific nested PCR was positive for at least one sample source for each of the 101 B. henselae-infected cats. Figure 2 represents the results using an area-proportional Venn diagram that demonstrates that 15/26 blood-negative cats had positive results following liquid culture (13.4%) (23).
FIG 2.
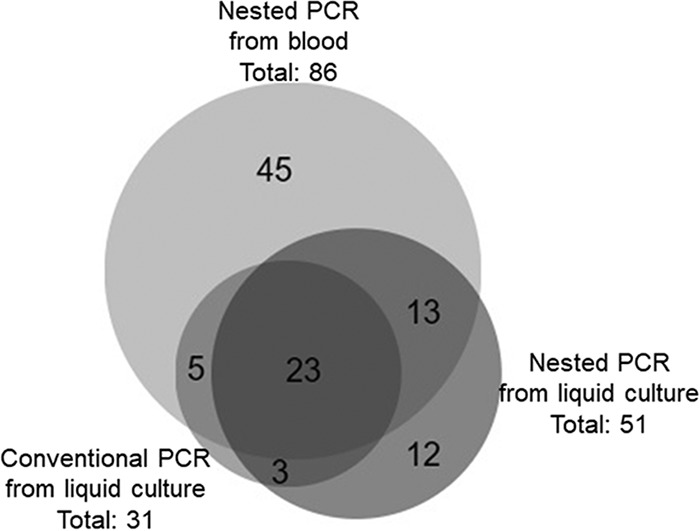
Area-proportional Venn diagram of Bartonella sp. PCR results from Brazilian cats.
By conventional PCR, Bartonella 16S-23S ITS DNA was amplified from only 31/112 (27.7%) cat BAPGM liquid culture DNA extractions. Eleven of 16 Gram-negative slant subculture isolates with suggestive Bartonella sp. morphology were Bartonella sp. PCR positive. From these 11 isolates (9.8% from total samples), six ITS amplicons of sufficient quality for DNA sequencing had 100% similarity with the B. henselae Brazil-1 strain (GenBank accession number DQ346666.1), and seven ftsZ amplicons of sufficient quality for DNA sequencing had 100% similarity with the B. henselae complete genome (GenBank accession number HG969191). Three isolates were the first samples of B. henselae isolated in Brazil to be deposited at the Adolfo Lutz Culture Collection in Brazil under accession numbers IAL 3714, 3715, and 3716.
By multiple logistic regression analysis, young cats (<1 year old) were 6 times more likely than adult cats to be PCR positive (aOR, 6.18; 95% confidence interval [CI], 1.24 to 38.77). Four cats less than and seven cats more than 1 year of age were PCR negative. Bloodstream infection was more prevalent in stray cats (95%, 80/84 cats) than in client-owned cats (75%, 21/28 cats). Stray cats were 13 times more likely to be PCR positive than the pet cats (aOR, 13.15; 95% CI, 2.71 to 87.66). Based on the same analysis, sex was not significantly associated with B. henselae bloodstream infection.
Compared to the ITS PCR, the ftsZ nested PCR from blood was a more sensitive assay for the amplification of B. henselae DNA, with an 85.1% (95% CI, 2.7 to 87.7) sensitivity and a negative predictive value of 42.3% (95% CI, 25.5 to 61.1), compared to ftsZ nested PCR from liquid culture with a 50.5% (95% CI, 40.9 to 60.0) sensitivity and a negative predictive value of 18.0% (95% CI, 10.4 to 29.5) and compared to conventional ITS PCR from liquid culture, with a sensitivity of 30.7% (95% CI, 22.5 to 40.3) and negative predictive value of 13.6% (95% CI, 7.8 to 22.7). The CHAID analysis also indicated that the ftsZ nested PCR from blood was the single assay most associated with Bartonella bloodstream infection, followed by the ftsZ nested PCR from liquid culture.
DISCUSSION
In this study, 90% of pet and stray cats from Brazil were PCR positive from blood, liquid blood culture, or slant tube subculture isolates. The sensitivity of Bartonella sp. PCR detection increased using liquid blood culture over PCR from DNA extracted directly from blood samples. Besides the high bloodstream infection detection, bacteremia was confirmed in only 9.8% by bacterial isolation. As in previous studies, liquid culture and nested PCR used together increased the analytical sensitivity (18, 19, 24, 25). If we had performed only the ITS conventional PCR assay, which in our lab is less sensitive than ftsZ nested PCR (50 GE versus 10 GE, respectively), for testing the cats in this study, Bartonella sp. bloodstream infection would have been detected in only 27.7% versus 45.5%. Bartonella PCR prevalence in cats has varied from 0% in Norway to 97% in Brazil, with a median worldwide prevalence of approximately 30.5% (6, 26). A high Bartonella prevalence was detected in this study, compared to most other Bartonella sp. cat prevalence studies reported from Brazil (Table 2) (5, 6, 8, 27–38). Discrepancy among studies may be related to differences in the analytical sensitivity of laboratory diagnostic methods used among different studies. As illustrated by this study, the reported prevalence is dependent upon the methods used to establish the presence or absence of bloodstream infection.
TABLE 2.
Epidemiology of Bartonella spp. in Brazilian cats as reflected by culture, serology, and PCR findingsa
| Publication yr | Region of Brazil | Cat origin | Wild felid | Prevalence positive/total (%) |
Bartonella species | Reference | ||
|---|---|---|---|---|---|---|---|---|
| Culture | Serology | PCR | ||||||
| 2007 | Southeast | D/S | NP | 32/200 (16) | NP | B. henselae | 27 | |
| 2010 | South | S | NP | NP | 8/47 (17)b | 5 B. henselae, 3 B. clarridgeiae | 28 | |
| 2010 | Southeast | S | NP | 25/37 (68) | 36/37 (97) | Bartonella spp. | 6 | |
| 2010 | Many regions | Small neotropical felids | NP | NP | 10/67 (15) | 10 B. henselae | 29 | |
| 2011 | Southeast | D/S | NP | 19/40 (47) | 17/40 (42) | B. henselae | 30 | |
| 2012 | Southeast | D/S | NP | NP | 2/26 (4.3) | 2 B. henselae | 31 | |
| 2012 | Northeastern | D | NP | NP | 9/200 (4.5) | 6 B. henselae, 3 B. clarridgeiae | 32 | |
| 2012 | Southeast | Neotropical felids | NP | 40/84 (48) | 2/109 (1.8) | 1 B. koehlerae | 33 | |
| 2013 | Midwest | D/S | NP | NP | 4/163 (2.5) | 3 B. henselae, 1 B. clarridgeiae | 38 | |
| 2014 | Southeast | S | NP | NP | 11/37 (30) | 6 B. henselae, 5 B. clarridgeiae | 34 | |
| 2014 | South | S | NP | NP | 12/47 (26)b | 8 B. henselae, 6 B. clarridgeiae | 8 | |
| 2015 | Southeast | D/S | NP | NP | 46/151 (30) | B. henselae and B. clarridgeiae | 35 | |
| 2015 | Midwest | D/S | NP | NP | 3/182 (1.6) | 3 B. clarridgeiae | 5 | |
| 2016 | South | D | NP | NP | 6/30 (20) | Bartonella spp. | 36 | |
| 2017 | Northeastern | D | NP | 6/40 (15) | 0/40 (0) | Bartonella spp. | 37 | |
Abbreviations: D, domestic; S, stray; NP, not performed.
Same population of cats tested in the two studies.
Another factor to consider in the context of bloodstream infection is that 63.4% of cats in this study were under 1 year of age. Studies have reported a higher seroprevalence in older cats, whereas bacteremia is higher in younger cats (39, 40), which is consistent with our findings where cats under 1 year of age were statistically more likely to have bloodstream infection (P < 0.01). Statistically, B. henselae DNA was more frequently amplified from stray cats than pet cats, which is also in agreement with findings from previous studies (41, 42).
In this current study, B. henselae species-specific nested ftsZ PCR was more sensitive than the conventional ITS PCR from liquid culture that we use. Pennisi et al. also found that nested PCR increases the detection sensitivity of Bartonella sp. infection (25). Our results reinforce the importance of multiple sample analyses to prevent false-negative results and erroneous epidemiologic and diagnostic conclusions (43).
Bartonella sp. DNA was detected in 76.8% of cat samples from DNA extracted directly from blood (without culture) using a nested PCR. It is possible that the amount of Bartonella sp. DNA in the initial blood sample was below the minimum detection threshold, and following culture, there were enough bacteria for detection due to enhancement of bacterial replication during the 10-day incubation in liquid medium. The nested PCR of liquid culture detected an additional 13.4% (15/112) of positive cats.
The lower number of liquid culture ftsZ nested PCR-positive samples (n = 51) than of blood (n = 86) may indicate the amplification of nonviable bacteria in blood that failed to grow in liquid culture. The fastidious characteristic of the genus, besides the difficulty in cultivating a wild strain, could be the reason for these numbers. Blood bacterial numbers would also have been affected by dilution in a large volume of culture medium (dilution effect). Since there was no increase in the amount of bacteria in the liquid culture medium, the concentration of bacteria in these samples was below the nested PCR level of detection (44). It could happen for other reasons such as antimicrobial treatment, besides the fastidious characteristic of the genus.
Table 3 estimates the number of bacterial DNA copies necessary in initial blood or liquid culture samples that would be required to reach the minimum detection threshold for the ITS and fstZ PCR assays in the LARDBI, Brazil, assuming that there was no bacterial multiplication in liquid culture. We estimated these numbers using the limit of detection for the conventional ITS PCR (25 GE per μl) and fstZ nested PCR (4 GE per μl). We also considered (i) the amount of blood used initially in each step—200 μl of whole blood and 500 μl of whole blood “diluted” in 2 ml of liquid culture medium, (ii) the amount of sample used in DNA extraction (200 μl of whole blood and 1 ml of liquid culture), (iii) the volume of resuspension at the end of extraction (200 μl for both), and (iv) the amount of DNA sample used in each PCR (2.5 μl in nested PCR and 5 μl in conventional PCR). Using these data, we calculated the limit of Bartonella sp. detection in each method assuming that there was no bacterial multiplication in culture. Importantly, PCR sensitivity can vary within laboratories due to differences in thermocyclers and between laboratories due to differences in primer design, DNA polymerases, thermocyclers, and other factors. Although unsettling for clinicians and diagnosticians, a negative Bartonella PCR result does not exclude the possibility of bloodstream infection.
TABLE 3.
Limit of Bartonella sp. detection in each method
| GE/μla in initial blood sample | Liquid culture |
Blood, nested PCR | |
|---|---|---|---|
| Conventional PCR | Nested PCR | ||
| Below 4 | Negative | Negative | Negative |
| From 4 to 9 | Negative | Negative | Positive |
| From 10 to 24 | Negative | Positive | Positive |
| Over 25 | Positive | Positive | Positive |
GE, genome equivalent; 1 GE/μl = 1,000 copies/ml.
From 101 positive samples, only 11 Bartonella sp. isolates were obtained. Sequence analysis revealed DNA sequence homology among the 6 isolates with the GenBank Brazil-1 B. henselae strain. This low number of isolates reflects the Bartonella species' fastidious nature: primary Bartonella isolation is difficult to achieve from reservoir and nonreservoir sick patients (19). All six isolates came from cats that were ftsZ nested PCR positive. Two of these came from cats that were positive only in blood or liquid culture prior to slant subculture.
In a previous study from our laboratory, we found a 3.2% rate of Bartonella sp. bloodstream infection in Brazilian blood donors (16/500, 15 for B. henselae and one for Bartonella clarridgeiae), using only the 16S-23S conventional PCR assay to screen BAPGM enrichment blood cultures and slant subculture isolates (2). Five of 6 B. henselae isolates from donors were obtained from enrichment blood culture conventional PCR-negative DNA amplifications. Importantly, only 3/16 Bartonella-infected human donors were seroreactive to B. henselae or Bartonella quintana by indirect immunofluorescent assay (IFA) and only 1/6 Bartonella enrichment culture PCR-positive donors was confirmed bacteremic by liquid subculture isolation (2). The Brazilian blood donors were recruited from the same region as the cats in this study, potentially explaining a particularly high environmental exposure rate to B. henselae. The data reported in this study reinforce the concept that there is no gold standard diagnostic test to confirm Bartonella sp. infection.
Conclusion.
By combining methods and samples, a higher prevalence of Bartonella sp. bloodstream infection was documented in cats from Southeastern Brazil. Our results support previous studies that recommend the use of multiple diagnostic modalities to detect Bartonella sp. bloodstream infection. As documentation of Bartonella bacteremia by isolation in humans is a very infrequent occurrence, our study reinforces the importance of using several tests in combination to prevent false-negative epidemiological and diagnostic test results.
ACKNOWLEDGMENTS
We thank Hanako Nancy Momma for collecting all samples used in this study and Ricardo Maggi for helpful review of the manuscript.
This project was partially funded by the Faculty Grant in Global Health to D.G.S. from the Johns Hopkins Center for Global Health and by grants to P.E.N.F.V. from FAEPEX-UNICAMP (Fundo de Apoio ao Ensino, à Pesquisa e à Extensão—UNICAMP) under protocols number 234/10 and 292/10.
In conjunction with Sushama Sontakke and North Carolina State University, Edward B. Breitschwerdt holds U.S. patent no. 7,115,385, media and methods for cultivation of microorganisms, which was issued on 3 October 2006. He is a cofounder, stockholder, and chief scientific officer for Galaxy Diagnostics, a company that provides advanced diagnostic testing for the detection of Bartonella species infections. The remaining authors have no competing interests.
REFERENCES
- 1.Mogollon-Pasapera E, Otvos L, Giordano A, Cassone M. 2009. Bartonella: emerging pathogen or emerging awareness? Int J Infect Dis 13:3–8. doi: 10.1016/j.ijid.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Pitassi LH, de Paiva Diniz PP, Scorpio DG, Drummond MR, Lania BG, Barjas-Castro ML, Gilioli R, Colombo S, Sowy S, Breitschwerdt EB, Nicholson WL, Velho PE. 2015. Bartonella spp. bacteremia in blood donors from Campinas, Brazil. PLoS Negl Trop Dis 9:e0003467. doi: 10.1371/journal.pntd.0003467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaiser PO, Riess T, O'Rourke F, Linke D, Kempf VA. 2011. Bartonella spp.: throwing light on uncommon human infections. Int J Med Microbiol 301:7–15. doi: 10.1016/j.ijmm.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Chomel BB, Boulouis HJ, Maruyama S, Breitschwerdt EB. 2006. Bartonella spp. in pets and effect on human health. Emerg Infect Dis 12:389–394. doi: 10.3201/eid1203.050931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braga IA, Dias IS, Chitarra CS, Amude AM, Aguiar DM. 2015. Molecular detection of Bartonella clarridgeiae in domestic cats from Midwest Brazil. Braz J Infect Dis 19:451–452. doi: 10.1016/j.bjid.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Souza A, Almeida D, Guterres A, Gomes R, Favacho A, Moreira N, Maia L, Rozental T, Torres Filho R, Cerqueira A, Lemos E, Almosny N. 2010. Bartonelose: análise molecular e sorológica em gatos do Rio de Janeiro—Brasil. Rev Bras Cie Vet 17:7–11. [Google Scholar]
- 7.Diniz P, Velho P, Pitassi L, Drummond M, Lania B, Barjas-Castro M, Sowy S, Breitschwerdt E, Scorpio D. 2016. Risk factors for Bartonella species infection in blood donors from southeast Brazil. PLoS Negl Trop Dis 10:e0004509. doi: 10.1371/journal.pntd.0004509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Staggemeier R, Pilger DA, Spilki FR, Cantarelli VV. 2014. Multiplex SYBR Green-real time PCR (qPCR) assay for the detection and differentiation of Bartonella henselae and Bartonella clarridgeiae in cats. Rev Inst Med Trop Sao Paulo 56:93–95. doi: 10.1590/S0036-46652014000200001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breitschwerdt EB. 2017. Bartonellosis, One Health and all creatures great and small. Vet Dermatol 28:96–e21. doi: 10.1111/vde.12413. [DOI] [PubMed] [Google Scholar]
- 10.Diaz MH, Bai Y, Malania L, Winchell JM, Kosoy MY. 2012. Development of a novel genus-specific real-time PCR assay for detection and differentiation of Bartonella species and genotypes. J Clin Microbiol 50:1645–1649. doi: 10.1128/JCM.06621-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breitschwerdt EB, Maggi RG, Cadenas MB, Diniz PP. 2009. A groundhog, a novel Bartonella sequence, and my father's death. Emerg Infect Dis 15:2080–2086. doi: 10.3201/eid1512.AD1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breitschwerdt EB. 2015. Did Bartonella henselae contribute to the deaths of two veterinarians? Parasit Vectors 8:317. doi: 10.1186/s13071-015-0920-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Velho PE, Pimentel V, Del Negro GM, Okay TS, Diniz PP, Breitschwerdt EB. 2007. Severe anemia, panserositis, and cryptogenic hepatitis in an HIV patient infected with Bartonella henselae. Ultrastruct Pathol 31:373–377. doi: 10.1080/01913120701696601. [DOI] [PubMed] [Google Scholar]
- 14.Woudstra C, Fach P, Chomel BB, Haddad N, Boulouis HJ. 2017. Draft genome sequences of 12 feline Bartonella henselae isolates. Genome Announc 5:e00075-17. doi: 10.1128/genomeA.00075-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brunt J, Guptill L, Kordick DL, Kudrak S, Lappin MR. 2006. American Association of Feline Practitioners 2006 panel report on diagnosis, treatment, and prevention of Bartonella spp. infections. J Feline Med Surg 8:213–226. doi: 10.1016/j.jfms.2006.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breitschwerdt EB, Kordick DL. 2000. Bartonella infection in animals: carriership, reservoir potential, pathogenicity, and zoonotic potential for human infection. Clin Microbiol Rev 13:428–438. doi: 10.1128/CMR.13.3.428-438.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson CA, Saha S, Mead PS. 2016. Cat-scratch disease in the United States, 2005–2013. Emerg Infect Dis 22:1741–1746. doi: 10.3201/eid2210.160115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duncan AW, Maggi RG, Breitschwerdt EB. 2007. A combined approach for the enhanced detection and isolation of Bartonella species in dog blood samples: pre-enrichment liquid culture followed by PCR and subculture onto agar plates. J Microbiol Methods 69:273–281. doi: 10.1016/j.mimet.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 19.Maggi RG, Duncan AW, Breitschwerdt EB. 2005. Novel chemically modified liquid medium that will support the growth of seven bartonella species. J Clin Microbiol 43:2651–2655. doi: 10.1128/JCM.43.6.2651-2655.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drummond MR, Pitassi LH, Lania BG, Dos Santos SR, Gilioli R, Velho PE. 2011. Detection of Bartonella henselae in defibrinated sheep blood used for culture media supplementation. Braz J Microbiol 42:430–432. doi: 10.1590/S1517-83822011000200003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bemis DA, Kania SA. 2007. Isolation of Bartonella sp. from sheep blood. Emerg Infect Dis 13:1565–1567. doi: 10.3201/eid1310.070570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawasato KH, de Oliveira LC, Velho PE, Yamamoto L, Del Negro GM, Okay TS. 2013. Detection of Bartonella henselae DNA in clinical samples including peripheral blood of immune competent and immune compromised patients by three nested amplifications. Rev Inst Med Trop Sao Paulo 55:1–6. doi: 10.1590/S0036-46652013000100001. [DOI] [PubMed] [Google Scholar]
- 23.Hulsen T, de Vlieg J, Alkema W. 2008. BioVenn—a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fenollar F, Sire S, Wilhelm N, Raoult D. 2005. Bartonella vinsonii subsp. arupensis as an agent of blood culture-negative endocarditis in a human. J Clin Microbiol 43:945–947. doi: 10.1128/JCM.43.2.945-947.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pennisi MG, La Camera E, Giacobbe L, Orlandella BM, Lentini V, Zummo S, Fera MT. 2010. Molecular detection of Bartonella henselae and Bartonella clarridgeiae in clinical samples of pet cats from Southern Italy. Res Vet Sci 88:379–384. doi: 10.1016/j.rvsc.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 26.Boulouis HJ, Chang CC, Henn JB, Kasten RW, Chomel BB. 2005. Factors associated with the rapid emergence of zoonotic Bartonella infections. Vet Res 36:383–410. doi: 10.1051/vetres:2005009. [DOI] [PubMed] [Google Scholar]
- 27.Loureiro VS, Hagiwara M. 2007. A survey of anti-Bartonella henselae antibodies in domiciliated cats in the city of São Paulo, state of São Paulo and its importance in public health. Rev Bras Cie Vet 14:39–42. [Google Scholar]
- 28.Staggemeier R, Venker CA, Klein DH, Petry M, Spilki FR, Cantarelli VV. 2010. Prevalence of Bartonella henselae and Bartonella clarridgeiae in cats in the south of Brazil: a molecular study. Mem Inst Oswaldo Cruz 105:873–878. doi: 10.1590/S0074-02762010000700006. [DOI] [PubMed] [Google Scholar]
- 29.Guimaraes AM, Brandão PE, Moraes W, Kiihl S, Santos LC, Filoni C, Cubas ZS, Robes RR, Marques LM, Neto RL, Yamaguti M, Oliveira RC, Catão-Dias JL, Richtzenhain LJ, Messick JB, Biondo AW, Timenetsky J. 2010. Detection of Bartonella spp. in neotropical felids and evaluation of risk factors and hematological abnormalities associated with infection. Vet Microbiol 142:346–351. doi: 10.1016/j.vetmic.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 30.Crissiuma A, Favacho A, Gershony L, Mendes-de-Almeida F, Gomes R, Mares-Guia A, Rozental T, Barreira J, Lemos E, Labarthe N. 2011. Prevalence of Bartonella species DNA and antibodies in cats (Felis catus) submitted to a spay/neuter program in Rio de Janeiro, Brazil. J Feline Med Surg 13:149–151. doi: 10.1016/j.jfms.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Bortoli CP, André MR, Seki MC, Pinto AA, Machado Se T, Machado RZ. 2012. Detection of hemoplasma and Bartonella species and co-infection with retroviruses in cats subjected to a spaying/neutering program in Jaboticabal, SP, Brazil. Rev Bras Parasitol Vet 21:219–223. doi: 10.1590/S1984-29612012000300008. [DOI] [PubMed] [Google Scholar]
- 32.Braga MDSCDO, Diniz PP, André MR, Bortoli CP, Machado RZ. 2012. Molecular characterisation of Bartonella species in cats from São Luís, state of Maranhão, north-eastern Brazil. Mem Inst Oswaldo Cruz 107:772–777. doi: 10.1590/S0074-02762012000600011. [DOI] [PubMed] [Google Scholar]
- 33.Filoni C, Catao-Dias JL, Cattori V, Willi B, Meli ML, Correa SH, Marques MC, Adania CH, Silva JC, Marvulo MF, Ferreira Neto JS, Durigon EL, de Carvalho VM, Coutinho SD, Lutz H, Hofmann-Lehmann R. 2012. Surveillance using serological and molecular methods for the detection of infectious agents in captive Brazilian neotropic and exotic felids. J Vet Diagn Invest 24:166–173. doi: 10.1177/1040638711407684. [DOI] [PubMed] [Google Scholar]
- 34.Andre MR, Baccarim Denardi NC, Marques de Sousa KC, Goncalves LR, Henrique PC, Grosse Rossi Ontivero CR, Lima Gonzalez IH, Cabral Nery CV, Fernandes Chagas CR, Monticelli C, Alexandre de Santis AC, Machado RZ. 2014. Arthropod-borne pathogens circulating in free-roaming domestic cats in a zoo environment in Brazil. Ticks Tick Borne Dis 5:545–551. doi: 10.1016/j.ttbdis.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 35.Andre MR, Dumler JS, Herrera HM, Goncalves LR, de Sousa KC, Scorpio DG, de Santis AC, Domingos IH, de Macedo GC, Machado RZ. 2016. Assessment of a quantitative 5′ nuclease real-time polymerase chain reaction using the nicotinamide adenine dinucleotide dehydrogenase gamma subunit (nuoG) for Bartonella species in domiciled and stray cats in Brazil. J Feline Med Surg 18:783–790. doi: 10.1177/1098612X15593787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malheiros J, Costa MM, do Amaral RB, de Sousa KC, Andre MR, Machado RZ, Vieira MI. 2016. Identification of vector-borne pathogens in dogs and cats from southern Brazil. Ticks Tick Borne Dis 7:893–900. doi: 10.1016/j.ttbdis.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 37.Fontalvo MC, Favacho ARM, Araujo AC, Santos NMD, Oliveira GMB, Aguiar DM, Lemos ERS, Horta MC. 2017. Bartonella species pathogenic for humans infect pets, free-ranging wild mammals and their ectoparasites in the Caatinga biome, northeastern Brazil: a serological and molecular study. Braz J Infect Dis 21:290–296. doi: 10.1016/j.bjid.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miceli NG, Gavioli FA, Gonçalves LR, André MR, Sousa VRF, Sousa KC, Machado RZ. 2013. Molecular detection of feline arthropod-borne pathogens in cats in Cuiabá, state of Mato Grosso, central-western region of Brazil. Rev Bras Parasitol Vet 22:385–390. doi: 10.1590/S1984-29612013000300011. [DOI] [PubMed] [Google Scholar]
- 39.Guptill L. 2010. Bartonellosis. Vet Microbiol 140:347–359. doi: 10.1016/j.vetmic.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 40.Guptill L. 2010. Feline bartonellosis. Vet Clin North Am Small Anim Pract 40:1073–1090. doi: 10.1016/j.cvsm.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 41.Guptill L, Wu CC, HogenEsch H, Slater LN, Glickman N, Dunham A, Syme H, Glickman L. 2004. Prevalence, risk factors, and genetic diversity of Bartonella henselae infections in pet cats in four regions of the United States. J Clin Microbiol 42:652–659. doi: 10.1128/JCM.42.2.652-659.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gurfield AN, Boulouis HJ, Chomel BB, Kasten RW, Heller R, Bouillin C, Gandoin C, Thibault D, Chang CC, Barrat F, Piemont Y. 2001. Epidemiology of Bartonella infection in domestic cats in France. Vet Microbiol 80:185–198. doi: 10.1016/S0378-1135(01)00304-2. [DOI] [PubMed] [Google Scholar]
- 43.Lynch T, Iverson J, Kosoy M. 2011. Combining culture techniques for Bartonella: the best of both worlds. J Clin Microbiol 49:1363–1368. doi: 10.1128/JCM.02403-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson IG. 1997. Inhibition and facilitation of nucleic acid amplification. Appl Environ Microbiol 63:3741–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]