ABSTRACT
The plasmid-located colistin resistance gene mcr-1 confers low-level resistance to colistin, a last-line antibiotic against multidrug-resistant Gram-negative bacteria. Current CLSI-EUCAST recommendations require the use of a broth microdilution (BMD) method with cation-adjusted Mueller-Hinton (CA-MH) medium for colistin susceptibility testing, but approximately 15% of all MCR-1 producers are classified as sensitive in that broth. Here we report on an improved calcium-enhanced Mueller-Hinton (CE-MH) medium that permits simple and reliable determination of mcr-1-containing Enterobacteriaceae. Colistin susceptibility testing was performed for 50 mcr-1-containing Escherichia coli and Klebsiella pneumoniae isolates, 7 intrinsically polymyxin-resistant species, K. pneumoniae and E. coli isolates with acquired resistance to polymyxins due to mgrB and pmrB mutations, respectively, and 32 mcr-1-negative, colistin-susceptible isolates of Acinetobacter baumannii, Citrobacter freundii, Enterobacter cloacae, E. coli, K. pneumoniae, and Salmonella enterica serovar Typhimurium. A comparison of the colistin MICs determined in CA-MH medium and those obtained in CE-MH medium was performed using both the BMD and strip-based susceptibility test formats. We validated the data using an isogenic IncX4 plasmid lacking mcr-1. Use of the CE-MH broth provides clear separation between resistant and susceptible isolates in both BMD and gradient diffusion assays; this is true for both mcr-1-containing Enterobacteriaceae isolates and those exhibiting either intrinsic or acquired colistin resistance. CE-MH medium is simple to prepare and overcomes current problems associated with BMD and strip-based colistin susceptibility testing, and use of the medium is easy to implement in routine diagnostic laboratories, even in resource-poor settings.
KEYWORDS: colistin susceptibility testing, mcr-1 resistance, calcium ions
INTRODUCTION
Polymyxins, which include polymyxin B and colistin, are pentacationic lipopeptide antibiotics produced by the bacterium Paenibacillus polymyxa (1). In analogy to polymyxin B, colistin selectively binds to the lipopolysaccharide (LPS) of Gram-negative bacteria. It competitively displaces divalent cations (such as Ca2+ and Mg2+) that bridge adjacent LPS molecules, and insertion of its lipopeptide moiety induces expansion of the outer membrane and, with time, loss of physical integrity of the phospholipid bilayer of the inner membrane. The increase in the permeability of the cell membranes leads to leakage of intracellular contents and ultimately bacterial death (2–6).
Colistin has been used for decades as an oral drug in veterinary medicine, particularly to treat infections associated with pathogenic Escherichia coli and Salmonella spp. in pig and veal calf herds (7, 8). With the emergence of multidrug-resistant (MDR) Gram-negative bacteria exhibiting resistance to third-generation cephalosporins and carbapenems, colistin is now also used as a last-line antibiotic for treatment of health care-associated infections (9, 10).
Resistance to colistin generally involves mutations in chromosomal genes. Intrinsic resistance to colistin in strains such as Morganella morganii, Proteus mirabilis, Proteus vulgaris, Providencia rettgeri, and Serratia marcescens strains is due to modification of lipopolysaccharide with amino sugars such as l-Ara4N or overexpression of outer membrane proteins. Acquired resistance to polymyxins in strains such as Klebsiella pneumoniae and E. coli strains involves mutations in the PmrAB/PhoPQ two-component sensing systems or alterations of the mgrB and pmrB genes (11–13).
Recently, a plasmid-borne mcr-1 gene conferring resistance to colistin that could be transmitted by horizontal gene transfer was identified (14). This new mechanism could contribute to increased occurrence of colistin resistance. The degree of resistance conferred by mcr-1 is lower than that of the other colistin resistance mechanisms encoded on the chromosome (15).
Current antimicrobial susceptibility testing for colistin is fraught with pitfalls, and a joint CLSI-EUCAST Polymyxin Breakpoints Working Group recently provided warnings regarding the credibility of methods used to determine the MICs of colistin-resistant isolates (16, 17). Currently, it is recommended that the broth microdilution (BMD) method should be used as a standard format for colistin antimicrobial susceptibility testing, but it has been claimed that BMD is time-consuming and requires trained technical staff for manual preparation of antibiotic solutions and assay assessment (18). Some of these difficulties are overcome by the rapid polymyxin NP test, but failure to detect polymyxin-resistant E. coli and Enterobacter sp. strains using this assay has been reported (19).
The difficulties with colistin resistance testing are associated with the physical properties of cationic lipopeptides, including their poor agar diffusion characteristics and the unidentified conditions required for the activity of MCR-1. Limited laboratory screening procedures using cation-adjusted Mueller-Hinton (CA-MH) medium for BMD have implications for public health and lead to underestimation of the true prevalence of mcr-1 and thus to possible treatment failures due to methodology-based nondetection of resistance. Therefore, there is a need for a simple assay that promotes reliable and reproducible susceptibility testing using different formats, in order to implement preventive measures (20).
Here we report on a novel medium for the improved determination of mcr-1-producing Enterobacteriaceae. The broth described here allows reliable monitoring of MCR-1-dependent activity and requires optimized levels of Ca2+ for the detection of colistin resistance in mcr-1-containing bacteria. The medium is easy to prepare and usable in several assay formats (e.g., microdilution and gradient diffusion) and therefore is a useful addition to protocols currently used for testing of colistin resistance.
(Konrad Gwozdzinski performed this work as a partial fulfillment of the requirements for a Ph.D. at the Faculty of Medicine, Justus Liebig University, Giessen, Germany, 2018. Saina Azarderakhsh performed this work as a partial fulfillment of the requirements for an M.Sc. at the Faculty of Biology, Philipps University of Marburg, Marburg, Germany, 2017.)
MATERIALS AND METHODS
Isolates used in the study.
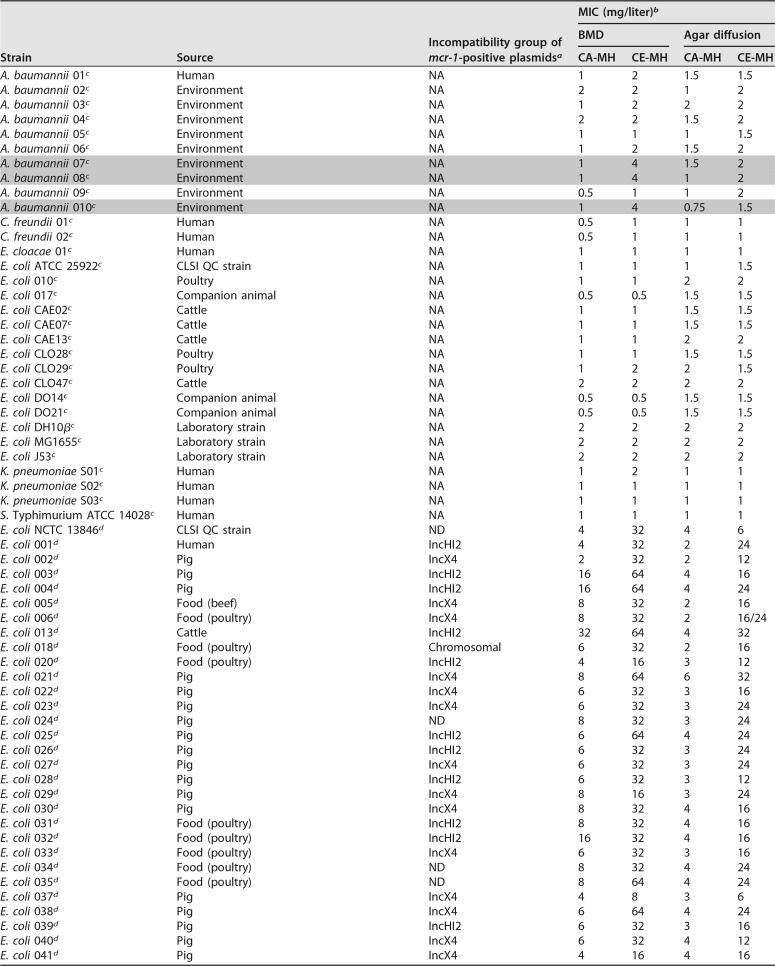
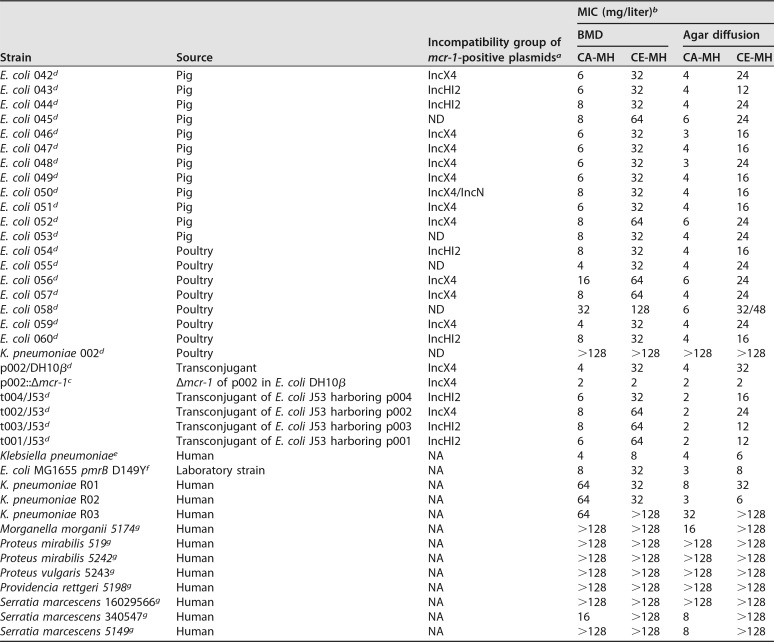
We included 50 mcr-1-containing isolates of E. coli and K. pneumoniae (21), 7 isolates of intrinsically colistin-resistant species, including M. morganii, P. mirabilis, P. vulgaris, P. rettgeri, and S. marcescens, 8 isolates of Acinetobacter baumannii, E. coli, or K. pneumoniae exhibiting acquired resistance to polymyxins, including 1 isolate of K. pneumoniae with a mutation in the mgrB gene and 1 isolate of E. coli with a mutated pmrB gene (D149Y), 32 mcr-1-negative and colistin-susceptible isolates representing diverse bacterial species, such as A. baumannii, Citrobacter freundii, Enterobacter cloacae, E. coli, K. pneumoniae, and Salmonella enterica serovar Typhimurium, and 5 E. coli J53-DH10β transconjugants with mcr-1 on either IncX4 or IncHI2 plasmids (22). Details of the relevant characteristics of the isolates are provided in Table 1.
TABLE 1.
Properties of isolates included in the study
a NA, not applicable; ND, not determined.
b Colistin MIC values were determined by both broth microdilution and strip-based methods, in CA-MH broth and CE-MH broth. False-susceptible isolates that were resistant with the test method (BMD in CE-MH broth) and susceptible with the reference method (BMD in CA-MH broth) are highlighted in gray.
c mcr-1-negative, colistin-susceptible isolates.
d mcr-1-positive isolates.
e Mutation in the mgrB gene involved in colistin resistance.
f Mutation in the pmrB gene (D149Y) involved in colistin resistance.
g Intrinsically polymyxin-resistant species.
Mutagenesis of the pmrB gene.
A mutation in the pmrB gene of E. coli MG1655 leading to activation of the arnBCADTEF-pmrE operon (23) was introduced through a plasmid-based portable multiplex automated genome engineering (pORTMAGE) approach, as described previously (24). The sequence of the oligonucleotide carrying a G445T substitution is shown in Table SA4 in the supplemental material. The pORTMAGE plasmid was maintained by addition of 100 mg/liter ampicillin (Sigma-Aldrich) to the medium. For verification of the mutated gene, whole-genome DNA was isolated using the PureLink genome DNA minikit (Thermo Fischer Scientific, Langenselbold, Germany), following the manufacturer's instructions. A Nextera XT library of the genome (Illumina, San Diego, CA, USA) was sequenced with a MiSeq system (using 2 by 300 cycles). Raw data were assembled using SPAdes (25), and comparison with the wild-type pmrB gene was performed using BLASTn.
Preparation of inactivated human serum samples.
For normal human serum (NHS) collection, a consenting adult volunteer donated blood. After clotting at room temperature, the NHS component was aseptically harvested and used fresh. NHS was heated for 30 min at 56°C in order to inactivate complement.
Preparation of calcium-enhanced Mueller-Hinton broth.
The medium devised here is based on Mueller-Hinton (MH) broth supplemented with calcium chloride dehydrate, i.e., calcium-enhanced Mueller-Hinton (CE-MH) broth. To prepare 1 liter of CE-MH medium, 23 g of MH broth powder (product no. 70192; Sigma-Aldrich, Darmstadt, Germany) was dissolved in 1 liter of distilled water and sterilized by autoclaving at 121°C for 15 min. After the solution was cooled to room temperature, 5 ml of a sterile-filtered 1 M stock solution of calcium chloride dihydrate (product no. 5239.3; Carl Roth, Karlsruhe, Germany) was added to the MH broth to achieve a final molarity of 5 mM in the medium. For solid media, 15 g/liter agar was added before autoclaving. Storage of MH agar plates supplemented with 5 mM calcium chloride dihydrate for up to 4 weeks at 4°C did not affect strip-based colistin susceptibility testing. The reference medium, CA-MH broth, was prepared from MH broth supplemented with Ca2+ and Mg2+, according to CLSI-EUCAST guidelines (16).
MIC determination.
BMD to determine the MICs of the isolates was performed according to the recommendations of the joint CLSI-EUCAST Polymyxin Breakpoints Working Group (16). A range of 0.25 to 128 mg/liter colistin sulfate (Sigma-Aldrich) was tested. Plain 96-well polystyrene microplates (Greiner, Frickenhausen, Germany) were used for broth microdilution experiments. Each isolate was examined for growth in either CA-MH or CE-MH medium. MIC determination was performed by visual inspection following overnight incubation of the plate at 37°C.
MIC determination by the strip-based method was performed in accordance with the manufacturer's instructions (product no. 01B10093; Liofilchem, Roseto degli Abruzzi, Italy). The MIC was read at 1 dilution step above the area where the growth inhibition area intersected with the antibiotic strip. When small colonies grew within the area of inhibition or a haze of growth occurred around the MIC endpoint, the highest MIC intersection was noted. For MIC determination including serum, CA-MH medium was supplemented with 20% heat-inactivated NHS.
Interpretation of MIC data.
We used the EUCAST breakpoint values for polymyxins as a reference (26). Enterobacterial isolates with colistin MICs of ≤2 mg/liter were categorized as susceptible, and those with MICs of >2 mg/liter were categorized as resistant.
Precision and reproducibility testing of CE-MH broth.
Three isolates (E. coli 010, E. coli CLO28, and mcr-1-expressing E. coli 051) were chosen at random and tested according to the requirements described in the CLSI document for development of in vitro susceptibility testing criteria and quality control (QC) parameters (27). Three replicates using individual inoculum preparations of the appropriate strains were tested for 5 consecutive test days, using the 15-replicate (3 by 5 days) plan. These data were used to assess the interassay and intra-assay precision and reproducibility of the MICs obtained in CE-MH medium with the BMD and agar-based methods. On each testing day, the CLSI-recommended E. coli ATCC 25922 and E. coli NCTC 13846 QC strains were included. All of the MICs determined were within the acceptable QC range.
Deletion of the mcr-1 gene from p002.
The mcr-1 deletion mutant of p002 (mcr-1-containing IncX4 plasmid of E. coli 002) (22) (Fig. SA2), p002::Δmcr-1, was generated using the primers listed in Table SA5, as described previously (28). Plasmids were maintained by the addition of 2 mg/liter colistin sulfate salt (Sigma-Aldrich) or 30 mg/liter kanamycin (Sigma-Aldrich) to the broth. The complete sequence of p002 was determined using long-read single-molecule real-time (SMRT) sequencing (Pacific Biosciences, Menlo Park, CA, USA) supplemented with short-read sequencing using the Illumina platform, as described previously (29). For sequencing of p002::Δmcr-1, plasmid DNA was isolated using the Qiagen plasmid maxi kit (Qiagen, Hilden, Germany), following the manufacturer's instructions. A Nextera XT library of the plasmid (Illumina) was sequenced on the NextSeq 500 platform (using 2 by 150 cycles). Raw data were assembled using SPAdes (25), and comparison of p002 with p002::Δmcr-1 was performed using BLASTn.
Statistical analysis.
In our study, CA-MH broth was the reference medium. Categorical agreement and essential agreement (EA) between CA-MH and CE-MH media for both BMD and strip-based methods were calculated according to ISO guidelines (30) and by using kappa scores (31). Categorical agreement is defined as the MIC from the test that is in the same susceptibility category as the currently used reference method. EA is defined as the MIC obtained for the test that is within ±1 twofold dilution of the currently used reference method. Statistical analysis was performed using the MedCalc software package (MedCalc, Mariakerke, Belgium) and Excel (Microsoft, Redmond, CA, USA).
Accession number(s).
The plasmid sequences of p002 and p002::Δmcr-1 were deposited in the GenBank database under accession numbers MF381176 and MF381175, respectively.
RESULTS
A preliminary analysis of the colistin MIC values revealed that mcr-1-containing Enterobacteriaceae isolates exhibited increased colistin resistance when grown in the presence of heat-inactivated NHS (see Table SA1 in the supplemental material). Because serum contains high levels of calcium ions (32), we assessed whether Ca2+ added to growth media could be used as an enhancing agent for the detection of mcr-1-containing Enterobacteriaceae. We determined that elevated concentrations of Ca2+ in the growth medium are optimal for the unambiguous detection of mcr-1-positive Gram-negative bacteria.
We devised a novel formulation, called calcium-enhanced Mueller-Hinton (CE-MH) broth, which contains 200 mg/liter Ca2+ ions (5 mM), and we compared it to the reference medium (CA-MH), employing microdilution and strip-based methods. The concentration of calcium used in CE-MH broth had no adverse effects on bacterial growth (Fig. SA1).
We examined 50 mcr-1-positive isolates by broth microdilution and observed a pronounced difference in MIC values between CA-MH broth and the CE-MH broth devised here. The colistin MICs ranged from 4 mg/liter to 8 mg/liter in CA-MH broth and increased to 16 to 32 mg/liter in CE-MH broth (Table 1). Polymyxin-sensitive isolates lacking mcr-1 remained susceptible to colistin when examined in CE-MH medium. The exceptions were 3 A. baumannii isolates that were classified as susceptible by the reference method and as resistant by BMD in CE-MH broth (Table 1); this was not observed when the agar diffusion format was used.
Whole-genome sequencing performed for the A. baumannii isolates revealed mutations in the pmrC gene in the isolates (pmrC I228V and R348K for A. baumannii 07, V231A and A401V for A. baumannii 08, and H36Q, A55V, and A401G for A. baumannii 010), compared with the colistin-susceptible strain A. baumannii ATCC 17978. This suggests improved detection of colistin-resistant A. baumannii with BMD performed in CE-MH medium.
We next examined the usefulness of the medium in an agar-based diffusion format (strip test). A significant increase in colistin MIC values was also noted with CE-MH medium, compared to the reference CA-MH medium, when strips were used on CA-MH agar plates (Fig. 1 and Table 1). Isolates that were intrinsically resistant to colistin, such as M. morganii, Proteus sp., P. rettgeri, and S. marcescens isolates, and isolates that had acquired resistance due to mutations in the mgrB gene (K. pneumoniae) or the pmrB gene (E. coli) also exhibited increased MIC values with CE-MH medium (Table 1). Thus, CE-MH medium clearly detects all classes of colistin-resistant bacteria, particularly mcr-1-containing isolates, regardless of the assay format (liquid or solid) used. However, isolates of A. baumannii remain a challenge when CE-MH medium is used in an agar diffusion format.
FIG 1.
Effects of an elevated calcium concentration on the colistin MICs of mcr-1-containing isolates, as determined in cation-adjusted Mueller-Hinton (CA-MH) broth and calcium-enhanced Mueller-Hinton (CE-MH) broth by the agar diffusion method. (A) Colistin MICs of representative colistin-susceptible and mcr-1-positive isolates. (B) Susceptibility testing of 50 isolates containing the mcr-1 gene. A significant upward shift in colistin MIC values was observed in CE-MH medium, compared to the reference CA-MH medium.
To directly compare the resistance levels of isolates harboring mcr-1 on IncX4 or IncHI2 plasmids, we used transconjugants of E. coli J53 harboring either plasmid. As before, we observed increased colistin MIC values for transconjugants grown in CE-MH medium, compared to CA-MH medium, as determined by both agar diffusion and microdilution methods. The colistin MIC values were independent of the incompatibility group of the plasmid used (Table 1).
To confirm the contribution of the mcr-1 gene to calcium-induced resistance to colistin, the mcr-1 gene was deleted from the IncX4 plasmid p002 to create the isogenic plasmid variant p002::Δmcr-1. The colistin MICs of E. coli DH10β containing the wild-type p002 or p002::Δmcr-1 were determined in CA-MH medium to be 4 mg/liter and 2 mg/liter, respectively. When tested in CE-MH medium, the MIC of the strain with p002 increased to 32 mg/liter, while that of the strain with p002::Δmcr-1 remained 2 mg/liter, indicating an essential role of the mcr-1 gene in calcium-induced colistin resistance (Table 2).
TABLE 2.
Colistin susceptibility testing results for E. coli DH10β harboring p002 or p002::Δmcr-1, as determined by broth microdilution in CA-MH broth and CE-MH broth
| Strain | MIC (mg/liter)a |
|
|---|---|---|
| CA-MH | CE-MH | |
| E. coli DH10β | 2 | 2 |
| p002 in DH10β | 4 | 32 |
| p002::Δmcr-1 in DH10β | 2 | 2 |
The MIC is defined as the lowest concentration of colistin that inhibits visible growth of the tested isolate, as observed with the unaided eye.
The comparison of the MIC values obtained in CA-MH and CE-MH media demonstrated almost identical essential and categorical agreements for colistin-susceptible isolates, ranging from slight to almost perfect agreement for colistin-resistant isolates using both BMD and agar-based methods (Table SA2). For colistin-susceptible isolates, the essential agreement (EA) values calculated for the BMD and strip-based methods were 91% (29/32 isolates [95% confidence interval [CI], 83 to 96%]) and 100% (32/32 isolates [95% CI, 96 to 100%]), respectively. For colistin-resistant isolates, the EA values for the BMD and strip-based methods were 24% (16/68 isolates [95% CI, 12 to 41%]) and 13% (9/68 isolates [95% CI, 5 to 34%]), respectively. Essential agreement values obtained for colistin-resistant isolates were significantly lower than the EA values for colistin-sensitive isolates as colistin MICs in CE-MH medium. The categorical agreement values (test results with correct susceptibility categorization) for colistin-resistant and colistin-susceptible isolates varied from 91% to 99% for the BMD method and from 87% to 100% for the gradient test method.
Precision and reproducibility results were within the acceptable ranges for 2 colistin-susceptible isolates and 1 colistin-resistant isolate of E. coli, as observed with BMD and agar-diffusion-based methods in CE-MH medium (Table SA3). For the QC strains, i.e., colistin-susceptible E. coli ATCC 25922 and colistin-resistant, mcr-1-positive E. coli NCTC 13846, the MICs determined with both BMD and gradient tests were within acceptable ranges, ranging from 0.5 to 1.5 mg/liter and from 6 to 32 mg/liter, respectively.
DISCUSSION
The simple and accurate phenotypic detection of colistin resistance mediated by the horizontally transferable mcr-1 gene in Enterobacteriaceae remains a challenge for routine microbiology laboratories determining antibiotic susceptibilities. The mcr-1 gene is highly conserved and is capable of conferring resistance to colistin but clearly only under defined conditions. Thus, even though the presence of the mcr-1 gene results in increased resistance to polymyxin antibiotics, many isolates exhibit colistin MICs of 2 mg/liter, just below the EUCAST clinical breakpoint, and therefore are categorized as sensitive despite harboring the plasmid-borne colistin resistance gene (15). This prompted a joint CLSI-EUCAST subcommittee to issue warnings related to the overall poor quality of colistin susceptibility testing (16). Thus, approaches to simplify and to improve MIC determinations for cationic antimicrobial peptides, e.g., colistin, are highly desirable.
Our studies were prompted by the observation that the presence of heat-inactivated serum increased the colistin MIC values of isolates harboring mcr-1. Because the concentration of Ca2+ ions in serum is approximately 2.5 mM (32), we used increasing concentrations of these ions to titrate and to devise the medium presented in this study (CE-MH medium). All of the mcr-1-containing isolates grown in CE-MH medium exhibited increased MIC values, compared to growth in the reference CA-MH medium. There was a clear separation of MIC values between resistant and susceptible bacteria when CE-MH medium was used.
The CE-MH broth used here has a very important property, i.e., it increases colistin MICs only in isolates that are resistant to polymyxins. Colistin-susceptible isolates that lack mcr-1 do not display increased MICs when grown in this medium, and the concentration of calcium used has no adverse effects on bacterial growth. The use of a strain harboring an IncX4 plasmid lacking the mcr-1 gene confirms the contribution of the gene to calcium-dependent resistance to colistin. The increase in the MIC values for isolates exhibiting intrinsic or acquired resistance to colistin suggests that the elevated level of Ca2+ has a general effect, probably due to positively charged modified lipopolysaccharides. The mechanisms underlying the calcium effect remain to be understood but, inasmuch as there is no increased resistance of colistin-sensitive isolates, Ca2+ does not antagonize the bactericidal effect of colistin by preventing binding of the peptide to the bacterial cell wall.
False-susceptible and resistant isolates constitute a serious problem for current colistin susceptible testing and consequently for adequate therapy (33). This study included a set of A. baumannii isolates that were classified as resistant with our test method (BMD in CE-MH medium) and as susceptible with the reference method (BMD in CA-MH medium) (Table 1). Genome analysis of these isolates demonstrated specific alterations in the sequence of the pmrCAB operon, compared to the colistin-sensitive A. baumannii ATCC 17978 strain, providing evidence for the efficacy of the CE-MH medium, compared to the reference broth. Further studies with a large and diverse panel of A. baumannii isolates will be required to validate these findings.
The essential and categorical agreements between MIC values obtained in CA-MH and CE-MH media with BMD and strip-based methods were highly congruent and ranged from 91% to 100% for colistin-susceptible isolates, indicating that, as an alternative to CA-MH broth, CE-MH medium could also be used for colistin susceptibility testing for Enterobacteriaceae. The novelty of this study is that CE-MH medium allows resistance determinations using both solid and liquid formats, without a loss of specificity. This could simplify MIC determinations, for instance by the use of strip tests, a methodology that is widely used in many laboratories and could be again implemented as a suitable method for colistin resistance testing, due to its improved performance with CE-MH agar plates versus CA-MH agar plates (Table 1).
One limitation of this medium is that it does not differentiate between MCR-1 producers and colistin-resistant isolates displaying other colistin resistance mechanisms, i.e., isolates with intrinsic or adaptive colistin resistance, and additional testing for the presence of mcr-1 by other methods is required. However, this limitation could be easily overcome by supplementing our CE-MH medium with a zinc-chelating agent, such as EDTA (34). A further limitation is that, unlike the rapid polymyxin NP test, which can determine colistin susceptibility in 2 h, the test formats described here require overnight incubation. The real advantage of our formulation is that it enables assays to be performed in extremely simple agar-diffusion-based formats, such as by employing Etests.
The Ca2+-enhanced medium described here will make surveillance studies easier and lead to the development of simpler assays for tracking of mcr-1-containing Enterobacteriaceae. Routine colistin resistance testing will improve our understanding of the true prevalence of mcr-1 and help us to devise guidelines and studies to limit its spread. Ultimately, standardized studies, ideally performed under the supervision of internationally authorized committees on antibiotic susceptibility testing, will be required to validate our assay formats, particularly with respect to the levels of skills required to implement the assay and its value in clinical settings and for patient management.
Supplementary Material
ACKNOWLEDGMENTS
We thank Christina Gerstmann and Natalia Lest (Institute of Medical Microbiology, Giessen, Germany) for excellent technical assistance.
This study was supported by grants to the German Center of Infection Research (DZIF), through the German Federal Ministry of Education and Research (BMBF) (grant 8000 701-3 [HZI] to T.C. and C.I. and grants TI06.001 and 8032808811 to T.C.). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
A European patent form, corresponding to this work, has been filed on behalf of the Justus Liebig University (Giessen, Germany).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01950-17.
REFERENCES
- 1.Conly JM, Johnston BL. 2006. Colistin: the phoenix arises. Can J Infect Dis Med Microbiol 17:267–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falagas ME, Kasiakou SK. 2005. Colistin: the revival of polymyxins for the management of multidrug-resistant Gram-negative bacterial infections. Clin Infect Dis 40:1333–1341. doi: 10.1086/429323. [DOI] [PubMed] [Google Scholar]
- 3.Falagas ME, Kasiakou SK, Tsiodras S, Michalopoulos A. 2006. The use of intravenous and aerosolized polymyxins for the treatment of infections in critically ill patients: a review of the recent literature. Clin Med Res 4:138–146. doi: 10.3121/cmr.4.2.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yahav D, Farbman L, Leibovici L, Paul M. 2012. Colistin: new lessons on an old antibiotic. Clin Microbiol Infect 18:18–29. doi: 10.1111/j.1469-0691.2011.03734.x. [DOI] [PubMed] [Google Scholar]
- 5.Li J, Nation RL, Milne RW, Turnidge JD, Coulthard K. 2005. Evaluation of colistin as an agent against multi-resistant Gram-negative bacteria. Int J Antimicrob Agents 25:11–25. doi: 10.1016/j.ijantimicag.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Loho T, Dharmayanti A. 2015. Colistin: an antibiotic and its role in multiresistant Gram-negative infections. Acta Med Indones 47:157–168. [PubMed] [Google Scholar]
- 7.European Medicines Agency. 2013. Use of colistin products in animals within the European Union: development of resistance and possible impact on human and animal health. EMA/755938/2012. European Medicines Agency, London, UK: http://www.ema.europa.eu/docs/en_GB/document_library/Report/2013/07/WC500146813.pdf. [Google Scholar]
- 8.Schwarz S, Johnson AP. 2016. Transferable resistance to colistin: a new but old threat. J Antimicrob Chemother 71:2066–2070. doi: 10.1093/jac/dkw274. [DOI] [PubMed] [Google Scholar]
- 9.Tängdén T, Giske CG. 2015. Global dissemination of extensively drug-resistant carbapenemase-producing Enterobacteriaceae: clinical perspectives on detection, treatment and infection control. J Intern Med 277:501–512. doi: 10.1111/joim.12342. [DOI] [PubMed] [Google Scholar]
- 10.Karaiskos I, Giamarellou H. 2014. Multidrug-resistant and extensively drug-resistant Gram-negative pathogens: current and emerging therapeutic approaches. Expert Opin Pharmacother 15:1351–1370. doi: 10.1517/14656566.2014.914172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen HD, Groisman EA. 2013. The biology of the PmrA/PmrB two-component system: the major regulator of lipopolysaccharide modifications. Annu Rev Microbiol 67:83–112. doi: 10.1146/annurev-micro-092412-155751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller AK, Brannon MK, Stevens L, Johansen HK, Selgrade SE, Miller SI, Høiby N, Moskowitz SM. 2011. PhoQ mutations promote lipid A modification and polymyxin resistance of Pseudomonas aeruginosa found in colistin-treated cystic fibrosis patients. Antimicrob Agents Chemother 55:5761–5769. doi: 10.1128/AAC.05391-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olaitan AO, Morand S, Rolain JM. 2014. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 15.Chew KL, La M-V, Lin RTP, Teo JWP. 2017. Colistin and polymyxin B susceptibility testing for carbapenem-resistant and mcr-positive Enterobacteriaceae: comparison of Sensititre, MicroScan, Vitek 2, and Etest with broth microdilution. J Clin Microbiol 55:2609–2616. doi: 10.1128/JCM.00268-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.European Committee on Antimicrobial Susceptibility Testing. 2016. Recommendations for MIC determination of colistin (polymyxin E), as recommended by the joint CLSI-EUCAST Polymyxin Breakpoints Working Group. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/General_documents/Recommendations_for_MIC_determination_of_colistin_March_2016.pdf.
- 17.Giske CG, Kahlmeter G. 2018. Colistin antimicrobial susceptibility testing: can the slow and challenging be replaced by the rapid and convenient? Clin Microbiol Infect 24:93–94. doi: 10.1016/j.cmi.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 18.Jayol A, Nordmann P, Lehours P, Poirel L, Dubois V. 2018. Comparison of methods for detection of plasmid-mediated and chromosomally encoded colistin resistance in Enterobacteriaceae. Clin Microbiol Infect 24:175–179. doi: 10.1016/j.cmi.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 19.Simar S, Sibley D, Ashcraft D, Pankey G. 2017. Evaluation of the rapid polymyxin NP test for polymyxin B resistance. J Clin Microbiol 55:3016–3020. doi: 10.1128/JCM.00934-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nordmann P, Jayol A, Poirel L. 2016. Rapid detection of polymyxin resistance in Enterobacteriaceae. Emerg Infect Dis 22:1038–1043. doi: 10.3201/eid2206.151840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imirzalioglu C, Falgenhauer L, Schmiedel J, Waezsada S, Gwozdzinski K, Roschanski N, Roesler U, Kreienbrock L, Schiffmann AP, Irrgang A, Käsbohrer A, Bauerfeind R, Domann E, Chakraborty T. 2017. Evaluation of a loop-mediated isothermal amplification-based assay for the rapid detection of plasmid-encoded colistin resistance gene mcr-1 in Enterobacteriaceae isolates. Antimicrob Agents Chemother 61:e02326-16. doi: 10.1128/AAC.02326-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falgenhauer L, Waezsada SE, Yao Y, Imirzalioglu C, Käsbohrer A, Roesler U, Michael GB, Schwarz S, Werner G, Kreienbrock L, Chakraborty T. 2016. Colistin resistance gene mcr-1 in extended-spectrum β-lactamase-producing and carbapenemase-producing Gram-negative bacteria in Germany. Lancet Infect Dis 16:282–283. doi: 10.1016/S1473-3099(16)00009-8. [DOI] [PubMed] [Google Scholar]
- 23.Phan MD, Nhu NTK, Achard MES, Forde BM, Hong KW, Chong TM, Yin WF, Chan KG, West NP, Walker MJ, Paterson DL, Beatson SA, Schembri MA. 2017. Modifications in the pmrB gene are the primary mechanism for the development of chromosomally encoded resistance to polymyxins in uropathogenic Escherichia coli. J Antimicrob Chemother 72:2729–2736. doi: 10.1093/jac/dkx204. [DOI] [PubMed] [Google Scholar]
- 24.Nyerges Á Csörgő B, Nagy I, Bálint B, Bihari P, Lázár V, Apjok G, Umenhoffer K, Bogos B, Pósfai G, Pál C. 2016. A highly precise and portable genome engineering method allows comparison of mutational effects across bacterial species. Proc Natl Acad Sci U S A 113:2502–2507. doi: 10.1073/pnas.1520040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.European Committee on Antimicrobial Susceptibility Testing. 2015. Breakpoint tables for interpretation of MICs and zone diameters. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_5.0_Breakpoint_Table_01.pdf.
- 27.Clinical and Laboratory Standards Institute. 2018. Development of in vitro susceptibility testing criteria and quality control parameters—5th ed. M23. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 28.Kostylev M, Otwell AE, Richardson RE, Suzuki Y. 2015. Cloning should be simple: Escherichia coli DH5α-mediated assembly of multiple DNA fragments with short end homologies. PLoS One 10:e0137466. doi: 10.1371/journal.pone.0137466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Falgenhauer L, Ghosh H, Doijad S, Yao Y, Bunk B, Spröer C, Kaase M, Hilker R, Overmann J, Imirzalioglu C, Chakraborty T. 2017. Genome analysis of the carbapenem- and colistin-resistant Escherichia coli isolate NRZ14408 reveals horizontal gene transfer pathways towards panresistance and enhanced virulence. Antimicrob Agents Chemother 61:e02359-16. doi: 10.1128/AAC.02359-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.International Organization for Standardization. 2007. Clinical laboratory testing and in vitro diagnostic test systems—susceptibility testing of infectious agents and evaluation of performance of antimicrobial susceptibility test devices, part 2: evaluation of antimicrobial susceptibility test devices. ISO 20776-2:2007 International Organization for Standardization, Geneva, Switzerland: https://www.iso.org/standard/41631.html. [Google Scholar]
- 31.Tang W, Hu J, Zhang H, Wu P, He H. 2015. Kappa coefficient: a popular measure of rater agreement. Shanghai Arch Psychiatry 27:62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fijorek K, Püsküllüoğlu M, Tomaszewska D, Tomaszewski R, Glinka A, Polak S. 2014. Serum potassium, sodium and calcium levels in healthy individuals: literature review and data analysis. Folia Med Cracov 54:53–70. [PubMed] [Google Scholar]
- 33.Matuschek E, Åhman J, Webster C, Kahlmeter G. 5 December 2017. Antimicrobial susceptibility testing of colistin: evaluation of seven commercial MIC products against standard broth microdilution for Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter spp. Clin Microbiol Infect doi: 10.1016/j.cmi.2017.11.020. [DOI] [PubMed] [Google Scholar]
- 34.Esposito F, Fernandes MR, Lopes R, Muñoz M, Sabino CP, Cunha MP, Silva KC, Cayô R, Martins WM, Moreno AM, Knöbl T, Gales AC, Lincopan N. 2017. Detection of colistin-resistant MCR-1-positive Escherichia coli using inhibition by EDTA and zeta potential assays. J Clin Microbiol 55:3454–3465. doi: 10.1128/JCM.00835-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.