Abstract
Educated natural killer (NK) cells possess inhibitory receptors specific for self major histocompatibility (MHC) class I molecules and kill cancer cells more efficiently than NK cells not possessing such receptors (from hereon referred to as hyporesponsive NK cells). The mechanism behind this functional empowerment through education has so far not been fully described. In addition, distinctive phenotypical markers of educated NK cells at the single cell level are lacking. We developed a refined version of the image mean square displacement (iMSD) method, called iMSD carpet analysis, and used it in combination with single-particle tracking to characterize the dynamics of the activating receptor NKp46 and the inhibitory receptor Ly49A on resting educated versus hyporesponsive murine NK cells. The majority of both NKp46 and Ly49A molecules were restricted to microdomains; however, individual NKp46 molecules resided in these domains for shorter periods and diffused faster on the surface of educated, compared to hyporesponsive, NK cells. In contrast, the movement of Ly49A was more constrained in educated NK cells compared to hyporesponsive NK cells. Either disrupting the actin cytoskeleton or adding cholesterol to the cells prohibited activating signaling, indicating that the dynamics of receptor movements within the cell membrane are critical for proper activation of NK cells. The faster and more dynamic movement of NKp46 in educated NK cells may facilitate a swifter response in interactions with target cells.
INTRODUCTION
Natural Killer (NK) cells are cytotoxic lymphocytes of the innate immune system that are essential for the control of infections and for tumor immunosurveillance (1, 2). NK cells are increasingly used in cancer therapy trials. Their capacity to recognize “missing-self”, the absence of endogenous major histocompatibility (MHC) class I allelic transcripts, is often exploited in therapeutic settings (3). By missing-self, NK cells can kill cells that lack MHC class I ligands for their inhibitory receptors (4). The murine Ly49 family of receptors, for which MHC class I molecules act as ligands, represents the most thoroughly studied inhibitory receptor family in mice. These receptors are the functional equivalent of human killer immunoglobulin receptors (KIRs). Stochastic expression of the genes encoding these receptors produces subsets of NK cells displaying different combinations of Ly49 family members. Thus individual NK cells have different MHC class I allelic specificity for which they perform “missing-self” killing. Not all NK cells possess an inhibitory receptor that recognizes a self MHC class I allele. NK cells undergo an educational process to ensure that only NK cells with self-specific inhibitory receptors are able to perform missing-self killing. NK cells without self-specific inhibitory receptors are hyporesponsive in the resting state (5, 6).
The mechanism behind the functional dichotomy between educated and hyporesponsive NK cells is not well understood. However, educated NK cells migrate in a more dynamic fashion (7) and form more stable conjugates with target cells compared to hyporesponsive cells (8). Only a few differences between these two cell types at the transcript and protein levels have been described. One difference is a higher frequency of cells expressing DNAX accessory molecule-1 (DNAM-1) in the educated human NK cell subset (9, 10). No cell surface proteins have however been described as being present solely on educated NK cells. Rather there is a difference in the frequency of DNAM-1 positive cells in the respective subpopulations of NK cells. Thus, no marker of education has so far been described at the single cell level.
The importance of the organization and degree of movement of receptors and signaling mediators within the cell membrane for immune cell regulation has gained intensified attention in recent years. Both the actin cytoskeleton and the lipid composition of the membrane itself can affect the diffusion of receptors within the membrane and lead to a heterogeneous localization pattern (11, 12). The limitation of free diffusion by the cytoskeleton is a major regulator of B and Natural Killer T cell responses (13–15). Membrane lipid composition can affect both receptor localization and conformation. Both of these mechanisms can regulate receptor signaling. Furthermore, manipulation of the membrane lipid composition strongly affects T cell responses (11). Less is known about how receptor dynamics regulate NK cell responses. Most studies have indicated that inhibitory NK cell receptors diffuse freely and accumulate spontaneously at the immune synapse, whereas some reported that the cytoskeleton limits diffusion (16–18). One study indicated that activating receptors are confined to membrane nanodomains in educated NK cells, whereas their diffusion is limited by the cytoskeleton in hyporesponsive NK cells (19). The regulation of missing-self killing may thus lie in receptor organization, rather than the presence or absence of a particular receptor.
Many different modes of molecular movements occur at the cell surface over a large range of time scales. Apart from free diffusion, movement can be constrained by crowding, compartmentalization by the cytoskeleton, lipid micro- or nanodomains (sometimes referred to as lipid rafts), or protein-protein interactions (20). Because all methods currently used for tracking the movements of proteins within membranes have limitations in the range of diffusion rates that can be accurately assessed, it is challenging to select a proper method to quantify the entire range of protein dynamics on cell membranes. Here, we used total internal reflection fluorescence (TIRF) microscopy to assess the dynamics of the activating receptor NKp46 and the inhibitory receptor Ly49A in NK cells. With a camera as the detector, large amounts of data can be collected in a short amount of time, allowing the assessment of more complex types of movement than merely free diffusion. We employed two different analytical methods to cover the full range of different possible types of movements of the studied receptors. Single-particle tracking (SPT) can reveal the behavior of single molecules and detect membrane compartmentalization, but does not detect fast diffusion because pinpointing precise localizations requires time for photons to accumulate (21, 22). In contrast, spatiotemporal image correlation spectroscopy (STICS) with image mean square displacement (iMSD) distinguishes and characterizes a broad range of dynamic processes, including fast motion (23, 24).
In this study, we utilized NK cell education as a model system to study differences in receptor dynamics between active and hyporesponsive immune cells. We used SPT and developed and utilized a novel STICS-iMSD analysis approach for rapid evaluation of dynamic molecular movement at the cell surface, which we termed iMSD carpet analysis. With these approaches, we investigated how the spatiotemporal dynamics of activating NKp46 and inhibitory Ly49A receptors were related to the educational status of the NK cell. Our results revealed that NKp46 exhibited a more dynamic movement pattern in educated NK cells compared to hyporesponsive cells. In contrast, Ly49A was more confined and diffused more slowly in educated NK cells than in hyporesponsive cells. Either disrupting the cytoskeleton or increasing the amount of cholesterol in the membrane prohibited activating signaling initiation in educated NK cells. These findings indicate that receptor dynamics within the membrane is tightly linked to NK cell function and plays a role in NK cell regulation.
RESULTS
NKp46 and Ly49A are transiently confined to microdomains on primary resting NK cells
Fluorescence Correlation Spectroscopy (FCS) has previously been used to characterize the diffusion of receptors at the NK cell surface (18, 19). However, this technique is limited in its ability to detect different types of movement, since it only reports the mean diffusion time of detected molecules through the focal volume. Quantitative temporal analysis of image stacks, on the other hand, provides a means to cover a larger range of the different types of dynamic movement which may be present at the cell surface. Compared to other imaging methods, TIRF microscopy yields a superior signal-to-noise ratio in combination with high temporal resolution. We thus decided to employ TIRF imaging to study the dynamics of activating and inhibitory NK cell receptors labeled with fluorescent antibodies. To determine whether the dynamic behavior of membrane proteins was affected by being bound by an antibody, we tracked the movement of fluorescently tagged proteins bound by antibodies. The diffusion of human leukocyte antigen (HLA) coupled to yellow fluorescent protein (YFP) was not affected by the addition of a specific antibody recognizing HLA in a cell line, as measured by Fluorescence Correlation Spectroscopy (FCS) (fig. S1A and S1B). HLA-YFP also diffused at a similar rate on both the top and the bottom of the cell (fig. S1A and S1B). Furthermore, in stably transfected CHO cell lines, spontaneous clustering, observed as brighter spots of green fluorescent protein (GFP)–tagged Ly49A (Ly49A-GFP), occurred both on unstained cells (Fig. 1A and 1B) and on cells co-stained with an antibody recognizing Ly49A (Fig. 1C and 1D). There was no difference in the size of Ly49A clusters detected by GFP labeling and those detected with antibodies (fig. S1E).
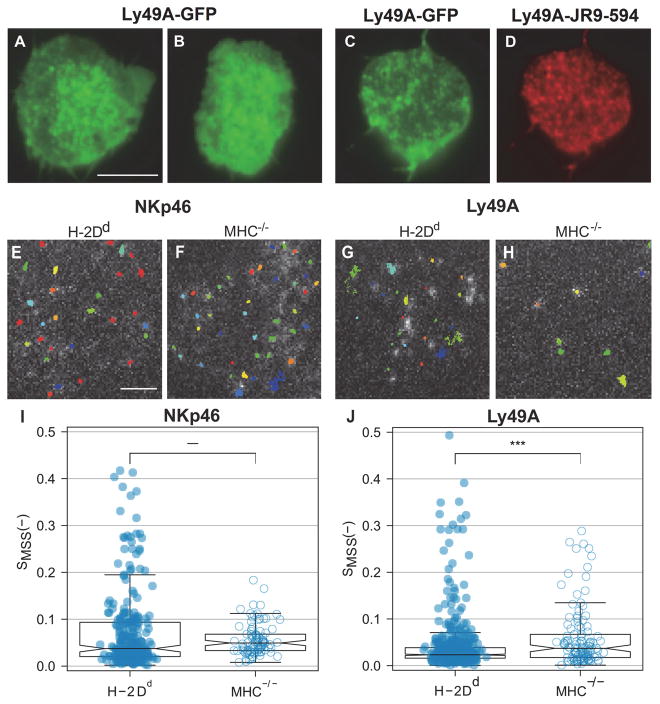
Fig. 1. Dynamic confinement of NKp46 and Ly49A at the NK cell surface.
(A–D) Representative fluorescence images showing transfected Ly49A-GFP in CHO cells. The images were merged from 6000 frames acquired at a 36 ms frame-to-frame rate. (A–B) Transfected Ly49A-GFP without antibody labeling (C) Transfected Ly49A-GFP incubated with an antibody that recognizes Ly49A (clone JR9.318 conjugated to abberior star 594). (D) The JR9.318-abberior star 594 labeling in the same cell as in panel C. (E–J) Single particle tracking (SPT) in freshly isolated H-2Dd compared to MHC−/− murine Ly49A+ NK cells captured at a 20 ms frame-to-frame rate. (E–H) Representative single particle tracks of NKp46 (E, F), and Ly49A (G, H) in H-2Dd and MHC−/− NK cells. (I, J) The degree of anomalous diffusion (Hurst parameter) for NKp46, median H-2Dd = 0.037 and MHC−/− = 0.049 (I), and Ly49A, median H-2Dd = 0.023 and MHC−/− = 0.037 (J), was calculated on 100 frames segments of trajectories. All the values calculated for a single trajectory were averaged. Each dot represents a single trajectory, H-2Dd NKp46 N=264, MHC−/− NKp46 = 94, H-2Dd Ly49A = 521, and MHC−/− Ly49A = 116. Data was pooled from 7 – 27 cells per group, from three to six independent experiments. The boxes represent medians (line inside box), the 95 % confidence intervals (notches), first and third quartiles (end of boxes), and the whiskers 1.5 times the interquartile ranges. P-values: *** ≤ 0.001 (two-sample Wilcoxon ranking test). Scale bars, 5 μm (A–D) and 2 μm (E–H).
TIRF image time series of the activating receptor NKp46 and the inhibitory receptor Ly49A were thereafter recorded on freshly isolated murine NK cells. NK cells were isolated from mice expressing the H-2Dd allele of MHC class I, the educating ligand for Ly49A (25), and from MHC−/− mice, which lack educating MHC class I ligands. All NK cells from MHC−/− mice are thus hyporesponsive. Accumulation of both NKp46 and Ly49A in certain regions was visible as distinct high-intensity spots in the compiled image stacks of Ly49A+ NK cells from MHC−/− mice, indicating a high degree of confinement of receptors or receptor clusters. Fluorescence intensity time traces at selected spots, however, showed a dynamic behavior, wherein receptors or receptor clusters disappeared and reappeared in certain spots over time (fig. S2A and S2B). The same pattern was observed in Ly49A+ NK cells from H-2Dd mice (fig. S2C and S2D).
The distinctly isolated fluorescent spots made it possible to analyze cells using SPT. We identified and tracked single NKp46 molecules (Fig. 1E and 1F) and single Ly49A molecules (Fig. 1G, 1H). We first addressed the question of the extent to which the particles were freely diffusing. The Hurst parameter, SMSS, quantifies the degree of anomalous diffusion, which is the deviation from Brownian (random) motion. An SMSS of 0.5 represents free diffusion, < 0.5 indicates confined movement, and > 0.5 indicates active transport. The calculation of the Hurst parameter necessitates trajectories longer than 100 movie frames, which may hide short-term confinements and changes in diffusion. However, it does not require a threshold or choice of initial parameters for the movement (26). In agreement with the visually observed clustering pattern, the majority of both the NKp46 and Ly49A molecules were generally restricted in their movement. Most tracks displayed an SMSS < 0.1, regardless of the educational status of the NK cell (Fig. 1I and 1J). There were no statistically significant differences in the degree of restriction to the movement of NKp46 between the two NK cell types (Fig. 1I). Ly49A molecules, however, had on average a lower SMSS in the educated H-2Dd NK cells, compared to the hyporesponsive MHC−/− NK cells (Fig. 1J). Notably, a portion of both receptors was considerably less constrained in their movement, especially NKp46 in the educated H-2Dd NK cells (Fig. 1I). This movement pattern was consistent with a transient confinement, wherein most receptors were confined within microdomains, but some were entering and leaving these domains. The transient nature of the confinement was more prominent for the NKp46 receptor in the educated H-2Dd NK cells, compared to the MHC−/− cells.
NKp46 resides in larger microdomains and diffuses faster than Ly49A in educated NK cells
We next studied the nature of the confined movement of the receptors in more detail. SPT defines both the diffusion coefficient at different time scales and the size of the domains limiting the diffusion. The size of the microdomains was similar for both receptors in H-2Dd NK cells (Fig. 2A and 2B). However, the domains containing NKp46 were substantially larger (Fig. 2A), and those containing Ly49A were smaller (Fig. 2B), in educated H-2Dd NK cells compared to their counterparts in hyporesponsive MHC−/− NK cells.
Fig. 2. NKp46 diffuses faster and cover larger areas on educated NK cells, whereas Ly49A diffuses slower and cover smaller areas in educated NK cells.
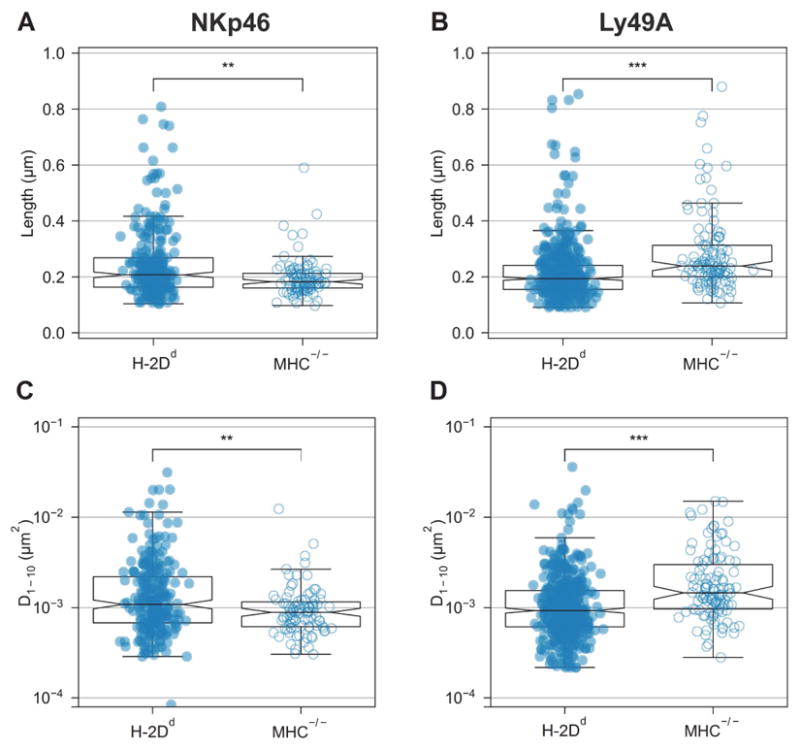
Single particle tracking (SPT) trajectories in freshly isolated H-2Dd compared to MHC−/− murine NK cells captured at 20 ms frame-to-frame rate. Each dot represents a single trajectory. (A–B) NKp46 microdomain median width: H-2Dd = 0.21, MHC−/− = 0.18 μm (A) and Ly49A microdomain median width: H-2Dd = 0.19 and MHC−/− = 0.24 μm (B). (C–D) NKp46 short range diffusion coefficient D1-10 median: H-2Dd = 1.1*10−3 and MHC−/− = 0.89*10−3 μm2/s (C) Ly49A short range diffusion coefficient D1-10 median: H-2Dd = 0.93*10−3 and MHC−/− = 1.5*10−3μm2/s (D). The tracks are the same as those depicted in Fig. 1. The boxes represent medians (line inside box), the 95 % confidence intervals (notches), first and third quartiles (end of boxes), and the whiskers 1.5 times the interquartile ranges. P-values: ** ≤ 0.01, *** ≤ 0.001 (two-sample Wilcoxon ranking test).
The short-range diffusion (D1-10) reported by SPT was, in general, very low, in line with the low values of the Hurst parameter (Fig. 2C, 2D). This very low diffusion rate indicates that the majority of the receptors are immobile or highly constrained in their movement. However, there were large heterogeneities among the tracks of individual molecules (note the log scale in Fig. 2C and 2D). Whereas most particles were nearly immobile, some diffused at a perceptible rate. NKp46 molecules, on average, diffused faster in H-2Dd NK cells compared to MHC−/− NK cells (Fig. 2C). Ly49A instead, on average, diffused faster in MHC−/− NK cells compared to H-2Dd NK cells (Fig. 2D). Because D1-10 indicates diffusion within a very short range, it describes diffusion within, as well as outside of, microdomains. The reported diffusion rate is thus an average of movement inside and outside microdomains, as well as across microdomain borders.
In conclusion, NKp46 on average both resided in larger domains and diffused faster in educated H-2Dd NK cells compared to hyporesponsive MHC−/− NK cells. The heterogeneity between tracks was also larger in educated H-2Dd NK cells, which, taken together with the heterogeneity in the Hurst parameter, suggests that NKp46 molecules are highly constrained and diffuse relatively slowly in hyporesponsive cells, whereas it exhibits a more dynamic pattern of movements upon NK cell education.
iMSD carpet analysis reveals shorter microdomain dwell times for both NKp46 and Ly49A in educated H-2Dd NK cells
SPT analysis detects the detailed movements of individual particles (in this case, receptor molecules or clusters) over time and hence provides a very precise account of movements of individual particles. However, the method is sensitive to labeling densities, sampling frequency, fluorophore blinking, and image noise, and the analysis is time-consuming and data power-intensive. To provide a fast and relatively simple method for detailed analysis of receptor movements that also encompasses a large range of time scales, we developed a new analysis method based on STICS. With STICS, dynamics can be extracted from a series of correlation images by means of iMSD analysis (21). In contrast to SPT, spatial separation of molecules is not necessary, therefore no signal thresholds need to be applied. Image correlation spectroscopy is therefore more resistant to noise than SPT analysis. It is also compatible with higher densities of labeled molecules.
In this study, we applied a global fit to the entire STICS series to increase the number of data points. Since the correlation function was radially symmetric for the data presented here, we simplified the fitting process by combining x and y into a single spatial component, r, by vector addition (Fig. 3A). This approach made it possible to model various modes of particle dynamics over different time scales. A pseudo-image (“the correlation carpet”) with the radial component, δ, as a single spatial component and the lag time, τ, as temporal coordinate, was generated (Fig. 3A, lower panel). We termed this analysis approach “iMSD carpet analysis” (see Materials and Methods for further details). We tested four models of movement: free diffusion, active transport, trapping, and confinement. All of these models, except the active transport model, have been previously described (27). To assess the reliability of the analysis method, we generated several series of simulated data and fitted them to the four models. Firstly, we generated simulated diffusion data with different numbers of molecules and noise variance. In each case, the diffusion model gave the best fit to the simulated data (fig. S3 and table S1). Secondly, we made simulations for each of the four types of movement and fitted these to the models using the iMSD carpet analysis. Two data sets with different signal to noise ratios (noise variance of 1 and 0.01) were simulated. In each case, the correct model gave the best fit (R2 value closest to one) to the simulated data (fig. S4 and table S2).
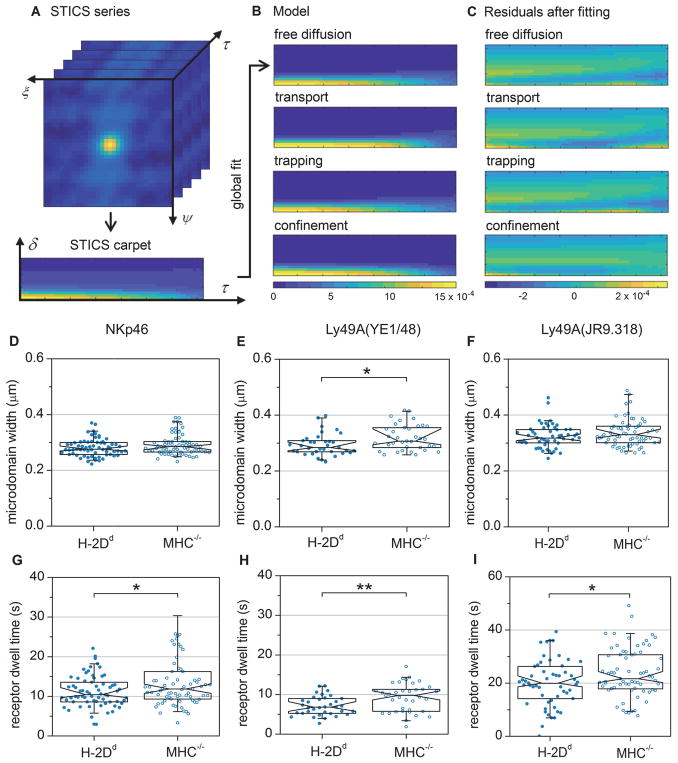
Fig. 3. iMSD carpet analysis reveals a smaller Ly49A domain size and shorter average microdomain dwell times for NKp46 and Ly49A in educated NK cells compared to hyporesponsive NK cells.
(A) The three-dimensional STICS series was calculated from a recorded image time series, followed by conversion into a two-dimensional carpet by combining the spatial lags, ξ and ψ, into a single radial component, δ. (B) The data was fitted with four models representing different types of particle dynamics. Panes show the fitted models of a sample data set. (C) The residuals of the fit of each model for the sample data set shown in (B). The color scale in B and C represents the correlation amplitude. (D–I) TIRF imaging and fitting of the confinement carpet iMSD model to freshly isolated Ly49A+ NK cells. The graphs show analyses of microdomain widths of (D) NKp46, (E) Ly49A (labelled with antibody YE1/48), and (F) Ly49A (labelled with antibody JR9.318). (G–I) Receptor dwell times within individual domains of (G) NKp46, (H) Ly49A (labelled with antibody YE/148), and (I) Ly49A (labelled with antibody JR9.318). Each dot represents one cell, N=67 for NKp46 in H-2Dd cells, 72 for NKp46 in MHC−/− cells, 53 for Ly49A in H-2Dd cells, 68 for Ly49A in MHC−/− cells (JR9.318), 35 for Ly49A in H-2Dd cells and 37 for Ly49A in MHC−/− cells (YE/148). Data were pooled from six (NKp46 antibody and JR9.318) or three (YE1/48) independent experiments. No more than one outlier per sample was excluded prior to analysis, using Grubbs’ test. The boxes represent median, first and third quartiles; the notch represent the 95 % confidence intervals. The whiskers show the 95 % ranges. P-values: * ≤ 0.05, ** ≤ 0.01 (Mann-Whitney’s test). Comparisons that are not marked were not statistically significantly different.
Using the iMSD carpet approach, we next fitted the results from the TIRF image series of freshly isolated Ly49A+ NK cells from H-2Dd and MHC−/− mice. An example of the best fits of the four models for a typical cell (Fig. 3B) and the residuals (Fig. 3C) are shown. Among the four models, the “confinement” model gave the best fit to the data in the majority of cases (table S3). This model represents confinement in isolated domains which receptors can enter and exit. The iMSD carpet analysis thus reported the same general dynamic pattern of receptor movements for the freshly isolated NK cells as the SPT analysis did, namely a movement characterized by transient confinement.
The “confinement” model fits the width of the domains with restricted diffusion, and the mean time receptors or receptor clusters are confined. When we applied the confinement model to the cell line transfected with Ly49A-GFP (Fig. 1A to 1D), antibody staining affected neither the best fit microdomain size (fig. S5A), nor the time the Ly49A-GFP resided in microdomains (fig. S5B). Thus, the labeling with antibodies did not change the detected characteristics of receptor movements in this case.
The microdomain widths resulting from fitting the confinement model to the time series of NKp46 and Ly49A in freshly isolated NK cells (Fig. 3D and 3E) were slightly larger than the microdomain sizes predicted from the SPT analysis (Fig. 2A and 2B). However, the confinement domain width from the iMSD model is not directly comparable with the domain diagonal length obtained by SPT, so this result was not unexpected (see Discussion for details). For NKp46, there was no statistically significant difference in the domain width for H-2Dd NK cells compared to MHC−/− NK cells (Fig. 3D). This outcome was in contrast to SPT, where a smaller domain size was detected in MHC−/− cells (Fig. 2A). This difference could be due to discrepancies in cluster detection between the two methods, most likely due to the fact that the domains constraining NKp46 movement reported by SPT were smaller than the diffraction limit. The average domain width constraining Ly49A movement was, however, smaller in H-2Dd NK cells compared to MHC−/− NK cells (Fig. 3E), in agreement with the SPT analysis (Fig. 2B). When Ly49A was labeled with a different antibody (JR9.318) that fluoresces in the deep red spectrum, there was a trend towards smaller microdomain widths in H-2Dd NK cells, albeit not statistically significant (Fig. 3F). This observation was in agreement with the suggestion that the diffraction limit influences the ability to distinguish individual domains with the iMSD analysis.
The iMSD carpet analysis uniquely determines the microdomain dwell time, i.e., how much time each receptor or receptor cluster spends within each isolated microdomain. In contrast to the microdomain width, this parameter is theoretically not affected by the optical resolution (see Materials and Methods for equations). For both receptors, the microdomain dwell time was shorter in H-2Dd, compared to MHC−/− NK cells (Fig. 3G to 3I). This result does not mean that the receptors spend less total time within microdomains, but rather that once within a microdomain, the receptor exits faster in H-2Dd compared to MHC−/− cells. The shorter microdomain dwell time supports a more agile receptor movement of NKp46 and Ly49A in educated H-2Dd cells, with more frequent exits of receptors from microdomains, in agreement with the larger heterogeneity in the Hurst parameter observed by SPT analysis.
Cytoskeleton destabilization decreases NKp46 confinement and disrupts activating signaling
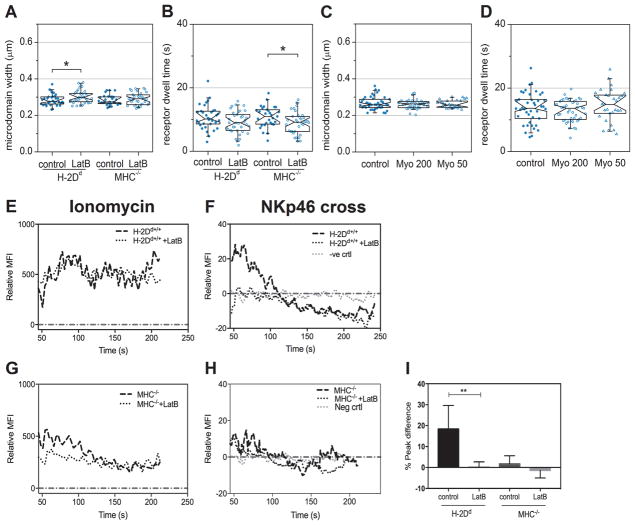
To evaluate whether the cytoskeleton influences receptor dynamics, we measured the movement of NKp46 and Ly49A in NK cells treated with the actin polymerization inhibitor Latrunculin B (LatB). For Ly49A, we observed no differences between treated and untreated NK cells, neither in the microdomain width (fig. S6A), nor for the microdomain dwell time (fig. S6B). However, for NKp46, the microdomain width increased in H-2Dd NK cells and the dwell time decreased in MHC−/− NK cells upon treatment (Fig. 4A and 4B), suggesting that NKp46 diffusion is restricted by the cytoskeleton in resting NK cells. For the hyporesponsive MHC−/− cells, this result is in line with previously published data suggesting that NKp46 diffuses faster following disruption of the actin cytoskeleton (19). Several studies have reported that activating receptors are actively transported into the immune cell synapse (28, 29). However, treatment with Myovin1, a reagent that inhibits active transport by the myosin V motor protein, had no effect on either the microdomain dwell times or the microdomain sizes (Fig. 4C and 4D for NKp46 and fig. S6C and S6D for Ly49A), indicating that receptors are not actively transported in cells without active immune synapses. These findings suggest that the actin cytoskeleton restricts the movement of NKp46, but not that of Ly49A, in resting NK cells.
Fig. 4. Actin depolymerization reduces NKp46 confinement and disrupts activating signaling.
TIRF imaging and fitting of the confinement carpet iMSD model to freshly isolated Ly49A+ NK cells. The graphs show analyses of NKp46 microdomain width (A) and microdomain dwell time (B) in NK cells treated with the actin polymerization inhibitor Latrunculin B (LatB), and NKp46 microdomain width (C) and microdomain dwell time (D) in H-2Dd NK cells treated with the myosin V inhibitor Myovin1 (Myo). MFI= Mean Fluorescence Intensity. Each dot represents one cell. N=34 H-2Dd control, 39 H-2Dd LatB, 32 MHC−/− control 34 MHC−/− LatB (A, B); N=42 for control, 45 for Myo 200, 29 for Myo 50, 42 for MHC−/− control, 46 for Myo 200, 29 for Myo 50 (C, D). Data were pooled from three independent experiments. No more than one outlier per group was detected and removed using Grubbs’ test. The boxes represent medians, first and third quartiles, the notches represent the 95 % confidence intervals. The whiskers show the 95 % range. P-values: * ≤ 0.05 (Mann-Whitney’s test). (E–H) Representative Fluo-4 fluorescence time traces indicating Ca2+ release in H-2Dd and MHC−/− NK cells after treatment with ionomycin stimulation (E, G) or cross-linking of NKp46 (F, H), normalized to median base line by subtraction, in H-2Dd and MHC−/− NK cells as indicated. (I) Peak difference in Ca2+ flux in resting NK cells following NKp46 cross-linking with or without pre-treatment with Latrunculin B in the indicated NK cells. For the negative control, NKp46 was not cross-linked. Bars show the mean with standard deviation. Data were pooled from 8 independent experiments. P-values: * ≤ 0.05 (Mann-Whitney’s test). Statistical tests were performed between control groups and treatment within each mouse from which the cells were harvested (H-2Dd control versus H-2Dd treated and MHC−/− control versus MHC−/− treated); comparisons that are not marked were not statistically significantly different.
Although the actin cytoskeleton is crucial for the so-called NK cell “lytic hit”, the execution of target cell killing (30), whether the cytoskeleton is important for transmission of activating signals from surface receptors into the NK cell has not, to our knowledge, been previously investigated. To specifically test the role of the actin cytoskeleton in proximal activating signal transduction, we analyzed Ca2+ release into the cytoplasm from internal stores following activating receptor cross-linking. Although educated H-2Dd NK cells retained the capacity to release Ca2+ from internal stores upon stimulation with the ionophore ionomycin after disrupting the cytoskeleton with LatB (Fig. 4E), cross-linking NKp46 completely abrogated Ca2+ release (representative Ca2+ release curve in Fig. 4F). This result suggests that the actin cytoskeleton plays a crucial role not only in the execution of killing, but also in the signaling that initiates it. In MHC−/− cells, LatB treatment affected neither the response to ionomycin (Fig. 4G), nor the lack of response to cross-linking NKp46 (Fig. 4H). The decrease in Ca2+ signaling after cross-linking of NKp46 in H-2Dd NK cells was statistically significant (Fig. 4I). Thus, conclusively, a preserved actin cytoskeleton was crucial for signal initiation after NKp46 cross-linking in murine NK cells on this setting.
Cholesterol addition decreases the microdomain dwell time and disrupts activating signaling
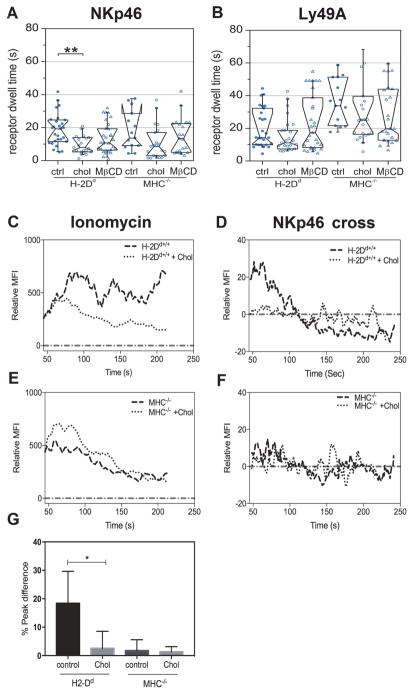
Cholesterol is one of the major constituents of the plasma membrane that also influences signaling in many biological systems. Cholesterol-rich domains are proposed to take part in the formation of signaling platforms necessary for the initiation of activating signals in immune cells (31, 32). To investigate the role of the membrane composition in NK cell receptor dynamics and signaling, we first performed iMSD carpet analysis on TIRF image series of freshly isolated NK cells in which cholesterol was added to or depleted from the cell membrane. A short incubation of the NK cells with a water-soluble form of cholesterol that readily integrates into the membrane considerably decreased the microdomain dwell time of NKp46 in H-2Dd NK cells but had no statistically significant effect on the microdomain dwell time in MHC−/− NK cells (Fig. 5A). Cholesterol addition did not alter the microdomain dwell time of Ly49A (Fig. 5B) or the microdomain sizes for either of the receptors in either cell type (fig. S7A and S7B). Depletion of membrane cholesterol with the cholesterol sequestering compound methyl-β-cyclodextrin (MβCD) had no statistically significant effect on receptor dynamics (Fig. 5A and 5B).
Fig. 5. The role of cholesterol in receptor dynamics and signal initiation.
(A) NKp46 microdomain dwell time and (B) Ly49A microdomain dwell time, as detected by antibodies (clone JR9), after adding cholesterol (chol), depleting cholesterol with methyl-β-cyclodextrin (MβCD), or in untreated control cells (ctrl). MFI= Mean Fluorescence Intensity. Each dot represents one cell. N= 27 H-2Dd ctrl, 21 H-2Dd chol, 29 H-2Dd MβCD, and 14 MHC−/− ctrl, 21 MHC−/− chol, 21 MHC−/− MβCD (A); N=27 H-2Dd ctrl, 24 H-2Dd chol, 28 H-2Dd MβCD, and 14 MHC−/− ctrl, 23 MHC−/− chol, 25 MHC−/− MβCD (B). Data were pooled from three independent experiments. No more than one outlier per group was detected and removed using Grubbs’ test. The boxes represent medians, first and third quartiles, the notches represent the 95 % confidence intervals. The whiskers show the 95 % range. P-values: * ≤ 0.05 (Mann-Whitney’s test). (C–F) representative Fluo-4 fluorescence time traces indicating Ca2+ release in H-2Dd and MHC−/− NK cells after stimulation with ionomycin (C, E) or crosslinking of NKp46 (D, F) normalized to median base line by subtraction, in the absence or presence of added cholesterol (+chol). Statistical tests were performed between control groups and treatment within each mouse (H-2Dd ctrl versus H-2Dd treated and MHC−/− ctrl versus MHC−/− treated); differences not marked in the figure were not statistically significantly different.
Next, we studied activating signal initiation in NK cells after incubation with cholesterol. Although H-2Dd NK cells to which cholesterol had been added were capable of releasing Ca2+ in response to ionomycin treatment (Fig. 5C), Ca2+ release upon cross-linking the receptor for NKp46 was completely abolished in these cells (representative curve in Fig. 5D), suggesting that the lipid composition of the plasma membrane has a strong impact on initiation of activating signaling in NK cells. In MHC−/− cells, cholesterol treatment had no effect on Ca2+ release in response to ionomycin stimulation (Fig. 5E), nor on NKp46 cross-linking (Fig. 5F). The difference in peak Ca2+ release was statistically significant in H-2Dd NK cells, whereas there was no statistically significant change in MHC−/− NK cells (Fig. 5G).
DISCUSSION
Several studies have focused on visualizing immune synapses during interactions of NK cells with target cells (16, 33). However, the functional outcome of such interactions is to a large extent determined by previous experiences of the NK cell. Education is a process which enables NK cells to perform missing-self killing. Education thus ought to leave observable imprints on the NK cells before they encounter their targets. However, there have only been a few reports of such phenotypic differences (7, 19, 34). This lack of observable features is problematic, both from the mechanistic understanding point-of-view, and also for pinpointing which cells would be most clinically useful, for instance in cancer therapy. Here we characterize such imprints of education at the level of spatiotemporal receptor dynamics by using ultrasensitive imaging and a newly developed analytic method, namely, iMSD carpet analysis.
In agreement with previous studies, both murine NKp46 and Ly49A were highly confined in microdomains in resting cells on a non-activating surface. However, the educational status of the NK cell clearly affected the receptor dynamics, which became most evident in a composite picture of how the parameters investigated were affected by education. Our iMSD carpet analysis resolved the dynamic nature of receptor transits into and out of microdomains. Compared to NKp46 in hyporesponsive MHC−/− NK cells, NKp46 in educated H-2Dd NK cells resided in larger microdomains, had a faster overall diffusion rate, and spent less time within each microdomain. Taken together, these results are indications of a more dynamic movement pattern following education, wherein NKp46 switches, from being constrained to moving freely, more frequently. Functionally, this dynamic movement pattern of NKp46 could allow the NK cell to more rapidly scan the membrane of a target cell, form activating receptor-ligand bonds, and initiate a cytotoxic response. Keeping activating receptors in check to prevent inappropriate cytotoxicity may be one of the mechanisms by which a hyporesponsive NK cell remains hyporesponsive. This hypothesis is in agreement with the observation that educated human NK cells, possessing the inhibitory receptor NKG2A, form more and larger conjugates than hyporesponsive NK cells that lack self-specific inhibitory receptors (7).
Ly49A was on the other hand less restricted than NKp46, as indicated by the larger confinement zones, in hyporesponsive NK cells. Because Ly49A binds to MHC class I in cis within the same cell membrane, cis interactions could potentially constrain Ly49A movement in the H-2Dd NK cells (35). Limiting the free movement of inhibitory receptors could thus potentially lower the NK cell activation threshold in addition to limiting the proportion of Ly49A molecules free to interact in trans (36, 37).
The actin cytoskeleton plays a crucial role in cytotoxic lymphocyte synapse formation and killing (12, 30). Our data showing that Ca2+ release is abrogated upon cross-linking of an activating receptor suggests that interaction with the actin cytoskeleton, or proteins tethered to the cytoskeleton, is an integral part also of activating signal initiation in NK cells, in line with what has been suggested for T cells (38).
Because cholesterol is generally thought to decrease the fluidity of the plasma membrane, it was at first surprising that cholesterol addition increased the dynamic movements of NKp46. One explanation for this behavior could be that addition of cholesterol increases the sizes of cholesterol-rich domains, thus allowing NKp46 to diffuse within these domains, rather than transiting between cholesterol-rich and cholesterol-depleted domains, where the transitions would reduce the diffusion rate. Only the educated NK cells showed a decreased microdomain dwell time after cholesterol addition. This could suggest that NKp46 localizes to different types of compartments depending on the educational status, which would be in line with the previously observed suggestion that activating receptor movement would be hindered by the cytoskeleton in hyporesponsive NK cells, but rather restricted by nanodomains in educated NK cells (19). An alternative explanation would be that the amount of cholesterol in the membrane differs between the two NK cell subsets.
iMSD carpet analysis is a promising approach for rapid screening of molecular dynamics in live cells across cellular disciplines. It is less sensitive to labeling concentrations and noise than is SPT. In SPT, fluorescence signals with insufficient signal-to-noise ratio fall below the localization and tracking threshold and can no longer contribute to the analysis. Hence, there is a sharp transition point at which SPT analysis is no longer possible. With fluorescence fluctuation-based approaches, however, every signal fluctuation, no matter how small, contributes to the final correlation function. Thus, there is no strict lower threshold. Spot variation (sv) FCS has been developed to identify sub- and superdiffusion, yet this approach is indirect and elaborate (39). Scanning FCS approaches and raster image correlation spectroscopy (RICS) can at least partly account for the large heterogeneity of biological systems by allowing for removal of immobile features (40). However, data interpretation using these methods can be tedious, especially in systems where multiple processes are taking place simultaneously. In comparison, the iMSD carpet analysis covers a wide range of temporal resolution and is relatively straightforward to apply.
The diffusion coefficients reported in the literature are often not directly comparable and can differ by as much as an order of magnitude even when detected by established methods, such as SPT, FCS, and fluorescence recovery after photobleaching (FRAP). This discrepancy is due to differences in the upper and lower limits of the temporal range detected by the different techniques (41). In comparison, the iMSD carpet analysis will report parameter values for the most prominent dynamic feature of the system, regardless of whether this feature is relatively fast diffusion, or a confined and comparably slow movement. The iMSD carpet analysis is thus able to capture a broader time scale of dynamics. However, in our hands it was not possible to fit more than one type of movement, for instance a combination of free diffusion and confinement, and still get reliable output parameter values. Therefore, the reported output values are an average of all movements taking place among the fluorescently labeled molecules and should thus be regarded somewhat qualitatively, rather than strictly quantitatively. For comparison between groups of cells or individual cells, this approach is nevertheless useful and informative. An alternative strategy that was not utilized here, but may be useful if the dynamics change on a larger scale, during a treatment for instance, would be to compare which model fits best in different cells, rather than comparing the output parameters of one model.
The analysis step is faster with iMSD compared to SPT, which is favorable for large-scale screening purposes. Potential applications of the iMSD carpet analysis include finding global differences between cell populations and identifying individual cells with an active phenotype for clinical or experimental purposes. As an extension of what was applied in this study, a cellular footprint could be divided into several smaller regions of interest instead of restricting analysis to a single part of the cell. The resulting map could reveal spatial heterogeneities in different parts of the cell with a resolution half the size of the region of interest. Theoretically, iMSD carpet analysis can also give information about cluster size beyond the diffraction limit. However, this approach is only possible if individual domains are spatially separated by a distance greater than the diffraction limit.
Taken together, our data indicate that the education of NK cells leaves an imprint on receptor dynamics that is distinguishable using TIRF-iMSD carpet analysis, at least at the cell population level. The differences in dynamic receptor movement patterns could, in the future, be used to identify the most active NK cells for use in cancer therapy. The data collection and analysis is rapid and can be used on naïve NK cells without the need for synapse formation with other cells. Studies testing this approach in other systems and cell lines should be conducted. Further, limiting activating receptor movement can be one of the mechanisms by which hyporesponsive NK cells remain hyporesponsive, and could potentially be investigated as a means to dampen immune responses.
MATERIALS AND METHODS
Mice
All mice were from a C57BL/6 (B6) background. Hyporesponsive NK cells were obtained from B6.Kb−/−Db−/− mice, called MHC−/− mice in this study. Educated NK cells were from B6.Kb−/−Db−/−Dd+/+ mice, here called H-2Dd mice (42). Both mouse strains were maintained and bred at the animal facility in the Department of Microbiology, Tumor and Cell biology, or at Comparative Medicine Wallenberg, both at the Karolinska Institutet, Sweden, according to Swedish laws and regulations (Ethical permit diarium numbers N418/12 and N70/15).
Transgene construction
The HLA-B*5101 (B51) open reading frame was amplified by PCR from plasmid IHW10033 (IHWG Genebank). The PCR product was cloned into a pCR3 vector and sequenced to ensure identity with the published sequence. The YFP open reading frame was inserted in frame at the C-terminus of B51 and the B51-YFP fusion gene was subcloned into a pRRLsin.PPTshCMV.pre (Lenti) vector. Lentiviruses were produced using standard techniques and used to infect the human cell line 721.221 which do not have HLA on their surface (43). Transduced cells were purified using flow sorting based on EYFP fluorescence. The 721.221 B51-YFP cells had also been transduced with KIR2DL1. KIR2DL1 was amplified by PCR from cDNA prepared from YTS 2DL1 cells (Cohen et al., 1999, Immunity, 10: 661–671) (kindly provided by D. Burshtyn) and cloned into a pRRLsin.PPT.pgk.pre vector. Lentiviruses were used to infect 721.221 B51-YFP cells and transduced cells were purified using a KIR2DL1 mAb (HP 3E4) and flow sorting. The Ly49A-GFP construct has been previously published [18].
Cell lines and transfection procedure
The human 721.221 cells were cultured at 37 °C, 7.5 % CO2 in Iscove’s Modified Dulbecco’s Medium (HyClone) supplemented with 10 % Fetal Bovine Serum (FBS), 5 % (500 U/ml) penicillin-streptomycin, 5 % non-essential amino acids, 5 % (5 mM) sodium pyruvate, and 0.5 % (2.5 mM) beta-mercaptoethanol.
The Chinese hamster ovary (CHO) cell line AA8 was cultured in Minimal Essential Medium Eagle, alpha modification 1X (HyClone) supplemented with 10 % FBS, 5 % (500 U/ml) penicillin-streptomycin, and 0.5 % (2.5 mM) β-mercaptoethanol. Cells were transfected using standard procedures with a 1:5 ratio mixture of Ly49A-GFP construct and Lipofectamine 2000 (Invitrogen). Cells were analyzed 2–4 days after transfection.
NK cell isolation from splenocytes
Spleens were mechanically homogenized for single cell suspension. Erythrocytes were lysed by incubating the cell suspension with 0.83% NH4Cl for 5 min on ice. Splenocyte single cell suspensions were obtained by filtration through a 40 μm nylon cell strainer (Corning). NK cells were subsequently enriched by MACS™ NK cell isolation kit mouse II (Miltenyi Biotec NordenAB, Sweden) according to the manufacturer’s instructions. The purity of NK cells isolated from splenocytes was 85 ± 5%.
Antibodies and preparation of cells for imaging and spectroscopy
Clone JR9.318, an antibody that recognizes Ly49A, was purified from hybridoma supernatants either in our lab or by MabTech (Nacka, Sweden) and conjugated to Abberior star 635-NHS dye (Abberior GmbH, Germany). The antibody recognizing NKp46 (clone 29A1.4, BioLegend), were conjugated to MFP488 (MoBiTec GmbH, Germany). For the experiments with transfected CHO cells, the JR9 antibody clone was conjugated to the fluorophore Abberior star 594-NHS (Abberior GmbH, Germany). Antibody-dye conjugations were performed according to the manufacturers’ protocols and purified using PD Minitrap G-25 columns (GE healthcare Bioscience, Sweden). The antibody recognizing Ly49A clone YE1/48.10.6-FITC was purchased (BioLegend). All antibodies were titrated for optimal staining for each batch, but were always in the range of 1–10 μg/ml final solution during staining.
Fc receptors were blocked in 16 % mouse serum in phospate buffered saline (PBS) before antibody labeling. NK cells were stained in solution, centrifuged (450 G for 3 minutes) and resuspended in a 1:1 mixture of transparent Roswell Park Memorial Institute medium 1640 (RPMI, PAA The Cell Culture Company) and PBS with 0.5 % FBS. Resuspended NK cells were transferred to poly-L-lysine–coated dishes with glass bottoms (MatTek Corporation, USA, P35G-1.5-14.C, and Greiner BioOne 627861). For actin inhibition experiments, NK cells were resuspended in transparent RPMI only, due to FBS interfering with the inhibitor (44). The actin inhibitor Latrunculin B (Molecular Probes, final concentration 2 μg/ml), and myosin inhibitor Myovin1 (Calbiochem, 475984, final concentration 50 or 200 μg/ml) was added to NK cells 10 min after cell transfer to coated dishes. Imaging started at least 10 min after inhibitor addition. Only Ly49A+ NK cells were imaged, as judged visually at the microscope. Similar experiments for cholesterol addition to the NK cell membrane were performed using 60 nM water-soluble cholesterol (Sigma Aldrich, C4951), for cholesterol addition and 10 mM methyl-β-cyclodextrin (Sigma Aldrich, C4555) for cholesterol removal. Compounds were added to cells in the well 30 minutes prior to imaging.
Total internal reflection fluorescence (TIRF) microscopy
For isolated NK cells and 721.221 cells, a wide-field fluorescence inverted microscope (Zeiss Elyra PS1) with Zen 2012 Black software was used, at the Advanced Light Microscopy facility at SciLifeLab, Stockholm, Sweden. The microscope was run in TIRF illumination mode using an alpha Plan-Apochromat 100x oil immersion objective (NA 1.46). Fluorescence was excited with 488 nm and 633 nm laser light and detected in two channels (bandpass filter 495–575 nm and long pass filter 655 nm) with an EMCCD camera (Andor iXon 897). Exposure times were either 10 or 20 ms, resulting in 20 or 36 ms frame-to-frame intervals. Recorded image series were saved as Zeiss LSM format and analyzed without further modification. Transfected CHO AA8 cells were imaged on a wide-field fluorescence inverted microscope (Zeiss 200M) with Zen 2 Blue software. The Zeiss 200M microscope was run in TIRF illumination mode using a 100x oil immersion objective. Fluorescence was excited with 488 nm and 561 nm light, for GFP and labelled antibody, respectively. Fluorescence was detected with a 74 HE GFP/mRFP filter and a CCD camera (Rolera em-c2) at 36 ms frame-to-frame intervals. Recorded image series were saved as Zeiss .czi format and converted to multi-tif in Fiji (ImageJ, version 2.0.0-rc-43/1.50g) prior to analysis. For all TIRF measurements, 6000 frames were acquired per cell and spectral channel, at a pixel size of 0.10 μm. To minimize photobleaching channels were imaged sequentially with the longer wavelength first (633 nm excitation). Cells were imaged at room temperature (22–24 °C).
Image mean square displacement analysis of images
TIRF images were analyzed in Matlab using a script with a graphical user interface. A square region of interest of 32 × 32 pixels (3.2 × 3.2 μm2) was placed within the cellular footprint, avoiding clusters of unusually high brightness. Cells that were out-of-focus or had a smaller footprint than the region of interest were excluded before analysis.
From the Gaussian distribution width of the STICS correlation function, the underlying type of motion can be identified after subtraction of the immobile fraction. From the recorded time series, I(x, y, t), a region of interest containing the cell adhesion area was extracted. First, the temporal average of all frames, <I(x, y, t)>t, was subtracted from each individual frame, i, in the series, Ii(x, y, t), followed by addition of the average intensity of the entire stack <I(x, y, t)>xyt,
| (1) |
This removed contributions of the immobile fraction to the correlation. The data was then corrected for bleaching by subtracting the average intensity of that frame, <Ji(x, y, t)>xy, followed again by addition of the average intensity of the entire stack, <I(x, y, t)>xyt,
| (2) |
Subsequently, the spatiotemporal correlation,
| (3) |
was calculated in a multi-tau fashion with 6 blocks of 12 linear delay times, τ. As the underlying dynamics was isotropic, the two spatial correlation coordinates, ξ and ψ, were combined into a single spatial coordinate, δ, by means of . This two-dimensional dataset, G(δ, τ), termed the correlation carpet, was then least-squares fitted with several equations representing different modes of particle dynamics as previously described in the Supporting Information of (45). Briefly, for fluorescence fluctuations, the correlation amplitude can be expressed as:
| (4) |
With the normalization factor given by the average number of particles, N, residing in the point spread function and the gamma factor which is 0.5 for a 2D Gaussian, the probability distribution, p, to find a particle at position ξ, ψ, time τ, and the point spread function, W. For particle movement in 2D, the probability distribution becomes:
| (5) |
For free diffusion with diffusion coefficient, D, the mean square displacement, σ2, is:
| (6) |
For active transport with average velocity, v, the mean square displacement is given by:
| (7) |
For square confinement within a compartment of width, L, it can be demonstrated that:
| (8) |
And for binding in 2D, the probability distribution yields:
| (9) |
Note that in equations (6) to (9) the point spread function, W, is included in the size parameters s0 and sB. Insertion of equation (5) in equation (4) and equations (6) to (9) in equation (5) yields the following model equations, which were used to fit our data.
Free diffusion:
| (10) |
Active transport:
| (11) |
Trapping:
| (12) |
Confinement:
| (13) |
With offset, A0, particle number, N, diffusion coefficient, D, apparent particle size, s0, velocity, v, trapping compartment width, L, trapping time, τC, cluster amplitude, AB, dwell time, τB, and apparent microdomain size, sB. The microdomain width was calculated as the square root of sB. Note that both the apparent particle size and the apparent microdomain width were broadened by the instrument point spread function. The average size of both can be obtained by subtracting the size of the PSF. Due to subtraction of the immobile fraction, a term G(δ, τ → ∞) should be subtracted from each model. Yet for all models, this term approaches zero except for the trapping model in which a spatial correlation remains that was subtracted. The fitting procedure was terminated if 1) the tolerance on the model value was <10−21, 2) the tolerance on the coefficient values was <10−21, 3) the maximum number of iterations exceeded 1000, or, 4) the maximum number of evaluations of the model exceeded 1000. The lower and upper boundaries for the fit parameters were chosen according to the technical limitations (the temporal and spatial resolution) of the measurement.
Simulation of different modes of dynamics
We simulated molecules exhibiting different modes of dynamics in a plane of 128 × 128 pixels using a custom script written in Matlab. In this plane, 1000 molecules were seeded at random positions and plotted by convoluting their position with a Gaussian of width 2.5 pixels followed by the addition of Gaussian noise (variance 0.01) to the image (Matlab function imnoise). For each run, we simulated a total of 6000 frames and assigned each pixel a size of 100 nm and each time step to be 36 ms, i.e., the same values as in the actual experiment. For the border of the simulated plane we applied reflective boundary conditions. To simulate free diffusion, for each molecule in each time step a random angle φ was assigned. Particle displacement was calculated as r = 4Dt. A displacement of Δx = rcosφ and Δy = rsinφ was then added to the current particle position. For the simulation of active transport, a random direction φ was assigned to each particle during the initial seeding process. In each time step of the simulation, the particle position was advanced in that direction by calculating displacement as r = vt and adding Δx = rcosφ and Δy = rsinφ to the x and y position, respectively. For trapping, the plane was subdivided into 16 × 16 compartments corresponding to a confinement width L of 8 pixels. Molecules inside these compartments followed the same rules as for the simulation of free diffusion. For confinement, an off (koff) and on rate (kon) was assigned. No displacement was added, instead, in each time step a random number between zero and one was generated for each particle. If the particle was in the on state that random number was compared to the off rate to decide if the particle was not drawn in the next frame, simulating exit from the domain. If in the off state, the random number was compared to the on rate to decide if the particle was added back into the next frame, simulating entering the domain. From the simulated stacks of images, regions of 32 × 32 pixels were extracted and analyzed with the same script and settings as the experimental data.
Single particle tracking (SPT) analysis
Image sequences from images which had sufficiently low labeling intensity to allow identification of single particles were treated using a home-written software in Igor Pro (Wavemetrics, Lake Oswego, OR) as described elsewhere (46) with slight modifications. Five images were grouped to increase the signal-to-noise ratio and reach single-molecule sensitivity. Fluorescent peaks were detected in each image and then linked over the complete image sequence to obtain trajectories. Diffusion coefficients, length of the confinement regions and moment scaling spectrum were evaluated on segments of the trajectory obtained with a sliding window of 100 frames. Each of the segmented trajectories was treated as an individual trajectory and computed to extract the initial diffusion coefficient, D1-10, the Hurst parameter SMSS and the length of displacement. The mean square displacement for each time interval, tlag was calculated with the following equation:
| (14) |
Where N is the number of frames and r the displacement; i and n are integers.
D1-10 was estimated with a linear fit of the 10 first time intervals of the MSD vs tlag curve:
| (15) |
Where D1-10 is the diffusion coefficient for the first 10 tlag and the noise is composed of the movement of the particle during the acquisition time and the error on the localization due to the experimental noise. The Hurst parameter was computed as described in detail in Ewers et. al. (47). The length of displacement was determined as the diagonal of the smallest rectangle containing 95 % of the particle position during a 100 frames segment. D1-10, SMSS, and the length of displacement of each single-molecule are calculated by averaging the values of all segments comprised in the trajectory. Box plots, including notches, were computed for each type of cells using R (R Foundation for Statistical Computing).
Ca2+ release assays
NK cells were isolated from splenocytes as described earlier and surface stained with antibodies against the following antigens: NKRP1-C (PK136), Ly49A (YE1/48), CD3 (145-2C11) all purchased from Biolegend and used at 2–5 μg/ml, and biotin-labelled antibody against NKp46 (29A1.4, Miltenyi Biotech) in 10 μg/ml concentration. Cells were maintained in Hanks buffered salt solution without Ca2+ and Mg2+ (HBSS) supplemented with 1 % FBS during the assay. The calcium loading dye Fluo-4 [Molecular Probes] dissolved in dimethyl sulfoxide was added to a final concentration of 2 μg/ml and cells were further incubated for 30 min on ice. Cells were washed once in HBSS 1 % FBS and resuspended in HBSS. Cells were incubated with 60 nM water-soluble cholesterol (Sigma Aldrich, C4951), 10 mM methyl-β-cyclodextrin, or 2 μg/ml Latrunculin B for 30 min at 37 °C. Cell suspensions were vortexed prior to sorting by flow cytometry (FACS Calibur, BD Bioscience). After acquiring the basal calcium fluorescence signal for 30 seconds, 10 μg/ml of streptavidin-coated beads (Miltenyi Biotec Norden, Sweden) were added to crosslink NKp46. As a positive control for Ca2+ release, each sample was treated with 5 μg/ml ionomycin (Sigma-Aldrich). Samples were vortexed, placed back in the flow cytometer, and Ca2+ events were recorded for 4 min. Data were analyzed with FlowJo™ software version 10.
Fluorescence correlation spectroscopy
Fluorescence correlation spectroscopy (FCS) was carried out on a Zeiss LSM 780 system with software ZEN 2012 black at the Advanced Light Microscopy facility at Science for Life laboratories, Stockholm, Sweden. The room was kept at a stable 22 °C. Excitation lasers were an Argon laser (for 488) and HeNe laser (for 633), used with a 40x water immersion objective (NA 1.2). A power series of measurements of freely diffusing Rhodamine 110 (20 nM) and Alexa 647 (2 nM) in PBS were performed to measure the size of the detection volume. Although the YFP fluorescence is optimal at 514 nm excitation, the 488 laser gave sufficient excitation. Living cells with intact membrane and rounded nucleus in differential interference contrast, with B51-YFP or Ly49A-GFP expression neither too high nor too low were selected for measurement. For the B51-YFP experiments, focus was positioned both on the upper and the lower membrane (order of top -> bottom was switched between cells) Laser powers for cells were 0.5 % for the first two experiments, and 0.05 % for the later two experiments. 7 repeats of 10 seconds were recorded.
FCS analysis
FCS analysis was carried out with the same graphical user interface and underlying mathematical models as described in [18]. Briefly, the diffusion of free fluorophores was fitted with a free 3D diffusion model with one triplet state and one diffusion component. From the known diffusion coefficients of Rhodamin 110 and Alexa 647 (3.3 × 10−10 and 3.3 × 10−10, respectively), the focal waist radius, ω, was calculated. FCS curves for single cells were pre-evaluated, and curve segments containing large clusters, movement of the whole cell or obvious bleaching were excluded before analysis. The cells were thereafter fitted to a 2D-membrane diffusion model. The density of moving entities, the diffusion coefficient (assuming only one mode of movement), and the brightness per moving entity were thereafter calculated, using ω. Finally cells with diffusion coefficients below 0.05 μm2/s (corresponding to >500 ms transit time of fluorescent entities through the focus) were excluded from analysis. See Bagawath-Singh et al for formulas and calculations [18].
Statistical analysis
Figures and statistical analyses for iMSD carpet analysis and FCS were done in Origin (version X), Prism (Version 6,0 GraphPad, La Jolla, CA, USA), and R (R Foundation for Statistical Computing). All statistical tests were done under the assumption that the inter-experimental variation was negligible. The significance for SPT measurements was determined by a two-sample Wilcoxon ranking test with the null hypothesis (H0) that population A is not different from population B. iMSD data sets were examined for a gaussian distribution and homogeneity of variance using Shapiro-Wilk’s normality test and maximum one outlier per sample were identified and removed using the Grubb’s test, after verifying that the value was due to technical anomalies in the measurements. For transfected AA8 cells, multiple comparisons were performed. Kruskal-Wallis one-way analysis of variance with Dunn’s multiple comparison correction was therefore applied. Data sets were analyzed with Mann-Whitney’s test in GraphPad Prism. P-values for statistical significance were p < 0.05 for *, p < 0.01 for **, and p < 0.001 for ***.
Supplementary Material
Movie M1. Single molecule tracking of NKp46 in H-2D2 NK cells
Movie M2. Single molecule tracking of Ly49A in H-2D2 NK cells
Fig. S1. Diffusion rate and cluster size at the cell surface are not affected by antibody binding.
Fig. S2. Dynamic clustering of NKp46 and Ly49A at the NK cell surface.
Fig. S3. Simulation of diffusion with different numbers of molecules and variance of the noise.
Fig. S4. Fitting of simulated data sets of the different modes of movement.
Fig. S5. Cluster dynamics of transfected Ly49A-GFP at the cell surface is not significantly affected by antibody binding.
Fig. S6. Carpet iMSD analysis of Ly49A after LatB and Myovin1 treatment.
Fig. S7. Carpet iMSD microdomain widths after cholesterol or MbCD treatment.
Table S1. Simulation of diffusion with different numbers of molecules and variance of the noise.
Table S2. R2 of fitting simulated data to the four different models, using carpet iMSD analisys
Table S3. Median R2 and percentage of cells with best fit for each model.
Editors Summary: Receptor dynamics shape NK cell function.
Natural killer (NK) cells are a subset of innate lymphoid cells that target virally-infected and malignant cells. An education process ensures that activating receptors promote NK cell activation in response to infected or abnormal cells and that inhibitory receptors prevent inappropriate NK cell activation by healthy cells. Cells that lack inhibitory receptors arehyporesponsive. Staaf et al. found that the activating receptor NKp46 and the inhibitory receptor Ly49A exhibited dynamic movements in the membrane that differed between educated and hyporesponsive NK cells. Although both receptors were generally restricted to membrane microdomains, NKp46 molecules spent less time in an individual microdomain and diffused faster on educated cells compared to hyporesponsive cells. Conversely, Ly49A molecules were overall more constrained and diffused more slowly on educated cells compared to hyporesponsive cells. Interfering with receptor dynamics reduced signaling by NKp46, indicating that the dynamic movements of these receptors are likely important determinants of NK cell responsiveness.
Acknowledgments
We thank H. Blom (facility manager at the Advanced Light Microscopy facility, SciLifeLab, Stockholm, Sweden) for assistance in establishing TIRF, access to the microscope, and good advice throughout the process, and S. Wennmalm (facility manager at the FCS facility, SciLifeLab, Stockholm, Sweden) for access to the FCS microscope and support for crucial control experiments. We also thank W. Held at the Ludwig Center for Cancer Research of the University of Lausanne for sharing the 721.221. KIR2DL1-B51-YFP cell line with our laboratory, R. Mehr for careful reading of the manuscript, and D. M. Jameson for proofreading.
Funding: S.J., E.S., and S.B.S. were supported by Vetenskapsrådet, Selma Andersons stipend fund, Hierta-Retzius stipend fund, Magnus Bergvalls stiftelse, Clas Groschinsky’s memorial foundation, Robert Lundberg’s Memorial Foundation, and The Swedish Foundation for International Cooperation in Research and Higher Education. J.P. was supported by Vinnova, the Swedish agency for innovation. E.G. and P.N.H. were supported by NIH grants P41-GM103540 and P50-GM076516.
Footnotes
Author contributions: S.J., E.S., and P.N.H. conceived the project. E.S. performed most of the TIRF experiments. P.N.H. conceived the iMSD carpet analysis and constructed the Matlab analysis script with input from E.G. and E.S. P.N.H. and E.S. analyzed the data for iMSD carpet analysis. P.N.H. performed the simulations of receptor movements. J.P. performed the SPT analysis. S.B.S. performed calcium release and cholesterol iMSD experiments. S.J. was involved in the design and analysis of iMSD and Ca release experiments and wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Tu MM, Mahmoud AB, Wight A, Mottashed A, Belanger S, Rahim MM, Abou-Samra E, Makrigiannis AP. Ly49 family receptors are required for cancer immunosurveillance mediated by natural killer cells. Cancer Res. 2014;74:3684–3694. doi: 10.1158/0008-5472.CAN-13-3021. [DOI] [PubMed] [Google Scholar]
- 2.Jost S, Altfeld M. Control of human viral infections by natural killer cells. Annual review of immunology. 2013;31:163–194. doi: 10.1146/annurev-immunol-032712-100001. [DOI] [PubMed] [Google Scholar]
- 3.Velardi A. Natural killer cell alloreactivity 10 years later. Current opinion in hematology. 2012;19:421–426. doi: 10.1097/MOH.0b013e3283590395. [DOI] [PubMed] [Google Scholar]
- 4.Kärre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319:675–678. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- 5.Fernandez NC, Treiner E, Vance RE, Jamieson AM, Lemieux S, Raulet DH. A subset of natural killer cells achieve self-tolerance without expressing inhibitory receptors specific for self MHC molecules. Blood. 2005 doi: 10.1182/blood-2004-08-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim S, Poursine-Laurent J, Truscott SM, Lybarger L, Song YJ, Yang L, French AR, Sunwoo JB, Lemieux S, Hansen TH, Yokoyama WM. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436:709–713. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 7.Forslund E, Sohlberg E, Enqvist M, Olofsson PE, MKJ, Önfelt B. Microchip-Based Single-Cell Imaging Reveals That CD56dimCD57-KIR-NKG2A+ NK Cells Have More Dynamic Migration Associated with Increased Target Cell Conjugation and Probability of Killing Compared to CD56dimCD57-KIR-NKG2A- NK Cells. J Immunol. 2015;195:3374–3381. doi: 10.4049/jimmunol.1500171. [DOI] [PubMed] [Google Scholar]
- 8.Thomas LM, Peterson ME, Long EO. Cutting edge: NK cell licensing modulates adhesion to target cells. J Immunol. 2013;191:3981–3985. doi: 10.4049/jimmunol.1301159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodridge JP, Önfelt B, Malmberg K-J. Newtonian cell interactions shape natural killer cell education. Immunol Rev. 2015;267:197–213. doi: 10.1111/imr.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enqvist M, Ask EH, Forslund E, Carlsten M, Abrahamsen G, Beziat V, Andersson S, Schaffer M, Spurkland A, Bryceson Y, Önfelt B. Coordinated expression of DNAM-1 and LFA-1 in educated NK cells. J Immunol. 2015;194:4518–4527. doi: 10.4049/jimmunol.1401972. [DOI] [PubMed] [Google Scholar]
- 11.Wu W, Shi X, Xu C. Regulation of T cell signalling by membrane lipids. Nat Rev Immunol. 2016;16:690–701. doi: 10.1038/nri.2016.103. [DOI] [PubMed] [Google Scholar]
- 12.Mattila PK, Batista FD, Treanor B. Dynamics of the actin cytoskeleton mediates receptor cross talk: An emerging concept in tuning receptor signaling. The Journal of cell biology. 2016;212:267–280. doi: 10.1083/jcb.201504137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu C, Miller H, Orlowski G, Hang H, Upadhyaya A, Song W. Actin reorganization is required for the formation of polarized BCR signalosomes in response to both soluble and membrane-associated antigens. J Immunol. 2012;188:3237–3246. doi: 10.4049/jimmunol.1103065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mattila PK, Feest C, Depoil D, Treanor B, Montaner B, Otipoby KL, Carter R, Justement LB, Bruckbauer A, Batista FD. The actin and tetraspanin networks organize receptor nanoclusters to regulate B cell receptor-mediated signaling. Immunity. 2013 doi: 10.1016/j.immuni.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 15.Torreno-Pina JA, Manzo C, Salio M, Aichinger MC, Oddone A, Lakadamyali M, Shepherd D, Besra GS, Cerundolo V, Garcia-Parajo MF. The actin cytoskeleton modulates the activation of iNKT cells by segregating CD1d nanoclusters on antigen-presenting cells. Proc Natl Acad Sci U S A. 2016;113:E772–781. doi: 10.1073/pnas.1514530113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pageon SV, Aquino G, Lagrue K, Kohler K, Endres RG, Davis DM. Dynamics of natural killer cell receptor revealed by quantitative analysis of photoswitchable protein. Biophys J. 2013;105:1987–1996. doi: 10.1016/j.bpj.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Standeven LJ, Carlin LM, Borszcz P, Davis DM, Burschtyn DN. The actin cytoskeleton controls the efficiency of killer-Ig-like receptor accumulation at inhibitory NK Cell Immune Synapses. J Immunol. 2004;173:5671–5625. doi: 10.4049/jimmunol.173.9.5617. [DOI] [PubMed] [Google Scholar]
- 18.Bagawath-Singh S, Staaf E, Stoppelenburg AJ, Spielmann T, Kambayashi T, Widengren J, Johansson S. Cytokines induce faster membrane diffusion of MHC class I and the Ly49A receptor in a subpopulation of Natural Killer Cells. Front Immunol. 2016;7 doi: 10.3389/fimmu.2016.00016. Article 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guia S, Jaeger BN, Piatek S, Mailfert S, Trombik T, Fenis A, Chevrier N, Walzer T, Kerdiles YM, Marguet D, Vivier E, Ugolini S. Confinement of activating receptors at the plasma membrane controls natural killer cell tolerance. Sci Signal. 2011;4:ra21. doi: 10.1126/scisignal.2001608. [DOI] [PubMed] [Google Scholar]
- 20.Kusumi A, Fujiwara TK, Chadda R, Xie M, Tsunoyama TA, Kalay Z, Kasai RS, Suzuki GN. Dynamic organizing principles of the plasma membrane that regulate signal transduction: Commemorating the fortieth anniversary of Singer and Nicolson’s fluid-mosaic model. Annu Rev Cell Dev Biol. 2012;28:215–250. doi: 10.1146/annurev-cellbio-100809-151736. [DOI] [PubMed] [Google Scholar]
- 21.Manzo C, Garcia-Paranjo M. A review of progress in single particle tracking: from methods to biophysical insights. Rep Prog Phys. 2015;78:124601, 124629pp. doi: 10.1088/0034-4885/78/12/124601. [DOI] [PubMed] [Google Scholar]
- 22.Saxon MJ, Jacobson K. Single-particle tracking: applications to membrane dynamics. Annu Rev Biophys Biomol Struct. 1997;26:373–399. doi: 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- 23.Herbert B, Constantino S, Wiseman PW. Spatiotemporal Image Correlation Spectroscopy (STICS) Theory, Verification,and Application to Protein Velocity Mapping in Living CHO Cells. Biophys J. 2005;88:3601–3614. doi: 10.1529/biophysj.104.054874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Rienzo C, Gratton E, Beltram F, Cardarelli F. Fast spatiotemporal correlation spectroscopy to deterimne protein lateral diffusion laws in live cell membranes. PNAS. 2013;110:12307–12312. doi: 10.1073/pnas.1222097110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olsson-Alheim MY, Sundbäck J, Kärre K, Sentman CL. The MHC class I molecule H-2Dp inhibits murine NK cells via the inhibitory receptor Ly49A. J Immunol. 1999;162:7010–7014. [PubMed] [Google Scholar]
- 26.Ewers H, Sbalzarini IF, Koumoutsakos P, Helenius A. Single-particle tracking of murine polyoma virus-like particles on live cells and artificial membranes. Proc Natl Acad Sci USA. 2005;102 doi: 10.1073/pnas.0504407102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Rienzo C, Gratton E, Beltram F, Cardarelli F. Fast spatiotemporal correlation spectroscopy to deterimne protein lateral diffusion laws in live cell membranes. PNAS. 2013;110:12307–12312. doi: 10.1073/pnas.1222097110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wulfing C, Sumen C, Sjaastad MD, Wu LC, Dustin ML, Davis MM. Costimulation and endogenous MHC ligands contribute to T cell recognition. Nat Immunol. 2002;3:42–47. doi: 10.1038/ni741. [DOI] [PubMed] [Google Scholar]
- 29.Mossman KD, Campi G, Groves JT, Dustin ML. Altered TCR signaling from geometrically repatterned immunological synapses. Science. 2005;310:1191–1193. doi: 10.1126/science.1119238. [DOI] [PubMed] [Google Scholar]
- 30.Mace EM, Orange JS. Lytic immune synapse function requires filamentous actin deconstruction by Coronin 1A. Proc Natl Acad Sci U S A. 2014;111:6708–6713. doi: 10.1073/pnas.1314975111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Field KA, Holowka D, Baird B. Compartmentalized activation of the high affinity immunoglobulin E receptor within membrane domains. The Journal of biological chemistry. 1997;272:4276–4280. doi: 10.1074/jbc.272.7.4276. [DOI] [PubMed] [Google Scholar]
- 32.Xavier R, Brennan T, Li Q, McCormack C, Seed B. Membrane compartmentation is required for efficient T cell activation. Immunity. 1998;8:723–732. doi: 10.1016/s1074-7613(00)80577-4. [DOI] [PubMed] [Google Scholar]
- 33.Oszmiana A, Williamson DJ, Cordoba SP, Morgan DJ, Kennedy PR, Stacey K, Davis DM. The Size of Activating and Inhibitory Killer Ig-like Receptor Nanoclusters Is Controlled by the Transmembrane Sequence and Affects Signaling. Cell Rep. 2016;15:1957–1972. doi: 10.1016/j.celrep.2016.04.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Höglund P, Brodin P. Current perspectives of natural killer cell education by MHC class I molecules. Nat Rev Immunol. 2010;10:724–734. doi: 10.1038/nri2835. [DOI] [PubMed] [Google Scholar]
- 35.Chalifour A, Scarpellino L, Back J, Brodin P, Devevre E, Gros F, Levy F, Leclercq G, Hoglund P, Beermann F, Held W. A Role for cis Interaction between the Inhibitory Ly49A receptor and MHC class I for natural killer cell education. Immunity. 2009;30:337–347. doi: 10.1016/j.immuni.2008.12.019. [DOI] [PubMed] [Google Scholar]
- 36.Stromqvist J, Johansson S, Xu L, Ohsugi Y, Andersson K, Muto H, Kinjo M, Hoglund P, Widengren J. A modified FCCS procedure applied to Ly49A-MHC class I cis-interaction studies in cell membranes. Biophys J. 2011;101:1257–1269. doi: 10.1016/j.bpj.2011.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doucey MA, Scarpellino L, Zimmer J, Guillamue P, Luescher IF, Bron C, Held W. Cis-association of Ly49A with MHC class I restrics natural killer cell inhibition. Nature Immunol. 2004;5:328–336. doi: 10.1038/ni1043. [DOI] [PubMed] [Google Scholar]
- 38.Kumari S, Depoil D, Martinelli R, Judokusumo E, Carmona G, Gertler FB, Kam LC, Carman CV, Burkhardt JK, Irvine DJ, Dustin ML. Actin foci facilitate activation of the phospholipase C-gamma in primary T lymphocytes via the WASP pathway. eLife. 2015;4 doi: 10.7554/eLife.04953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klotzsch E, Schütz GJ. A critical survey of methods to detect plasma membrane rafts. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120033. doi: 10.1098/rstb.2012.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ries J, Yu SR, Burkhardt M, Brand M, Schwille P. Modular scanning FCS quantifies receptor-ligand interactions in living multicellular organisms. Nature Methods. 2009;6:643–645. doi: 10.1038/nmeth.1355. [DOI] [PubMed] [Google Scholar]
- 41.Bates IR, Wiseman PW, Hanrahan JW. Investigating membrane protein dynamics in living cells. Biochem Cell Biol. 2006;84:825–831. doi: 10.1139/o06-189. [DOI] [PubMed] [Google Scholar]
- 42.Johansson S, Johansson M, Rosmaraki E, Vahlne G, Mehr R, Salmon-Divon M, Lemonnier F, Karre K, Hoglund P. Natural killer cell education in mice with single or multiple major histocompatibility complex class I molecules. J Exp Med. 2005;201:1145–1155. doi: 10.1084/jem.20050167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shimizu Y, DeMars R. Production of human cells expressing individual transferred HLA-A,-B,-C genes using an HLA-A,B,-C null human cell line. J Immunol. 1989;142:3320–3328. [PubMed] [Google Scholar]
- 44.Wakatsuki T, Schwab B, Thompson NC, Elson EL. Effects of cytochalasin D and latrunculin B on mechanical properties of cells. J Cell Sci. 2001;114:1025–1036. doi: 10.1242/jcs.114.5.1025. [DOI] [PubMed] [Google Scholar]
- 45.Di Rienzo C, Gratton E, Beltram F, Cardarelli F. Fast spatiotemporal correlation spectroscopy to determine protein lateral diffusion laws in live cell membranes. Proc Natl Acad Sci U S A. 2013;110:12307–12312. doi: 10.1073/pnas.1222097110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Veya L, Piguet J, Vogel H. Single-Molecule Imaging Deciphers the Relation between Mobility and Signaling of a Prototypical G Protein-Coupled Receptor in Living Cells. J Biol Chem. 2015;290:27723–27735. doi: 10.1074/jbc.M115.666677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ewers H, Smith AE, Sbalzarini IF, Lilie H, Koumoutsakos P, Helenius A. Single-particle tracking of murine polyoma virus-like particles on live cells and artificial membranes. Proc Natl Acad Sci U S A. 2005;102:15110–15115. doi: 10.1073/pnas.0504407102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie M1. Single molecule tracking of NKp46 in H-2D2 NK cells
Movie M2. Single molecule tracking of Ly49A in H-2D2 NK cells
Fig. S1. Diffusion rate and cluster size at the cell surface are not affected by antibody binding.
Fig. S2. Dynamic clustering of NKp46 and Ly49A at the NK cell surface.
Fig. S3. Simulation of diffusion with different numbers of molecules and variance of the noise.
Fig. S4. Fitting of simulated data sets of the different modes of movement.
Fig. S5. Cluster dynamics of transfected Ly49A-GFP at the cell surface is not significantly affected by antibody binding.
Fig. S6. Carpet iMSD analysis of Ly49A after LatB and Myovin1 treatment.
Fig. S7. Carpet iMSD microdomain widths after cholesterol or MbCD treatment.
Table S1. Simulation of diffusion with different numbers of molecules and variance of the noise.
Table S2. R2 of fitting simulated data to the four different models, using carpet iMSD analisys
Table S3. Median R2 and percentage of cells with best fit for each model.