Abstract
Summary
Nuclear magnetic resonance (NMR) spectroscopy, along with X-ray crystallography and cryoelectron microscopy, is one of the three major tools that enable the determination of atomic-level structural models of biological macromolecules. Of these, NMR has the unique ability to follow important processes in solution, including conformational changes, internal dynamics and protein–ligand interactions. As a means for facilitating the handling and analysis of spectra involved in these types of NMR studies, we have developed PINE-SPARKY.2, a software package that integrates and automates discrete tasks that previously required interaction with separate software packages. The graphical user interface of PINE-SPARKY.2 simplifies chemical shift assignment and verification, automated detection of secondary structural elements, predictions of flexibility and hydrophobic cores, and calculation of three-dimensional structural models.
Availability and implementation
PINE-SPARKY.2 is available in the latest version of NMRFAM-SPARKY from the National Magnetic Resonance Facility at Madison (http://pine.nmrfam.wisc.edu/download_packages.html), the NMRbox Project (https://nmrbox.org) and to subscribers to the SBGrid (https://sbgrid.org). For a detailed description of the program, see http://www.nmrfam.wisc.edu/pine-sparky2.htm.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Numerous different groups and institutions have developed the software packages in current use in the field of biomolecular NMR, and many utilize different nomenclatures, data input procedures, and even computer operating systems. These differences have impeded research progress, particularly by non-experts. The Integrative NMR package (Lee et al., 2016a) offered a partial solution by integrating NMRFAM-SPARKY (Lee et al., 2015) with APES for peak picking (Shin et al. 2008), PINE for automated assignment (Bahrami et al. 2009), ARECA (Dashti et al. 2016) for validation of peak assignments; TALOS-N for shift based torsion angle restraints (Shen and Bax, 2013), CS-Rosetta (Shen et al., 2008), for structure determination from chemical shifts, AUDANA (Lee et al., 2016b) and PONDEROSA-C/S (Lee et al., 2014) for automated structure determination from NOE spectra, and data visualization by NDP-PLOT and an enhanced mode of the PyMOL software package (The PyMOL Molecular Graphics System, Version 1.7.4 Schrödinger, LLC.).
However, as part of this package, the original PINE-SPARKY (Lee et al., 2009) was cumbersome. Users had to pick peaks from NMR spectra, generate a set of peak list files, open a web browser, visit the PINE web page and submit generated peak list files one-by-one for each experiment. To import and verify chemical shifts, the user had to wait for an email notice, download and unpack the compressed results, use the PINE2SPARKY converter to apply PINE probabilistic assignments to SPARKY projects, and use PINE-SPARKY extensions to create the actual assignment labels. Only after following these steps could the user carry out further analysis, such as validation of chemical shift referencing by LACS (Wang et al., 2005), secondary structure determination by PECAN (Eghbalnia et al., 2005), analysis of chemical shifts by reference to the PACSY database (Lee et al., 2012) or 3 D structure determination by CS-Rosetta (Shen et al., 2008).
PINE-SPARKY.2, which comes as a plug-in to NMRFAM-SPARKY, integrates all of these tasks and provides, in addition, easy-to-use visual analysis tools based on probability theory. PINE-SPARKY.2 incorporates a new server and various programs written in CGI/Perl, PYTHON, and BASH scripts that integrate PINE, PECAN, LACS, PACSY, TALOS-N, and CS-ROSETTA. Typing the two-letter-code he calls up the user manual, which is also available on-line (http://www.nmrfam.wisc.edu/pine-sparky2.htm).
2 Implementation
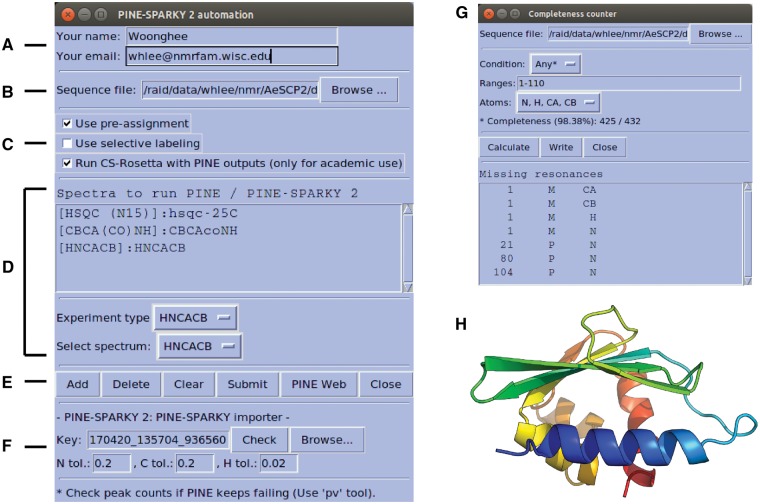
PINE-SPARKY.2 can be launched from the automated assignment sub-menu of NMRFAM menu or by typing the two-letter-code ep in the NMRFAM-SPARKY. This opens a graphical user interface that makes all the features of NMRFAM-SPARKY simultaneously accessible. Users provide name and email information (Fig. 1A), which the plug-in uses to interact with the PINE web server. The PINE web server sends an email containing a URL where the results can be retrieved. Users can provide a sequence file directly to the plugin (Fig. 1B), otherwise the sequence in the Sequence Entry plug-in (two-letter-code sq), will be imported.
Fig. 1.
Graphical user interface (GUI) for PINE-SPARKY.2 (A)–(F) input; see text). (G) Completeness Counter. (H) Example of a structural model obtained from assigned chemical shifts by CS-Rosetta
The PINE-SPARKY.2 plug-in offers three options (Fig. 1C): (i) Use pre-assignment: This option is used to restrain already assigned resonances. (ii) Use selective labeling: This option allows specification of the amino acid types expected in a spectrum. (iii) Run CS-Rosetta with PINE outputs: This option executes 3 D structure calculations using the CS-Rosetta server (http://csrosetta.bmrb.wisc.edu/csrosetta hosted by BMRB).
PINE-SPARKY.2 supports 19 different NMR experiments. The user specifies the NMR experiments with spectral data to be analyzed by clicking the Add button from the spectrum list (Fig. 1DE). Peaks need to be identified in the spectra to be analyzed, and this can be accomplished with the automated peak-picking program APES (two-letter-code ae). Then, an assignment job is submitted to the PINE web server (Supplementary Fig. S1) by clicking the Submit button. A unique Key identifier generated by the PINE-SPARKY importer (Fig. 1F) handles cross-talk between PINE-SPARKY.2 and the PINE web server. PINE-SPARKY.2 checks the status file from the URL associated with the Key, and the PINE web server updates the status of the PINE job in the status file. Predictions of secondary structures (PECAN), referencing errors (LACS), hydrophobicities (PACSY), torsion angles and flexibilities (TALOS-N), and 3 D structures (CS-ROSETTA) are executed sequentially by BASH and PYTHON scripts based on the chemical shifts with the highest probabilities given by PINE (described more fully in the manual). By clicking the Check button, the PINE-SPARKY importer retrieves the results. Replacement of previously downloaded results can be accomplished by clicking the Browse button before clicking on Check. The PINE-SPARKY importer automatically sets up visualization of the PINE results and incorporates them into the current project. It asks a series of interactive yes/no questions to determine whether the user wants to (i) download the results in the PINE sub-directory under working directory; (ii) visualize secondary structures determined by the PECAN algorithm (Supplementary Fig. S2A); (iii) visualize PINE probabilities for spin system assignments (Supplementary Fig. S2B); (iv) visualize chemical shift referencing analysis by the LACS algorithm (Supplementary Fig. S2C);(v) visualize hydrophobic core residues predicted from PACSY database (Supplementary Fig. S2D; Lee et al., 2012); (vi) visualize RCI S2 (random coil index order parameter; Berjanskii and Wishart, 2005); and/or (vii) generate PINE probabilistic labels and accept the most probable ones with P > 0.5 (Supplementary Fig. S3). Then the Completeness Counter (two-letter-code cm) can be used to find unassigned resonances (Fig. 1G).
As a test of PINE-SPARKY.2, we used data from three multidimensional NMR spectra (2 D 1 H, 15 N-HSQC, 3 D CBCA(CO)NH and HNCACB) from the small (110 amino acid residue) protein AeSCP-2 (BMRB Entry 16662) as inputs for automated assignment and fed the assignment results into CS-Rosetta for structure determination from chemical shifts alone. We compared the resulting structure (Fig. 1H) with that determined manually from NOE data (PDB ID 2KSH; Singarapu et al., 2010). Following superposition, the pairwise backbone RMSD for the two structures was 1.21 Å and the all-heavy-atom RMSD was 2.14 Å (Supplementary Fig. S4; see the Supplementary Material for details).
Supplementary Material
Acknowledgements
PINE-SPARKY.2 utilizes the CS-Rosetta web server service provided by BioMagResBank (https://csrosetta.bmrb.wisc.edu/csrosetta); we are grateful to Jon Wedell for its maintenance.
Funding
Supported by the United States National Institutes of Health (P41GM103399).
Conflict of Interest: none declared.
References
- Bahrami A. et al. (2009) Probabilistic interaction network of evidence algorithm and its application to complete labeling of peak lists from protein NMR spectroscopy. PLOS Comput. Biol., 5, e1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berjanskii M.V., Wishart D.S. (2005) A simple method to predict protein flexibility using secondary chemical shifts. J. Am. Chem. Soc., 127, 14970–14971. [DOI] [PubMed] [Google Scholar]
- Dashti H. et al. (2016) Probabilistic validation of protein NMR chemical shift assignments. J. Biomol. NMR, 64, 17.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eghbalnia H.R. et al. (2005) Protein energetic conformational analysis from NMR chemical shifts (PECAN) and its use in determining secondary structural elements. J. Biomol. NMR, 32, 71–81. [DOI] [PubMed] [Google Scholar]
- Lee W. et al. (2009) PINE-SPARKY: graphical interface for evaluating automated probabilistic peak assignments in protein NMR spectroscopy. Bioinformatics, 25, 2085–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. et al. (2012) PACSY, a relational database management system for protein structure and chemical shift analysis. J. Biomol. NMR, 54, 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. et al. (2014) PONDEROSA-C/S: client–server based software package for automated protein 3D structure determination. J. Biomol. NMR, 60, 73–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. et al. (2015) NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics, 31, 1325–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. et al. (2016a) Integrative NMR for biomolecular research. J. Biomol. NMR, 64, 307–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. et al. (2016b) The AUDANA algorithm for protein 3D structure determi-nation from NMR NOE data. J. Biomol. NMR, 65, 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Bax A. (2013) Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J Biomol NMR, 56, 227–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y. et al. (2008) Consistent blind protein structure generation from NMR chemical shift data. Proc. Natl. Acad. Sci., 105, 4685–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J. et al. (2008) Structural proteomics by NMR spectroscopy. Expert Rev. Proteomics, 5, 589–601. [DOI] [PubMed] [Google Scholar]
- Singarapu K.K. et al. (2010) Differences in the structure and dynamics of the apo- and palmitate-ligated forms of Aedes aegypti sterol carrier protein 2 (AeSCP-2). J. Biol. Chem., 285, 17046–17053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. et al. (2005) Linear analysis of carbon-13 chemical shift differences and its application to the detection and correction of errors in referencing and spin system identifications. J. Biomol. NMR, 32, 13–22. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.