Abstract
Motivation
Across biology, we are seeing rapid developments in scale of data production without a corresponding increase in data analysis capabilities.
Results
Here, we present Aether (http://aether.kosticlab.org), an intuitive, easy-to-use, cost-effective and scalable framework that uses linear programming to optimally bid on and deploy combinations of underutilized cloud computing resources. Our approach simultaneously minimizes the cost of data analysis and provides an easy transition from users’ existing HPC pipelines.
Availability and implementation
Data utilized are available at https://pubs.broadinstitute.org/diabimmune and with EBI SRA accession ERP005989. Source code is available at (https://github.com/kosticlab/aether). Examples, documentation and a tutorial are available at http://aether.kosticlab.org.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Aether
Data accumulation is exceeding Moore’s law, which only still progresses due to advances in parallel chip architecture (Esmaeilzadeh et al., 2013). Fortunately, the shift away from in-house computing clusters to cloud infrastructure has yielded approaches to computational challenges in biology that both make science more reproducible and eliminate time lost in high-performance computing queues (Beaulieu-Jones and Greene, 2017; Garg et al., 2011); however, existing off-the-shelf tools built for cloud computing often remain inaccessible, cumbersome, and in some instances, costly.
Solutions to parallelizable compute problems in computational biology are increasingly necessary; however, batch job-oriented cloud computing ystems, such as Amazon Web Services (AWS) Batch, Google preemptible Virtual Machines (VMs), Apache Spark and MapReduce implementations are either closed source, restrictively licensed, or locked in their own ecosystems making them inaccessible to many bioinformatics labs (Shvachko et al., 2010; Yang et al., 2007). Other approaches for bidding on cloud resources exist, but they neither provide implementations nor interface with a distributed batch job process with a backend implementation of all necessary networking (Andrzejak et al., 2010; Tordsson et al., 2012; Zheng et al., 2015).
Our proposed tool, Aether, leverages a linear programming (LP) approach to minimize cloud compute cost while being constrained by user needs and cloud capacity, which are parameterized by the number of cores, RAM, and in-node solid-state drive space. Specifically, certain types of instances are allocated to large web service providers (e.g. Netflix) and auctioned on a secondary market when they are not fully utilized (Zheng et al., 2015). Users bid amongst each other for use of this already purchased but unused compute time at extremely low rates (up to 90% off the listed price; https://aws.amazon.com/ec2/pricing/). However, this market is not without its complexities. For instance, significant price fluctuations, up to an order of magnitude, could lead to early termination of multi-hour compute jobs (Fig. 1A). Clearly, bidding strategies must be dynamic to overcome such hurdles.
Fig. 1.
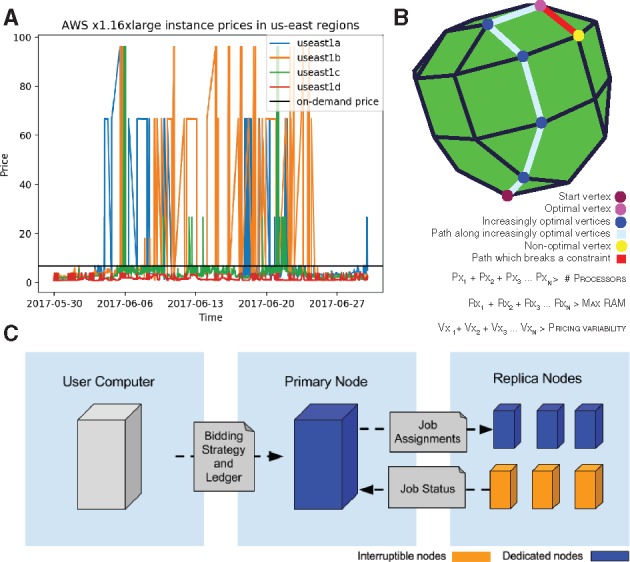
Overview of Aether. (A) Pricing history of an x1.16xlarge EC2 Instance showcasing variability of an order of magnitude, in both directions, for spot prices. (B) Simplified example showing three constraints on a sample bidding approach minimizing an objective function cTx considering cost according to a system of constraints represented as inequalities. x1, x2, … xn represent the number of specific types of compute nodes to solve for. Each inequality represents a constraint and adds another dimension to the space which the simplex algorithm needs to traverse vertices in to find ideal solution. The green line represents the optimal solution. (C) General overview of Aether
Aether consists of bidder and batch job processing command line tools that query instance metadata from the vendor application programming interface (APIs) to formulate the LP problem. LP is an optimization method that simultaneously solves a large system of equations to determine the best outcome of a scenario that can be described by linear relationships. The Aether bidder, described in detail in the Supplementary Methods, generates and solves a system of 140 inequalities using the simplex algorithm (Fig. 1B). For the purposes of reproducibility, an implementation of the bidder using CPLEX is also provided as an optional command line flag.
Subsequently, the replica nodes specified by the LP result are placed under the control of a primary node, which assigns batch processing jobs over transmission control protocol, monitors for any failures, gathers all logs, sends all results to a specified cloud storage location, and terminates all compute nodes once processing is complete (Fig. 1C). Additionally, Aether is able to distribute compute across multiple cloud providers. Sample code for this is provided with the Aether implementation although it was not utilized in our reported tests due to cost feasibility. Our implementation runs on any Unix-like system; we ran our pipeline and cost analysis using AWS but have provided code to spin up compute nodes on either Microsoft Azure or on a user's local physical clusters.
To test our bidding approach and batch job pipeline at scale, we used our framework to de novo assemble and annotate 1572 metagenomic, longitudinal samples from the stool of 222 infants in Northern Europe (Supplementary Fig. S1; Bäckhed et al., 2015; Kostic et al., 2015; Vatanen et al., 2016; Yassour et al., 2016). The sequencing data within datasets from the DIABIMMUNE consortium ranged from 4680 to 22, 435, 430 reads/sample with a median of 19, 020, 036 reads/sample. Assemblies were performed with MEGAHIT and annotations were done with PROKKA (Li et al., 2015; Seemann, 2014).
Metagenomic data, typically shotgun DNA sequencing of microbial communities, is difficult to analyze because of the enormous amounts of compute required to naively assemble short sequence reads into large contiguous spans (contigs) of DNA. To accomplish our assemblies, our bidding algorithm suggested that the optimal strategy would be to spin up 30TB of RAM across underutilized compute nodes. Our networked batch job processing module utilized these nodes for 13 h and yielded an assembly and annotation cost of ∼US$0.30 per sample (Supplementary Fig. S2). Theoretically, the pipeline can complete in the time it takes for the longest sub-process (i.e. assembly in this case) to finish (∼7 h). Spinning up the same nodes for this long without a bidding approach would cost ∼US$1.60 per sample (Supplementary Fig. S3). In order for on-site hardware to achieve the same cost efficiency as our pipeline, one would have to carry out on the order of 1 million assemblies over the lifespan of the servers, a practically insurmountable task (Supplementary Fig. S2). Such efficiency in both time and cost at scale is unprecedented. In fact, due to resource paucity, computational costs have forced the field of metagenomics to rely on algorithmic approaches that utilize mapping back to reference genomes rather than de novo methods (Truong et al., 2015).
Additional testing of Aether showed marginally better relative cost savings (compared to the assembly example) when tasked with aligning braw reads to the previously assembled genomes with BWA-MEM (Li, 2009; http://arxiv.org/abs/1303.3997); this is not surprising as shorter computational tasks are less sensitive to the risks of early spot instance termination. Additionally, in simulated runs of the bidder incorporating pricing history from periods where ask prices were approximately an order of magnitude higher than normal on the east coast of the United States (Fig. 1A), Aether suggested utilization of different instance types that would have resulted in similar cost and time to completion as our actual run. To allow users to make optimal usage of these benefits, the ability to simulate bidding for different timeframes is included as a feature. By not having to potentially re-run analysis pipelines (due to being outbid on compute during runtime), we claim that utilizing Aether leads to a reduction of market inefficiencies. We have both qualitatively and empirically compared Aether to existing AWS tools such as AWS Batch and Spot Fleet Pricing (Supplementary Fig. S1.3 and Supplementary Methods). Additionally, where empirical validation of benefit was possible, we have iterated on previous work and incorporated strategies such as basing a subset of constraints on service level agreements (Andrzejak et al., 2010). Future directions include training the bidding algorithm to predict its own effect on pricing variability when being utilized at massive scale as well as distributing compute nodes across datacenters when enough resources are being spun up to strongly influence the market.
To our knowledge, this is the first implementation of a bidding algorithm for cloud compute resources that is tied both to an easy-to-use front-end as well as a distributed backend that allows for spinning up purchased compute nodes across multiple providers. Conceivably, this tool can be applied to any number of disciplines, bringing cost-effective cloud computing into the hands of scientists in fields beyond biology.
Supplementary Material
Acknowledgements
We thank Thomas Lane and Chengwei Luo for their feedback and review of the manuscript. We thank Tommi Vatanen for helping us with data access.
Funding
This work was funded by National Institutes of Health/National Human Genome Reasearch Institute (NIH/NHGRI) T32 HG002295, PI: Park, Peter J (J.M.L.); an AWS Research Credits for Education Grant (J.M.L. and A.D.K.); a Microsoft Azure for Research Grant (B.T.T. and C.J.P.), NIH National Institute of Environmental Health Sciences (NIEHS) R00 ES023504 (C.J.P.); NIEHS R21 ES025052 (C.J.P.); National Science Foundation (NSF) Big Data Spoke grant (C.J.P.), a Smith Family Foundation Award for Excellence in Biomedical Research (A.D.K.); and an American Database Association (ADA) Pathway to Stop Diabetes Initiator Award (A.D.K.).
Conflict of Interest: none declared.
References
- Andrzejak A. et al. (2010) Decision model for cloud computing under SLA constraints. In: 2010 IEEE International Symposium on Modeling, Analysis and Simulation of Computer and Telecommunication Systems. pp. 257–266. IEEE.
- Bäckhed F. et al. (2015) Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microb., 17, 852. [DOI] [PubMed] [Google Scholar]
- Beaulieu-Jones B.K., Greene C.S. (2017) Reproducibility of computational workflows is automated using continuous analysis. Nat. Biotechnol., 35, 342–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmaeilzadeh H. et al. (2013) Power challenges may end the multicore era. Commun. ACM, 56, 93–102. [Google Scholar]
- Garg S.K. et al. (2011) Environment-conscious scheduling of HPC applications on distributed Cloud-oriented data centers. J. Parallel Distrib. Comput., 71, 732–749. [Google Scholar]
- Kostic A.D. et al. (2015) The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microb., 17, 260–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D. et al. (2015) MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics, 31, 1674–1676. [DOI] [PubMed] [Google Scholar]
- Li H. et al. (2009). Fast and accurate short read alignment with Burrows Wheeler transform. Bioinformatics, 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics, 30, 2068–2069. [DOI] [PubMed] [Google Scholar]
- Shvachko K. et al. (2010) The Hadoop distributed file system. In: 2010 IEEE 26th Symposium on Mass Storage Systems and Technologies (MSST). pp. 1–10, IEEE.
- Tordsson J. et al. (2012) Cloud brokering mechanisms for optimized placement of virtual machines across multiple providers. Future Gener. Comput. Syst., 28, 358–367. [Google Scholar]
- Truong D.T. et al. (2015) MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods, 12, 902–903. [DOI] [PubMed] [Google Scholar]
- Vatanen T. et al. (2016) Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell, 165, 1551. [DOI] [PubMed] [Google Scholar]
- Yang H.-C. et al. (2007) Map-reduce-merge: simplified relational data processing on large clusters. In: Proceedings of the 2007 ACM SIGMOD International Conference on Management of Data, SIGMOD ’07. New York, NY, USA, pp. 1029–1040. ACM.
- Yassour M. et al. (2016) Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci. Transl. Med., 8, 343ra81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L. et al. (2015) How to bid the cloud. ACM SIGCOMM Comput. Commun. Rev., 45, 71–84. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


