Abstract
Summary
GeoBoost is a command-line software package developed to address sparse or incomplete metadata in GenBank sequence records that relate to the location of the infected host (LOIH) of viruses. Given a set of GenBank accession numbers corresponding to virus GenBank records, GeoBoost extracts, integrates and normalizes geographic information reflecting the LOIH of the viruses using integrated information from GenBank metadata and related full-text publications. In addition, to facilitate probabilistic geospatial modeling, GeoBoost assigns probability scores for each possible LOIH.
Availability and implementation
Binaries and resources required for running GeoBoost are packed into a single zipped file and freely available for download at https://tinyurl.com/geoboost. A video tutorial is included to help users quickly and easily install and run the software. The software is implemented in Java 1.8, and supported on MS Windows and Linux platforms.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Locations of infected hosts (LOIH) are critical pieces of metadata required for exploring the spread and evolutionary dynamics of pathogens such as viruses. This information is often retrieved from GenBank, a public database of nucleotide sequences which is maintained by the National Center of Biotechnology Information (NCBI) (Benson et al., 2013). Researchers have used the geospatial metadata in virus GenBank records for a wide range of public health studies. For instance, the LOIH of viruses based on GenBank metadata have been used to map the global spread of viruses (Messina et al., 2014), investigate the environmental predictors of virus diffusion (Magee et al., 2015), and trace the origin of infectious disease outbreaks (Wallace and Fitch, 2008).
Currently, the extraction of the LOIH of viruses is performed manually and requires a significant investment of time and effort. The designated field for storing the LOIH of viruses in GenBank records is the country field, which despite its name, may contain geospatial metadata of varying degrees of specificity. For instance, the country field of the GenBank record with accession no. CY058987 (https://www.ncbi.nlm.nih.gov/nuccore/CY058987) contains the province-level metadata ‘China: Hubei’. Due to the nature of virus nomenclature, additional geospatial metadata may often be found in the strain and isolate fields of GenBank records. In the aforementioned GenBank record, the strain field contains the location ‘Wuhan’ embedded in the strain name ‘A/Wuhan/390/2005’. Thus, researchers often need to manually integrate locations from different GenBank record fields to retrieve the most specific spatial resolution. Other times, the geospatial metadata available in GenBank is not sufficient for a given study (Scotch et al., 2011). For instance, a researcher modeling the spread of the rabies virus within an US state would likely need at least the county-level LOIH of all virus samples, but GenBank may not contain such precise information. In such cases, researchers often search full-text publications linked to GenBank for more specific information. Moreover, depending on the type of study, they may also need to normalize each extracted LOIH to its latitude/longitude coordinates (e.g. for continuous phylogeography) or to a standardized string representation (e.g. for discrete phylogeography). The ambiguity of locations (e.g. Paris can be in France or Texas, USA) makes this task especially challenging.
We present GeoBoost, a knowledge-driven framework for automatically extracting, integrating and normalizing the LOIH of viruses from GenBank records and related full-text articles. It builds upon our prior work in this area (Tahsin et al., 2016), including additional features to enhance its usability and performance. To the best of our knowledge, this is the only publicly accessible software available for this task. Related work has been performed to extract, and often normalize, different forms of GenBank metadata (Carter and Gatherer, 2016; Chen and Sarkar, 2011; Gratton et al., 2017; Sarkar, 2010). For instance, the Tempus et Locus (TeL) software was developed to extract GenBank sequences containing the date of collection and/or location of sampling of the sequence (Carter and Gatherer, 2016). However, GeoBoost is the only software we know of which attempts to enhance existing geographic metadata in GenBank by extracting and integrating geographic information from related full-text publications.
In addition to outputting the most specific and most probable LOIH of a virus, GeoBoost also assigns a probability score to each possible LOIH of the virus based on its specificity (e.g. Phoenix is more specific than Arizona) and likelihood of being correct. Researchers can then use these scores for selection of geographic locations to build precise models of virus spread. Additionally, if no further location information can be found, it will also indicate this ‘failure’, saving researchers additional time in ruling out locations. GeoBoost is easy to install and run, and could accelerate bioinformatics research that incorporates the LOIH of viruses.
2 Materials and methods
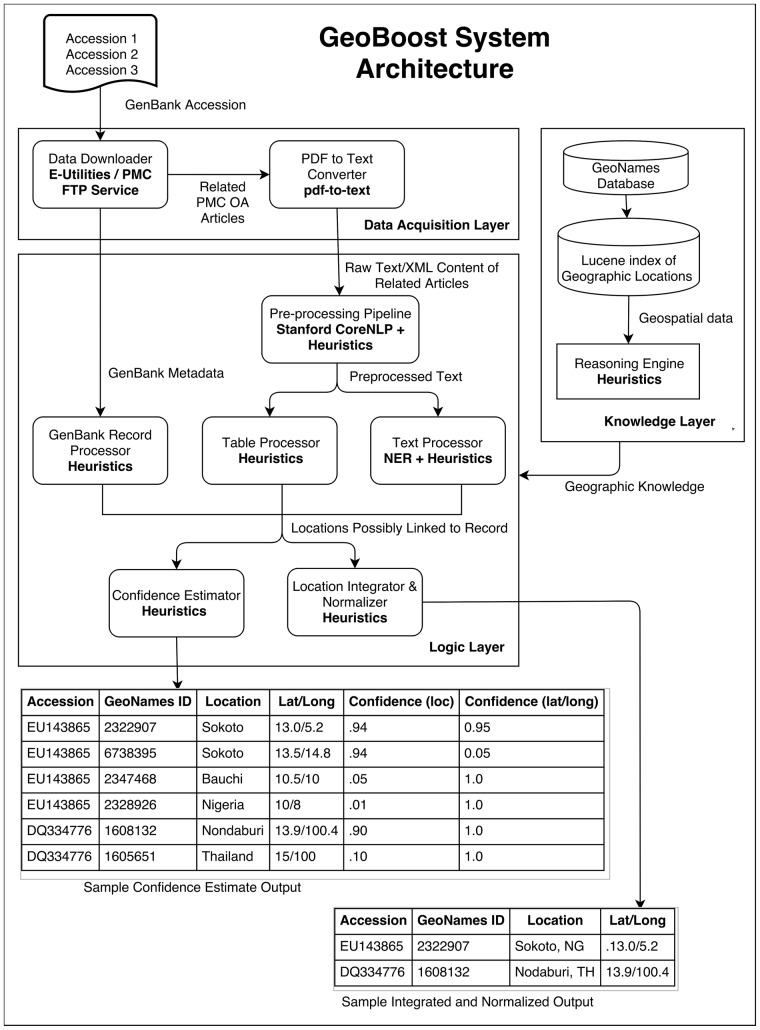
GeoBoost (see Fig. 1) is a knowledge-driven framework and central to its function is a knowledge base (KB) of geographic locations. Our KB is primarily based on the GeoNames.org database, which contains geospatial data for over 10 million geographic locations. Given a list of GenBank accession numbers, GeoBoost uses the Entrez Programming Utilities (Sayers, 2008) to download relevant metadata from the corresponding GenBank records. It also downloads all PubMed Central (PMC) Open Access articles linked to each record in both PDF and XML format, if available, and converts the PDF files to text files using the pdf-to-text software (http://www.foolabs.com/xpdf/home.html). It then uses knowledge-driven heuristics to extract and integrate geospatial metadata from relevant fields in each GenBank record, until a user-provided sufficiency criterion is satisfied. For instance, if the sufficiency criterion is ‘ADM1’ (i.e. states or provinces of a country), GeoBoost will stop searching once it finds ADM1-level or more specific geographic information. If GeoBoost fails to find sufficient geospatial metadata for a record even after analyzing all relevant fields, it proceeds to search the free-text and tabular content of linked articles. If GeoBoost is not given a sufficiency criterion, it searches all available sources for the most specific LOIH.
Fig. 1.
GeoBoost System Architecture. Given a user-provided list of GenBank accession numbers corresponding to viruses, the Logic Layer uses the geographic knowledge provided by the Knowledge Layer, and the GenBank metadata and PMC OA articles downloaded by the Data Acquisition layer to output: (i) the most probable, integrated, normalized location of infected host (LOIH) of each virus (integrated and normalized output), and (ii) the probability scores of each possible LOIH of each virus (confidence estimate output). More detailed information about GeoBoost architecture is provided in Online Appendix A
When searching the tabular content of an article associated with a GenBank record, GeoBoost uses simple rules to analyze the structure and content of each table and extract possible links between GenBank records and geographic locations. When searching the free-text content of a related article, GeoBoost applies a Named Entity Recognition system (Weissenbacher et al., 2015) for detecting geographic location mentions in text, and uses rule-based heuristics to determine which of the extracted locations are more likely to be linked to the record.
After extracting locations from all possible sources, GeoBoost integrates and normalizes them, and outputs the most probable and most specific LOIH of each virus (P location | GenBank record). For instance, if it extracted ‘USA’ from the GenBank record, ‘Paris’ from the free-text content of a related article, and ‘Texas’ from the tabular content of a related article, it would output ‘Paris, Texas, USA’, given there is more evidence for the later than for ‘Paris, France’. GeoBoost normalizes each LOIH to its corresponding GeoNames ID and latitude/longitude coordinates based on rule-based heuristics. It also assigns probability scores to each possible LOIH of a virus using a complex set of heuristics that assigns higher scores to more accurate and more specific locations. The probability score assignment process is performed in two stages. In the first stage, GeoBoost assigns probability scores to every location extracted by the pipeline from either the GenBank record or linked article. In the next stage, it assigns probability scores to all possible latitude/longitude pairs associated with each candidate location in GeoNames. Probability scores assigned in both steps add up to 1.0.
To estimate the performance of GeoBoost, we used two different manually annotated sets of GenBank records, created through our prior work (Tahsin et al., 2016). The first set (Flu) included 5728 GenBank records corresponding to influenza viruses. We annotated the LOIH of the viruses in this set based on information in the GenBank records and 60 full-text PMC articles linked to these records. The second set (Non-Flu) included 100 GenBank records corresponding to six different non-influenza viruses. We annotated the LOIH of the viruses in this set based on information in the GenBank records and 10 full-text PMC articles linked to these records. For accuracy, we calculated the percentage of the records for which the top ranked latitude/longitude coordinates outputted by GeoBoost was within 50 miles of the manually annotated latitude/longitude coordinates. We also calculated the time taken by GeoBoost to process each record using a JVM heap size of 1GB, a download speed of ∼100 Mbps, and the Windows 10 Operating System. We measured the accuracy and time taken by GeoBoost for each dataset under two different settings: (i) when configured to download and extract information from related PMC OA articles along with GenBank metadata (default configuration), (ii) when configured to use GenBank metadata only. This allowed us to assess the added benefit of extracting and integrating information from related articles, in addition to using GenBank metadata.
3 Results
In Table 1, we show the results of our evaluation. Under default configuration, GeoBoost had a high level of accuracy for both test sets (81% for Flu and 80% for Non-Flu), and took less than 5 s to process each record (1.59 s for Flu and 4.65 s for Non-Flu). When configured to exclude information from related PMC OA articles, GeoBoost’s accuracy fell by 11% for the Flu set and 25% for the Non-Flu set, demonstrating the value of extracting and integrating additional information from related articles.
Table 1.
Performance evaluation of GeoBoost relative to manually annotated gold standard
| Test set | Accuracy |
Time per record |
||
|---|---|---|---|---|
| Including PMC OA (%) | Excluding PMC OA (%) | Including PMC OA | Excluding PMC OA | |
| Flu_Set | 81 | 70 | 1.59s | 1.01s |
| Non-Flu_Set | 80 | 55 | 4.65s | 1.40s |
| Average | 80.5 | 62.5 | 3.12s | 1.21s |
Supplementary Material
Acknowledgements
We would like to thank Robert Rivera, Rachel Beard, Mari Firago and Dr. Garrick Wallstrom for their contributions. R.R., R.B. and M.F. annotated data for the test set. R.R. and G.W. contributed to confidence estimate process. R.R. reviewed the paper.
Funding
Research reported here was supported by the NIAID of the NIH under Award Number R01 AI117011 to G.G. and M.S. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Conflict of Interest: none declared.
References
- Benson D.A. et al. (2013) GenBank. Nucleic Acids Res., 41, D36–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter A.R., Gatherer D. (2016) Tempus et Locus: a tool for extracting precisely dated viral sequences from GenBank, and its application to the phylogenetics of primate erythroparvovirus 1 (B19V). bioRxiv doi: 10.1101/061697. [Google Scholar]
- Chen E.S., Sarkar I.N. (2011) Towards structuring unstructured genbank metadata for enhancing comparative biological studies. AMIA Jt. Summits. Transl. Sci. Proc., 2011, 6–10. [PMC free article] [PubMed] [Google Scholar]
- Gratton P. et al. (2017) A world of sequences: can we use georeferenced nucleotide databases for a robust automated phylogeography? J. Biogeogr., 44, 475–486. [Google Scholar]
- Magee D. et al. (2015) Combining phylogeography and spatial epidemiology to uncover predictors of H5N1 influenza A virus diffusion. Arch. Virol., 160, 215–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina J.P. et al. (2014) Global spread of dengue virus types: mapping the 70 year history. Trends Microbiol., 22, 138–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar I.N. (2010) Leveraging biomedical ontologies and annotation services to organize microbiome data from Mammalian hosts. AMIA Annu. Symp. Proc., 2010, 717–721. [PMC free article] [PubMed] [Google Scholar]
- Sayers E. (2008) E-utilities quick start. In: Entrez Programming Utilities Help. National Center for Biotechnology Information, Bethesda, MD. [Google Scholar]
- Scotch M. et al. (2011) Enhancing phylogeography by improving geographical information from GenBank. J. Biomed. Inform., 44) (suppl 1, S44–S47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahsin T. et al. (2016) A high-precision rule-based extraction system for expanding geospatial metadata in GenBank records. J. Am. Med. Informatics Assoc., 23, 934–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace R.G., Fitch W.M. (2008) Influenza A H5N1 immigration is filtered out at some international borders. PLoS One, 3, e1697.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenbacher D. et al. (2015) Knowledge-driven geospatial location resolution for phylogeographic models of virus migration. Bioinformatics, 31, i348–i356. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.