Abstract
Interleukin (IL)-11 is a multifunctional cytokine that was traditionally recognized for its hematopoietic and anti-inflammatory functions, but has recently been shown also to be involved in tumorigenesis. IL-11 signaling is initiated by binding of the cytokine to the IL-11 receptor (IL-11R), which is not directly involved in signaling but required for IL-11 binding to the signal-transducing receptor glycoprotein (gp) 130. In classic signaling, IL-11 binds to the membrane-bound IL-11R to initiate signal transduction. Additionally, IL-11 signaling can be initiated via soluble IL-11R, known as trans-signaling, and this pathway only requires the three extracellular domains of the IL-11R, but not stalk, transmembrane, or intracellular region. Here, we analyzed the role of the IL-11R stalk region, a 55 amino acid stretch connecting the extracellular domains with the transmembrane helix, in classic IL-11 signaling with the help of cytokine-dependent cell lines. We showed that the stalk region is crucial for IL-11 signaling via the membrane-bound IL-11R. Using different deletion variants, we found that a minimal length of 23 amino acid residues is required for efficient signal transduction. We further found that classic IL-11 signaling depended solely on the length, but not the sequence, of the IL-11R stalk region, suggesting that the stalk functions as a spacer in the signaling complex. We previously described the IL-11R stalk region as determinant of proteolysis and regulator of IL-11 trans-signaling. The results presented here reveal an additional function in classic IL-11 signaling, highlighting the importance of the IL-11R stalk in IL-11 signaling.
Keywords: interleukin, STAT3, cytokine, proliferation, signal transduction, interleukin-11, interleukin-11 receptor, gp130, stalk region
Introduction
Interleukin (IL)-11 is a cytokine which had initially been described as a hematopoietic and anti-inflammatory factor (1–3). Recent studies, however, connect dysregulated IL-11 signaling with tumorigenesis, especially in the gastrointestinal tract (4–7). For signal transduction, IL-11 has to first bind to its nonsignaling α-receptor, the IL-11 receptor (IL-11R)2. This complex then recruits two molecules of the signal-transducing β-receptor glycoprotein (gp) 130, which dimerize and thus initiate intracellular signaling cascades (8, 9). IL-11 mainly induces the Janus kinase/signal transducers and activators of transcription (Jak/STAT) pathway, but has also been shown to activate the mitogen-activated protein kinase/extracellular regulated kinase (MAPK/Erk), phosphoinositide 3-kinase (PI3K), and yes-associated protein (YAP) pathways (10–12).
IL-11 is a member of the IL-6 family of cytokines, which is characterized by their α-helical fold in an up-up-down-down topology and their common use of gp130 as signal-transducing receptor. This family also contains IL-6, IL-27, leukemia inhibitory factor (LIF), oncostatin M (OSM), ciliary neurotrophic factor (CNTF), and cardiotrophin-like cytokine (CLC), which all signal via different receptor complexes that contain one or two molecules gp130. Intriguingly, IL-11 and IL-6 are the only known cytokines that initiate signal transduction via a gp130 homodimer; all other cytokines utilize heterodimers of gp130 in combination with a second signal-transducing receptor (10). A prerequisite for gp130 dimerization is binding of IL-11 to the IL-11R, which is not directly involved in signal transduction but is required for formation of the signaling complex, as IL-11 alone cannot bind to gp130 (13, 14). Although gp130 is ubiquitously expressed, the IL-11R is only found on a limited number of cell types, which restricts the cells that can respond to IL-11.
The IL-11R consists of three extracellular domains (D1–D3), which are connected to a transmembrane region and an intracellular part through a 55 amino acid residue–long stalk region (15). For binding of IL-11, the fibronectin-type III (FNIII)–like domains D2 and D3, which harbor the cytokine-binding module (CBM), are essential, whereas the Ig-like D1 domain has been reported to be negligible for signaling (16, 17). The stalk region is also not directly involved in signal transduction, because recombinant soluble IL-11R, which only consists of the three extracellular domains, is able to bind IL-11 and induce signal transduction (17, 18). We have recently shown that the IL-11R can be cleaved off the cell surface by ectodomain shedding, and that the liberated soluble IL-11R ectodomain is able to bind IL-11 and induce signaling via the so-called trans-signaling pathway (19). However, the function of the stalk region in signaling via the membrane-bound IL-11R, referred to as classic signaling, has not been determined.
It has been hypothesized that the stalk region is required for positioning of the extracellular domains, because cytokines bind to the domains D2 and D3 of gp130 (20), which are separated from the membrane by three FNIII-like domains (D4–D6). Previous modeling approaches suggested that gp130 is flexible and adapts its overall structure upon binding of the IL-11/IL-11R complex (8), and that the domains D4, D5, and D6 can span a distance between 83 and 96 Å, depending on their conformation (21).
It was previously shown that IL-6 classic signaling, which requires a signaling complex that is similar to that of IL-11, depends on an IL-6R stalk region which spans 83.6 Å (22 amino acid residues) (21). However, the crystal structure of IL-11 revealed differences from IL-6 in terms of structure and receptor-binding site features, suggesting that IL-11 and IL-6 engage gp130 differently (22). To elucidate these potential differences further, we deleted different stretches of the IL-11R stalk region and analyzed whether and how these deletions influence classic IL-11 signaling. We found that deletion of the majority of the stalk prevents signaling, and that the minimum required length for a full response to IL-11 is 23 amino acid residues, corresponding to ∼87.4 Å. We further show that the signaling capacity depends solely on the length, but not the sequence, of the stalk region.
Results
Deletion of the IL-11R stalk region prevents classic IL-11 signaling
We have previously shown that the length of the stalk region of the IL-6R is critical for IL-6 classic signaling (21). Like IL-6, IL-11 signals via a gp130 homodimer and requires the preceding binding to a nonsignaling α-receptor. Because the crystal structure of IL-11 suggests differences in the binding of the cytokine to gp130 compared with IL-6, we sought to analyze whether the stalk region of the IL-11R serves a similar purpose as the stalk of the IL-6 in the correct positioning of the extracellular domains toward gp130, and whether there is also a critical length of the stalk region that is needed to allow IL-11 classic signaling.
To analyze the role of the IL-11R stalk region in classic IL-11 signaling, we made use of the cytokine-dependent cell line Ba/F3-gp130 (23), which has been stably transduced with IL-11R (termed Ba/F3–gp130–IL-11R) or variants thereof. Ba/F3-gp130 cell lines, which have been transduced with biologically active variants of the IL-11R, do not express IL-11 themselves but are responsive to IL-11 (15). We further used the synthetic cytokine Hyper–IL-6 (Hy–IL-6), which consists of IL-6 connected by a linker to the soluble IL-6R. Hyper–IL-6 induces dimerization of gp130 independently of the IL-11R and hence serves as positive control. Because the individual cell lines might respond differently to cytokine treatment and especially to stimulation with IL-11, all cell lines will at least proliferate when stimulated with Hyper–IL-6, albeit the amount of proliferation might not be identical for all cell lines, which are all derived individually.
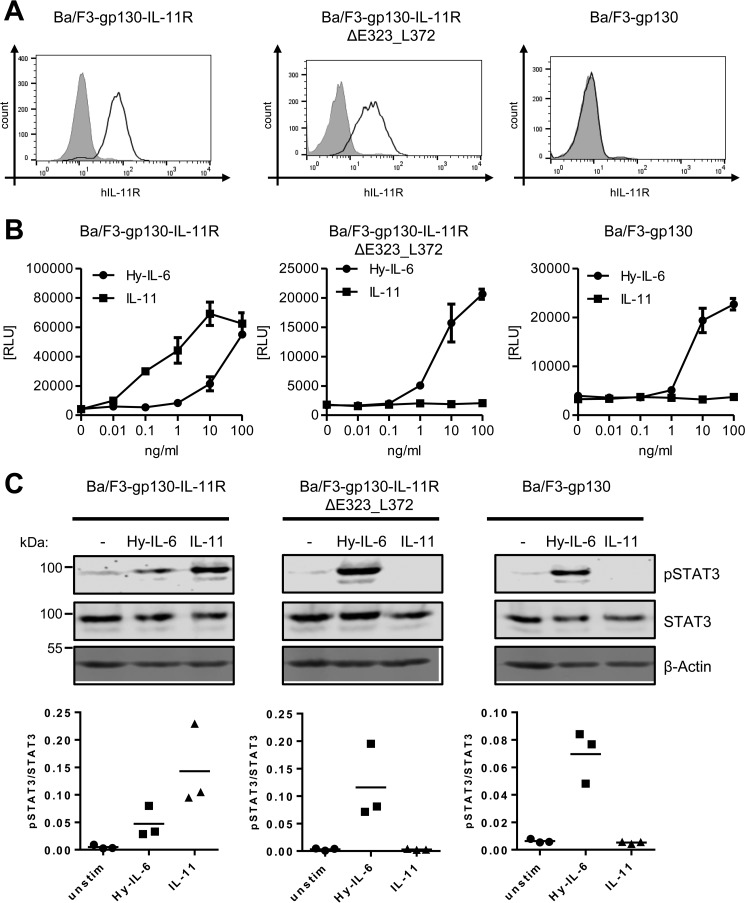
To investigate a potential role for the stalk region in classic signaling, we first deleted the amino acid residues Glu-323 to Leu-372 (IL-11RΔE323_L372), which constitute the majority of the stalk. A deletion of the entire stalk prevented transport of the receptor to the cell surface (data not shown), and was therefore not further analyzed in this study. We generated stably transduced Ba/F3–gp130–IL-11RΔE323_L372 cells and analyzed the amount of IL-11R at the cell surface via flow cytometry. The ΔE323_L372 deletion, which results in a stalk region of only five amino acid residues, did not compromise the IL-11R, as the deletion variant was detected at the cell surface in comparable amount as the WT receptor, whereas the signal was absent in the parental Ba/F3-gp130 cells (Fig. 1A). To analyze the biological activity of IL-11RΔE323_L372, we determined proliferation in response to different concentrations of IL-11 or Hy–IL-6. As shown previously, Ba/F3-gp130 cells respond to Hy–IL-6 but not to IL-11, whereas proliferation of Ba/F3–gp130–IL-11R cells can be induced by both cytokines (15) (Fig. 1B, left panel). In contrast, Ba/F3–gp130–IL-11RΔE323_L372 responded to stimulation by Hy–IL-6, but not IL-11 (Fig. 1B, middle panel), thus mimicking Ba/F3-gp130 cells (Fig. 1B, right panel). As a second readout for the biological activity of the IL-11R variants, we examined phosphorylation of the transcription factor STAT3, which is induced by IL-11. Consistent with the cytokine-dependent proliferation, we detected activation of STAT3 in Ba/F3–gp130–IL-11R cells upon stimulation with IL-11 and Hy–IL-6, but only with Hy–IL-6 in Ba/F3–gp130–IL-11RΔE323_L372 and Ba/F3-gp130 cells (Fig. 1C). These results indicate that the stalk region of the IL-11R is required for classic IL-11 signaling.
Figure 1.
IL-11R stalk is essential for classic signaling. A, cell surface expression of IL-11R variants on Ba/F3–gp130–IL-11R, Ba/F3–gp130–IL-11RΔE323_L372, and Ba/F3-gp130 cells was assessed by flow cytometry. B, equal amounts of Ba/F3–gp130–IL-11R, Ba/F3–gp130–IL-11RΔE323_L372, and Ba/F3-gp130 cells were stimulated with increasing amounts (0–100 ng/ml) of either IL-11 or Hy–IL-6 for 48 h. Concentration-dependent proliferation was determined as described in “Experimental Procedures.” Error bars represent S.D. of triplicates from one representative experiment. C, STAT3 phosphorylation of Ba/F3–gp130–IL-11R, Ba/F3–gp130–IL-11RΔE323_L372, and Ba/F3-gp130 cells in response to 10 ng/ml IL-11 or 10 ng/ml Hy–IL-6 stimulation for 15 min was determined by Western blotting. Total STAT3 was determined to ensure equal protein loading.
The signaling capacity of the IL-11R depends on the length of its stalk region
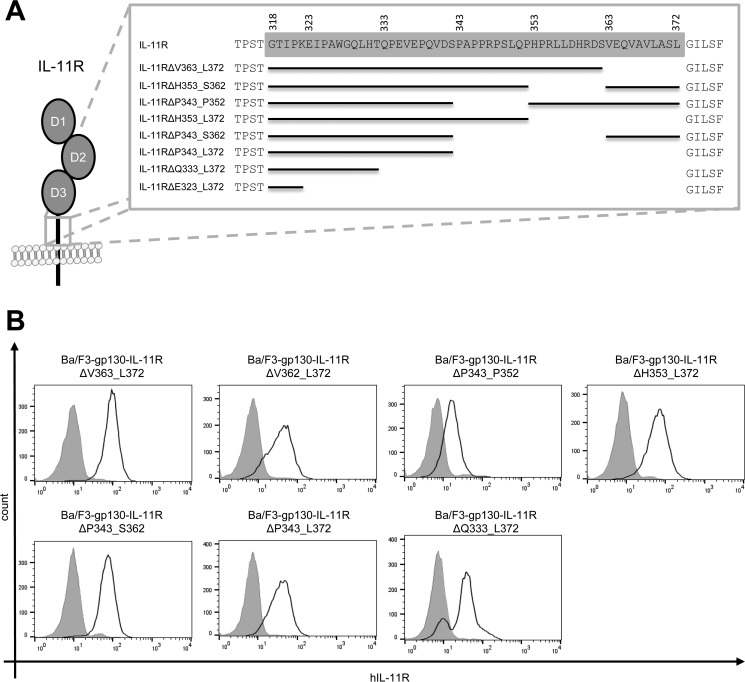
Having shown that the IL-11RΔE323_L372 variant lacks biological activity, we sought to further analyze which part and which length of the stalk region is necessary for classic signaling. As depicted in Fig. 2A, we therefore generated seven additional deletion variants of the IL-11R: IL-11RΔV363_L372, IL-11RΔH353_S362, and IL-11RΔP343_P352 all lacked 10 amino acid residues; IL-11RΔH353_L372 and IL-11RΔP343_S362 lacked 20 amino acid residues; and IL-11RΔP343_L372 and IL-11RΔQ333_L372 had 30 and 40 amino acid residues deleted, respectively. To be able to analyze IL-11–dependent signal transduction, we transduced all seven IL-11R variants stably in Ba/F3-gp130 cells, which were all efficiently transported to the cell surface as measured by flow cytometry (Fig. 2B). However, amounts of IL-11R at the cell surface varied between the individual cell lines, which could possibly influence their response toward stimulation with IL-11.
Figure 2.
Deletions in the IL-11R stalk do not compromise cell surface expression. A, schematic overview of IL-11R stalk deletions. Amino acid residues of the stalk region (spanning Gly-318 to Leu-372) are highlighted in gray. Missing stretches in the scheme indicate deletions. B, cell surface expression of the above-depicted IL-11R deletion variants stably transduced into Ba/F3-gp130 cells.
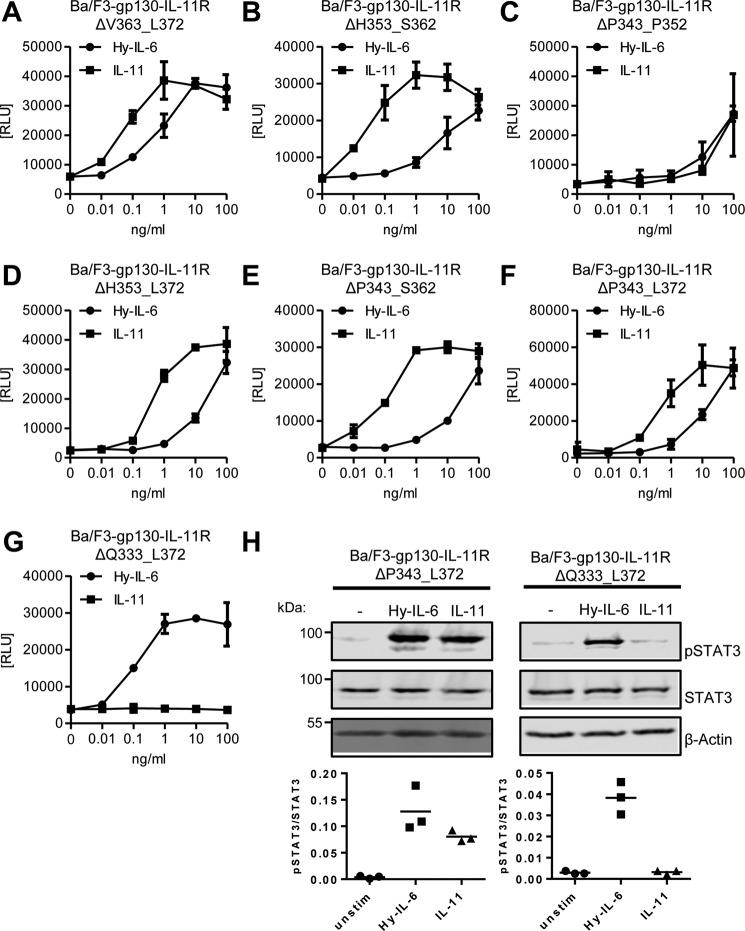
We then analyzed IL-11–dependent proliferation of all these cell lines. Deletion of 10 amino acid residues in the variants IL-11RΔV363_L372, IL-11RΔH353_S362, and IL-11RΔP343_P352 did not alter response to IL-11 (Fig. 3, A–C). Furthermore, deletion of 20 amino acid residues in IL-11RΔH353_L372 or IL-11RΔP343_S362 did not impair IL-11–induced proliferation (Fig. 3, D and E), and also removal of 30 amino acid residues was tolerated, because Ba/F3–gp130–IL-11RΔP343_L372 proliferated in a dose-dependent manner in response to IL-11 that was indistinguishable from the other cell lines tested (Fig. 3F). In contrast, deletion of 40 amino acid residues in the IL-11RΔQ333_L372 mutant led to a complete loss of IL-11–induced proliferation (Fig. 3G), which we previously also observed with largest deletion variant of 50 amino acid residues (IL-11RΔE323_L372) (Fig. 1C). Notably, these cells still responded normally to Hy–IL-6, ensuring that the cells were in principle able to proliferate when gp130 was activated by a ligand. Additionally, we analyzed STAT3 phosphorylation in Ba/F3–gp130–IL-11RΔP343_L372 and Ba/F3–gp130–IL-11RΔQ333_L372 in response to stimulation with IL-11 and Hy–IL-6. In line with the other data, deletion of 30 amino acid residues did not compromise phosphorylation upon IL-11 stimulation, whereas deletion of 40 amino acid residues rendered the cells unresponsive toward IL-11 treatment and Hy–IL-6 stimulation was intact in both cell lines (Fig. 3H). These results show that deletion of 30 amino acid residues, resulting in a stalk with a length of 25 amino acid residues, is sufficient for classic IL-11 signaling, whereas 15 remaining amino acid residues are not sufficient, indicating that classic signaling depends on the length of the stalk region.
Figure 3.
Classic IL-11 signaling depends on the stalk length. A–G, equal amounts of the different Ba/F3-gp130 cell lines were stimulated with increasing amounts (0–100 ng/ml) of either IL-11 or Hy–IL-6 for 48 h. Concentration-dependent proliferation was determined as described in “Experimental Procedures.” The analyzed cell line is given above the respective diagram. Error bars represent S.D. of triplicates from one representative experiment. H, STAT3 phosphorylation of Ba/F3–gp130–IL-11RΔP343_L372 and Ba/F3–gp130–IL-11RΔQ333_L372 cells in response to 10 ng/ml IL-11 or 10 ng/ml Hy–IL-6 stimulation for 15 min was determined via Western blotting. Total STAT3 was detected to ensure equal protein loading. The ratio of pSTAT3/STAT3 was determined from three independent experiments.
A stalk length of 23 amino acid residues is the minimal requirement for classic IL-11 signaling
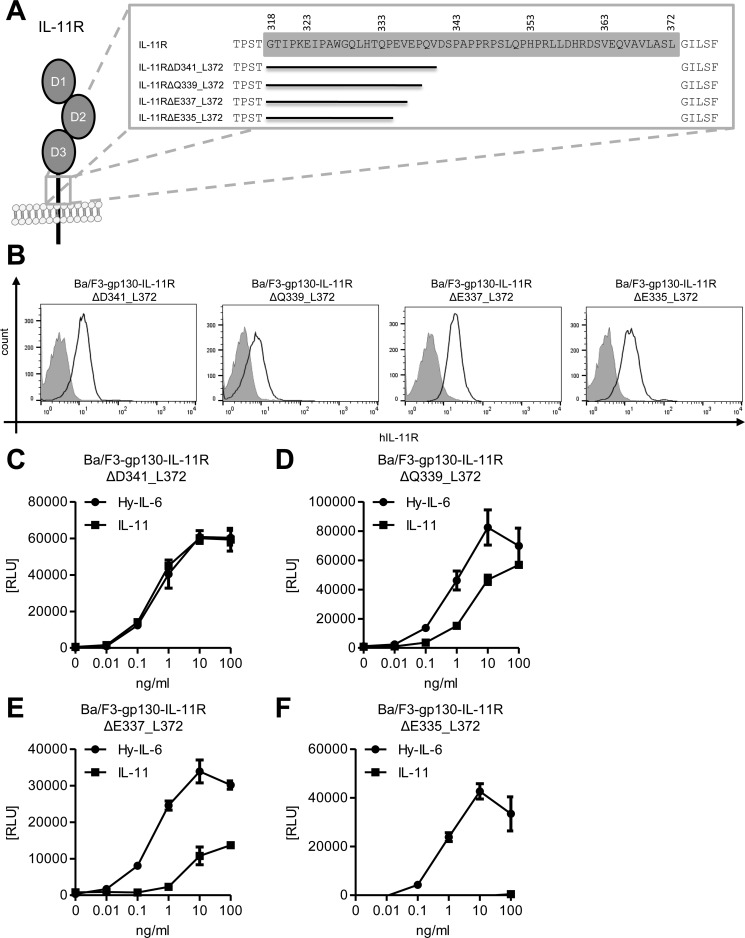
To narrow down the essential stalk length further, we made four additional stalk deletion variants, whose length was between IL-11RΔP343_L372 and IL-11RΔQ333_L372. These variants, which each differ by 2 amino acid residues in length, are depicted in Fig. 4A, and result in stalk lengths between 23 and 17 amino acid residues. As seen for all receptor variants before, these deletions also did not compromise transport of the IL-11R to the cell surface when we measured IL-11R expression in stably transduced Ba/F3-gp130 cell lines via flow cytometry (Fig. 4B).
Figure 4.
A stalk region of 23 amino acid residues is required for classic signaling. A, schematic overview of the four additional IL-11R stalk deletion variants. Amino acid residues of the stalk are highlighted in gray. Missing stretches in the scheme indicate deletions. B, cell surface expression of the above depicted IL-11R deletion variants stably transduced into Ba/F3-gp130 cells. C–F, equal amounts of the different Ba/F3-gp130 cell lines were stimulated with increasing amounts (0–100 ng/ml) of either IL-11 or Hy–IL-6 for 48 h. Concentration-dependent proliferation was determined as described in “Experimental Procedures.” The analyzed cell line is given above the respective diagram. Error bars represent S.D. of triplicates from one representative experiment.
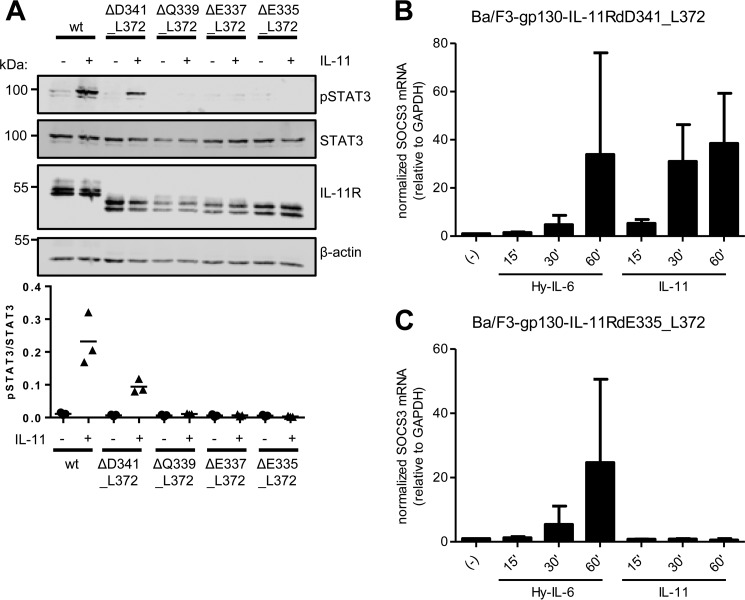
We further analyzed IL-11–induced proliferation of all four cell lines. Interestingly, the stalk region of 23 amino acid residues in length present in Ba/F3–gp130–IL-11RΔD341_L372 was sufficient to provoke a full response to IL-11 (Fig. 4C). In contrast, Ba/F3-gp130 cell lines expressing deletion variants with a stalk length of either 21 (IL-11RΔQ339_L372) or 19 amino acid residues (IL-11RΔE337_L372) still responded to IL-11, but the induced proliferation was distinctly lower than the response caused by Hy–IL-6 treatment (Fig. 4, D and E). Further decreasing the stalk length to 17 amino acid residues (IL-11RΔE335_L372) completely abrogated IL-11–induced proliferation (Fig. 4F). We again complemented our analyses with STAT3 phosphorylation assays in the four Ba/F3–gp130–IL-11R cell lines. In line with our previous results, we could detect robust, distinct phosphorylation of STAT3 upon IL-11 stimulation in Ba/F3–gp130–IL-11R and Ba/F3–gp130–IL-11RΔD341_L372 cells (Fig. 5A). In contrast, we observed no phosphorylation of STAT3 in Ba/F3–gp130–IL-11RΔQ339_L372, Ba/F3–gp130–IL-11RΔE337_L372, and Ba/F3–gp130–IL-11RΔE335_L372 cells (Fig. 5A). Furthermore, we analyzed induction of gene expression of SOCS3 in response to stimulation with Hy–IL-6 and IL-11 after 15, 30, and 60 min. As shown in Fig. 5B, SOCS3 expression was strongly induced by Hy–IL-6 and IL-11 in Ba/F3–gp130–IL-11RΔD341_L372 cells, showing again that this IL-11R deletion variant is biologically active. In contrast, although we detected an increase in SOCS3 in Ba/F3–gp130–IL-11RΔE335_L372 cells treated with Hy–IL-6, no SOCS3 induction was observed when the same cells were treated with IL-11 (Fig. 5C), confirming that the IL-11RΔE335_L372 variant lacks biological activity.
Figure 5.
IL-11-induced STAT3 phosphorylation depends on IL-11R stalk length. A, Ba/F3–gp130–IL-11R, Ba/F3–gp130–IL-11RΔD341_L372, Ba/F3–gp130–IL-11RΔQ339_L372, Ba/F3–gp130–IL-11RΔE337_L372, and Ba/F3–gp130–IL-11RΔE335_L372 cells were either stimulated with 10 ng/ml IL-11 for 15 min or left untreated. STAT3 phosphorylation was determined by Western blotting. Total STAT3 and IL-11R were visualized within the cell lysate. β-actin was determined to ensure equal protein loading. The ratio of pSTAT3/STAT3 was determined from three independent experiments. B, Ba/F3–gp130–IL-11RΔD341_L372 cells were stimulated with either 10 ng/ml Hy–IL-6, 10 ng/ml IL-11 for the indicated time points, or left untreated. The relative amounts of SOCS3 mRNA were quantified by quantitative real-time PCR. Values were normalized to GAPDH (n = 3 independent experiments, three technical replicates per experiment, mean ± S.D.). C, the experiment was performed with Ba/F3–gp130–IL-11RΔE335_L372 cells as described in B.
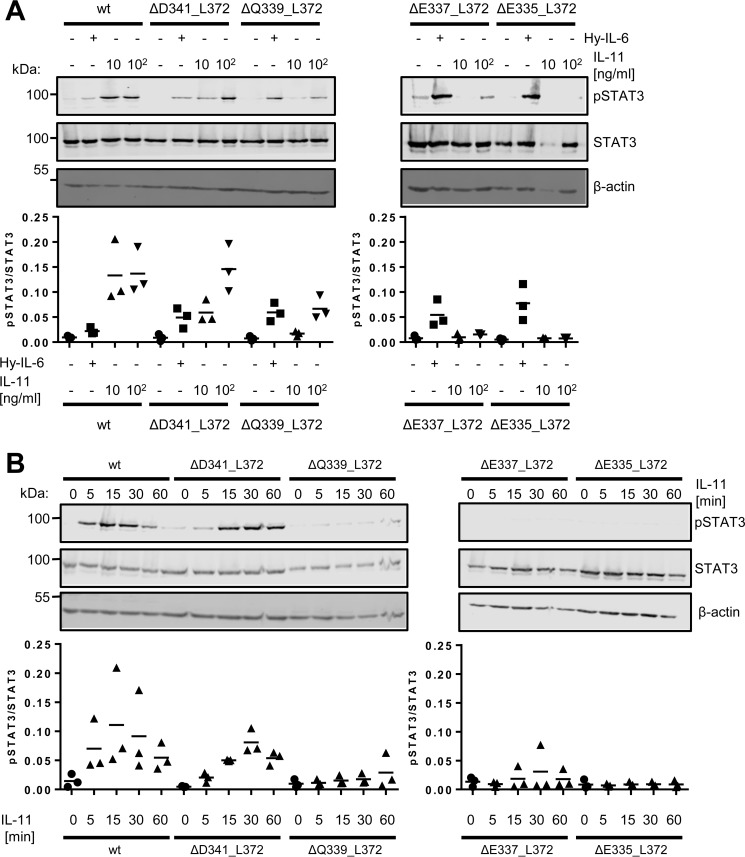
To analyze whether the decreased proliferation that we observed in the 21 and 19 amino acid residue–long stalk variants IL-11RΔQ339_L372 and IL-11RΔE337_L372 was also mirrored in STAT3 phosphorylation, we stimulated the cell lines with 10 ng/ml and 100 ng/ml IL-11 (Fig. 6A). Although no difference between the two concentrations was detected for Ba/F3–gp130–IL-11R cells, an increase in STAT3 phosphorylation upon stimulation with 100 ng/ml IL-11 compared with 10 ng/ml could be observed in Ba/F3–gp130–IL-11RΔD341_L372 cells. This shows that 10 ng/ml were already sufficient to activate the IL-11R in both cells lines, but full activation of the IL-11RΔD341_L372 was only achieved with the higher concentration of IL-11. In contrast, no induction of STAT3 phosphorylation was detected in the Ba/F3–gp130–IL-11RΔQ339_L372 and Ba/F3–gp130–IL-11RΔE337_L372 cell lines with 10 ng/ml IL-11 (Fig. 6A). However, stimulation with 100 ng/ml IL-11 revealed a weak STAT3 phosphorylation (Fig. 6A). These results are in accordance with the proliferation data (Fig. 4, D and E) and underline the finding that these receptor variants are not inactive per se, but rather show a decrease in signaling capability. Accordingly, the Ba/F3–gp130–IL-11RΔE335_L372 cell line showed practically no induction of STAT3 phosphorylation, even with 100 ng/ml IL-11 (Fig. 6A). Furthermore, when we measured phosphorylation of STAT3 over a time period of 60 min, we observed a normal STAT3 phosphorylation in Ba/F3–gp130–IL-11R cells, which declined stepwise in lockstep with the shortening of the stalk region of the IL-11R variants (Fig. 6B).
Figure 6.
Influence of cytokine concentration and duration of stimulation on IL-11–induced STAT3 phosphorylation. A, Ba/F3–gp130–IL-11R, Ba/F3–gp130–IL-11RΔD341_L372, Ba/F3–gp130–IL-11RΔQ339_L372, Ba/F3–gp130–IL-11RΔE337_L372, and Ba/F3–gp130–IL-11RΔE335_L372 cells were stimulated with either 10 ng/ml Hy–IL-6, 10 ng/ml IL-11, 100 ng/ml IL-11, or left untreated for 15 min. STAT3 phosphorylation was determined by Western blotting, and total STAT3 was visualized to ensure equal protein loading. The ratio of pSTAT3/STAT3 was determined from three independent experiments. B, Ba/F3–gp130–IL-11R, Ba/F3–gp130–IL-11RΔD341_L372, Ba/F3–gp130–IL-11RΔQ339_L372, Ba/F3–gp130–IL-11RΔE337_L372, and Ba/F3–gp130-IL–11RΔE335_L372 cells were stimulated with 10 ng/ml IL-11 for different time periods (0–60 min). STAT3 phosphorylation was determined by Western blotting, and total STAT3 was visualized to ensure equal protein loading. The ratio of pSTAT3/STAT3 was determined from three independent experiments.
In summary, our results indicate that for a full induction of IL-11 classic signaling, the IL-11R stalk needs to be at least 23 amino acid residues in length. No signaling can be induced if the stalk is 17 amino acid residues or shorter, albeit the receptor is still present at the cell surface. Interestingly, a stalk length between 17 and 23 amino acid residues results in decreased, but not fully abrogated, classic signaling, arguing for a certain degree of flexibility.
IL-11 signaling does not primarily depend on the sequence of the IL-11R stalk
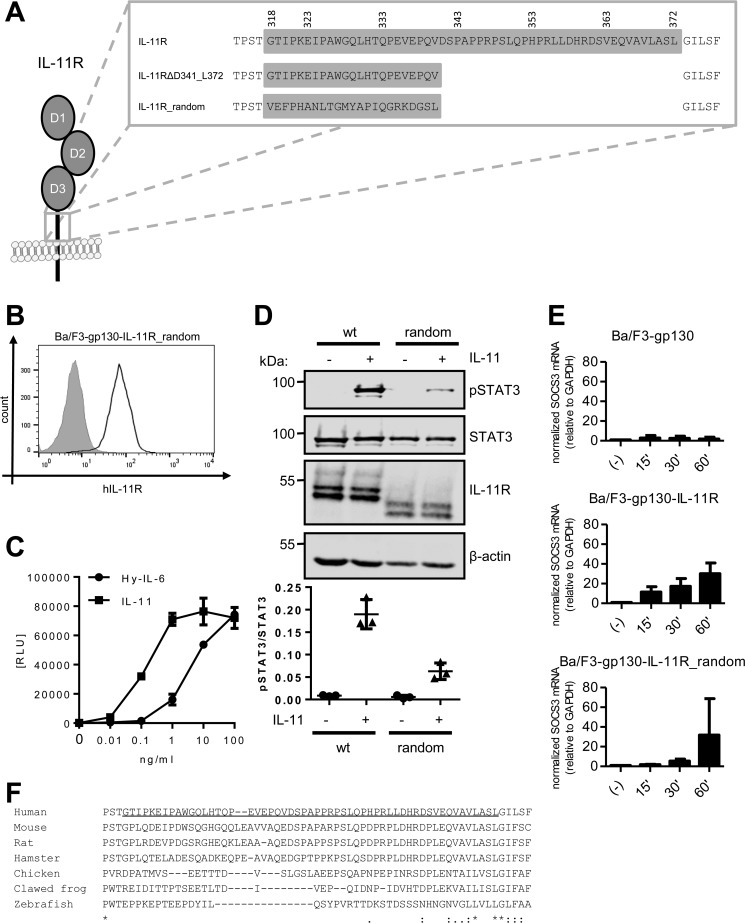
Finally, we sought to analyze whether the efficient formation of the signaling complex solely depends upon the length of the IL-11R stalk, or whether the amino acid residues within the stalk are directly involved. To this end, we made use of an IL-11R variant, where we inserted a random 23 amino acid residue sequence (IL-11R_random, Fig. 7A), as we have identified this as the minimal length required for efficient IL-11 signaling (Fig. 4C). The sorting and transport of the IL-11R to the cell surface of stably transduced Ba/F3-gp130 cells was independent of the sequence of the stalk region, as judged by efficient cell surface localization of the IL-11R_random variant detected via flow cytometry (Fig. 7B). Furthermore, we found that cells expressing this IL-11R variant (Ba/F3–gp130–IL-11R_random) showed IL-11–dependent proliferation as well as proliferation induced by Hy–IL-6 (Fig. 7C). Additionally, Ba/F3–gp130–IL-11R_random cells were also able to induce phosphorylation of STAT3 upon IL-11 stimulation, albeit less than cells expressing the IL-11R WT receptor (Fig. 7D). This was further confirmed by quantitative real-time PCR of SOCS3 expression in response to IL-11 treatment. Although Ba/F3-gp130 cells did not respond to IL-11 at all, SOCS3 expression increased over time in Ba/F3–gp130–IL-11R cells and was even detectable after only 15 min of stimulation (Fig. 7E). In contrast, Ba/F3–gp130–IL-11R_random cells responded slower to IL-11 treatment, although they reached the same amount of SOCS3 mRNA after 60 min as the Ba/F3–gp130–IL-11R cells (Fig. 7E). Together, these results indicate that classic IL-11 signaling depends on a certain length of the IL-11R stalk region, but that the actual sequence of this stalk is not of critical importance. However, although the IL-11R_random variant was biologically active, its capability to transmit IL-11–induced signals was reduced compared with the WT IL-11R. These findings support the hypothesis that the IL-11R stalk is required for positioning the extracellular domains in a correct distance to the CBM of gp130, which then allows the formation of the signaling complex.
Figure 7.
Classic IL-11 signaling is independent of the stalk sequence. A, stalk sequences of IL-11R, IL-11RΔD341_L372, and IL-11R_random. B, cell surface expression of IL-11R_random was assessed by flow cytometry. C, equal amounts of Ba/F3–gp130–IL-11R_random cells were stimulated with increasing amounts (0–100 ng/ml) of either IL-11 or Hy–IL-6 for 48 h. Concentration-dependent proliferation was determined as described in “Experimental Procedures.” D, STAT3 phosphorylation of Ba/F3–gp130–IL-11R_random in response to 10 ng/ml IL-11 or 10 ng/ml Hy–IL-6 stimulation for 15 min was determined by Western blotting. Total STAT3 was determined to ensure equal protein loading. The ratio of pSTAT3/STAT3 was determined from three independent experiments. E, Ba/F3-gp130, Ba/F3–gp130–IL-11R and Ba/F3–gp130–IL-11R_random cells were stimulated with 10 ng/ml IL-11 for the indicated time points. The relative amounts of SOCS3 mRNA were quantified as described in the legend to Fig. 5B. Values were normalized to GAPDH (n = 3 independent experiments, three technical replicates per experiment, mean ± S.D.). F, sequence alignment of the amino acid residues of the IL-11R stalk sequences from human (Homo sapiens), mouse (Mus musculus), rat (Rattus norvegicus), hamster (Mesocricetus auratus), chicken (Gallus gallus), clawed frog (Xenopus tropicalis), and zebrafish (Danio rerio). The predicted stalk region of the human IL-11R has been underlined.
Discussion
IL-11 signal transduction is mediated via a complex of IL-11R and a gp130 homodimer. It is well known that the extracellular domains D2 and D3 of the IL-11R are responsible for binding of IL-11 to its receptor, which is a prerequisite for gp130 homodimerization and initiation of signal transduction. In contrast, the IL-11R stalk region is not directly involved in signal transduction, because IL-11 can bind to membrane-bound as well as soluble forms of the IL-11R, and both complexes efficiently recruit a gp130 homodimer (17–19). Accordingly, the intracellular and the transmembrane region also are dispensable for IL-11 classic signaling (17–19), but the intracellular region has been shown to be required for the correct sorting of the IL-11R in polarized cells and the transcytotic activity of the IL-11R (24).
Besides IL-11, gp130 mediates signal transduction of multiple other cytokines, which all differ in their receptor-binding sites (10). This variety of interactions has been proposed to result in slightly different orientations of cytokines and the gp130 CBM (25), and possibly also different overall structures of the signaling complexes. Among these cytokines, IL-11 and IL-6 stand out because they are the only cytokines which utilize a gp130 homodimer (10). This has led to the assumption that the signaling complexes of both cytokines are equal, and current knowledge is that dimerization of gp130 by IL-11 and IL-6 leads to the same intracellular signaling cascades (26). Conversely, monoclonal antibodies directed against gp130 have been described which specifically inhibit IL-11 or IL-6 signaling (27). Furthermore, the recently solved crystal structure of IL-11 revealed differences in the gp130 interaction site compared with that of IL-6, indicating that the two cytokines might bind to gp130 in a different manner (22). However, we have previously shown that chimeric receptors of the IL-11R and IL-6R are indistinguishable in their biological activity (15), pointing toward similar formation of the signaling complexes.
Here, we found that the IL-11R stalk region is required for classic signaling, as deletion of the majority of the stalk (deleting 50 of the 55 amino acid residues) prevented signal transduction, albeit the receptor was still expressed and transported to the cell surface. Furthermore, we showed that a stalk region of at least 23 amino acid residues is required for efficient signal transduction. Interestingly, an IL-11R variant with a random stalk of the same length was also biologically active, although its signaling capacity was reduced compared with the IL-11R with the full-length stalk region. Overall, these results indicate that the exact amino acid sequence of the stalk region is not crucial for IL-11 signaling and further support the proposed role for the stalk region as a spacer to position the CBM of the IL-11R in a correct distance in relation to the two gp130 receptors. Importantly, the required stalk length for IL-11 classic signaling is similar to what we have reported previously for the IL-6R (21). We cannot rule out completely the influence of the amount of the IL-11R at the cell surface, as this varied between the individual Ba/F3-gp130 cell lines expressing the different IL-11R deletion variants. In conclusion, our data support the theory that the orientation of IL-11 and IL-6 in relation to gp130, and hence the overall geometry of the signaling complexes of both cytokines, are similar, although our results do not exclude subtle differences in the binding interfaces. Nevertheless, there are no indications that these gp130 dimers differ in a way that would result in differences in the intracellular signaling cascades. Whether qualitative or quantitative differences in terms of signaling exist between IL-11 classic and trans-signaling is also unknown to date.
However, our results raise the question of why the IL-11R stalk length exceeds the minimal requirement for signal transduction by 22 amino acid residues. Comparison of the stalk regions of the IL-11R across different species revealed no conservation regarding the amino acid sequence (Fig. 7F), further supporting our finding that the sequence is not directly involved in signal transduction. Indeed, the predicted stalk region e.g. in the IL-11R from zebrafish is 13 amino acid residues shorter than the human orthologue, illustrating that functional IL-11R does not require the length of 55 amino acid residues found in the human IL-11R. Intriguingly, the human stalk region spans 209 Å (3.8 Å per amino acid), whereas the cytokine-binding region of gp130 is only in a distance of 96 Å separated from the plasma membrane (31 Å for each of the three FNIII-like domains). This indicates a huge amount of flexibility for the final formation of the IL-11/IL-11R/gp130 signaling complex. The minimal functional stalk determined in this study spans only 87.4 Å, similar to the minimal stalk region of the IL-6R (21), indicating that the gp130 molecules have to bend toward the plasma membrane to reach the IL-11/IL-11R complex. Indeed, structural data revealed a bended, C-shaped conformation of gp130 (28), which is in accordance with our results here.
Interestingly, even the shortest stalk region shown in Fig. 7F is longer than the minimal length identified in this study, which indicates that the IL-11R stalk might have further functions in addition to the correct spacing between the membrane and the gp130 receptors. We have previously shown that the stalk also acts as a determinant of proteolysis (19), but it is also possible that this region has further, yet unknown, functions which have to be explored in further studies.
Experimental procedures
Cells and reagents
Ba/F3-gp130 cells were obtained from Immunex (Seattle, WA) (23). Retroviral transduction of Ba/F3-gp130 cells was performed as described previously (29). Cells were cultured in DMEM high-glucose culture medium (Gibco, Thermo Fisher Scientific), supplemented with 10% fetal bovine serum, penicillin (60 mg/liter), and streptomycin (100 mg/liter) in the presence of 10 ng/ml Hyper–IL-6 or, if the cells were transduced with biologically active variants of the IL-11R, in the presence of 10 ng/ml IL-11 instead, as they do not produce IL-11 endogenously. Hyper–IL-6 and IL-11 were produced as described previously (24, 30). Anti-phosphoSTAT3 (pTyr705), anti-STAT3 (124H6), and anti-Myc (71D10) antibodies were obtained from Cell Signaling Technology, the anti-IL-11R (N-20) and anti-β-actin (C-4) antibodies were from Santa Cruz Biotechnology (Dallas, TX). IRDye-conjugated anti-mouse and anti-rabbit antibodies were purchased from LI-COR Biosciences (Lincoln, NE), and the Alexa Fluor 488–conjugated anti-rabbit antibody from Thermo Fisher Scientific.
Construction of plasmids
pcDNA3.1 expression plasmids for Myc-tagged hIL-11R have been described previously (15). Deletions within the IL-11R stalk have been constructed by splicing by overlapping extensions PCR as described previously (19). The IL-11R deletion variants IL-11RΔV363_L372, IL-11RΔH353_S362, and IL-11RΔP343_P352 have been described previously (19). The random sequence has been designed based on natural distribution of amino acids using the ExPASy RandSeq tool (31) and was constructed by PCR. All variants were subcloned into pMOWS by digestion of pcDNA3.1 plasmids with PmeI (29). The generated constructs were verified via Sanger sequencing (GATC Biotech, Konstanz, Germany).
Flow cytometry
To analyze cell surface expression of IL-11R variants, ∼500,000 cells were washed with PBS and stained with α-Myc-antibody diluted 1:100 in FACS buffer (1% BSA in PBS) for 1 h, washed two times and incubated with Alexa Fluor 488–conjugated anti-rabbit antibody for 1 h in the dark. After two more washing steps, cells were analyzed on a FACSCanto II (BD Biosciences). The obtained data were analyzed with FlowJo software (Ashland, OR).
Ba/F3-gp130 cell viability assay
Cytokine-dependent proliferation of Ba/F3-gp130–derived cell lines in response to IL-11 or Hyper–IL-6 stimulation was analyzed using the CellTiter-Blue Viability Assay (Promega, Madison, WI) according to the manufacturer's instructions as described previously (32). Because Ba/F3 cells proliferate dose dependently in response to cytokines and undergo apoptosis without an appropriate stimulus, the cell viability is used as readout for the proliferation of the different cell lines. Data are shown as relative light units (RLU).
Stimulation of cells and Western blotting
For analysis of STAT3 phosphorylation, the different Ba/F3-gp130–derived cell lines were starved in serum-free medium for 3 h and stimulated with 10 ng/ml Hyper–IL-6 or 10 ng/ml IL-11 for 15 min. Afterward, cells were harvested by centrifugation and directly boiled in Laemmli buffer. Cell lysates were separated by SDS-PAGE and blotted onto nitrocellulose membranes, which were blocked with Odyssey Blocking Buffer (LI-COR Biosciences) for 1 h at room temperature. Then, membranes were treated with primary antibody at 4 °C overnight and, after washing, with IRDye-conjugated secondary antibodies for 1 h in the dark at room temperature. Signals were analyzed with an Odyssey Fc and the ImageStudio software (LI-COR Biosciences).
Quantitative real-time PCR
Ba/F3-gp130 cell lines were stimulated with 10 ng/ml IL-11 or Hy–IL-6 for the indicated time points. Total RNA was isolated from 2 × 106 cells using the NucleoSpin RNA II kit (Macherey-Nagel, Düren, Germany) according to manufacturer's instructions. Concentration and purity of the extracted RNA was analyzed using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). For gene expression analysis, 1 μg total RNA was reversely transcribed into cDNA with Oligo-dT15 primers and the RevertAid reverse transcriptase (Thermo Fisher Scientific). cDNA transcribed from 50 ng RNA was used as template for each quantitative real-time PCR reaction using the Power SYBR Green PCR Master Mix (Thermo Fisher Scientific) according to manufacturer's instructions on a LightCycler® 480 System (Roche). Samples were measured in triplicates and normalized to GAPDH expression. Relative gene expression was calculated using the 2−ΔCt method. Primer sequences were mSOCS3_fwd: GCTCCAAAAGCGAGTACCAGC, mSOCS3_rev: AGTAGAATCCGCTCTCCTGCAG, mGAPDH_fwd: AAGGTCATCCCAGAGCTGAA, mGAPDH_rev: CTGCTTCACCACCTTCTTGA.
Sequence alignment
For the multiple sequence alignment the online tool Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/)3 (33) was used. The alignment was performed with the full sequences of IL-11 receptors from the following species: Homo sapiens (UniProt: Q14626), Mus musculus (UniProt: Q64385), Rattus norvegicus (UniProt: Q99MF4), Mesocricetus auratus (UniProt: A0A1U8C6K8), Xenopus tropicalis (UniProt: A4IHR2), Gallus (UniProt: A0A1D5PDZ2), Danio rerio (UniProt: A8WH84).
Data presentation
Unless stated otherwise, all experiments were performed at least three times with similar outcome. Western blots and flow cytometry data are shown from one representative experiment, and quantification of three independent experiments is presented next to the respective Western blots. Cell viability data are shown from one representative experiment (mean ± S.D., n = 3 biological replicates).
Author contributions
J. L. investigation; J. L. methodology; J. L. and C. G. writing-original draft; J. L. and C. G. writing-review and editing; C. G. conceptualization; C. G. supervision; C. G. funding acquisition.
Acknowledgments
We thank Stefan Düsterhöft for critical reading of the manuscript.
This work was funded by Deutsche Forschungsgemeinschaft Grants SFB877 project A10 (Bonn, Germany) and the Cluster of Excellence “Inflammation at Interfaces.” The authors declare that they have no conflicts of interest with the contents of this article.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- IL-11R
- interleukin 11-receptor
- gp
- glycoprotein
- YAP
- yes-associated protein
- CBM
- cytokine-binding module.
References
- 1. Paul S. R., Bennett F., Calvetti J. A., Kelleher K., Wood C. R., O'Hara R. M. Jr., Leary A. C., Sibley B., Clark S. C., and Williams D. A. (1990) Molecular cloning of a cDNA encoding interleukin 11, a stromal cell-derived lymphopoietic and hematopoietic cytokine. Proc. Natl. Acad. Sci. U.S.A. 87, 7512–7516 10.1073/pnas.87.19.7512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Orazi A., Cooper R. J., Tong J., Gordon M. S., Battiato L., Sledge G. W. Jr., Kaye J. A., Kahsai M., and Hoffman R. (1996) Effects of recombinant human interleukin-11 (Neumega rhIL-11 growth factor) on megakaryocytopoiesis in human bone marrow. Exp. Hematol. 24, 1289–1297 [PubMed] [Google Scholar]
- 3. Trepicchio W. L., Wang L., Bozza M., and Dorner A. J. (1997) IL-11 regulates macrophage effector function through the inhibition of nuclear factor-kappaB. J. Immunol. 159, 5661–5670 [PubMed] [Google Scholar]
- 4. Xu D. H., Zhu Z., Wakefield M. R., Xiao H., Bai Q., and Fang Y. (2016) The role of IL-11 in immunity and cancer. Cancer Lett. 373, 156–163 10.1016/j.canlet.2016.01.004 [DOI] [PubMed] [Google Scholar]
- 5. Ernst M., Najdovska M., Grail D., Lundgren-May T., Buchert M., Tye H., Matthews V. B., Armes J., Bhathal P. S., Hughes N. R., Marcusson E. G., Karras J. G., Na S., Sedgwick J. D., Hertzog P. J., and Jenkins B. J. (2008) STAT3 and STAT1 mediate IL-11-dependent and inflammation-associated gastric tumorigenesis in gp130 receptor mutant mice. J. Clin. Invest. 118, 1727–1738 10.1172/JCI34944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Putoczki T. L., Thiem S., Loving A., Busuttil R. A., Wilson N. J., Ziegler P. K., Nguyen P. M., Preaudet A., Farid R., Edwards K. M., Boglev Y., Luwor R. B., Jarnicki A., Horst D., Boussioutas A., et al. (2013) Interleukin-11 is the dominant IL-6 family cytokine during gastrointestinal tumorigenesis and can be targeted therapeutically. Cancer Cell 24, 257–271 10.1016/j.ccr.2013.06.017 [DOI] [PubMed] [Google Scholar]
- 7. Ernst M., and Putoczki T. L. (2014) Molecular pathways: IL11 as a tumor-promoting cytokine-translational implications for cancers. Clin. Cancer Res. 20, 5579–5588 10.1158/1078-0432.CCR-13-2492 [DOI] [PubMed] [Google Scholar]
- 8. Matadeen R., Hon W. C., Heath J. K., Jones E. Y., and Fuller S. (2007) The dynamics of signal triggering in a gp130-receptor complex. Structure 15, 441–448 10.1016/j.str.2007.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stahl N., Boulton T. G., Farruggella T., Ip N. Y., Davis S., Witthuhn B. A., Quelle F. W., Silvennoinen O., Barbieri G., and Pellegrini S., et al. (1994) Association and activation of Jak-Tyk kinases by CNTF-LIF-OSM-IL-6 beta receptor components. Science 263, 92–95 10.1126/science.8272873 [DOI] [PubMed] [Google Scholar]
- 10. Garbers C., Hermanns H. M., Schaper F., Müller-Newen G., Grötzinger J., Rose-John S., and Scheller J. (2012) Plasticity and cross-talk of interleukin 6-type cytokines. Cytokine Growth Factor Rev. 23, 85–97 10.1016/j.cytogfr.2012.04.001 [DOI] [PubMed] [Google Scholar]
- 11. Eulenfeld R., Dittrich A., Khouri C., Müller P. J., Mütze B., Wolf A., and Schaper F. (2012) Interleukin-6 signalling: More than Jaks and STATs. Eur. J. Cell Biol. 91, 486–495 10.1016/j.ejcb.2011.09.010 [DOI] [PubMed] [Google Scholar]
- 12. Taniguchi K., Wu L. W., Grivennikov S. I., de Jong P. R., Lian I., Yu F. X., Wang K., Ho S. B., Boland B. S., Chang J. T., Sandborn W. J., Hardiman G., Raz E., Maehara Y., Yoshimura A., Zucman-Rossi J., Guan K. L., and Karin M. (2015) A gp130-Src-YAP module links inflammation to epithelial regeneration. Nature 519, 57–62 10.1038/nature14228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hilton D. J., Hilton A. A., Raicevic A., Rakar S., Harrison-Smith M., Gough N. M., Begley C. G., Metcalf D., Nicola N. A., and Willson T. A. (1994) Cloning of a murine IL-11 receptor alpha-chain; requirement for gp130 for high affinity binding and signal transduction. EMBO J. 13, 4765–4775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nandurkar H. H., Hilton D. J., Nathan P., Willson T., Nicola N., and Begley C. G. (1996) The human IL-11 receptor requires gp130 for signalling: Demonstration by molecular cloning of the receptor. Oncogene 12, 585–593 [PubMed] [Google Scholar]
- 15. Nitz R., Lokau J., Aparicio-Siegmund S., Scheller J., and Garbers C. (2015) Modular organization of interleukin-6 and interleukin-11 α-receptors. Biochimie 119, 175–182 10.1016/j.biochi.2015.11.005 [DOI] [PubMed] [Google Scholar]
- 16. Schleinkofer K., Dingley A., Tacken I., Federwisch M., Müller-Newen G., Heinrich P. C., Vusio P., Jacques Y., and Grötzinger J. (2001) Identification of the domain in the human interleukin-11 receptor that mediates ligand binding. J. Mol. Biol. 306, 263–274 10.1006/jmbi.2000.4387 [DOI] [PubMed] [Google Scholar]
- 17. Pflanz S., Tacken I., Grötzinger J., Jacques Y., Minvielle S., Dahmen H., Heinrich P. C., and Müller-Newen G. (1999) A fusion protein of interleukin-11 and soluble interleukin-11 receptor acts as a superagonist on cells expressing gp130. FEBS Lett. 450, 117–122 10.1016/S0014-5793(99)00477-9 [DOI] [PubMed] [Google Scholar]
- 18. Baumann H., Wang Y., Morella K. K., Lai C. F., Dams H., Hilton D. J., Hawley R. G., and Mackiewicz A. (1996) Complex of the soluble IL-11 receptor and IL-11 acts as IL-6-type cytokine in hepatic and nonhepatic cells. J. Immunol. 157, 284–290 [PubMed] [Google Scholar]
- 19. Lokau J., Nitz R., Agthe M., Monhasery N., Aparicio-Siegmund S., Schumacher N., Wolf J., Möller-Hackbarth K., Waetzig G. H., Grötzinger J., Müller-Newen G., Rose-John S., Scheller J., and Garbers C. (2016) Proteolytic cleavage governs interleukin-11 trans-signaling. Cell Rep. 14, 1761–1773 10.1016/j.celrep.2016.01.053 [DOI] [PubMed] [Google Scholar]
- 20. Pflanz S., Kurth I., Grötzinger J., Heinrich P. C., and Müller-Newen G. (2000) Two different epitopes of the signal transducer gp130 sequentially cooperate on IL-6-induced receptor activation. J. Immunol. 165, 7042–7049 10.4049/jimmunol.165.12.7042 [DOI] [PubMed] [Google Scholar]
- 21. Baran P., Nitz R., Grötzinger J., Scheller J., and Garbers C. (2013) Minimal interleukin 6 (IL-6) receptor stalk composition for IL-6 receptor shedding and IL-6 classic signaling. J. Biol. Chem. 288, 14756–14768 10.1074/jbc.M113.466169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Putoczki T. L., Dobson R. C., and Griffin M. D. (2014) The structure of human interleukin-11 reveals receptor-binding site features and structural differences from interleukin-6. Acta Crystallogr. D Biol. Crystallogr. 70, 2277–2285 10.1107/S1399004714012267 [DOI] [PubMed] [Google Scholar]
- 23. Gearing A. J., Beckett P., Christodoulou M., Churchill M., Clements J., Davidson A. H., Drummond A. H., Galloway W. A., Gilbert R., and Gordon J. L. (1994) Processing of tumour necrosis factor-α precursor by metalloproteinases. Nature 370, 555–557 10.1038/370555a0 [DOI] [PubMed] [Google Scholar]
- 24. Monhasery N., Moll J., Cuman C., Franke M., Lamertz L., Nitz R., Görg B., Häussinger D., Lokau J., Floss D. M., Piekorz R., Dimitriadis E., Garbers C., and Scheller J. (2016) Transcytosis of IL-11 and apical redirection of gp130 is mediated by IL-11α receptor. Cell Rep. 16, 1067–1081 10.1016/j.celrep.2016.06.062 [DOI] [PubMed] [Google Scholar]
- 25. Boulanger M. J., and Garcia K. C. (2004) Shared cytokine signaling receptors: Structural insights from the gp130 system. Adv. Protein Chem. 68, 107–146 10.1016/S0065-3233(04)68004-1 [DOI] [PubMed] [Google Scholar]
- 26. Garbers C., and Scheller J. (2013) Interleukin-6 and interleukin-11: Same but different. Biol. Chem. 394, 1145–1161 10.1515/hsz-2013-0166 [DOI] [PubMed] [Google Scholar]
- 27. Gu Z. J., Wijdenes J., Zhang X. G., Hallet M. M., Clement C., and Klein B. (1996) Anti-gp130 transducer monoclonal antibodies specifically inhibiting ciliary neurotrophic factor, interleukin-6, interleukin-11, leukemia inhibitory factor or oncostatin M. J. Immunol. Methods 190, 21–27 10.1016/0022-1759(95)00232-4 [DOI] [PubMed] [Google Scholar]
- 28. Xu Y., Kershaw N. J., Luo C. S., Soo P., Pocock M. J., Czabotar P. E., Hilton D. J., Nicola N. A., Garrett T. P., and Zhang J.-G. (2010) Crystal structure of the entire ectodomain of gp130: insights into the molecular assembly of the tall cytokine receptor complexes. J. Biol. Chem. 285, 21214–21218 10.1074/jbc.C110.129502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ketteler R., Glaser S., Sandra O., Martens U. M., and Klingmüller U. (2002) Enhanced transgene expression in primitive hematopoietic progenitor cells and embryonic stem cells efficiently transduced by optimized retroviral hybrid vectors. Gene Ther. 9, 477–487 10.1038/sj.gt.3301653 [DOI] [PubMed] [Google Scholar]
- 30. Fischer M., Goldschmitt J., Peschel C., Brakenhoff J. P., Kallen K. J., Wollmer A., Grötzinger J., and Rose-John S. (1997) I. A bioactive designer cytokine for human hematopoietic progenitor cell expansion. Nat. Biotechnol. 15, 142–145 10.1038/nbt0297-142 [DOI] [PubMed] [Google Scholar]
- 31. Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., and Bairoch A. (2003) ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788 10.1093/nar/gkg563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Garbers C., Thaiss W., Jones G. W., Waetzig G. H., Lorenzen I., Guilhot F., Lissilaa R., Ferlin W. G., Grötzinger J., Jones S. A., Rose-John S., and Scheller J. (2011) Inhibition of classic signaling is a novel function of soluble glycoprotein 130 (sgp130), which is controlled by the ratio of interleukin 6 and soluble interleukin 6 receptor. J. Biol. Chem. 286, 42959–42970 10.1074/jbc.M111.295758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]