Abstract
DNA interstrand cross-links (ICLs) block the progress of the replication and transcription machineries and can weaken chromosomal stability, resulting in various diseases. FANCD2–FANCI-associated nuclease (FAN1) is a conserved structure-specific nuclease that unhooks DNA ICLs independently of the Fanconi anemia pathway. Recent structural studies have proposed two different mechanistic features for ICL unhooking by human FAN1: a specific basic pocket that recognizes the terminal phosphate of a 1-nucleotide (nt) 5′ flap or FAN1 dimerization. Herein, we show that despite lacking these features, Pseudomonas aeruginosa FAN1 (PaFAN1) cleaves substrates at ∼3-nt intervals and resolves ICLs. Crystal structures of PaFAN1 bound to various DNA substrates revealed that its conserved basic Arg/Lys patch comprising Arg-228 and Lys-260 recognizes phosphate groups near the 5′ terminus of a DNA substrate with a 1-nt flap or a nick. Substitution of Lys-260 did not affect PaFAN1's initial endonuclease activity but significantly decreased its subsequent exonuclease activity and ICL unhooking. The Arg/Lys patch also interacted with phosphates at a 3-nt gap, and this interaction could drive movement of the scissile phosphates into the PaFAN1-active site. In human FAN1, the ICL-resolving activity was not affected by individual disruption of the Arg/Lys patch or basic pocket. However, simultaneous substitution of both FAN1 regions significantly reduced its ICL-resolving activity, suggesting that these two basic regions play a complementary role in ICL repair. On the basis of these findings, we propose a conserved role for two basic regions in FAN1 to guide ICL unhooking and to maintain genomic stability.
Keywords: DNA endonuclease, protein–nucleic acid interaction, enzyme mechanism, molecular biology, DNA repair, protein structure, bacterial FAN1, basic pocket, DNA interstrand cross-link, FANCD2–FANCI-associated nuclease, ICL unhooking
Introduction
A class of bifunctional exogenous (platinum derivatives, mitomycin C, psoralen, and nitrogen mustards) and endogenous (aldehydes) chemicals can covalently link the bases of two complementary DNA strands (1–3). The resulting DNA interstrand cross-links (ICLs)2 are highly cytotoxic as they block the progress of the replication and transcription machineries. If not properly repaired, ICLs can induce chromosomal instability, resulting in various pathologies (1, 4).
To counteract this threat, several modes of ICL repair operate in vertebrates mainly in the context of replication (5–10). One of these is the Fanconi anemia (FA)-dependent ICL repair pathway (9, 10), in which one or two replication forks approach the ICL and stall some distance away from it. Activation of the FA core complex then leads to ubiquitylation of the FANCD2–FANCI (ID) complex (11, 12) and recruitment of ERCC1-XPF and possibly other endo- or exonucleases to make incisions on either side of the ICL in one strand to unhook the cross-link (13–15). Replication of one of the two strands is then completed by translesion DNA synthesis (TLS), and the second strand is repaired by homologous recombination.
Several additional endo- and exonucleases have been implicated in ICL repair, but their exact roles in ICL repair are not well understood (16). Among these, the FANCD2–FANCI-associated nuclease FAN1 is a structure-specific endo- and exonuclease that cleaves various substrates, including 5′ flaps, replication forks, splayed arms, duplex DNA, and ssDNA (17–20). FAN1 contains a UBZ domain that interacts with the ID complex and was initially proposed to be an ICL-unhooking nuclease in the FA pathway. However, a number of studies suggested that the cross-link repair function of FAN1 is not within the FA pathway as indicated: (i) homologs of FAN1 have been found in bacteria, archaea, and unicellular eukaryotes that lack FA proteins (19, 21–23); (ii) mutations in FAN1 do not cause FA, but instead a kidney disorder called karyomegalic interstitial nephritis (KIN) (24); and (iii) N-terminal UBZ domain is dispensable for ICL repair but is required for genomic maintenance, suggesting that the interaction of FAN1 with ID serves a function outside of ICL repair (25–27). Nevertheless, the sensitivity of FAN1-deficient cells to ICL-forming agents implicates FAN1 in ICL resolution independently of the FA pathway.
In addition to KIN (24), mutations in FAN1 have been associated with predisposition to colorectal and pancreatic cancer as well as autism and schizophrenia (28–31), raising the possibility that defects in ICL repair may be associated with a wide variety of diseases. In vitro studies showed that human FAN1 can degrade ICL-containing oligonucleotides past the lesion in replication-like intermediates, thereby unhooking the ICLs (32–34). FAN1 incises substrates 2–5 nucleotides into the duplex from the ss/ds junction of DNA with a 5′ flap or a nick, followed by 5′ to 3′ exonucleolytic cleavage of the duplex even in presence of an ICL (17–20, 23, 32–34).
Recently, several crystal structures of FAN1 bound to DNA have been reported, and mechanisms of how they degrade ICL-containing DNA substrates have been proposed (23, 33–35). To date, these are the only structures of an ICL-repair nuclease bound to DNA. Interestingly, two very different models for endonuclease activity have been proposed (33–35). Although the monomer structure of Homo sapiens FAN1 (HsFAN1) is similar in both structures, there are fundamental differences in quaternary structures and the proposed mechanisms of how FAN1 resolves ICLs. In one group of structures, HsFAN1 acts as a monomer and binds to the 5′-terminal phosphate of a flap or a nicked DNA via a basic pocket clustered with positively charged residues (33). HsFAN1 unhooks ICLs using successive DNA cleavage steps in 3-nt intervals, using the basic pocket, which is 3 nts to the 5′ side from the active site, as a guide for the incisions.
In the second set of structures, HsFAN1 forms a “head to tail” dimer in the presence of DNA (34). The authors suggest that FAN1 dimer scans, latches, and unwinds DNA to pull the ssDNA to the active site of a FAN1. Mutations of the residues at the dimeric interface abolished the nuclease activity of the HsFAN1, raising the possibility that the dimerization is important for its activity.
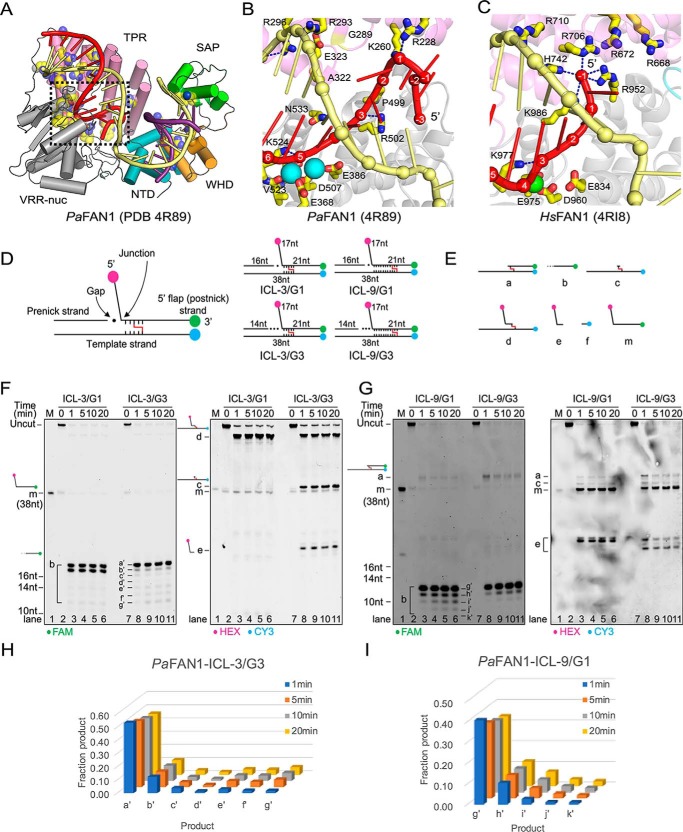
Bacterial FAN1 homologs share a high similarity with HsFAN1 (Fig. 1A) (23, 35). Pseudomonas aeruginosa FAN1 (PaFAN1) shares 34% sequence similarity with HsFAN1, and the root mean square deviation between the two structures is 3.1 Å for 456 Cα atoms. PaFAN1 functions as a monomer and makes an initial endonucleolytic incision near an ss/dsDNA junction followed by exonucleolytic cleavage in the duplex region of the substrate. However, bacterial FAN1 exhibited several features distinct from HsFAN1. First, PaFAN1 produces at least two endonucleolytic products as it incises the phosphate of the 5th or 6th nt in the duplex region of an ss/ds DNA junction (23). Second, PaFAN1 does not have a specific basic pocket that is critical for the proposed “3-nt exonuclease” mechanism (Fig. 1, B and C) (33). Third, PaFAN1 lacks the basic motif in the SAP domain required for dimerization in one of the structures of the human nuclease (Fig. 1A) (34). These features raise the question if PaFAN1 can unhook ICLs and, if so, by which mechanism.
Figure 1.
ICL-unhooking activity of PaFAN1. A, overall structure of PaFAN1 bound to 5′ flap DNA (PDB code 4R89). B, close-up view of the 5′ flap-binding region (red) of PaFAN1. Two Mn2+ ions are shown as cyan spheres. C, corresponding region in HsFAN1 (PDB code 4RI8). D, structures of substrates used in the experiment. Substrates containing a 1- or 3-nt gap were labeled with three fluorophores, HEX (pink), Cy3 (blue), and FAM (green). Gap region is marked with filled circles. ICL-unhooking activity was tested toward four substrates containing ICL (represented as red line) 3 or 9 nt downstream from the ss/ds junction (right, ICL-3/G1, ICL-3/G3, ICL-9/G1, and ICL-9/G3). E, structures of predicted products. F and G, denaturing PAGE of ICL unhooking reactions with the DNA substrates (25 nm) and PaFAN1 (250 nm) for the indicated times. A 10-fold excess of enzyme to DNA substrate was used to clearly detect the cleaved products b and a′–g′. Fluorescence signals were scanned using two wavelengths of 488 and 532 nm for FAM (left panel) and HEX or CY3 (right panel), respectively. H and I, graph showing the quantified FAM-labeled products generated at different time points in reactions with ICL-3/G3 (H) or ICL-9/G1 (I).
To address these issues, we examined the ICL-unhooking activity and the structural mechanism of DNA binding and incision of PaFAN1. We first show that PaFAN1 efficiently unhooks ICLs independently of the position of the ICLs from the ss/ds junction. We determined the structures of PaFAN1 bound to 5′ flap DNAs with various gaps that mimic initial endonucleolytic cleavage product. Although PaFAN1 does not have the basic pocket, the phosphate-binding patch made of the conserved basic residues (Arg/Lys patch) is located 3 nt distant from the active site. Mutation of this patch did not alter the initial endonuclease activity, but it significantly decreased the subsequent nuclease and ICL-unhooking activity of PaFAN1. We propose that PaFAN1 can make multiple incisions on a substrate at a first site and cleaves DNA in 3-nt intervals. By comparison, ICL-resolving activity of HsFAN1 was only significantly decreased upon disruption of both the Arg/Lys patch and the basic pocket suggesting a complementary role of the two basic regions in maintaining the nuclease function. Based on these findings, we provide insights into how FAN1 might have evolved to have two basic regions to guide its nuclease activity and maintain genomic stability.
Results
Substrate specificity of PaFAN1
In a previous study, we have used a DNA substrate with a 4-nucleotide 5′ flap (23). However, because HsFAN1 most efficiently binds to a DNA with a 1-nt 5′ flap and a 4–8-nt 3′ flap, we examined the activity of PaFAN1 on a 1-nt 5′ flap DNA (33). PaFAN1 exhibited slightly better activity toward the 5′ flap of 1 nt relative to the 5′ flap of 4 nt (Fig. S1, A–D). However, in contrast to HsFAN1, addition of a 3′ flap to the 5′ flap DNA substrate decreased the nuclease activity of PaFAN1. We thus used the DNA substrate with a 1-nt 5′ flap in most nuclease assays in this study, except for the ICL-unhooking assay. We used the PaFAN1 with an N-terminal His tag throughout the study. We opted not to remove the tag because a small fraction of PaFAN1 undergoes internal cleavage during the removal of a His tag. The His tag, which is disordered in the structure, did not affect the nuclease activity of PaFAN1, consistent with the N terminus being far away from the active and DNA-binding sites (Fig. S1, E and F).
PaFAN1 efficiently incises ICL lesion
To examine whether the bacterial FAN1 homolog shares the ICL-resolving activity of mammalian FAN1, we examined the in vitro ICL-unhooking activity of PaFAN1 using various DNA ICL substrates (Fig. 1D). We first analyzed whether PaFAN1 can unhook the ICL near the ss/ds junction. We used DNA substrates containing a nitrogen-like ICL, which is free of duplex distortion (15, 36, 37), with the ICL located at the 3rd nucleotide of the 5′ flap strand and the 5th nucleotide from the junction template strand (ICL-3). In this DNA, the 3′ overhang of the 38-nt template strand is paired with the 16-nt pre-nick strand to form a 5′ flap structure with a gap of 1 nt to the junction (ICL-3/G1, Fig. 1D). The ICL substrates are labeled with HEX and FAM fluorophores at 5′ and 3′ ends of the 5′ flap strand, respectively, and with Cy3 at the 5′ end of the template strand.
Incubation of the ICL-3/G1 substrate with PaFAN1 yielded two FAM-labeled ssDNA oligonucleotides (product b), indicating that the incision took place 3′ to the ICL lesion (Fig. 1, E and F, left panel, lanes 2–6), consistent with incisions 5 and 6 nt into the duplex with a native flap substrate (18, Fig. 1, E and F, left panel, lanes 2–6). However, no further cleavage of the FAM-labeled ssDNA was observed, presumably because the flap fragment linked via the ICL interferes the subsequent nuclease activity. PaFAN1 did not cleave 5′ to the ICL, and no flap fragments (products a and e) were released, supporting previous observations that HsFAN1 did not release the flap when the ICL is within 4 nt from the ss/ds junction (Fig. 1F, right panel, lanes 2–6) (32). Instead, a single ICL-containing incision product of ICL-3/G1 labeled with HEX and CY3 was formed, which corresponds to product d resulting from incision 3′ to the ICL.
Because PaFAN1 makes an initial endonucleolytic incision followed by the exonucleolytic cleavage on the ICL-3/G1 substrate, we asked whether a gap formed by endonucleolytic cleavage would affect the subsequent ICL-unhooking activity of PaFAN1. We increased gap length between pre-nick duplex and the junction to 3 nt (ICL-3/G3) and tested whether this would affect the position of the initial incision. The presence of a longer gap did indeed alter the incision pattern of ICL substrates. PaFAN1 cleaved the ICL-3/G3 substrate (Fig. 1F, lanes 7–11, products b–e) both 5′ and 3′ of the ICL lesion as evidenced by formation of a fragment c with a single unhooked ICL, a fragment that incised 5′ to ICL (product e) and 3′ to ICL (product d) and produced seven FAM-labeled fragments (product b, 12–18 nt). The cleavage pattern 3′ to the ICL (product b) suggests that PaFAN1 successively incised seven nucleotides with a predominant 18-nt fragment and several less prominent shorter fragments (Fig. 1, F and H). The result also suggests that the 3′ end of the pre-nick strand can be used a reference point for cleavage, consistent with our previous study (23).
We then asked how far PaFAN1 can traverse into the duplex past the junction to unhook the ICL (Fig. 1, G and I). A previous study showed that HsFAN1 can cleave the ICL positioned at the 3rd to 6th nt from the terminal phosphate of a 1-nt 5′ flap (33). For this purpose, we used a substrate in which the ICL is positioned at the 9th nt of the 5′ flap strand and the 11th nt of the template strand from the junction (ICL-9). The appearance of HEX/CY3 bands assigned to structures a and e as well as FAM-labeled b indicated that PaFAN1 cleaved the 5′ as well as the 3′ side of the ICL-9/G1, demonstrating that PaFAN1 can traverse at least 9 nt from the junction (Fig. 1G, lanes 2–6). The g′ (12 nt) product is the most intense incision product past the ICL with the amount of subsequent smaller products decreasing continuously (Fig. 1I, ICL-9/G1). When the gap was increased to 3 nt (ICL-9/G3), the amount of product was decreased, but PaFAN1 efficiently incised this substrate on the 5′ and 3′ side of ICL as well (Fig. 1G, lanes 7–11, products a, e, and b). Interestingly, product e, formed by incision on the 5′ side of the ICL, displays different mobility for ICL-9/G1 and ICL-9/G3 (Fig. 1G, right panel), suggesting that the length of the gap determines the position of the first incision of PaFAN1 in the duplex.
Structures of PaFAN1 bound to a 5′ flap DNA show two different registers of scissile phosphate positioning in the active site
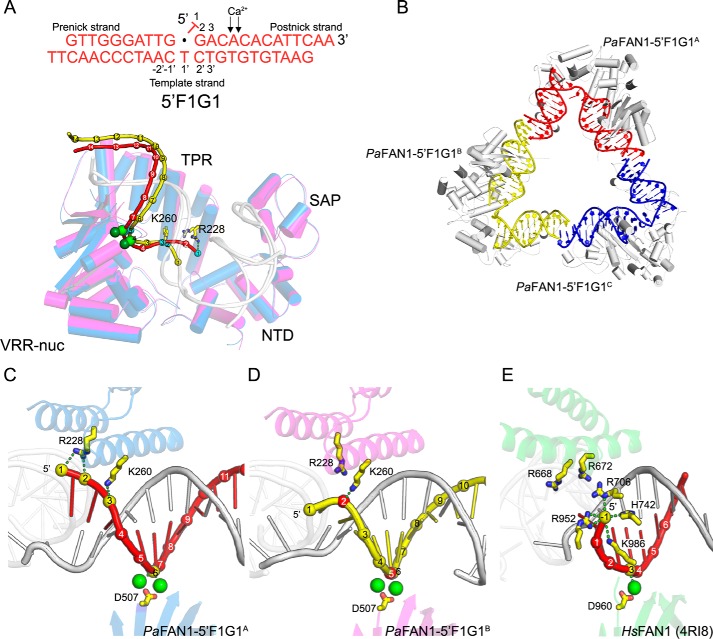
PaFAN1 makes two main initial incision products at the 5th or 6th position in the 5′ flap strand from the ss/ds junction. The position of the incision may be an important feature in determining how the protein unhooks the ICL. Although a specific basic pocket of HsFAN1 is not present in bacterial FAN1, our previously reported structure of PaFAN1 bound to DNA with a flap of 4 nt (PDB code 4R8A) showed that the phosphate groups at the ss/ds junction interact with Arg-228, Arg-257, and Lys-260 (23). However, the structure did not provide a clue on how PaFAN1 can catalyze multiple cleavage events. To gain insight into this unique ability, we determined the structure with PaFAN1 bound to its preferred DNA substrate (a 1-nt 5′ flap structure with a 1-nt gap (5′ F1G1)) in the presence of Ca2+ ion at 3.4 Å (Fig. 2, A–D, and Fig. S2A). The DNA in the crystal contained a 10-nt “pre-nick” strand (G10 to G1), a 14-nt “post-nick” strand (T1 to A14), and a 24-nt “template” strand (G12′ to T12′; Fig. 2A). The PaFAN1 structure is similar to that bound to DNA with a 5′ flap of 4 nt (PDB code 4R8A) with a root mean square deviation of 0.82 Å for 535 Cα atoms. The crystal contained three PaFAN1–5′F1G1 complexes (referred as PaFAN1–5′F1G1A, PaFAN1–5′F1G1B, and PaFAN1–5′F1G1C) in an asymmetric unit that can be classified into two different types of structures (Fig. 2B). Whereas the structures of PaFAN1–5′F1G1A and PaFAN1–5′F1G1B contained two Ca2+ ions at their active sites, PaFAN1–5′F1G1C had only one Ca2+ ion. The DNA molecules are linked through a pseudo-continuous helix formed through bp formed by the 2-nt overhangs (Fig. 2B). The interaction between the pre-nick duplex and NTD is virtually identical in the three molecules. However, the post-nick duplex exhibited two different modes of binding to the CTD. Arg-228 and Lys-260 in the TPR domain form a patch (hereafter referred as “Arg/Lys patch”) located 3 nt away from the active site, which interacts with the phosphates at the 5′ end of the post-nick strand. In PaFAN1–5′F1G1A, Arg-228 interacts with phosphates of T1 and G2, and Lys-260 interacts with phosphate of A3. PaFAN1–5′F1G1C also shows interaction between Lys-260 and phosphate of A3, whereas Arg-228 is retracted from the 5′ terminus of the post-nick (Fig. 2, A and C). In another mode, Lys-260 interacts with phosphate of G2 in PaFAN1–5′F1G1B (Fig. 2D). As a result, the scissile phosphate coordinated by a Ca2+ ion is also shifted by 1 nt with the active site located at the +5 phosphate in PaFAN1–5′F1G1B versus the +6 phosphate in PaFAN1–5′F1G1A and PaFAN1–5′F1G1C. The structure suggests that Arg-228 and Lys-260 contribute to the different distribution of the interaction between the post-nick strand and PaFAN1, in which the register of the post-nick strands differs by 1 nt, explaining how the multiple cleavage events can be mediated by PaFAN1. In the Arg/Lys patch, a basic residue was found to be highly conserved across the species at the position equivalent to Lys-260, whereas Arg-228 is only conserved among prokaryotic FAN1s (Fig. S3A). Lys-260 and Arg-228 are equivalent to Arg-668 and Arg-672, respectively, in the aligned structure of HsFAN1 (PDB code 4RI8, Fig. 2E, and Fig. S3B). However, in functional aspects, His-742, Arg-706, and Arg-952 of HsFAN1 are more similar to Lys-260 and Arg-228 as they primarily contact the 5′ phosphate.
Figure 2.
Crystal structure of PaFAN1–5′F1G1 in the presence of Ca2+ ions. A, DNA substrate used for crystallization (top). Superimposed structures of PaFAN1–5′F1G1A (blue) and PaFAN1–5′F1G1B (magenta) were bound to the 5′F1G1 DNA in the asymmetric unit (bottom). Two structures were aligned through their NTD. Each Ca2+ ion is shown as green and light green spheres for PaFAN1–5′F1G1A and PaFAN1–5′F1G1B, respectively. Pre-nick and template strands are colored in gray. Phosphate groups of post-nick strand DNA molecules (PaFAN1–5′F1G1A (red) and PaFAN1–5′F1G1B (yellow)) are shown as spheres and numbered from the 5′ end. Phosphate groups that interact with the Arg/Lys patch or metal ions are colored in cyan. B, three PaFAN1–DNA complexes in the asymmetric unit. C, close-up view of the active site and Arg/Lys patch of PaFAN1–5′F1G1A. D, PaFAN1–5′F1G1B, and E, HsFAN1–DNA (PDB code 4RI8). Phosphate groups that interact with the Arg/Lys patch or metal ion are colored in yellow (C and E) or red (D). See also Fig. S2 and S3.
Structure of PaFAN1 bound to a 5′ flap DNA with a 3-nt gap
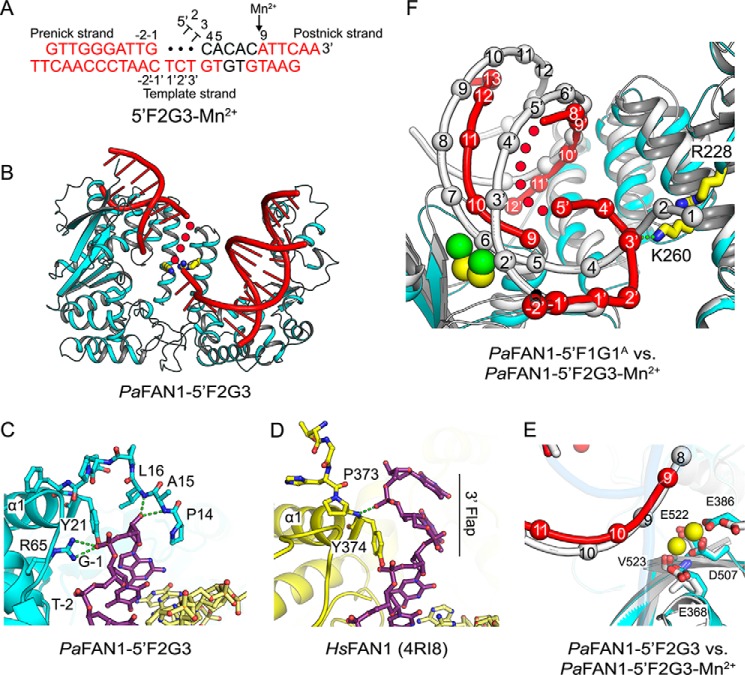
PaFAN1 makes an initial endonucleolytic cleavage followed by exonucleolytic incisions toward a flap or a nicked DNA substrate. To understand the effect of a gap produced by endonucleolytic cleavage on the subsequent exonuclease activity of PaFAN1, we determined the structure of PaFAN1 bound to a DNA with a 3-nt gap and a 2-nt 5′ flap (5′F2G3) at 2.7 Å (Fig. 3, A–E). With this structure, we wanted to test the hypothesis that the 5′-terminal phosphate of the 5′ flap strand interacts with the Arg/Lys patch. Although the structure clearly shows electron density for most of the DNA, a part of the gap region is disordered. DNA bound to PaFAN1 forms a continuous helix through base pairing of the 2-bp overhangs of consecutive DNA molecules that are related through a crystallographic symmetry (Fig. S2, B and C).
Figure 3.
Crystal structure of PaFAN1–5′F2G3 in the presence or absence of Mn2+. A, scheme of 5′F2G3 DNA used for crystallization. Disordered bases are colored in black, and unpaired bases at a gap are shown as filled circles. Scissile phosphates are indicated by a black arrow. B, overall structure of the PaFAN1–5′F2G3. C, close-up view of the interaction between the N-terminal loop of PaFAN1 and the pre-nick duplex. D, N-terminal loop of HsFAN1 and the 3′ flap. See also Fig. S1. E, close-up view of the active site in the PaFAN1–5′F2G3 in the presence (colored as in Fig. 1B) or absence (protein and DNA in gray) of Mn2+ ions. F, superposition of the PaFAN1–5′F2G3-Mn2+ (protein, DNA, and Mn2+ ions are colored in cyan, red, and yellow, respectively) and PaFAN1–5′F1G1A (protein and DNA in gray; Ca2+ ions in green) structures. Structures are aligned through their NTD. The gap region is flipped and interacts with the Arg/Lys patch. See also Fig. S2.
The structure shows two notable features compared with the PaFAN1 bound to 5′F1G1 or the 5′ flap DNA (PDB code 4R8A) structure. First, the main chain of Ala-15 and Leu-16 at the N-terminal loop, which was disordered in other structures, becomes ordered. As a result, residues Pro-14 to Tyr-21 along with Leu-195 form a pocket in which the 3′-terminal nucleotide of the pre-nick strand lodges (Fig. 3C). The backbone amide of Leu-16 interacts with the 3′ hydroxyl group of the ribose and the side chains of Tyr-21 and Arg-65 interact with the phosphate group of the 3′ end of the pre-nick strand. Thus, the presence of a 3′ flap on the pre-nick strand could disrupt the interaction with the N-terminal loop, consistent with the observation that addition of a 3′ flap decreased the nuclease activity of PaFAN1 (Fig. S1, B–D). This is markedly different from HsFAN1, which binds the 3′ flap with an extended conformation through residues Tyr-374, Val-577, and Arg-581. No residues at the N-terminal region of HsFAN1 block the path of the 3′ flap and the DNA binding affinity can be enhanced by the addition of a 3′ flap (33). Therefore, the interaction between the N-terminal loop and 3′ hydroxyl group of the pre-nick strand in PaFAN1 may be an important reference point for determining the position of cleavage (23).
Second, in the PaFAN1–5′F2G3 structure, the DNA region ranging from the active site to the junction of the pre-nick duplex undergoes a conformational change when a 3-nt gap is introduced between the pre- and post-nick duplexes (Fig. 3, A and B). Superposition of the PaFAN1–5′F2G3 structure onto the PaFAN1–5′F1G1A structure by aligning the NTD shows that the post-nick duplex undergoes a shift by 17–18 Å, whereas the pre-nick duplexes are aligned very well. Movement of the post-nick duplex locates the scissile phosphate at the active site, such that A9 (6th nt from the ss/ds junction) is positioned at the active site, which mimics the exonuclease-processed state (Fig. 3, A, E, and F). The distance between catalytic residue Asp-507 and scissile phosphate is 4.4 Å away from that of the PaFAN1–5′F1G1A structure.
To further understand the conformational changes during the activation process of PaFAN1, we determined the structure of PaFAN1–5′F2G3 with Mn2+ at 3.2 Å (Fig. 3, A, E, and F). Crystals soaked in a buffer containing the 5 mm MnCl2 for 14 min showed clear density for two Mn2+ ions (yellow spheres in Fig. 3, E and F). In the presence of Mn2+ ions, the pre-nick duplex retained almost the same conformation in the two structures, but the post-nick duplex near the active site is translated by 4–5.5 Å. Furthermore, the distance between the phosphates of A9 and T10 increased from 6.6 to 7.5 Å. Most remarkably, the 3-nt gap region becomes ordered and interacts with the Arg/Lys patch (Fig. S2D). The Mn2+ ion binding may have induced a local conformational change in the single-stranded gap in the PaFAN1–5′F2G3 crystal. A gap region containing T1′ to G8′ in the template strand interacts with the Arg/Lys patch via T3′ phosphate group and Lys-260 thereby effectively reducing the distance between the post-nick and pre-nick duplexes (Fig. 3, E and F). The T3′ residue of the backbone underwent the largest movement of 18.5 Å compared with that of PaFAN1–5′F1G1A. This interaction scrunches the three nucleotides in the gap, facilitating the translocation of the post-nick duplex toward the NTD, positioning the scissile phosphate of the post-nick into the active site. Based on this structure, we propose that the binding of the gap to the Arg/Lys patch can be used as a driving force to reposition the post-nick strand for successive exonuclease activities by PaFAN1 following the initial endonuclease incision.
Mutation of the Arg/Lys patch decreased ICL-unhooking activity of PaFAN1
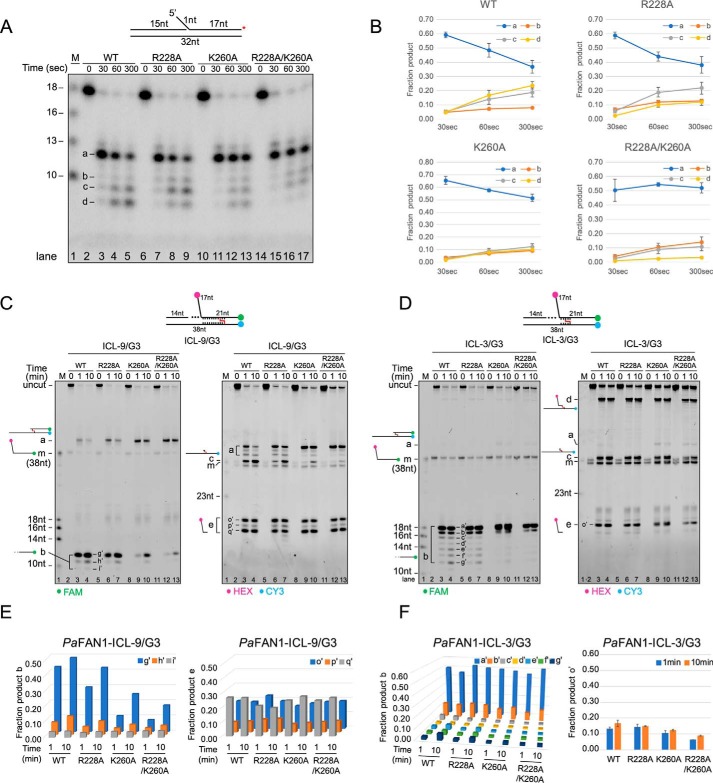
To assess the importance of the Arg/Lys patch for PaFAN1 activity, we generated three mutants, R228A, K260A, and R228A/K260A, and examined their nuclease and ICL unhooking activities (Fig. 4, A–F). For the nuclease activity assays, a substrate with a 1-nt 5′ flap was used. PaFAN1 generated multiple initial endonucleolytic products at the 5th and 6th phosphodiester from the junction, which were then processed further by exonucleolytic incisions (Fig. 4, A and B). Mutation of R228A did not alter the incision pattern. However, although the K260A and R228A/K260A proteins showed initial endonuclease activity (product a) similar to that of the WT PaFAN1, the exonuclease activity (product b to d) of these two mutants was slightly but reproducibly decreased. This result suggests that Lys-260 is important for exonucleolytic activity, likely through an interaction with terminal phosphate(s) of the initially cleaved DNA substrate, which could facilitate the translocation of the post-nick DNA with respect to the active site.
Figure 4.
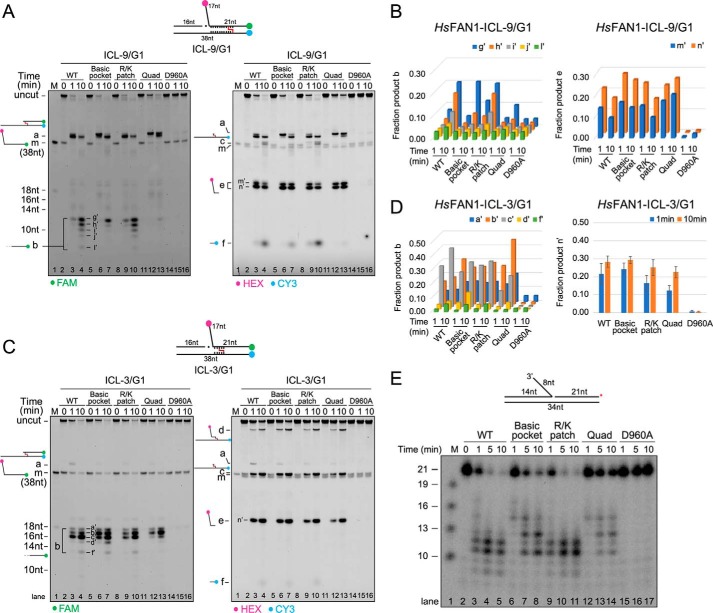
Nuclease and ICL-unhooking activity of the WT and the mutants of PaFAN1. A, time-dependent nuclease activity of WT (50 nm, lanes 2–5) and mutant PaFAN1 (50 nm, lanes 6–9; R228A, lanes 10–13; K260A, lanes 14–17; R228A/K260A) toward a 1-nt 5′ flap DNA (10 nm). DNA was labeled at 3′ end of a 5′ flap strand (red asterisk on top). Initial cleavage products are shown at 12- and 13-nt positions, which are further cleaved by exonuclease activity. B, quantified fraction product in each reaction. Data are represented as means ± S.D. C, time-dependent ICL-unhooking activity of WT PaFAN1 (250 nm, lanes 2–4) and mutants (250 nm, lanes 5–7; R228A, lanes 8–10; K260A, lanes 11–13; R228A/K260A) toward ICL-3/G3 (25 nm, top). Fluorescence signals were scanned using two wavelengths of 488 and 532 nm for FAM (left panel) and HEX or CY3 (right panel), respectively. D, time-dependent ICL-unhooking activity of various PaFAN1 proteins toward ICL-9/G3. E, graph showing the quantified fraction product “b” (left) or “e” (right) in C and F in reaction in D.
We next tested the ICL unhooking by these mutants (Fig. 4, C–F). The K260A and R228A/K260A mutants exhibited significantly decreased ICL-unhooking activity on the ICL-9/G3 substrate, which requires successive incisions to unhook the ICL, relative to that of the WT nuclease. Although product “c” representing the unhooking of the ICL is lost in the K260A and R228A/K260A mutants, formation of product “a” representing the incision of the flap is formed in all mutants (Fig. 4, C and D). Importantly, in both the ICL-9/G3 and ICL-3/G3 substrates, formation of the exonuclease products (Fig. 4, C and D, left panel, product b) is significantly reduced in K260A and R228A/K260A mutants. This result strongly supports an idea that Lys-260 is important for the recognition of the terminal phosphate of the initially cleaved product and subsequent 3-nt exonuclease activity.
To examine whether the Arg/Lys patch similarly directs incision in eukaryotic FAN1, we generated HsFAN1 with a mutation in Arg/Lys patch (R668A/R672A) and with a mutation in the previously identified basic pocket shown to be involved in 5′ flap binding (R706A/R952A). We also made the Arg/Lys patch/basic pocket quadruple mutant (R668A/R672A/R706A/R952A) and the nuclease-dead (D960A) mutant and performed ICL-unhooking analyses (Fig. 5, A–D). HsFAN1 exhibited a similar ICL-unhooking pattern toward ICL-3/G1, ICL-3/G3, ICL-9/G1, and ICL-9/G3 substrate, and we describe the data with the ICL-3/G1 and ICL-9/G1 substrate, for which it has higher activity, in detail. The basic pocket mutant was reported to have ∼400-fold decreased DNA-binding affinity compared with WT HsFAN1 (33). Both the Arg/Lys patch and the basic pocket mutants of HsFAN1 retained the unhooking activity of the ICL-3/G1 and ICL-9/G1 substrates similar to that of the WT nuclease (Fig. 5, A–D). The combined Arg/Lys patch/basic pocket quadruple mutant displayed clearly decreased exonuclease activity of the ICL-9/G1 substrate. The amount of product e (initial endonucleolytic cleavage product) by the quadruple mutant is similar to that of the WT HsFAN1, whereas the amount of product b was significantly reduced (Fig. 5, A, lane 13, and B). However, neither the basic pocket mutant nor the quadruple mutant did exhibit a pronounced decrease in exonuclease activity toward the ICL-3/G1. Subsequently, we examined the nuclease activity of various HsFAN1 proteins toward a nicked DNA with a 3′ flap (Fig. 5E). Although the basic pocket mutant showed a slightly reduced nuclease activity, a more significant decrease in activity was observed in the Arg/Lys patch/basic pocket quadruple mutation. A significant amount of the “14-nt” product remained, and the formation of the 12- and 11-nt product is decreased in this mutant. Based on these results, we conclude that the basic pocket plays an important role in degradation of the substrates by HsFAN1, and the Arg/Lys patch and basic pocket have a combined effect in substrate recognition and cleavage toward specific ICL substrates. However, it is clear that in addition to these features, other factors contribute to the ICL unhooking.
Figure 5.
Nuclease and ICL-unhooking activity of the WT and the mutants of HsFAN1. A, time-dependent ICL-unhooking activity of various HsFAN1 proteins (250 nm) toward ICL-9/G1 (25 nm). Denaturing PAGE with reactions containing WT (lanes 2–4), basic pocket (R706A/R952A, lanes 5–7), Arg/Lys patch (R668A/R672A, lanes 8–10), quadruple (R706A/R952A/R668A/R672A, lanes 11–13), and nuclease-dead (D960A, lanes 14–16) mutants. Fluorescence signals were scanned using two wavelengths and shown in the same way as in Fig. 4C. See also Fig. S3. B, bar graph showing the quantified fraction product b (left) or e (right) in A. C, time-dependent ICL-unhooking activity of various HsFAN1 proteins toward ICL-3/G1. D, bar graph showing the quantified fraction product b (left) or e (right) in C. E, time-dependent nuclease activity of WT (lanes 2–5) and mutant HsFAN1 (lanes 6–8; basic pocket, lanes 9–11; Arg/Lys patch, lanes 12–14; quadruple, lanes 15–17; D960A) toward a nicked DNA with an 8-nt 3′ flap. DNA was labeled at 3′ end of a nicked strand (red asterisk on top).
Disruption of initial cleavage inhibits gap generation and further exonucleolytic processing
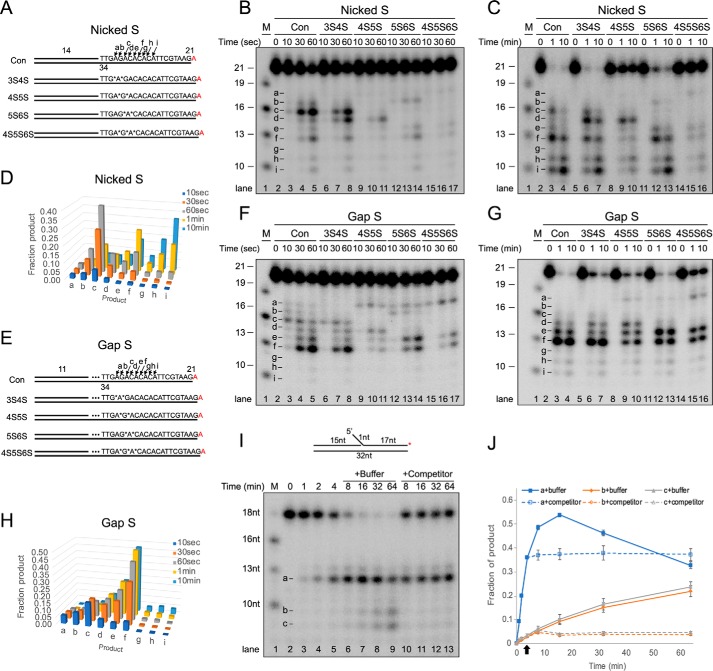
We then examined the importance of the gap formed upon initial endonucleolytic incision for the exonucleolytic processing by PaFAN1. We substituted several phosphodiester bonds of a 5′ flap strand with phosphorothioate bonds, which have been shown to be resistant to nucleolytic cleavage by metal-catalyzed endonucleases (38). We generated nicked DNA substrates (nicked S) with phosphorothioate bonds at incision sites of nucleotides 3/4 and 4/5 (3S4S), 4/5 and 5/6 (4S5S), 5/6 and 6/7 (5S6S), and 4/5, 5/6, and 6/7 (4S5S6S) (Fig. 6, A–D). PaFAN1 binds to the four phosphorothioate-substituted substrates with similar affinity as that of the unsubstituted substrate (Fig. S4, A–F). PaFAN1 initially cleaved the 4/5 and 5/6 nt from the junction, generating a 16-nt fragment via endonuclease activity (Fig. 6B, lanes 2–5, product c). PaFAN1 also produced 13- and 10-nt fragments (Fig. 6B, lanes 4 and 5, products f and i), suggesting that PaFAN1, like HsFAN1, incises DNA in 3-nt steps. However, PaFAN1 generated other fragments with various sizes at lower levels (Fig. 6B, products b, d, e, and h). These products are likely formed because PaFAN1 can recognize multiple phosphates at or near the 5′ terminus.
Figure 6.
Nuclease activity of PaFAN1 toward the phosphorothioate-substituted DNA. See also Fig. S4. A, structures of nicked DNA with a 14-nt pre-nick and a 21-nt post-nick duplex (Nicked S). Phosphodiester bonds are substituted with phosphorothioate bond at 3/4 and 4/5 nt (3S4S), at 4/5 and 5/6 nt (4S5S), at 5/6 and 6/7 nt (5S6S), and at 4/5/6/7 nt (4S5S6S) and marked by asterisks. Cleavage sites of substrates are marked by arrows with letters. DNA molecules were labeled at 3′ end by conjugating [α-32P]ATP, which is shown in red. B and C, denaturing PAGE of reactions with DNA substrates (10 nm) and PaFAN1 (10 nm, B) or PaFAN1 (50 nm, C) for indicated times. D, quantified reaction products in B and C. E, gapped DNA (Gap S) substrates with an 11-nt pre-nick, a 3-nt gap, and a 21-nt post-nick duplex. F and G, denaturing PAGE of reactions with DNA substrates (10 nm) and PaFAN1 (10 nm, F) or PaFAN1 (50 nm, G) for the indicated times. H, quantified reaction products in F and G. I, processivity analysis of PaFAN1. The 32P-labeled substrate with a 1-nt flap was incubated with PaFAN1 for different time points (lanes 2–5). At 4 min, an excess of unlabeled substrate (lanes 10–13) or a mock buffer (lanes 6–9) was added, and the reaction was further incubated. J, graph showing the fractions of products, a–c, in I. Arrow indicates a time when a buffer or a competitor was added.
Biochemical data and the PaFAN1–5′F1G1 structure suggest that substitution of phosphorothioate bonds at the nucleotides 4/5 and 5/6 would protect the substrate from initial endonucleolytic cleavage by PaFAN1. Indeed, PaFAN1 exhibited dramatically decreased nuclease activity on the 4S5S and 4S5S6S substrates (Fig. 6, B, lanes 9–11 and 15–17, and C, lanes 8–10 and 14–16). When the 4/5 and 5/6 sites are blocked, the 4S5S substrate is eventually incised between the 6th and 7th nucleotide (Fig. 6C, lane 10, product d) followed by incision between the 8th and 9th nucleotide (Fig. 6C, lanes 9 and 10, product f). The 3S4S substrate is efficiently cleaved at nucleotide 5/6 and 6/7 to form products c and d (Fig. 6, B, lanes 7 and 8, and C, lanes 6 and 7). Our data show that PaFAN1 only approximately follows the 3-nt step exonuclease rule and appears to more flexible with respect to the incision site than HsFAN1. Furthermore, the overall reduced nuclease activity on the 4S5S or in 4S5S6S substrate shows that the initial endonucleolytic incision is critical for the subsequent exonuclease steps.
We carried out the same set of experiments with an analogous substrate with a 3-nt gap (Gap S, Fig. 6, E–H), to examine whether a gap affected the cleavage pattern by PaFAN1. PaFAN1 cleaved at nucleotides 5/6, 7/8, and 8/9 downstream from the junction on the 3-nt gapped DNA, exhibiting a pattern similar to that on the nicked S DNA (Fig. 6, F and G, product c, e, and f). More e and f products are formed in a gapped DNA compared with a nicked substrate (Fig. 6F, lanes 4 and 5). Introduction of a gap facilitated the cleavage of the 4S5S and 4S5S6S substrates (Fig. 6G, lanes 9, 10, 15, and 16). This is probably because PaFAN1 initially incised at the “a” site of Gap S, which enables subsequent incisions at b and e, which differ by 3 nt (Fig. 6, B, F, lanes 10, 11, 16, and 17, C, and G, lanes 9, 10, 15, and 16). PaFAN1 produced nearly equal amounts of d, e, and f for 4S5S (Fig. 6G, lane 9 and 10), and equal amounts of e and f for the 5S6S (lanes 12 and 13) and 4S5S6S (lanes 15 and 16) substrates. The data suggest that whereas PaFAN1 prefers exonuclease activity in 3-nt steps, it can cleave adjacent phosphodiester bonds if the preferred incision site is blocked.
We next examined whether PaFAN1 dissociated after the initial incision and rebound to a substrate for the next cleavage or was processed continuously without dissociation. A 32P-labeled DNA substrate was incubated with PaFAN1 for different times, and subsequently, a 100-fold excess of unlabeled substrate competitor was added to the mixture at 4 min. Although addition of buffer did not seem to affect the incision reaction (Fig. 6, I, lanes 6–9, and J), addition of unlabeled competitor substrate clearly led to a decrease in endo- and exonuclease product substrate cleavage (lanes 10–13). If PaFAN1 incised a substrate in a strictly processive binding mode, we would expect the competitor to have less of an effect on nuclease activity. Thus, the data suggest that PaFAN1 is likely to dissociate from a substrate after initial cleavage and to rebind for a subsequent incision (Fig. 7). However, we do not rule out the possibility that processive binding can occur and may be detected under different assay conditions because both NTD and CTD quite specifically interact with the pre-nick and post-nick duplex, respectively.
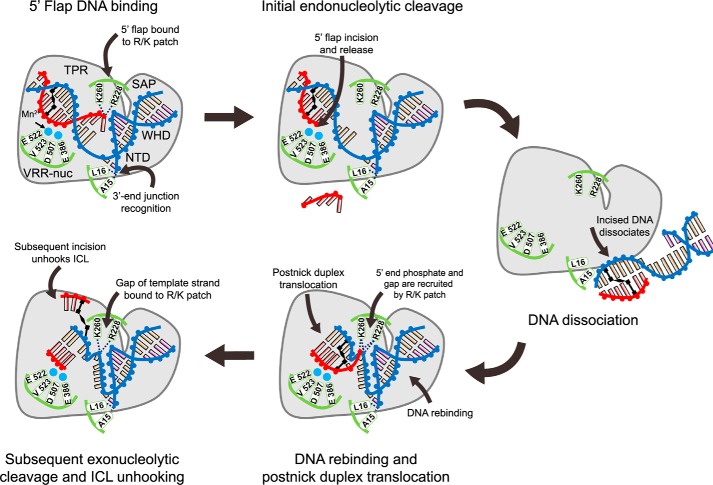
Figure 7.
Model for the ICL unhooking by PaFAN1. DNA strands are labeled as blue (template strand and pre-nick strand), red (5′ flap strand), and black for ICL-coupled bases. Mn2+ ions are shown in cyan spheres, and key residues for DNA recognition and cleavage are in green. (i) Recognition of a DNA substrate with a 5′ flap and an ICL by PaFAN1: pre-nick duplex by NTD, post-nick duplex by CTD, 3′-end junction by N-terminal loop, and 5′ flap by the Arg/Lys patch; (ii) 5′ flap is removed by initial endonucleolytic cleavage; (iii) the product is released from PaFAN1; (iv) PaFAN1 rebounds to the cleaved fragment and locates the next scissile phosphate group at the active site through the recognition of the phosphate of a gap by a Arg/Lys patch; (v) Subsequent exonucleolytic cleavages eventually unhook an ICL. The key features of PaFAN1 for ICL unhooking are marked: (a) 5′-terminal phosphates by a Arg/Lys patch (Arg-228 and Lys-260), (b) the pre-nick region by the N-terminal loop, and (c) a phosphate group at the gap by a Arg/Lys patch.
Discussion
Mammalian FAN1 repairs distorted helical structures caused by ICLs or other damage, thereby controlling the progression of stalled replication forks (17–20, 25–27). Although mammalian FAN1 can bind to the proximal site of replisome stalling via interaction with the ID complex, the nuclease also directly recognizes the substrate DNA and processes replication-like structures and ICLs (26, 32–35). Recent structural studies of HsFAN1 proposed two different mechanistic models for ICL unhooking by FAN1: the “3-nt exonuclease” or the “scanning and unwinding” model (33, 34). The key feature for the first model is a basic pocket that recognizes the 5′-terminal phosphate of substrates (33) and for the second model is the dimeric assembly of HsFAN1 in the presence of DNA (34). Puzzlingly, bacterial PaFAN1 does not possess any of these features, even though it shares overall structural similarity with HsFAN1 (23, 35). Thus, we were interested in examining whether bacterial FAN1 can resolve ICLs and, if so, how it achieves this task.
In this study, we demonstrated that bacterial FAN1 efficiently resolves ICL, and thus either a dimeric assembly or a specific basic pocket is not absolutely required for unhooking ICLs by FAN1 family members (Fig. 1, F–I). Although PaFAN1 did not release the 5′ flap when the ICL was too close to the ss/ds junction (example, within 3 nt), the nuclease could unhook ICL substrates at various positions located up to 9 nt from the junction. Structures of PaFAN1 bound to various DNA molecules suggest that the highly conserved Arg-228 and Lys-260 (Arg/Lys patch) play important roles in ICL unhooking by bacterial FAN1. Arg-228 and Lys-260 are 3 nt away from the active site and recognize the terminal phosphate groups of the 1-nt 5′ flap. A similar interaction was also observed in the HsFAN1–DNA with a 2-nt 5′ flap structure (4RIC) (33). In this structure, the 5′-terminal phosphate of a nick bypasses the basic pocket and interacts with two basic residues, Arg-668 and Arg-672, that correspond to Arg-228 and Lys-260, respectively (Fig. 1, B and C, and Fig. S3B). Thus, the Arg/Lys patch in bacterial FAN1 may perform a role equivalent to the basic pocket in recognizing the 5′ phosphate groups of the 5′ flap or a nicked strand. Lys-260 also interacts with a phosphate group in the gap, which suggests that a gap formed by the initial cleavage may contribute to the subsequent cleavage by PaFAN1. The importance of the interaction between the Arg/Lys patch and DNA was evident from the observation that mutation of Lys-260 decreased the exonuclease and ICL-unhooking activities of PaFAN1. However, this mutation did not affect the initial endonuclease activity of PaFAN1 toward a flap DNA substrate.
Substitution of two or three phosphate groups with phosphorothioates near the initial cleavage site almost completely inhibited the incision, although the addition of a gap facilitated cleavage (Fig. 6, A–H). These results suggest that the gap produced by the initial endonuclease activity is important for the subsequent exonuclease reactions. Thus, the interaction between Lys-260 and either the 5′-terminal phosphate or a phosphate group in the gap might be important for positioning the next cleavage site of the previously incised DNA substrate at the active site.
Consistent with the geometry of the Arg/Lys patch and active site, the primary products of PaFAN1 are fragments with 3-nt intervals, similar to those formed by HsFAN1 through the “3-nt step exonuclease” mechanism (Fig. 6, B and C). Although we cannot decisively conclude whether PaFAN1 follows processive mechanism in ICL resection or whether it dissociates after initial cleavage and rebinds to the substrate, two lines of evidence suggest that the nuclease can act through a distributive binding mechanism. First, in the crystal structure of PaFAN1–5′F1G1, PaFAN1 adopts two different binding states with the DNA substrate, in which the register of the post-nick duplex to the C-terminal domain is shifted by 1 nt (Fig. 2, A–D). Second, the observation that the presence of excess unlabeled substrate competitor inhibits successive incision events suggests that the nuclease is likely to be released from the substrate after initial incision and rebound for the next cleavage (Fig. 6, I and J). In mammalian FAN1, the presence of a UBZ domain may facilitate the association of the substrate DNA with the nuclease after the first cleavage.
However, a processive mode of binding has been observed in several nucleases that bind the 5′-terminal phosphate through a basic binding motif and drive the 5′ strand into the active site. λ exonuclease uses an electrostatic ratchet mechanism to unwind and attract the 5′ strand to the positively charged active site (39). Xrn1 exoribonuclease and hExo1 also contain a basic patch that recognizes the 5′-terminal phosphate of DNA substrates at or near the active site (40–43). Thus, further studies, including single molecule analyses, are required to more accurately determine the cleavage mechanism of substrates by PaFAN1.
Several minor products other than fragments with 3-nt intervals were observed, presumably because PaFAN1 can recognize multiple phosphates near the 5′ terminus rather than a single phosphate. Furthermore, if cleavage of the main site is blocked, PaFAN1 can cleave adjacent phosphodiester bonds (Fig. 6, A–D). As a result, the cleavage pattern by PaFAN1 does not strictly obey the 3-nt exonucleolytic rule. The 3-nt nuclease mechanism contributed by the Arg/Lys patch of PaFAN1 appears to be critical for unhooking ICLs because EXO1 and FEN1 that possess flap endonuclease and 5′ to 3′ exonuclease activities toward the flap substrates are unable to traverse and unhook ICLs (32).
Despite the significance of the Arg/Lys patch, mutation of Arg-228 or Lys-260 only affected the exonuclease but not the endonuclease activity of PaFAN1. Although the Arg/Lys patch is involved in recognizing the terminal phosphates of a 1-nt flap or a nick, the primary interaction of a substrate with PaFAN1 is achieved via four different domains: for the pre-nick duplex by the N-terminal winged-helix domain and the SAP domain, and for the post-nick duplex by the TPR and virus-type replication and repair-nuclease domains (23, 33–35). Thus, we speculate that the extensive interactions between the four different domains and the phosphate backbone of the substrate DNA are sufficient for the initial endonucleolytic cleavage.
One of the major roles of FAN1 in the maintenance of chromosomal stability is to recover stalled replication forks (25, 44, 45). In mammalian replication, the nuclease activity of FAN1 must be tightly controlled via several factors such as FANCD2 and Mre11 (44). A recent study showed that FAN1 is recruited to proliferating cell nuclear antigen, which then participates in the replication of a G4 quadruplex, implicating a role for FAN1 in replication of DNA with specific structures (45). Based on the structural and functional similarity between HsFAN1 and PaFAN1, it would be reasonable to assume that bacterial FAN1 is also involved in the progression of stalled replication forks. However, in contrast to HsFAN1, addition of a 3′ flap had an inhibitory effect on the nuclease activity of PaFAN1 because the N-terminal end of PaFAN1 blocks the path of the 3′ end of the pre-nick strand. This difference in substrate structures suggests that bacterial FAN1 may be involved in replication or metabolism of specific DNA structures.
To our surprise, mutation of the basic pocket did not significantly affect the ICL-unhooking activity of HsFAN1. Although mutation of the basic pocket of HsFAN1 resulted in a slight decrease in exonuclease activity toward ICL-9/G1, the activity toward the ICL-3/G1 substrate was not affected. We therefore surmised that the unchanged unhooking activity of the basic pocket mutant toward the ICL-3/G1 substrate is because the Arg/Lys patch can replace the function of the basic patch to some degree. Indeed, quadruple mutation of the Arg/Lys patch and the basic pocket reduced the exonuclease and ICL-unhooking activity to a greater extent (Fig. 5, A and C). Although the precise biological function of FAN1 is unclear, various disease-associated FAN1 mutations and functional analyses suggest that the nuclease activity of FAN1 is essential for ICL repair and the regulation of replication progress, and thereby maintenance of genome stability (25–27). The Arg/Lys patch and basic pocket in mammalian FAN1 may perform complementary roles as a dual safeguard to ensure interaction with the terminal phosphate, which is critical for the subsequent exonuclease and ICL-resolving activities. Thus, we propose that bacterial FAN1 evolved a basic pocket to bootstrap the substrate-binding function of the ancestral enzyme for processing severe DNA damage.
Experimental procedures
The gene encoding PaFAN1 (Uniprot ID Q9I2N0) was cloned, expressed in Escherichia coli Rosetta (DE3), and purified as described previously (23). Briefly, Rosetta (DE3) cells containing pET28a vector with the full-length PaFAN1 gene were grown in LB medium. PaFAN1 was induced by adding 0.5 mm isopropyl β-d-thiogalactopyranoside at A600 of 0.5, and cells were further incubated at 18 °C for 18 h. Cells were harvested, disrupted, and centrifuged, and the supernatant was loaded onto a Ni+-nitrilotriacetic acid affinity column and eluted with 130 mm imidazole. Eluents were further purified using anion-exchange chromatography and dialyzed against a buffer containing 20 mm Bistris propane-HCl (pH 7.0), 200 mm NaCl, and 5 mm DTT. Proteins were concentrated up to 20 mg/ml and stored at −80 °C. Each mutation was introduced by the PCR-based method and cloned into pET28a. For biochemical assay, human FAN1 (residues 330–1017; referred as HsFAN1 throughout the text) was used. PaFAN1 and HsFAN1 mutants were purified using the same protocol as those of WT PaFAN1.
PAGE-purified oligonucleotides were purchased from Bioneer Co. (Daejeon, Korea) for crystallization. Each oligonucleotide was mixed with an equimolar amount of an oligonucleotide with complementary sequence and annealed by heating at 95 °C for 5 min either in water or in SSC buffer (150 mm NaCl, 15 mm sodium citrate). DNAs were lyophilized and stored at −20 °C.
Oligonucleotides for nuclease activity assays were purchased from Bioneer Co. (Daejeon, Korea) or donated from the Bank of Modified Nucleic Acid Systems (BMNAS, POSTECH, Pohang, Korea). The 5′ flap strands were 3′-end-labeled using 2.5 μCi of [α-32P]ATP (PerkinElmer Life Sciences) and terminal deoxynucleotidyltransferase (Roche Applied Science) and then annealed to the template strand together with the 3′-nicked strand as described above.
ICL-containing oligonucleotides were synthesized as described previously (15, 36, 37). Briefly, oligonucleotides ICLSS1 and ICLSS2 or ICLSS3 and ICLSS4 (Table S1), containing fluorescent labels at one or both ends and an ICL precursor residue G[caret] (G[caret] = 7-deaza-(2,3-diacetoxy-propyl)-2′-deoxyguanosine), were annealed, and ICLs formed by oxidation with NaIO4 were followed by double-reductive amination with 1,2-dimethylethylenediamine and purified by gel electrophoresis and extraction of the ICL from a gel slice by electroelution. The resulting cross-linked oligonucleotides ICL-3 and ICL-9 only differ in the position of the ICL relative to the ss/dsDNA junction. To form the corresponding flap structures with a 1- or 3-nucleotide gap at the junction, ICL_G1 and ICL_G3 were annealed, yielding ICL3/G1, ICL3/G3, ICL9/G1, and ICL9/G3, respectively (Table S2).
PaFAN1–DNA complexes were prepared by mixing protein and DNA at 2:3 molar ratio and incubating at 4 °C for 30 min. Crystals of PaFAN1–DNA complexes were grown by hanging drop vapor diffusion method at 18 °C. PaFAN1–5′F2G3 co-crystals were obtained in a buffer containing 0.1 m HEPES (pH 7.2), 25% (w/v) polyethylene glycol (PEG) 8000, and 30 mm Gly-Gly-Gly. Crystals were cryo-protected using crystallization solution containing LiCl, gradually increasing the concentration of LiCl up to 3 m, and crystals were flash-frozen in liquid nitrogen. For Mn2+-bound PaFAN1–5′F2G3 co-crystals, the crystals were soaked into the reservoir buffer supplemented with 5 mm MnCl2 for 14 min and cryo-protected by gradually increasing LiCl concentrations up to 3 m.
PaFAN1–5′F1G1 co-crystals were obtained by same method, using the buffer containing 0.1 m Bistris propane (pH 7.6), 12% (w/v) PEG 8000, 0.2 m sodium acetate trihydrate, 10 mm calcium chloride dehydrate, and 1% (w/v) PEG 3350. Crystals were cryo-protected by the crystallization solution containing glycerol, gradually increasing the concentration of glycerol up to 30% (v/v). All data were collected at −170 °C at 0.9795 Å on Beamline 5C at the Pohang Accelerator Laboratory (PAL) and processed using the HKL2000 package (46). All data were scaled in the P212121 space group. To improve the usable resolution and quality of the resulting electron density maps, we used the Karplus CC* (Pearson correlation coefficient)-based data cutoff approach (Table 1) (47).
Table 1.
Statistics for data collection and refinement
| 5′F2G3 | 5′F2G3-Mn2+ | 5′F1G1 | |
|---|---|---|---|
| Data collection | |||
| Space group | P212121 | P212121 | P212121 |
| Cell dimensions | |||
| a, b, c (Å) | 80.9, 104.8, 105.6 | 80.8, 104.8, 105.6 | 140.3, 143.3, 172.7 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50–2.7 (2.75–2.7)a | 50–3.2 (3.26–3.2) | 50–3.4 (3.46–3.4) |
| Measured reflections | 356,808 | 99,109 | 557,812 |
| Unique reflections | 24,796 | 15,512 | 46,804 |
| Completeness (%) | 99.1 (98.4) | 99.9 (100) | 96.6 (91.1) |
| Average (I/σ) | 45.3 (1.5) | 16.2 (1.4) | 26.6 (1.4) |
| Rmerge (%) | 0.092 (2.416) | 0.098 (0.981) | 0.161 (1.908) |
| CC1/2 | 0.999 (0.646) | 0.998 (0.661) | 0.999 (0.292) |
| Redundancy | 14.4 (14.6) | 6.4 (6.5) | 12.0 (10.7) |
| Wilson B-factor (Å2) | 78.1 | 90.1 | 128.4 |
| Refinement | |||
| Resolution (Å) | 33.71–2.7 | 46.99–3.2 | 33.58–3.4 |
| No. of reflections | 24,874 | 15,341 | 463,39 |
| Rwork/Rfree (%)b | 19.3/24.3 | 18.2/23.8 | 22.1/28.0 |
| No. of atoms | |||
| Protein | 4444 | 4444 | 13231 |
| DNA | 734 | 779 | 2928 |
| Water/Mn2+/Ca2+ | 9/0/0 | 1/2/0 | 0/0/5 |
| B-factors | |||
| Protein | 84.5 | 94.4 | 156.9 |
| DNA | 139.3 | 159.1 | 191.4 |
| Water/Mn2+/Ca2+ | 61.6/0/0 | 63.6/118.9/0 | 0/0/143.1 |
| Root mean square deviations | |||
| Bond lengths (Å) | 0.009 | 0.013 | 0.013 |
| Bond angles (°) | 1.079 | 1.155 | 1.426 |
| Clash score | 8.4 | 7.1 | 17.2 |
| Ramachandran plot | |||
| Most favored (%) | 96.5 | 94.1 | 96.1 |
| Allowed (%) | 3.5 | 5.3 | 3.6 |
| Outlier (%) | 0 | 0.6 | 0.3 |
a Values in parentheses are for the highest shell.
b Rfree was calculated with 5% of the reflections.
All structures were determined by molecular replacement method using PaFAN1 structure (PDB code 4R8A) as the search model for phase calculation. Initial phases were obtained for PaFAN1–5′F2G3 (also for PaFAN1–5′F1G1-Ca2+ and PaFAN1–5′F2G3-Mn2+) co-crystal form using the program PHASER (48). Successive rounds of model building and refinement were performed using the programs COOT (49), PHENIX (48), and Refmac5 (50, 51), rigid body refinement, TLS and NCS refinement, individual B-value refinement, and positional and simulated annealing refinement. Detailed refinement statistics are listed in Table 1.
Nuclease activity assays were performed as described previously (23). 10 nm DNA substrates 32P-labeled at the 3′ end were mixed with 10 or 50 nm protein in a total 20-μl reaction buffer (25 mm Tris-HCl (pH 7.4), 25 mm NaCl, 5 mm β-mercaptoethanol, 0.1 mg/ml BSA) supplemented with 5 mm MgCl2 and incubated at 37 °C for the indicated time course. After incubation, the reaction was terminated by adding same volume of 2× reaction stop buffer (95% formamide, 18 mm EDTA, 0.025% SDS, 0.01% bromphenol blue) followed by boiling for 5 min at 100 °C. Products were analyzed by 15% denaturing PAGE containing 7 m urea in 1× TBE buffer. Images were detected and processed by Typhoon-9400 (GE Healthcare) and MultiGauge version 3.0 (Fuji film).
In vitro ICL-unhooking assays were performed in the same condition as the nuclease activity assay described above, but 25 nm fluorescently labeled ICL-containing oligonucleotides were mixed with 250 nm PaFAN1 or HsFAN1 for the indicated incubation time. Products were resolved by 15% denaturing PAGE containing 7 m urea in 1× TBE buffer, and images were detected by Typhoon-9400 (GE Healthcare) using lasers with 488 or 532 nm for FAM or HEX (or Cy3), respectively. Because of the overlapped range of the emission filter for HEX and Cy3, it was not possible to separate the detection of two fluorophores.
For DNA-mobility shift assay, 5 nm 32P-labeled DNA at the 5′ end of template strand was mixed with increasing amounts of proteins in a total 10 μl of reaction buffer (25 mm Tris-HCl (pH 7.4), 25 mm NaCl, 5 mm β-mercaptoethanol, 0.1 mg/ml BSA, 5% glycerol) supplemented with 10 mm CaCl2. Binding reactions were incubated on ice for 30 min followed by adding 2 μl of 6× DNA loading buffer (0.25% bromphenol blue, 0.25% xylene cyanol FF, 30% glycerol) and resolved by 10% native PAGE at 4 °C in 0.5× TBE buffer. Images were detected and processed by Typhoon-9400 (GE Healthcare) and MultiGauge version 3.0 (Fuji film).
To examine the distributive or processive mechanism, 20 nm DNA substrate labeled at 3′ end was mixed with 2 nm protein and incubated at 30 °C for different time points. At 4 min, 100-fold excess of unlabeled substrate was added to the reaction mixture and further incubated. Reaction was terminated, and each product was quantified as described above.
MultiGauge version 3.0 (Fuji film) was used to quantify in vitro biochemical experimental data. For nuclease activity assay, ICL-unhooking assay, and processivity assay, the fraction product was obtained by dividing intensity of the product by intensity of the whole lane. For DNA mobility shift assay, the fraction bound was calculated by dividing the protein-bound DNA by total DNA. Statistical parameters were expressed as average from duplicated experiments.
Author contributions
H. J. and Y. C. data curation; H. J., O. D. S., and Y. C. formal analysis; H. J. and Y. C. validation; H. J. and Y. C. visualization; H. J., U. R., and Y. C. methodology; H. J., O. D. S., and Y. C. writing-original draft; U. R., O. D. S., and Y. C. resources; G. L., O. D. S., and Y. C. conceptualization; G. L., O. D. S., and Y. C. supervision; Y. C. software; Y. C. funding acquisition; Y. C. investigation; Y. C. project administration.
Supplementary Material
Acknowledgment
Sequence alignment tool PROMALS3D (52) server is available in the Grishin Lab, UT Southwestern Medical Center (http://prodata.swmed.edu/promals3d/promals3d.php).
This work was supported by National Research Foundation of Korea (NRF), funded by the Korea government (Ministry of Education, Science, and Technology), Grants 2015R1A2A1A05001694, 2017M3A9F6029733, and NRF-2013M3A6A4044580, BK21 Program from the Ministry of Education (to Y. C.), National Institutes of Health Grant CA165911, and Korean Institute for Basic Science Grant IBS-R022-A1-2017 (to O. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S4 and Tables S1–S2.
The atomic coordinates and structure factors (codes 5Y7Q, 5Z6W, and 5Y7G) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- ICL
- inter-strand cross-link
- FAN1
- FANCD2–FANCI associated nuclease 1
- FA
- Fanconi-anemia
- TLS
- translesion synthesis
- UBZ
- ubiquitin-binding zinc finger
- KIN
- karyomegalic interstitial nephritis
- FAM
- carboxyfluorescein
- HEX
- 6-carboxy-2′,4,4′,5′,7,7′-hexachlorofluorescein
- Cy3
- cyanine 3
- SAP
- SAF-A/B, Acinus, and PIAS
- NTD
- N-terminal domain
- CTD
- C-terminal domain
- TPR
- tetratricopeptide repeat
- PDB
- Protein Data Bank
- nt
- nucleotide
- Bistris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- ssDNA
- single-stranded DNA
- dsDNA
- double-stranded DNA.
References
- 1. Deans A. J., and West S. C. (2011) DNA interstrand cross-link repair and cancer. Nat. Rev. Cancer 11, 467–480 10.1038/nrc3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schärer O. D. (2005) DNA interstrand cross-links: natural and drug-induced DNA adducts that induce unique cellular responses. Chembiochem 6, 27–32 10.1002/cbic.200400287 [DOI] [PubMed] [Google Scholar]
- 3. Huang Y., and Li L. (2013) DNA cross-linking damage and cancer–a tale of friend and foe. Transl. Cancer Res. 2, 144–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kottemann M. C., and Smogorzewska A. (2013) Fanconi anaemia and the repair of Watson and Crick DNA cross-links. Nature 493, 356–363 10.1038/nature11863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang J., Liu S., Bellani M. A., Thazhathveetil A. K., Ling C., de Winter J. P., Wang Y., Wang W., and Seidman M. M. (2013) The DNA translocase FANCM/MHF promotes replication traverse of DNA interstrand cross-links. Mol. Cell 52, 434–446 10.1016/j.molcel.2013.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang S., Liu K., Xiao L., Yang L., Li H., Zhang F., Lei L., Li S., Feng X., Li A., and He J. (2016) Characterization of a novel DNA glycosylase from S. sahachiroi involved in the reduction and repair of azinomycin B induced DNA damage. Nucleic Acids Res. 44, 187–197 10.1093/nar/gkv949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rolseth V., Krokeide S. Z., Kunke D., Neurauter C. G., Suganthan R., Sejersted Y., Hildrestrand G. A., Bjørås M., and Luna L. (2013) Loss of Neil3, the major DNA glycosylase activity for removal of hydantoins in single-stranded DNA, reduces cellular proliferation and sensitizes cells to genotoxic stress. Biochim. Biophys. Acta 1833, 1157–1164 10.1016/j.bbamcr.2012.12.024 [DOI] [PubMed] [Google Scholar]
- 8. Semlow D. R., Zhang J., Budzowska M., Drohat A. C., and Walter J. C. (2016) Replication-dependent unhooking of DNA interstrand cross-links by the NEIL3 glycosylase. Cell 167, 498–511 10.1016/j.cell.2016.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Räschle M., Knipscheer P., Knipsheer P., Enoiu M., Angelov T., Sun J., Griffith J. D., Ellenberger T. E., Schärer O. D., and Walter J. C. (2008) Mechanism of replication-coupled DNA interstrand cross-link repair. Cell 134, 969–980 10.1016/j.cell.2008.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang J., Dewar J. M., Budzowska M., Motnenko A., Cohn M. A., and Walter J. C. (2015) DNA interstrand cross-link repair requires replication-fork convergence. Nat. Struct. Mol. Biol. 22, 242–247 10.1038/nsmb.2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smogorzewska A., Matsuoka S., Vinciguerra P., McDonald E. R. 3rd, Hurov K. E., Luo J., Ballif B. A., Gygi S. P., Hofmann K., D'Andrea A. D., and Elledge S. J. (2007) Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell 129, 289–301 10.1016/j.cell.2007.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim J. M., Kee Y., Gurtan A., and D'Andrea A. D. (2008) Cell cycle-dependent chromatin loading of the Fanconi anemia core complex by FANCM/FAAP24. Blood 111, 5215–5222 10.1182/blood-2007-09-113092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Knipscheer P., Räschle M., Smogorzewska A., Enoiu M., Ho T. V., Schärer O. D., Elledge S. J., and Walter J. C. (2009) The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science 326, 1698–1701 10.1126/science.1182372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Klein Douwel D., Boonen R. A., Long D. T., Szypowska A. A., Räschle M., Walter J. C., and Knipscheer P. (2014) XPF-ERCC1 acts in unhooking DNA interstrand cross-links in cooperation with FANCD2 and FANCP/SLX4. Mol. Cell 54, 460–471 10.1016/j.molcel.2014.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hodskinson M. R., Silhan J., Crossan G. P., Garaycoechea J. I., Mukherjee S., Johnson C. M., Schärer O. D., and Patel K. J. (2014) Mouse SLX4 is a tumor suppressor that stimulates the activity of the nuclease XPF-ERCC1 in DNA cross-link repair. Mol. Cell 54, 472–484 10.1016/j.molcel.2014.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang J., and Walter J. C. (2014) Mechanism and regulation of incisions during DNA interstrand cross-link repair. DNA Repair 19, 135–142 10.1016/j.dnarep.2014.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kratz K., Schöpf B., Kaden S., Sendoel A., Eberhard R., Lademann C., Cannavó E., Sartori A. A., Hengartner M. O., and Jiricny J. (2010) Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand cross-linking agents. Cell 142, 77–88 10.1016/j.cell.2010.06.022 [DOI] [PubMed] [Google Scholar]
- 18. Liu T., Ghosal G., Yuan J., Chen J., and Huang J. (2010) FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science 329, 693–696 10.1126/science.1192656 [DOI] [PubMed] [Google Scholar]
- 19. MacKay C., Déclais A. C., Lundin C., Agostinho A., Deans A. J., MacArtney T. J., Hofmann K., Gartner A., West S. C., Helleday T., Lilley D. M., and Rouse J. (2010) Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell 142, 65–76 10.1016/j.cell.2010.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Smogorzewska A., Desetty R., Saito T. T., Schlabach M., Lach F. P., Sowa M. E., Clark A. B., Kunkel T. A., Harper J. W., Colaiácovo M. P., and Elledge S. J. (2010) A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand cross-link repair. Mol. Cell 39, 36–47 10.1016/j.molcel.2010.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yoshikiyo K., Kratz K., Hirota K., Nishihara K., Takata M., Kurumizaka H., Horimoto S., Takeda S., and Jiricny J. (2010) KIAA1018/FAN1 nuclease protects cells against genomic instability induced by interstrand cross-linking agents. Proc. Natl. Acad. Sci. U.S.A. 107, 21553–21557 10.1073/pnas.1011081107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang Y., Leung J. W., Lowery M., Matsushita N., Wang Y., Shen X., Huong D., Takata M., Chen J., and Li L. (2014) Modularized functions of the Fanconi anemia core complex. Cell Rep. 7, 1849–1857 10.1016/j.celrep.2014.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gwon G. H., Kim Y., Liu Y., Watson A. T., Jo A., Etheridge T. J., Yuan F., Zhang Y., Kim Y., Carr A. M., and Cho Y. (2014) Crystal structure of a Fanconi anemia-associated nuclease homolog bound to 5′ flap DNA: basis of interstrand cross-link repair by FAN1. Genes Dev. 28, 2276–2290 10.1101/gad.248492.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhou W., Otto E. A., Cluckey A., Airik R., Hurd T. W., Chaki M., Diaz K., Lach F. P., Bennett G. R., Gee H. Y., Ghosh A. K., Natarajan S., Thongthip S., Veturi U., Allen S. J., et al. (2012) FAN1 mutations cause karyomegalic interstitial nephritis, linking chronic kidney failure to defective DNA damage repair. Nat. Genet. 44, 910–915 10.1038/ng.2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lachaud C., Moreno A., Marchesi F., Toth R., Blow J. J., and Rouse J. (2016) Ubiquitinated Fancd2 recruits Fan1 to stalled replication forks to prevent genome instability. Science 351, 846–849 10.1126/science.aad5634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thongthip S., Bellani M., Gregg S. Q., Sridhar S., Conti B. A., Chen Y., Seidman M. M., and Smogorzewska A. (2016) Fan1 deficiency results in DNA interstrand cross-link repair defects, enhanced tissue karyomegaly, and organ dysfunction. Genes Dev. 30, 645–659 10.1101/gad.276261.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lachaud C., Slean M., Marchesi F., Lock C., Odell E., Castor D., Toth R., and Rouse J. (2016) Karyomegalic interstitial nephritis and DNA damage-induced polyploidy in Fan1 nuclease-defective knock-in mice. Genes Dev. 30, 639–644 10.1101/gad.276287.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Seguí N., Mina L. B., Lázaro C., Sanz-Pamplona R., Pons T., Navarro M., Bellido F., López-Doriga A., Valdés-Mas R., Pineda M., Guinó E., Vidal A., Soto J. L., Caldés T., Durán M., et al. (2015) Germline mutations in FAN1 cause hereditary colorectal cancer by impairing DNA repair. Gastroenterology 149, 563–566 10.1053/j.gastro.2015.05.056 [DOI] [PubMed] [Google Scholar]
- 29. Smith A. L., Alirezaie N., Connor A., Chan-Seng-Yue M., Grant R., Selander I., Bascuñana C., Borgida A., Hall A., Whelan T., Holter S., McPherson T., Cleary S., Petersen G. M., Omeroglu A., et al. (2016) Candidate DNA repair susceptibility genes identified by exome sequencing in high-risk pancreatic cancer. Cancer Lett. 370, 302–312 10.1016/j.canlet.2015.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bettencourt C., Hensman-Moss D., Flower M., Wiethoff S., Brice A., Goizet C., Stevanin G., Koutsis G., Karadima G., Panas M., Yescas-Gómez P., García-Velázquez L. E., Alonso-Vilatela M. E., Lima M., Raposo M., et al. (2016) DNA repair pathways underlie a common genetic mechanism modulating onset in polyglutamine diseases. Ann. Neurol. 79, 983–990 10.1002/ana.24656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ionita-Laza I., Xu B., Makarov V., Buxbaum J. D., Roos J. L., Gogos J. A., and Karayiorgou M. (2014) Scan statistic-based analysis of exome sequencing data identifies FAN1 at 15q13.3 as a susceptibility gene for schizophrenia and autism. Proc. Natl. Acad. Sci. U.S.A. 111, 343–348 10.1073/pnas.1309475110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pizzolato J., Mukherjee S., Schärer O. D., and Jiricny J. (2015) FANCD2-associated nuclease 1, but not exonuclease 1 or flap endonuclease 1, is able to unhook DNA interstrand cross-links in vitro. J. Biol. Chem. 290, 22602–22611 10.1074/jbc.M115.663666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang R., Persky N. S., Yoo B., Ouerfelli O., Smogorzewska A., Elledge S. J., and Pavletich N. P. (2014) DNA repair. Mechanism of DNA interstrand cross-link processing by repair nuclease FAN1. Science 346, 1127–1130 10.1126/science.1258973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhao Q., Xue X., Longerich S., Sung P., and Xiong Y. (2014) Structural insights into 5′ flap DNA unwinding and incision by the human FAN1 dimer. Nat. Commun. 5, 5726 10.1038/ncomms6726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jin H., and Cho Y. (2017) Structural and functional relationships of FAN1. DNA Repair 56, 135–143 10.1016/j.dnarep.2017.06.016 [DOI] [PubMed] [Google Scholar]
- 36. Angelov T., Guainazzi A., and Schärer O. D. (2009) Generation of DNA interstrand cross-links by post-synthetic reductive amination. Org. Lett. 11, 661–664 10.1021/ol802719a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mukherjee S., Guainazzi A., and Schärer O. D. (2014) Synthesis of structurally diverse major groove DNA interstrand cross-links using three different aldehyde precursors. Nucleic Acids Res. 42, 7429–7435 10.1093/nar/gku328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vosberg H. P., and Eckstein F. (1982) Effect of deoxynucleoside phosphorothioates incorporated in DNA on cleavage by restriction enzymes. J. Biol. Chem. 257, 6595–6599 [PubMed] [Google Scholar]
- 39. Zhang J., McCabe K. A., and Bell C. E. (2011) Crystal structures of λ exonuclease in complex with DNA suggest an electrostatic ratchet mechanism for processivity. Proc. Natl. Acad. Sci. U.S.A. 108, 11872–11877 10.1073/pnas.1103467108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jinek M., Coyle S. M., and Doudna J. A. (2011) Coupled 5′ nucleotide recognition and processivity in Xrn1-mediated mRNA decay. Mol. Cell 41, 600–608 10.1016/j.molcel.2011.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chang J. H., Xiang S., Xiang K., Manley J. L., and Tong L. (2011) Structural and biochemical studies of the 5′ → 3′ exoribonuclease Xrn1. Nat. Struct. Mol. Biol. 18, 270–276 10.1038/nsmb.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Orans J., McSweeney E. A., Iyer R. R., Hast M. A., Hellinga H. W., Modrich P., and Beese L. S. (2011) Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell 145, 212–223 10.1016/j.cell.2011.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Myler L. R., Gallardo I. F., Zhou Y., Gong F., Yang S. H., Wold M. S., Miller K. M., Paull T. T., and Finkelstein I. J. (2016) Single-molecule imaging reveals the mechanism of Exo1 regulation by single-stranded DNA binding proteins. Proc. Natl. Acad. Sci. U.S.A. 113, E1170–E1179 10.1073/pnas.1516674113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chaudhury I., Stroik D. R., and Sobeck A. (2014) FANCD2-controlled chromatin access of the Fanconi-associated nuclease FAN1 is crucial for the recovery of stalled replication forks. Mol. Cell. Biol. 34, 3939–3954 10.1128/MCB.00457-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Porro A., Berti M., Pizzolato J., Bologna S., Kaden S., Saxer A., Ma Y., Nagasawa K., Sartori A. A., and Jiricny J. (2017) Author Correction: FAN1 interaction with ubiquitylated PCNA alleviates replication stress and preserves genomic integrity independently of BRCA2. Nat. Commun. 8, 2285 10.1038/s41467-017-02130-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 47. Karplus P. A., and Diederichs K. (2012) Linking crystallographic model and data quality. Science 336, 1030–1033 10.1126/science.1218231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 50. Murshudov G. N., Vagin A. A., and Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 10.1107/S0907444996012255 [DOI] [PubMed] [Google Scholar]
- 51. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., and Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 10.1107/S0907444911001314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pei J., Kim B. H., and Grishin N. V. (2008) PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 36, 2295–2300 10.1093/nar/gkn072 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.